Abstract
The cleavage of amyloid precursor protein (APP) by β-site APP cleaving enzyme 1 (BACE1) is the rate limiting step in the generation of β-amyloid (Aβ) during Alzheimer’s disease (AD) pathogenesis. In AD brains, BACE1 is abnormally accumulated in the endocytic compartments, where the acidic pH is optimal for its activity. However mechanisms that regulate the endosome-to-TGN retrieval of BACE1 under physiological conditions remain unclear. Here we show the Par polarity complex containing Par3 and aPKC facilitates BACE1 retrograde trafficking from the endosomes to the TGN. We further show that Par3 functions through aPKC-mediated phosphorylation of BACE1 on Ser498. Finally, we found that Ser498 phosphorylation promotes the interaction between BACE1 and PACS1, which is necessary for the retrograde trafficking of BACE1 to the TGN. In human AD brains, there is a significant decrease in Ser498 phosphorylation of BACE1 suggesting that defective phosphorylation-dependent retrograde transport of BACE1 is important in the AD pathogenic process. Together, our studies provide mechanistic insight into a novel role for Par3 and aPKC in regulating the retrograde endosome-to-TGN trafficking of BACE1, and shed light on the mechanisms of AD pathogenesis.
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disease characterized by the presence of β-amyloid (Aβ) plaques and neurofibrillary tangles containing hyperphosphorylated tau. Aβ is generated by β- and γ-secretase cleavage of the single-transmembrane amyloid precursor protein (APP). Importantly, the cleavage of APP by the β-secretase, β-site APP cleaving enzyme 1 (BACE1), is the rate-limiting step of the Aβ generation pathway (Das et al., 2013; Thinakaran and Koo, 2008). Since both APP and BACE1 are normally expressed at high levels in neurons, their convergence within the cell would lead to large amounts of Aβ generation. Thus, mechanisms must exist to spatially segregate these two proteins at physiological states.
One key process in ensuring APP and BACE1 segregation is through modulating BACE1 trafficking. BACE1 is a Type I transmembrane protein that is normally delivered from the trans-Golgi network (TGN) to the plasma membrane through the secretory pathway. It is then internalized into endosomal compartments where the acidic environment is optimal for its enzymatic activity (Cole and Vassar, 2007; Tan and Evin, 2012). From the endosomes, BACE1 can be retrogradely transported to the TGN for re-insertion into the plasma membrane where its activity remains low due to the neutral environment. Since abnormally enlarged endosomal compartments and accumulation of Aβ in these compartments are some of the earliest pathogenic changes occurring in the AD brain (Cataldo et al., 2004; Ginsberg et al., 2010; Nixon, 2005), it is believed that defective BACE1 retrograde trafficking, which leads to its accumulation in endosomal compartments and convergence with APP, is a key pathogenic process during AD progression (Das et al., 2013; Kinoshita et al., 2003; Rajendran et al., 2006; Udayar et al., 2013; Ye and Cai, 2014; Zhang et al., 2015). Indeed, in AD neurons, defects have been observed in a number of proteins involved in the endosome-to-TGN retrograde trafficking pathway, including components of the retromer complex (Buggia-Prevot and Thinakaran, 2014; Muhammad et al., 2008; Reitz, 2015; Sullivan et al., 2011; Wen et al., 2011). However, the mechanisms regulating BACE1 retrograde endosome-to-TGN trafficking is still unclear.
Par3 is a large scaffolding molecule that is among a group of evolutionarily conserved proteins essential for cell polarization during animal development (Goldstein and Macara, 2007). Previous studies have implicated Par3 in trafficking processes such as vesicle exocytosis (Ahmed and Macara, 2017; Balklava et al., 2007; Das et al., 2014; Lalli, 2009; Zuo et al., 2009). We recently showed that Par3 regulates APP trafficking through the endocytic adaptor Numb (Sun et al., 2016). Here we show an unexpected role for the polarity protein Par3 in regulating BACE1 endosome-to-TGN retrograde trafficking in hippocampal neurons. We show that Par3 facilitates BACE1 retrograde trafficking to the trans-Golgi network (TGN) by recruiting atypical PKC (aPKC), which promotes BACE1 phosphorylation at Ser498. Phosphorylation of BACE1 increases its interaction with PACS1 and enhances its retrograde trafficking to the TGN. In the absence of Par3, there is a significant increase in the accumulation of BACE1 in the late endosome/lysosome compartments. Taken together, our studies provide mechanistic insights into a novel role for Par3 in regulating BACE1 retrograde trafficking and shed light on the mechanisms of AD pathogenesis.
Materials and Methods
Plasmids and Reagents
All plasmids encoding full length Par3b, Par3 shRNAs, and PKCζ have been previously described (Chen and Macara, 2005; McCaffrey and Macara, 2009; Zhang and Macara, 2008). Human BACE1-EGFP was a generous gift from Dr. Bradley T. Hyman (Harvard Medical School). BACE1 Ser498 mutants were generated by Quikchange (Stratagene) using WT human BACE1 as the template. All FLAG and mRFP tagged full length BACE1 constructs were generated by subcloning from the human BACE1-EGFP. For knockdown of PACS1, the following oligonucleotides against different regions of rat PACS1 were synthesized, annealed, and ligated into the pSUPER and pSUPER-GFP vectors. PACS1 shRNA#1: forward, 5′-gatccccGGACAAAGATCTTAACTCAttcaagagaTGAGTTAAGATCTTTGTCCtttttggaaa-3′, reverse, 5′-agcttttccaaaaaGGACAAAGATCTTAACTCAtctcttgaaTGAGTTAAGATCTTTGTCCggg-3′; PACS1 shRNA#2: forward 5′-gatccccGATGACAGCTTGACTGAAAttcaagagaTTTCAGTCAAGCTGTCATCtttttggaaa-3′, reverse, 5′-agcttttccaaaaaGATGACAGCTTGACTGAAAtctcttgaaTTTCAGTCAAGCTGTCATCggg-3′; PACS1 shRNA#3: forward 5′-gatccccGAAACAGACACTTTAGAAAttcaagagaTTTCTAAAGTGTCTGTTTCtttttggaaa-3′, reverse, 5′-agcttttccaaaaaGAAACAGACACTTTAGAAAtctcttgaaTTTCTAAAGTGTCTGTTTCggg-3′. GFP-Rab5 was generously provided by Dr. James Casanova (University of Virginia), and GFP-Rab7 was a gift from Dr. John Brumell (University of Toronto). LAMP1-mRFP was a gift from Dr. Qian Cai (Rutgers University). Rabbit anti-PACS1 polyclonal antibody and the HA-tagged PACS1 construct were generous gifts from Dr. Gary Thomas (University of Pittsburgh). Atypical PKC pseudosubstrate inhibitor was from Invitrogen and used at a final concentration of 2 ng/μl. Cycloheximide (Sigma) was used at 50 μM final concentration.
Primary hippocampal and cortical neurons and transfection
Hippocampal or cortical neuron cultures were prepared from embryonic day 18 rats as described previously (Bernard and Zhang, 2015; Wu et al., 2012). Hippocampal neurons were transfected using a CalPhos mammalian transfection kit (Clontech) (Sun et al., 2013) at DIV 4–5. Cortical neurons were infected with lentivirus at DIV 0.
Co-immunoprecipitation and Western Blotting
For co-immunoprecipitation experiments, HEK293 or N2a cells expressing different plasmid constructs or mouse brains were lysed on ice in buffer containing 25 mM Hepes, 150 mM NaCl, 10 mM MgCl2, 1% Nonidet P-40, and 10 mM DTT and supplemented with protease inhibitor cocktail (Sigma-Aldrich), phosphatase inhibitor cocktail (Sigma-Aldrich), 10 mM β-glycerophosphate, and 10 mM NaF. Lysates were cleared by centrifugation at 13,000 g for 10 min at 4°C. Cleared lysates were incubated with ant i-FLAG monoclonal antibody M2 (2 μg), anti-PACS1 polyclonal antibody or anti-BACE1 monoclonal antibody D10E5 (2 μg) for 1.5 h at 4°C, respectively, followed by incubation with 20 μl of Protein G sepharose 4B conjugate (Cat. No. 10-1241, ThermoFisher) preblocked with 5% BSA in lysis buffer for another 3 hours. Beads were washed three times with lysis buffer. Bound proteins were eluted with 3x Laemmli sample buffer and subjected to SDS-PAGE and Western blot analysis.
For Western blot analysis, the primary antibodies used were rabbit anti-Par3 antibody (1:5000; Cat. No. 07-330, Millipore), rabbit anti-BACE1 antibody (1:2000, D10E5, Cell Signaling), rabbit anti-phospho-BACE pSer498 antibody (1:2000, PA5-12549, ThermoFisher), rabbit anti-PACS1 antibody (1:2000, a generous gift from Dr. Gary Thomas), mouse anti-GAPDH antibody (1:8000, 6C5, Millipore), mouse anti-FLAG antibody (1:2000, M2, Sigma- Aldrich) and rabbit anti-GFP antibody (1:1000, A-11122, Life-technologies). The secondary antibodies used were Horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit antibodies (1:5000 Jackson ImmunoResearch Laboratories, West Grove, PA). Proteins were visualized by enhanced chemiluminescence and imaged using a Syngene G:BOX iChemi XR system and GeneSnap software (Version 7.09.a; Syngene USA, Frederick, MD).
Immunocytochemistry
Hippocampal neurons or N2a cells were fixed in 4% paraformaldehyde with 4% sucrose in PBS for 15 min at room temperature, then permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. Primary antibodies used include rabbit polyclonal BACE1 antibody (Laird et al., 2005; Ye and Cai, 2014) (1:100, a generous gift from Dr. Huaibin Cai), 6E10 (1:100), FLAG (mouse monoclonal, 1:500, Sigma; rabbit polyclonal, 1:100, Cell Signaling Technology), TGN (mouse monoclonal, 1:800, Thermo Scientific; rabbit polyclonal, 1:800, Abcam). Following washes with PBS, Alexa Fluor 405, 488, or 594- conjugated secondary antibodies (Invitrogen) diluted in 5% goat serum were incubated with the neurons at room temperature for 1 h. Neurons were then washed with PBS and mounted using VECTASHIELD (Vector Laboratories, Burlingame, CA). For Lysotracker staining, neurons were live labeled with 50nM Lysotracker Deep Red for 5 min and then imaged live with the 635nm laser.
Image Acquisition and Quantification
Confocal images were obtained using an Olympus FV1000 confocal microscope with a 60× water immersion objective (1.0 numerical aperture) with sequential-acquisition setting. Co-localization measurements were performed using NIH ImageJ with the Colocalization plug-in. Live imaging of hippocampal neurons were performed as previously described (Wu et al., 2017). Briefly, neurons were transferred into a 37°C heate d chamber. Cells were visualized with 488 nm excitation for eGFP and 543 nm for mRFP. Time-lapse images were collected under 5% of laser intensity with a 100 μm pinhole size. A total of 200 to 500 frames were captured. The stacks of images were imported into ImageJ. A membranous organelle was classified as immobile if it was stationary for the entire recording period; a motile one was counted only if the displacement was at least 10 μm. For tracing anterograde or retrograde movement of different vesicles in live neurons, single and separated axons were selected and kymographs were generated by ImageJ with extra plug-ins. The height of kymographs represents time (200–500 frames or 5–10 min), while the width represents the length (μm) of the axon imaged. Vesicle movement was counted and quantified by unpaired Student’s t-test.
Statistical analysis
All experiments were repeated at least three times. Unpaired Student’s t-test or ANOVA were used to calculate the p values. Error bars represent the S.E.M. of the samples.
Results
Par3 regulates BACE1 retrograde trafficking from endosomes to TGN
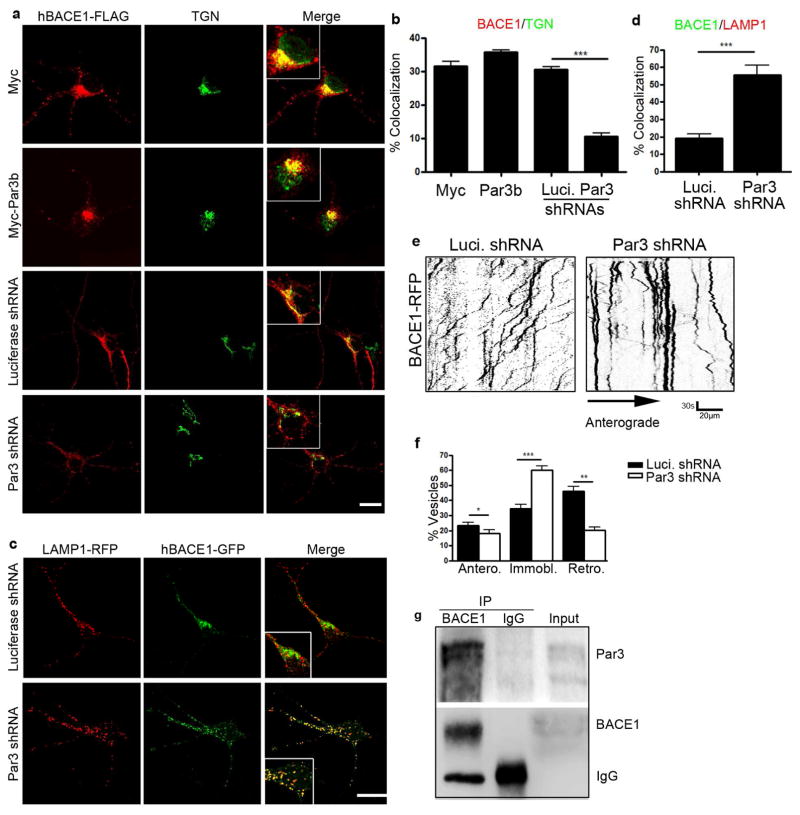
To explore a potential role for Par3 in BACE1 trafficking, we examined the subcellular distribution of BACE1 by co-immunostaining with a marker for the trans-Golgi network (TGN) (Wang et al., 2012) in hippocampal neurons expressing a control shRNA targeting luciferase or Par3 shRNA (Zhang and Macara, 2006). Consistent with previous reports (He et al., 2005; Vassar et al., 1999; Wang et al., 2012), BACE1 is normally predominantly localized to the TGN (Fig. 1a, Fig. S1 and S2). In Par3-depleted neurons, we observed a significant decrease in BACE1 co-localization with the TGN (Fig. 1 and Fig. S1 and S2). Similar results were observed in N2a cells (Fig. S3). When Par3 was overexpressed in hippocampal neurons or N2a cells, a slight increase in the TGN localization of exogenous BACE1 was observed (Fig. 1a and Fig. S3). Together, these results suggest there is a defect in the retrograde transport of BACE1 from the endosomes to the TGN in neurons depleted of Par3.
Figure 1. Par3 forms a complex with BACE1 and regulates its trafficking.
(a) Hippocampal neurons were transfected with indicated constructs. At DIV11 neurons were immunostained for FLAG (red) and TGN (green). Merge shows the overlap between BACE1 and TGN. Scale bar: 10μm. (b) Quantification of colocalization of BACE1 and TGN. (c) Hippocampal neurons were transfected with indicated constructs together with hBACE1-GFP and LAMP1-mRFP and imaged live on DIV11. Scale bar: 10μm. (d) Quantification of colocalization of hBACE1-GFP and LAMP1-mRFP. Data were expressed as Mean ± SEM with Student’s t test, *p<0.05; ** p<0.01; *** p<0.001. n=10–15. (e) Hippocampal neurons were transfected with indicated constructs together with hBACE1-RFP. Live images were captured on DIV11 and kymographs were generated to trace BACE1 trafficking in axons. Scale bar: 30s/20μm. (f) Quantification of the mobility of BACE1 containing vesicles (anterograde, immobile and retrograde). Data were expressed as Mean ± SEM with Student’s t test: *p<0.05; ** p<0.01; *** p<0.001. Data were analyzed from 10–15 neurons and 1000–2000 vesicles. (g) Endogenous BACE1 was immunoprecipitated from mouse cortex and the precipitated complex was analyzed by Western blotting for Par3 and BACE1.
To determine the identity of the BACE1 vesicles in Par3-depleted neurons, we examined the colocalization of BACE1 with different endosome markers. We found a significant increase in BACE1 colocalization with the late endosome/lysosome marker LAMP1 in Par3-depleted hippocampal neurons (Fig. 1c and d). No significant changes in colocalization with the early endosome marker Rab5 were observed (Fig. S4). To confirm that BACE1 is targeted to the late endosome/lysosome pathway, we treated N2a cells with cycloheximide (CHX) to inhibit protein synthesis. BACE1 turnover was significantly enhanced in the absence of Par3 (Fig. S5), which further shows that it is targeted to the lysosome pathway, since BACE1 is mainly degraded by the lysosomes (Koh et al., 2005). Together, these results suggest that depletion of Par3 targets BACE1 to late endosomes/lysosomes and inhibits BACE1 retrograde trafficking to the TGN.
We next aimed to examine BACE1 trafficking in live neurons. In neuronal processes, endosomes generated in the periphery are retrogradely transported back to the cell body where they may be targeted for degradation in mature lysosomes. Alternatively, cargos are diverted to TGN for signaling and/or recycling back to the plasma membrane (Lu and Hong, 2014; Yap and Winckler, 2012). The retromer complex, which is a key player in endosome-to-TGN trafficking, associates with their cargos in the periphery and regulates the long-range axonal transport of endocytic vesicles (Bhalla et al., 2012). To examine BACE1 trafficking in axons of live neurons, we performed time-lapse imaging in hippocampal neurons expressing BACE1-RFP. As seen in Figure 1e and f, axonal retrograde trafficking of BACE1 was significantly reduced in Par3-depleted neurons as compared with control neurons. This was accompanied by a concomitant increase in stationary vesicles. A slight but statistically significant decrease in anterogradely trafficking BACE1 vesicles was also observed (Fig. 1f), which is consistent with data showing that endocytosed BACE1 undergo bidirectional axonal trafficking (Buggia-Prevot et al., 2013).
The Par3/aPKC complex promotes phosphorylation of BACE1 on Ser498
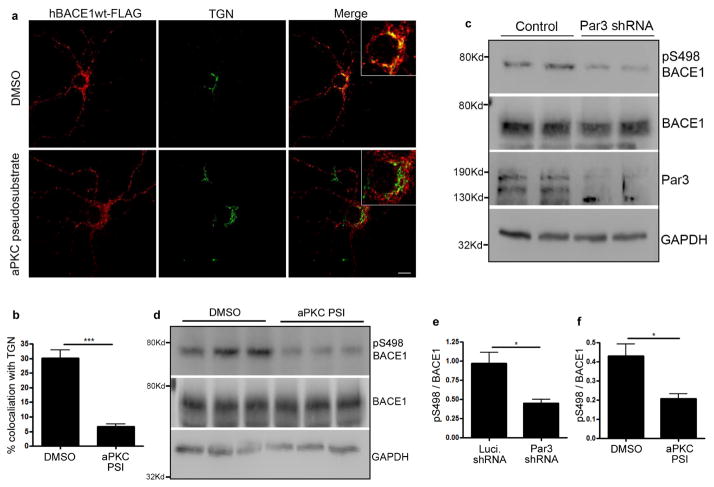
Next, we aimed to understand the mechanism by which Par3 regulates BACE1 trafficking. First, we performed co-immunoprecipitation between endogenous Par3 and BACE1 from mouse brain lysates. Par3 was found in the immunoprecipitates of BACE1, suggesting that Par3 and BACE1 form an endogenous complex (Fig. 1g). Since Par3 often functions to recruit substrates for aPKC (Goldstein and Macara, 2007), we wondered whether aPKC could be functioning downstream. To test this idea, we treated hippocampal neurons with an aPKC pseudosubstrate inhibitor (PSI) (Hao et al., 2010). The inhibitor caused BACE1 to redistribute away from the TGN similar to Par3-depleted neurons (Fig. 2a and b), indicating that aPKC is involved in this process.
Figure 2. The Par3/aPKC complex promotes BACE1 phosphorylation on Ser498.
(a) Hippocampal neurons were transfected with BACE1-FLAG and immunostained for FLAG (red) and TGN (green). Scale bar: 10 μm. (b) Quantification of colocalization of BACE1-FLAG and TGN. Data were expressed as Mean ± SEM with Student’s t test, ***p<0.001, n=15. (c) Primary hippocampal neurons were infected with Luciferase shRNA or Par3 shRNA expressing lentivirus at DIV 0. On DIV 7 neurons were lysed and analyzed by Western blotting. (d) Primary hippocampal neurons were treated with aPKC pseudosubstrate inhibitor (PSI) (2μg/ml) at DIV 10 and lysed on DIV 13. Lysates were analyzed by Western blotting to detect total BACE1 and phospho-Ser498 BACE1 levels. (e) Quantification of pSer498 normalized to total BACE1 from blots in (c). (f) Quantification of pSer498 normalized to total BACE1 from blots in (d). Data were expressed as Mean ± SEM with Student’s t test, *p<0.05; n=6.
We then wanted to determine the mechanism by which aPKC is involved in BACE1 trafficking. Since aPKC is a Ser/Thr kinase, we wondered whether aPKC might promote phosphorylation of BACE1. BACE1 is a single-transmembrane protein with a short cytoplasmic tail. There is a single serine site (Ser498) on the cytoplasmic tail, which is known to be phosphorylated (Walter et al., 2001). However it remains unclear how this phosphorylation is regulated in physiological and pathological processes. To see whether the Par3/aPKC complex regulates BACE1 phosphorylation on Ser498, we first examined Ser498 phosphorylation in hippocampal neurons depleted of Par3, using a phospho-specific antibody against the Ser498 site. The specificity of this antibody was tested using the S498A mutant (Fig. S6). We observed a significant decrease in p-Ser498 in hippocampal neurons infected with lentivirus expressing Par3 shRNA (Fig. 2c and e). To see whether aPKC is involved, we treated hippocampal neurons with the aPKC PSI. Neurons treated with the aPKC PSI show a significant decrease in p-Ser498 as compared with vehicle controls (Fig. 2d and f). Total BACE1 showed a slight and statistically insignificant decrease in Par3 shRNA expressing or aPKC PSI treated cells (Fig. S7). Taken together, these results show that the Par3/aPKC complex functions to promote Ser498 phosphorylation on BACE1.
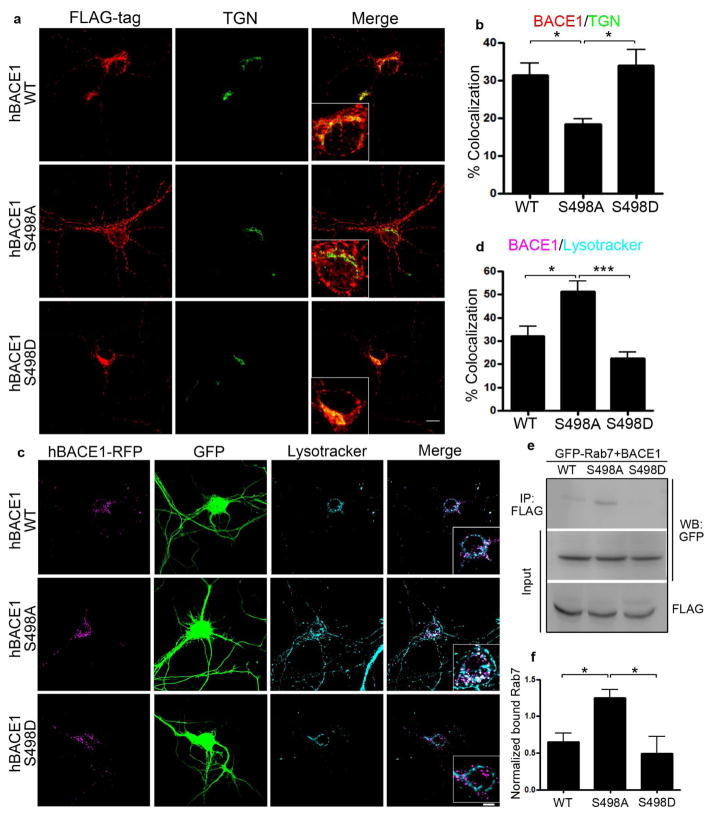
To determine whether Ser498 phosphorylation of BACE1 is downstream of Par3/aPKC in regulating BACE1 trafficking, we first expressed BACE1 Ser498 mutants in hippocampal neurons. Consistent with previous studies in non-neuronal cells (He et al., 2005; Wahle et al., 2005; Walter et al., 2001), BACE1 S498A showed a redistribution away from the TGN, while the S498D mutant predominantly localized to the TGN in hippocampal neurons (Fig. 3a and b). We next examined the colocalization of BACE1 mutants with organelle markers using live staining. The S498A mutant showed significantly increased colocalization with Lysotracker (Fig. 3c and d), suggesting that de-phosphorylation of the Ser498 site increases BACE1 targeting to the late endosome/lysosome pathway. To further confirm this, we performed co-immunoprecipitation between BACE1 mutants and Rab7. BACE1 S498A mutant showed significantly increased interaction with Rab7 (Fig. 3e and f), further showing that the S498A mutation increases BACE1 targeting to the late endosome/lysosome pathway.
Figure 3. Ser498 phosphorylation regulates BACE1 retrograde trafficking.
(a) Hippocampal neurons were transfected with indicated BACE1 constructs and immunostained for FLAG (red) and TGN (green). Scale bar: 10 μm. (b) Quantification of the colocalization of BACE1 and TGN. (c) Hippocampal neurons were transfected with indicated BACE1 constructs (magenta) and live stained with Lysotracker deep red (cyan) for 5 min. Merge shows the overlap between BACE1 and Lysotracker. Scale bar: 10 μm. (d) Quantification of the colocalization of BACE1 and Lysotracker. *p<0.05, ***p<0.001, n=10–15. (e) N2a cells were transfected with indicated BACE1 mutation constructs and GFP-Rab7. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoprecipitated complexes were analyzed by Western blot. (f) Quantification of blots in (e), normalized to GFP-Rab7 input, *p<0.05, n=3.
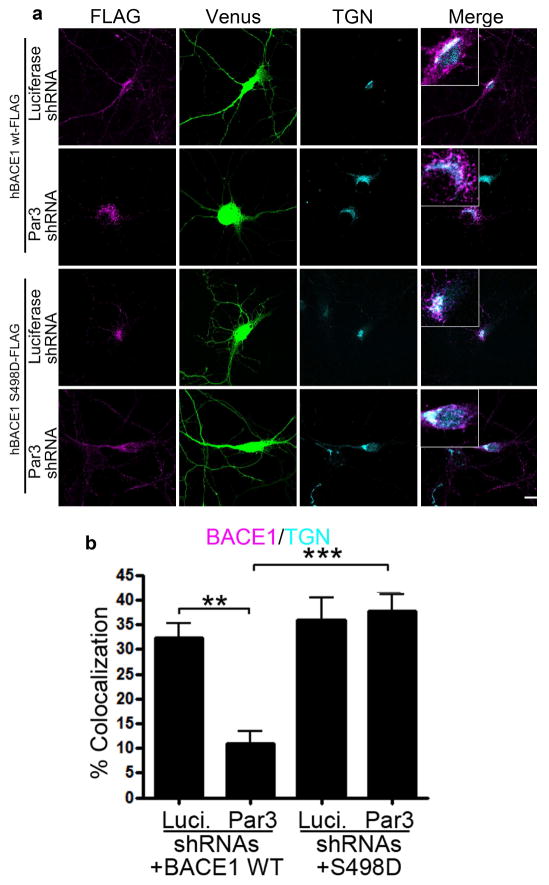
To see if Ser498 phosphorylation is downstream of Par3/aPKC in regulating BACE1 trafficking, we co-expressed different BACE1 mutants in hippocampal neurons depleted of Par3. BACE1 S498D was able to restore the TGN localization in Par3-depleted neurons (Fig. 4). Together, these results suggest that Ser498 phosphorylation of BACE1 is downstream of Par3/aPKC in regulating BACE1 trafficking.
Figure 4. Ser498 phosphorylation is downstream of Par3 in regulating BACE1 trafficking.
(a) Hippocampal neurons were transfected with indicated constructs. At DIV11, neurons were immunostained for FLAG (magenta) and TGN (cyan). Venus (green) indicates positive neurons. (b) Quantification of BACE1-FLAG and TGN colocalization. Data were expressed as Mean ± SEM with Student’s t test: ** p<0.01; *** p<0.001, n=10–15. Scale bar: 10μm.
Ser498 phosphorylation regulates BACE1 trafficking through modulating its interaction with PACS1
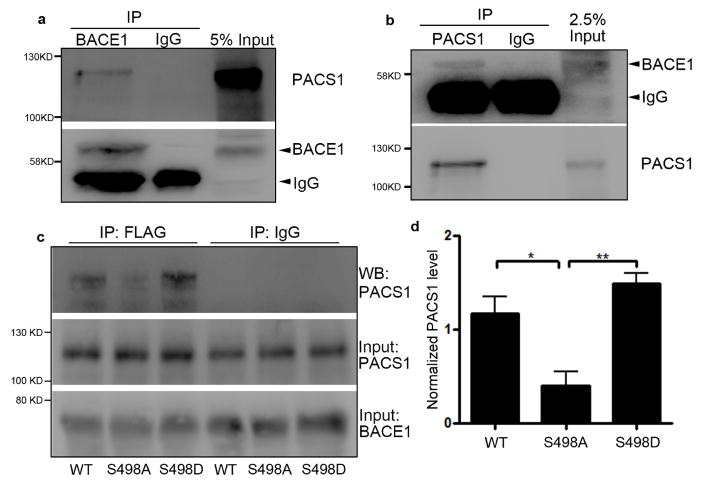
We next aimed to elucidate the mechanism by which Par3/aPKC-mediated phosphorylation of BACE1 on Ser498 regulates its trafficking. The Ser498 site lies in an acidic cluster dileucine motif in its cytoplasmic tail (He et al., 2002; He et al., 2003; Huse et al., 2000). One protein that binds to acidic cluster motifs and directs retrograde endosome-to-TGN trafficking is the phosphofurin acidic cluster sorting protein 1 (PACS1) (Wan et al., 1998). Thus, we tested the possibility that PACS1 is involved in the Par3/aPKC mediated retrograde trafficking of BACE1. First, we performed co-immunoprecipitation of endogenous BACE1 and PACS1 from mouse brain lysates. We were able to detect PACS1 in the immunoprecipitates of BACE1 (Fig. 5a), and vice versa (Fig. 5b). This suggests that BACE1 and PACS1 form an endogenous complex.
Figure 5. Ser498 phosphorylation of BACE1 promotes its interaction with PACS1.
(a) Endogenous BACE1 was immunoprecipitated from mouse cortex and the precipitated complex was analyzed by Western blotting for PACS1 and BACE1. (b) Endogenous PACS1 was immunoprecipitated from mouse cortex and the precipitated complex was analyzed by Western blotting for BACE1 and PACS1. (c) HEK293 cells were transfected with FLAG-tagged BACE1 constructs and PACS1-HA, lysed and immunoprecipitation was performed by anti-FLAG antibody. Precipitated complex was analyzed by Western blotting with the indicated antibodies. (d) Quantification of co-precipitated PACS1 normalized to input from blots in (c). Data were expressed as Mean ± SEM with Student’s t test: *p<0.05,** p<0.01, n=3.
To determine whether Ser498 regulates the interaction between BACE1 and PACS1, we co-expressed HA-tagged PACS1 with FLAG-tagged BACE1 Ser498 mutants in HEK293 cells and performed immunoprecipitation using FLAG antibody. The amount of PACS1 co-precipitated was detected by Western blotting. The S498A mutation significantly reduced the amount of PACS1 pulled down by BACE1, as compared with the WT and S498D BACE1 (Fig. 5c and d). This suggests that Ser498 phosphorylation promotes the interaction between BACE1 and PACS1.
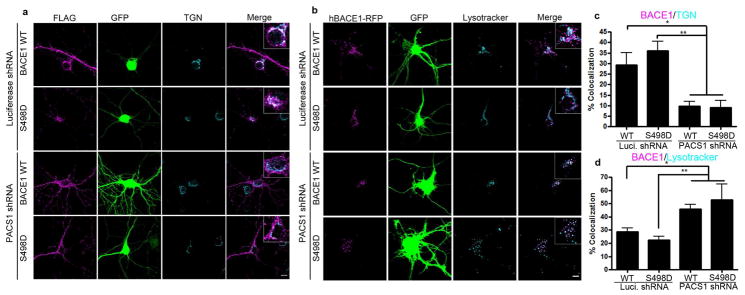
Finally, we examined whether PACS1 is necessary for the Ser498 phosphorylation-mediated retrograde trafficking of BACE1 to the TGN. We used short hairpin RNAs (shRNAs) to deplete endogenous PACS1 in hippocampal neurons. The efficiency of the PACS1 shRNAs were verified in both Rat2 cells and N2a cells (Fig. S8). Depletion of PACS1 caused wild type BACE1 to redistribute away from the TGN (Fig. 6a and c) and to the late endosomes/lysosomes (Fig. 6b and d). Similarly, the phosphomimetic S498D mutant also showed decreased localization to the TGN and increased targeting to the late endosomes/lysosomes when PACS1 was depleted (Fig. 6), suggesting that PACS1 is necessary for Ser498 phosphorylation-mediated retrograde trafficking of BACE1 to the TGN. Taken together, these results suggest that Ser498 phosphorylation-mediated binding of BACE1 to PACS1 is necessary for the retrograde trafficking of BACE1 from the endosomes to the TGN.
Figure 6. Depletion of PACS1 causes a redistribution of BACE1 constructs to the late endosomes/lysosomes.
(a) Hippocampal neurons were transfected with indicated constructs. At DIV10, neurons were immunostained for FLAG (magenta) and TGN (cyan). Merge shows the overlap between BACE1 and TGN. Venus (green) indicates positive neurons. (b) Hippocampal neurons were transfected with indicated BACE1 constructs (magenta) and live stained with Lysotracker Deep Red (cyan) for 5 min. Merge shows the overlap between BACE1 and Lysotracker. Venus (green) indicates positive neurons. (c) Quantification of BACE1-FLAG and TGN colocalization. (d) Quantification of BACE1-RFP and Lysotracker Deep Red colocalization. Data were expressed as Mean ± SEM with Student’s t test: *p<0.05; ** p<0.01; n=10–15. Scale bar: 10μm.
Ser498 phosphorylation of BACE1 is reduced in human AD brains
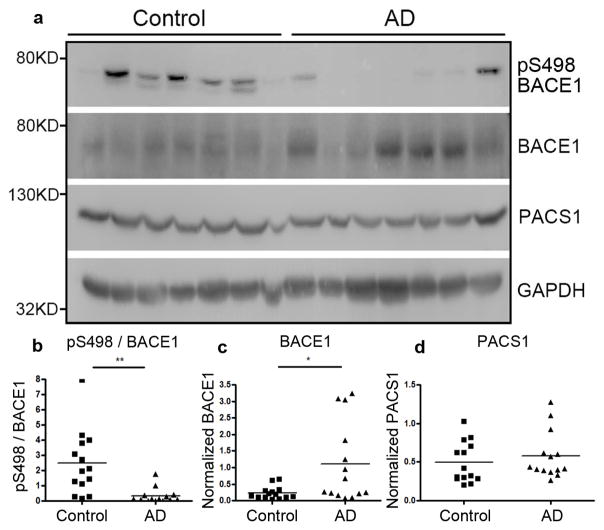
We wanted to determine whether the Par3/aPKC mediated BACE1 phosphorylation at Ser498 is important for AD progression in vivo. Our recent studies found that there is a significant decrease in Par3 protein levels in the temporal cortex of AD patients (Sun et al., 2016). If abnormalities in Par3/aPKC regulated Ser498 phosphorylation are important for AD progression, one would predict to see a decrease in Ser498 phosphorylation in AD patients. Thus, we performed Western blot on the temporal cortex of AD patients and healthy age matched control subjects. In support of our hypothesis, there was a drastic decrease in Ser498 phosphorylation of BACE1 in AD subjects (Fig. 7a and b). In addition, we observed a significant increase in BACE1 expression in AD subjects as compared with healthy controls (Fig. 7a and c), which is consistent with previous studies showing elevated levels of BACE1 mRNA (Coulson et al., 2010) and protein (Fukumoto et al., 2002; Harada et al., 2006; Holsinger et al., 2002; Li et al., 2004; Tyler et al., 2002; Yang et al., 2003) in the AD brain. Total PACS1 levels were not significantly changed (Fig. 7a and d). Together, these results suggest that decreased Ser498 phosphorylation of BACE1 is involved in AD progression.
Figure 7. Ser498 phosphorylation of BACE1 is reduced in Alzheimer’s brains.
(a) Lysates from the temporal lobes of AD patients or healthy age-matched controls were analyzed by Western blotting. (b) Quantification of Ser498 phoshphorylation normalized to total BACE1. (c) Quantification of total BACE1 levels normalized to GAPDH. (d) Quantification of PACS1 levels normalized to GAPDH, n=14, *p<0.05; ** p<0.01 by Student’s t-test.
Discussion
During AD progression, the amount of Aβ generation depends on the subcellular trafficking of BACE1 and its convergence with APP in different vesicular compartments. In this study, we show that Par3 and aPKC are novel regulators of BACE1 trafficking in hippocampal neurons. We found that Par3 forms a complex with BACE1, and the Par3/aPKC complex functions to promote BACE1 phosphorylation at Ser498, which increases its interaction with PACS1 and facilitates BACE1 retrograde trafficking to the TGN (Fig. S9). In the absence of Par3 or when aPKC activity is inhibited, we observed a reduction in Ser498 phosphorylation and defective retrograde trafficking of BACE1 from the endosomes to the TGN.
Defective endosome-to-TGN retrograde trafficking is characteristic of the AD brain. Anomalies in a number of proteins involved in this retrograde trafficking pathway have been observed in AD brains. For example, components of the retromer complex, Vps26 and Vps35, are significantly downregulated in the AD brain (Small et al., 2005). SorLA, a Vps10 family sorting receptor that facilitates endosome-to-TGN trafficking, is also downregulated in the AD (Dodson et al., 2006; Scherzer et al., 2004), and population-based studies have confirmed the association of gene variants of SorLA and sporadic AD (Lee et al., 2007; Rogaeva et al., 2007). Defects in the retromer complex or SorLA increase Aβ generation in cellular and mouse models of AD (Herskowitz et al., 2012; Wen et al., 2011), and both Vps26 and Vps35 are known to be involved in retrograde BACE1 trafficking (He et al., 2005; Wang et al., 2012). Our results show that loss of Par3 or inhibition of aPKC blocks retrograde endosome-to-TGN trafficking of BACE1, thereby increasing the amount of BACE1 in the endocytic compartments. This will increase the chance for BACE1 to converge with APP and facilitates Aβ generation, as Aβ has been reported to be generated in early endosomes (Kinoshita et al., 2003; Rajendran et al., 2006; Udayar et al., 2013), recycling endosomes (Das et al., 2013; Das et al., 2016), as well as late endosomes/multi-vesicular bodies and lysosomes (Edgar et al., 2015; Sannerud et al., 2016; Tam et al., 2014; Tang et al., 2015). Lysosomal defects that are wide-spread in the AD brain likely exacerbate the BACE1 and Aβ accumulations in these degradative organelles (Gowrishankar et al., 2015) or in late endocytic organelles destined for fusion with lysosomes (Takahashi et al., 2002; Ye and Cai, 2014). Intriguingly, Aβ40 is also generated in the TGN after retrograde trafficking from the endosomes in non-neuronal cells; however, the convergence with BACE1 is believed to occur in the endosomes (Choy et al., 2012). It will be important to better understand the physiologically relevant intracellular sites of APP and BACE1 convergence and Aβ generation, which will help elucidate the mechanisms of AD progression.
Significant evidence points to a role for BACE1 axonal trafficking in Aβ generation. Previous studies have shown that Aβ is released near presynaptic sites (Buxbaum et al., 1998; Lazarov et al., 2002; Sheng et al., 2002; Sokolow et al., 2012). Presynaptic Aβ is likely generated through BACE1 processing of APP in the axon, as abnormal BACE1 accumulation is found in the dystrophic axons near presynaptic terminals in mouse and human AD brains (Kandalepas et al., 2013; Zhang et al., 2009). Recent studies show that defective axonal retrograde trafficking of BACE1 causes it to accumulate in late endocytic vesicles and increases Aβ generation at presynaptic terminals (Feng et al., 2017; Ye et al., 2017). Our data show that loss of Par3 disrupts BACE1 axonal retrograde trafficking and causes an increase in stalled BACE1-containing vesicles in the axon (Fig. 1). It is plausible that this disrupted BACE1 axonal trafficking will cause an increase in presynaptic Aβ generation in Par3-depleted neurons.
Interestingly, our recent studies show that Par3 is decreased in AD patient brains. We found that Par3 regulates APP trafficking and intracellular Aβ generation in neurons (Sun et al., 2016). Here we found Par3 is also involved in the trafficking of BACE1; however, Par3 showed distinct effects on APP and BACE1. While Par3 regulates APP trafficking through the endocytic adaptor Numb (Sun et al., 2016), the effects of Par3 on BACE1 is through PACS1. This suggests that the observed phenotype of Par3 depletion is not due to a global disruption of vesicular trafficking. The distinct effects of Par3 on APP versus BACE1 trafficking are consistent with previous studies showing APP is internalized through the clathrin-dependent pathway while BACE1 is internalized through a clathrin-independent pathway (Sannerud et al., 2011).
We found that Par3/aPKC-dependent phosphorylation of BACE1 on Ser498 is important for its retrieval from the endosomes to the TGN (Fig. 4), which is consistent with previous reports on the role of Ser498 phosphorylation in BACE1 trafficking (He et al., 2005; Wahle et al., 2005; Walter et al., 2001). Interestingly, while previous studies in HEK293 cells and HeLa cells found that the non-phosphorylatable S498A mutant is retained in early endosomes (He et al., 2005; Wahle et al., 2005; Walter et al., 2001), we found that in hippocampal neurons, S498A is largely targeted to the late endosome/lysosome pathway. The differences in cell types and/or lysosome turnover rate likely account for the differential targeting of the S498A mutant. We further show that PACS1 binds BACE1 in a phosphorylation-dependent manner and promotes BACE1 retrograde trafficking (Fig. 6). It remains to be determined whether PACS1 functions coordinately with other proteins known to regulate the endosome-to-TGN trafficking of BACE1, such as Golgi-localized γ-ear-containing ARF-binding (GGA) proteins (He et al., 2005; Scott et al., 2006; Tesco et al., 2007; Wahle et al., 2005; Walker et al., 2012), the retromer complex (Wang et al., 2012), Sortilin and SorLA (Finan et al., 2011; Spoelgen et al., 2006), and/or proteins that are known to regulate the axonal retrograde trafficking of BACE1, such as the dynein adaptor Snapin (Feng et al., 2017; Ye and Cai, 2014; Ye et al., 2017).
BACE1 is post-translationally modified through phosphorylation, acetylation, S-palmitoylation and glycosylation on both the intracellular and extracellular domains (Kandalepas and Vassar, 2012). Here we show that phosphorylation at Ser498 is significantly reduced in the human AD brain as compared with healthy age-matched controls, suggesting that reduced phosphorylation of Ser498 contributes to AD progression. Interestingly, phosphorylation of Thr252 in the extracellular domain of BACE1 by Cdk5 increases its activity, and levels of Thr252 phosphorylation is increased in the AD patients (Song et al., 2015). Other post-translational modifications, such as glycosylation and acetylation, also affect BACE1 trafficking, activity and/or expression levels (Kizuka et al., 2015; Kizuka et al., 2016; Ko and Puglielli, 2009). A comprehensive understanding of abnormal post-translational modifications of BACE1 occurring in the AD brain will facilitate the development of therapies aimed at targeting BACE1 activity in AD patients.
BACE1 has primarily been examined in the context of neurodegeneration and AD. Here we show an unexpected mechanistic link between BACE1 and a polarity protein complex essential for all stages of animal development. This raises the possibility that BACE1 also plays important physiological roles during development. Indeed, studies have shown physiological roles for BACE1 during brain development (Cole and Vassar, 2008; Muller and Zheng, 2012; Soldano and Hassan, 2014), as well as myelination of the central and peripheral nervous system (Hu et al., 2006; Willem et al., 2006); however, the underlying molecular mechanisms still remain unclear. It will be interesting to see whether their interactions with the Par polarity complex are important for normal nervous system development.
Conclusions
In summary, our studies reveal that the Par complex, a key player in cell polarization during development, plays an important role in BACE1 trafficking. Par3 functions through directing distinct trafficking pathways for APP (Sun et al., 2016) and BACE1. Par3 and its binding partner aPKC function by promoting the phosphorylation of BACE1 at Ser498. Phosphorylation at this site is necessary for the retrograde trafficking of BACE1 to the TGN through its interaction with PACS1. During AD pathogenesis, Par3 protein level is decreased in the brain (Sun et al., 2016), along with a decrease in BACE1 Ser498 phosphorylation (Fig. 7). This leads to defective endosome-to-TGN retrograde trafficking of BACE1 and an increase in intracellular Aβ generation (Sun et al., 2016). Together, our results reveal an unexpected link between the Par complex and the retrograde trafficking of BACE1 and shed light on the mechanisms of Alzheimer’s disease pathogenesis.
Supplementary Material
Highlights.
Par3 and aPKC regulate BACE1 endosome-to-TGN trafficking.
Par3/aPKC complex promotes Ser498 phosphorylation of BACE1.
Ser498 phosphorylation facilitates BACE1 interaction with PACS1.
PACS1 is necessary for the endosome-to-TGN trafficking of BACE1.
Ser498 phosphorylation of BACE1 is decreased in the Alzheimer’s brain.
Acknowledgments
We would like to thank Dr. Bradley T. Hyman (Harvard Medical School) for the human BACE1-EGFP plasmid, Dr. Qian Cai (Rutgers University) for LAMP1-mRFP, Drs. John Brumell (University of Toronto), Ira Mellman (Genentech), James Casanova, Chan Choo Yap and Bettina Winckler (University of Virginia) for different Rab plasmids, Dr. Huaibin Cai (NIH) for the BACE1 antibody, and Dr. Gary Thomas (University of Pittsburgh) for the PACS1-HA construct and PACS1 antibody. We would like to thank Laura Bernard for technical assistance in the preparation of primary neuronal cultures. We would also like to thank the Harvard Brain Tissue Resource Center, the NIH NeuroBioBank and the Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona for human brain samples, and members of the Zhang laboratory for critical reading of the manuscript. This work was supported by National Institutes of Health grants NS065183, NS089578 and startup funds from Rutgers Robert Wood Johnson Medical School. The Banner Sun Health Research Institute Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research.
Abbreviations list
- AD
Alzheimer’s disease
- aPKC
atypical protein kinase C
- APP
amyloid precursor protein
- BACE1
β-APP cleaving enzyme 1
- DIV
days in vitro
- PACS1
phosphofurin acidic cluster sorting protein 1
- Par3
partitioning defective 3
- TGN
trans Golgi network
Footnotes
Competing interests
The authors declare they have no competing financial interests.
Authors’ contributions
MS and HZ designed the experiments. MS performed the experiments. MS and HZ analyzed the data. MS and HZ wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed SM, I, Macara G. The Par3 polarity protein is an exocyst receptor essential for mammary cell survival. Nat Commun. 2017;8:14867. doi: 10.1038/ncomms14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol. 2007;9:1066–1073. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- Bernard LP, Zhang H. MARK/Par1 Kinase Is Activated Downstream of NMDA Receptors through a PKA-Dependent Mechanism. PLoS One. 2015;10:e0124816. doi: 10.1371/journal.pone.0124816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla A, Vetanovetz CP, Morel E, Chamoun Z, Di Paolo G, Small SA. The location and trafficking routes of the neuronal retromer and its role in amyloid precursor protein transport. Neurobiol Dis. 2012;47:126–134. doi: 10.1016/j.nbd.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggia-Prevot V, Fernandez CG, Udayar V, Vetrivel KS, Elie A, Roseman J, Sasse VA, Lefkow M, Meckler X, Bhattacharyya S, George M, Kar S, Bindokas VP, Parent AT, Rajendran L, Band H, Vassar R, Thinakaran G. A function for EHD family proteins in unidirectional retrograde dendritic transport of BACE1 and Alzheimer’s disease Abeta production. Cell Rep. 2013;5:1552–1563. doi: 10.1016/j.celrep.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggia-Prevot V, Thinakaran G. Sorting the role of SORLA in Alzheimer’s disease. Sci Transl Med. 2014;6:223fs228. doi: 10.1126/scitranslmed.3008562. [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Thinakaran G, Koliatsos V, O’Callahan J, Slunt HH, Price DL, Sisodia SS. Alzheimer amyloid protein precursor in the rat hippocampus: transport and processing through the perforant path. J Neurosci. 1998;18:9629–9637. doi: 10.1523/JNEUROSCI.18-23-09629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Petanceska S, Terio NB, Peterhoff CM, Durham R, Mercken M, Mehta PD, Buxbaum J, Haroutunian V, Nixon RA. Abeta localization in abnormal endosomes: association with earliest Abeta elevations in AD and Down syndrome. Neurobiol Aging. 2004;25:1263–1272. doi: 10.1016/j.neurobiolaging.2004.02.027. [DOI] [PubMed] [Google Scholar]
- Chen X, I, Macara G. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- Choy RW, Cheng Z, Schekman R. Amyloid precursor protein (APP) traffics from the cell surface via endosomes for amyloid beta (Abeta) production in the trans-Golgi network. Proc Natl Acad Sci U S A. 2012;109:E2077–2082. doi: 10.1073/pnas.1208635109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SL, Vassar R. The Alzheimer’s disease beta-secretase enzyme, BACE1. Mol Neurodegener. 2007;2:22. doi: 10.1186/1750-1326-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SL, Vassar R. BACE1 structure and function in health and Alzheimer’s disease. Curr Alzheimer Res. 2008;5:100–120. doi: 10.2174/156720508783954758. [DOI] [PubMed] [Google Scholar]
- Coulson DT, Beyer N, Quinn JG, Brockbank S, Hellemans J, Irvine GB, Ravid R, Johnston JA. BACE1 mRNA expression in Alzheimer’s disease postmortem brain tissue. J Alzheimers Dis. 2010;22:1111–1122. doi: 10.3233/JAD-2010-101254. [DOI] [PubMed] [Google Scholar]
- Das A, Gajendra S, Falenta K, Oudin MJ, Peschard P, Feng S, Wu B, Marshall CJ, Doherty P, Guo W, Lalli G. RalA promotes a direct exocyst-Par6 interaction to regulate polarity in neuronal development. J Cell Sci. 2014;127:686–699. doi: 10.1242/jcs.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U, Scott DA, Ganguly A, Koo EH, Tang Y, Roy S. Activity-induced convergence of APP and BACE-1 in acidic microdomains via an endocytosis-dependent pathway. Neuron. 2013;79:447–460. doi: 10.1016/j.neuron.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U, Wang L, Ganguly A, Saikia JM, Wagner SL, Koo EH, Roy S. Visualizing APP and BACE-1 approximation in neurons yields insight into the amyloidogenic pathway. Nat Neurosci. 2016;19:55–64. doi: 10.1038/nn.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson SE, Gearing M, Lippa CF, Montine TJ, Levey AI, Lah JJ. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:866–872. doi: 10.1097/01.jnen.0000228205.19915.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar JR, Willen K, Gouras GK, Futter CE. ESCRTs regulate amyloid precursor protein sorting in multivesicular bodies and intracellular amyloid-beta accumulation. J Cell Sci. 2015;128:2520–2528. doi: 10.1242/jcs.170233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng T, Tammineni P, Agrawal C, Jeong YY, Cai Q. Autophagy-mediated Regulation of BACE1 Protein Trafficking and Degradation. J Biol Chem. 2017;292:1679–1690. doi: 10.1074/jbc.M116.766584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan GM, Okada H, Kim TW. BACE1 retrograde trafficking is uniquely regulated by the cytoplasmic domain of sortilin. J Biol Chem. 2011;286:12602–12616. doi: 10.1074/jbc.M110.170217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto H, Cheung BS, Hyman BT, Irizarry MC. Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch Neurol. 2002;59:1381–1389. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Mufson EJ, Counts SE, Wuu J, Alldred MJ, Nixon RA, Che S. Regional selectivity of rab5 and rab7 protein upregulation in mild cognitive impairment and Alzheimer’s disease. J Alzheimers Dis. 2010;22:631–639. doi: 10.3233/JAD-2010-101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B, I, Macara G. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar S, Yuan P, Wu Y, Schrag M, Paradise S, Grutzendler J, De Camilli P, Ferguson SM. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc Natl Acad Sci U S A. 2015;112:E3699–3708. doi: 10.1073/pnas.1510329112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Du Q, Chen X, Zheng Z, Balsbaugh JL, Maitra S, Shabanowitz J, Hunt DF, Macara IG. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical Pins. Curr Biol. 2010;20:1809–1818. doi: 10.1016/j.cub.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Tamaoka A, Ishii K, Shoji S, Kametaka S, Kametani F, Saito Y, Murayama S. Beta-site APP cleaving enzyme 1 (BACE1) is increased in remaining neurons in Alzheimer’s disease brains. Neurosci Res. 2006;54:24–29. doi: 10.1016/j.neures.2005.10.001. [DOI] [PubMed] [Google Scholar]
- He X, Chang WP, Koelsch G, Tang J. Memapsin 2 (beta-secretase) cytosolic domain binds to the VHS domains of GGA1 and GGA2: implications on the endocytosis mechanism of memapsin 2. FEBS Lett. 2002;524:183–187. doi: 10.1016/s0014-5793(02)03052-1. [DOI] [PubMed] [Google Scholar]
- He X, Li F, Chang WP, Tang J. GGA proteins mediate the recycling pathway of memapsin 2 (BACE) J Biol Chem. 2005;280:11696–11703. doi: 10.1074/jbc.M411296200. [DOI] [PubMed] [Google Scholar]
- He X, Zhu G, Koelsch G, Rodgers KK, Zhang XC, Tang J. Biochemical and structural characterization of the interaction of memapsin 2 (beta-secretase) cytosolic domain with the VHS domain of GGA proteins. Biochemistry. 2003;42:12174–12180. doi: 10.1021/bi035199h. [DOI] [PubMed] [Google Scholar]
- Herskowitz JH, Offe K, Deshpande A, Kahn RA, Levey AI, Lah JJ. GGA1-mediated endocytic traffic of LR11/SorLA alters APP intracellular distribution and amyloid-beta production. Mol Biol Cell. 2012;23:2645–2657. doi: 10.1091/mbc.E12-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger RM, McLean CA, Beyreuther K, Masters CL, Evin G. Increased expression of the amyloid precursor beta-secretase in Alzheimer’s disease. Ann Neurol. 2002;51:783–786. doi: 10.1002/ana.10208. [DOI] [PubMed] [Google Scholar]
- Hu X, Hicks CW, He W, Wong P, Macklin WB, Trapp BD, Yan R. Bace1 modulates myelination in the central and peripheral nervous system. Nat Neurosci. 2006;9:1520–1525. doi: 10.1038/nn1797. [DOI] [PubMed] [Google Scholar]
- Huse JT, Pijak DS, Leslie GJ, Lee VM, Doms RW. Maturation and endosomal targeting of beta-site amyloid precursor protein-cleaving enzyme. The Alzheimer’s disease beta-secretase. J Biol Chem. 2000;275:33729–33737. doi: 10.1074/jbc.M004175200. [DOI] [PubMed] [Google Scholar]
- Kandalepas PC, Sadleir KR, Eimer WA, Zhao J, Nicholson DA, Vassar R. The Alzheimer’s beta-secretase BACE1 localizes to normal presynaptic terminals and to dystrophic presynaptic terminals surrounding amyloid plaques. Acta Neuropathol. 2013;126:329–352. doi: 10.1007/s00401-013-1152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandalepas PC, Vassar R. Identification and biology of beta-secretase. J Neurochem. 2012;120(Suppl 1):55–61. doi: 10.1111/j.1471-4159.2011.07512.x. [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Fukumoto H, Shah T, Whelan CM, Irizarry MC, Hyman BT. Demonstration by FRET of BACE interaction with the amyloid precursor protein at the cell surface and in early endosomes. J Cell Sci. 2003;116:3339–3346. doi: 10.1242/jcs.00643. [DOI] [PubMed] [Google Scholar]
- Kizuka Y, Kitazume S, Fujinawa R, Saito T, Iwata N, Saido TC, Nakano M, Yamaguchi Y, Hashimoto Y, Staufenbiel M, Hatsuta H, Murayama S, Manya H, Endo T, Taniguchi N. An aberrant sugar modification of BACE1 blocks its lysosomal targeting in Alzheimer’s disease. EMBO Mol Med. 2015;7:175–189. doi: 10.15252/emmm.201404438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizuka Y, Nakano M, Kitazume S, Saito T, Saido TC, Taniguchi N. Bisecting GlcNAc modification stabilizes BACE1 protein under oxidative stress conditions. Biochem J. 2016;473:21–30. doi: 10.1042/BJ20150607. [DOI] [PubMed] [Google Scholar]
- Ko MH, Puglielli L. Two endoplasmic reticulum (ER)/ER Golgi intermediate compartment-based lysine acetyltransferases post-translationally regulate BACE1 levels. J Biol Chem. 2009;284:2482–2492. doi: 10.1074/jbc.M804901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh YH, von Arnim CA, Hyman BT, Tanzi RE, Tesco G. BACE is degraded via the lysosomal pathway. J Biol Chem. 2005;280:32499–32504. doi: 10.1074/jbc.M506199200. [DOI] [PubMed] [Google Scholar]
- Laird FM, Cai H, Savonenko AV, Farah MH, He K, Melnikova T, Wen H, Chiang HC, Xu G, Koliatsos VE, Borchelt DR, Price DL, Lee HK, Wong PC. BACE1, a major determinant of selective vulnerability of the brain to amyloid-beta amyloidogenesis, is essential for cognitive, emotional, and synaptic functions. J Neurosci. 2005;25:11693–11709. doi: 10.1523/JNEUROSCI.2766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli G. RalA and the exocyst complex influence neuronal polarity through PAR-3 and aPKC. J Cell Sci. 2009;122:1499–1506. doi: 10.1242/jcs.044339. [DOI] [PubMed] [Google Scholar]
- Lazarov O, Lee M, Peterson DA, Sisodia SS. Evidence that synaptically released beta-amyloid accumulates as extracellular deposits in the hippocampus of transgenic mice. J Neurosci. 2002;22:9785–9793. doi: 10.1523/JNEUROSCI.22-22-09785.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Schupf N, Manly J, Lantigua R, Stern Y, Rogaeva E, Wakutani Y, Farrer L, St George-Hyslop P, Mayeux R. The association between genetic variants in SORL1 and Alzheimer disease in an urban, multiethnic, community-based cohort. Arch Neurol. 2007;64:501–506. doi: 10.1001/archneur.64.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Lindholm K, Yang LB, Yue X, Citron M, Yan R, Beach T, Sue L, Sabbagh M, Cai H, Wong P, Price D, Shen Y. Amyloid beta peptide load is correlated with increased beta-secretase activity in sporadic Alzheimer’s disease patients. Proc Natl Acad Sci U S A. 2004;101:3632–3637. doi: 10.1073/pnas.0205689101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Hong W. From endosomes to the trans-Golgi network. Semin Cell Dev Biol. 2014;31:30–39. doi: 10.1016/j.semcdb.2014.04.024. [DOI] [PubMed] [Google Scholar]
- McCaffrey LM, I, Macara G. The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes Dev. 2009;23:1450–1460. doi: 10.1101/gad.1795909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad A, Flores I, Zhang H, Yu R, Staniszewski A, Planel E, Herman M, Ho L, Kreber R, Honig LS, Ganetzky B, Duff K, Arancio O, Small SA. Retromer deficiency observed in Alzheimer’s disease causes hippocampal dysfunction, neurodegeneration, and Abeta accumulation. Proc Natl Acad Sci U S A. 2008;105:7327–7332. doi: 10.1073/pnas.0802545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller UC, Zheng H. Physiological functions of APP family proteins. Cold Spring Harb Perspect Med. 2012;2:a006288. doi: 10.1101/cshperspect.a006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA. Endosome function and dysfunction in Alzheimer’s disease and other neurodegenerative diseases. Neurobiol Aging. 2005;26:373–382. doi: 10.1016/j.neurobiolaging.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci U S A. 2006;103:11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C. The role of the retromer complex in aging-related neurodegeneration: a molecular and genomic review. Mol Genet Genomics. 2015;290:413–427. doi: 10.1007/s00438-014-0939-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, St George-Hyslop P. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud R, Declerck I, Peric A, Raemaekers T, Menendez G, Zhou L, Veerle B, Coen K, Munck S, De Strooper B, Schiavo G, Annaert W. ADP ribosylation factor 6 (ARF6) controls amyloid precursor protein (APP) processing by mediating the endosomal sorting of BACE1. Proc Natl Acad Sci U S A. 2011;108:E559–568. doi: 10.1073/pnas.1100745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud R, Esselens C, Ejsmont P, Mattera R, Rochin L, Tharkeshwar AK, De Baets G, De Wever V, Habets R, Baert V, Vermeire W, Michiels C, Groot AJ, Wouters R, Dillen K, Vints K, Baatsen P, Munck S, Derua R, Waelkens E, Basi GS, Mercken M, Vooijs M, Bollen M, Schymkowitz J, Rousseau F, Bonifacino JS, Van Niel G, De Strooper B, Annaert W. Restricted Location of PSEN2/gamma-Secretase Determines Substrate Specificity and Generates an Intracellular Abeta Pool. Cell. 2016;166:193–208. doi: 10.1016/j.cell.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer CR, Offe K, Gearing M, Rees HD, Fang G, Heilman CJ, Schaller C, Bujo H, Levey AI, Lah JJ. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61:1200–1205. doi: 10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- Scott GK, Fei H, Thomas L, Medigeshi GR, Thomas G. A PACS-1, GGA3 and CK2 complex regulates CI-MPR trafficking. EMBO J. 2006;25:4423–4435. doi: 10.1038/sj.emboj.7601336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng JG, Price DL, Koliatsos VE. Disruption of corticocortical connections ameliorates amyloid burden in terminal fields in a transgenic model of Abeta amyloidosis. J Neurosci. 2002;22:9794–9799. doi: 10.1523/JNEUROSCI.22-22-09794.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SA, Kent K, Pierce A, Leung C, Kang MS, Okada H, Honig L, Vonsattel JP, Kim TW. Model-guided microarray implicates the retromer complex in Alzheimer’s disease. Ann Neurol. 2005;58:909–919. doi: 10.1002/ana.20667. [DOI] [PubMed] [Google Scholar]
- Sokolow S, Luu SH, Nandy K, Miller CA, Vinters HV, Poon WW, Gylys KH. Preferential accumulation of amyloid-beta in presynaptic glutamatergic terminals (VGluT1 and VGluT2) in Alzheimer’s disease cortex. Neurobiology of Disease. 2012;45:381–387. doi: 10.1016/j.nbd.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldano A, Hassan BA. Beyond pathology: APP, brain development and Alzheimer’s disease. Curr Opin Neurobiol. 2014;27:61–67. doi: 10.1016/j.conb.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Song WJ, Son MY, Lee HW, Seo H, Kim JH, Chung SH. Enhancement of BACE1 Activity by p25/Cdk5-Mediated Phosphorylation in Alzheimer’s Disease. PLoS One. 2015;10:e0136950. doi: 10.1371/journal.pone.0136950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoelgen R, von Arnim CA, Thomas AV, Peltan ID, Koker M, Deng A, Irizarry MC, Andersen OM, Willnow TE, Hyman BT. Interaction of the cytosolic domains of sorLA/LR11 with the amyloid precursor protein (APP) and beta-secretase beta-site APP-cleaving enzyme. J Neurosci. 2006;26:418–428. doi: 10.1523/JNEUROSCI.3882-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CP, Jay AG, Stack EC, Pakaluk M, Wadlinger E, Fine RE, Wells JM, Morin PJ. Retromer disruption promotes amyloidogenic APP processing. Neurobiol Dis. 2011;43:338–345. doi: 10.1016/j.nbd.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Asghar SZ, Zhang H. The polarity protein Par3 regulates APP trafficking and processing through the endocytic adaptor protein Numb. Neurobiol Dis. 2016 doi: 10.1016/j.nbd.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Bernard LP, Dibona VL, Wu Q, Zhang H. Calcium phosphate transfection of primary hippocampal neurons. J Vis Exp. 2013:e50808. doi: 10.3791/50808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi RH, Milner TA, Li F, Nam EE, Edgar MA, Yamaguchi H, Beal MF, Xu HX, Greengard P, Gouras GK. Intraneuronal Alzheimer A beta 42 accumulates in multivesicular bodies and is associated with synaptic pathology. American Journal of Pathology. 2002;161:1869–1879. doi: 10.1016/s0002-9440(10)64463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam JH, Seah C, Pasternak SH. The Amyloid Precursor Protein is rapidly transported from the Golgi apparatus to the lysosome and where it is processed into beta-amyloid. Mol Brain. 2014;7:54. doi: 10.1186/s13041-014-0054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Evin G. Beta-site APP-cleaving enzyme 1 trafficking and Alzheimer’s disease pathogenesis. J Neurochem. 2012;120:869–880. doi: 10.1111/j.1471-4159.2011.07623.x. [DOI] [PubMed] [Google Scholar]
- Tang W, Tam JH, Seah C, Chiu J, Tyrer A, Cregan SP, Meakin SO, Pasternak SH. Arf6 controls beta-amyloid production by regulating macropinocytosis of the Amyloid Precursor Protein to lysosomes. Mol Brain. 2015;8:41. doi: 10.1186/s13041-015-0129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesco G, Koh YH, Kang EL, Cameron AN, Das S, Sena-Esteves M, Hiltunen M, Yang SH, Zhong Z, Shen Y, Simpkins JW, Tanzi RE. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron. 2007;54:721–737. doi: 10.1016/j.neuron.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinakaran G, Koo EH. Amyloid precursor protein trafficking, processing, and function. J Biol Chem. 2008;283:29615–29619. doi: 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler SJ, Dawbarn D, Wilcock GK, Allen SJ. alpha- and beta-secretase: profound changes in Alzheimer’s disease. Biochem Biophys Res Commun. 2002;299:373–376. doi: 10.1016/s0006-291x(02)02635-9. [DOI] [PubMed] [Google Scholar]
- Udayar V, Buggia-Prevot V, Guerreiro RL, Siegel G, Rambabu N, Soohoo AL, Ponnusamy M, Siegenthaler B, Bali J, Simons M, Ries J, Puthenveedu MA, Hardy J, Thinakaran G, Rajendran L. A paired RNAi and RabGAP overexpression screen identifies Rab11 as a regulator of beta-amyloid production. Cell Rep. 2013;5:1536–1551. doi: 10.1016/j.celrep.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Wahle T, Prager K, Raffler N, Haass C, Famulok M, Walter J. GGA proteins regulate retrograde transport of BACE1 from endosomes to the trans-Golgi network. Mol Cell Neurosci. 2005;29:453–461. doi: 10.1016/j.mcn.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Walker KR, Kang EL, Whalen MJ, Shen Y, Tesco G. Depletion of GGA1 and GGA3 mediates postinjury elevation of BACE1. J Neurosci. 2012;32:10423–10437. doi: 10.1523/JNEUROSCI.5491-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Fluhrer R, Hartung B, Willem M, Kaether C, Capell A, Lammich S, Multhaup G, Haass C. Phosphorylation regulates intracellular trafficking of beta-secretase. J Biol Chem. 2001;276:14634–14641. doi: 10.1074/jbc.M011116200. [DOI] [PubMed] [Google Scholar]
- Wan L, Molloy SS, Thomas L, Liu G, Xiang Y, Rybak SL, Thomas G. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- Wang CL, Tang FL, Peng Y, Shen CY, Mei L, Xiong WC. VPS35 regulates developing mouse hippocampal neuronal morphogenesis by promoting retrograde trafficking of BACE1. Biol Open. 2012;1:1248–1257. doi: 10.1242/bio.20122451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Tang FL, Hong Y, Luo SW, Wang CL, He W, Shen C, Jung JU, Xiong F, Lee DH, Zhang QG, Brann D, Kim TW, Yan R, Mei L, Xiong WC. VPS35 haploinsufficiency increases Alzheimer’s disease neuropathology. J Cell Biol. 2011;195:765–779. doi: 10.1083/jcb.201105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willem M, Garratt AN, Novak B, Citron M, Kaufmann S, Rittger A, DeStrooper B, Saftig P, Birchmeier C, Haass C. Control of peripheral nerve myelination by the beta-secretase BACE1. Science. 2006;314:664–666. doi: 10.1126/science.1132341. [DOI] [PubMed] [Google Scholar]
- Wu Q, V, DiBona L, Bernard LP, Zhang H. The polarity protein partitioning-defective 1 (PAR-1) regulates dendritic spine morphogenesis through phosphorylating postsynaptic density protein 95 (PSD-95) J Biol Chem. 2012;287:30781–30788. doi: 10.1074/jbc.M112.351452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Sun M, Bernard LP, Zhang H. Postsynaptic density 95 (PSD-95) serine 561 phosphorylation regulates a conformational switch and bidirectional dendritic spine structural plasticity. J Biol Chem. 2017 doi: 10.1074/jbc.M117.782490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LB, Lindholm K, Yan R, Citron M, Xia W, Yang XL, Beach T, Sue L, Wong P, Price D, Li R, Shen Y. Elevated beta-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat Med. 2003;9:3–4. doi: 10.1038/nm0103-3. [DOI] [PubMed] [Google Scholar]
- Yap CC, Winckler B. Harnessing the power of the endosome to regulate neural development. Neuron. 2012;74:440–451. doi: 10.1016/j.neuron.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Cai Q. Snapin-mediated BACE1 retrograde transport is essential for its degradation in lysosomes and regulation of APP processing in neurons. Cell Rep. 2014;6:24–31. doi: 10.1016/j.celrep.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Feng T, Tammineni P, Chang Q, Jeong YY, Margolis DJ, Cai H, Kusnecov A, Cai Q. Regulation of Synaptic Amyloid-beta Generation through BACE1 Retrograde Transport in a Mouse Model of Alzheimer’s Disease. J Neurosci. 2017;37:2639–2655. doi: 10.1523/JNEUROSCI.2851-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, I, Macara G. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
- Zhang H, I, Macara G. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 RhoGAP and the Rho GTPase. Dev Cell. 2008;14:216–226. doi: 10.1016/j.devcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XM, Cai Y, Xiong K, Cai H, Luo XG, Feng JC, Clough RW, Struble RG, Patrylo PR, Yan XX. beta-Secretase-1 elevation in transgenic mouse models of Alzheimer’s disease is associated with synaptic/axonal pathology and amyloidogenesis: implications for neuritic plaque development. European Journal of Neuroscience. 2009;30:2271–2283. doi: 10.1111/j.1460-9568.2009.07017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Song M, Liu X, Su Kang S, Duong DM, Seyfried NT, Cao X, Cheng L, Sun YE, Ping Yu S, Jia J, Levey AI, Ye K. Delta-secretase cleaves amyloid precursor protein and regulates the pathogenesis in Alzheimer’s disease. Nat Commun. 2015;6:8762. doi: 10.1038/ncomms9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Guo W, Lipschutz JH. The exocyst protein Sec10 is necessary for primary ciliogenesis and cystogenesis in vitro. Mol Biol Cell. 2009;20:2522–2529. doi: 10.1091/mbc.E08-07-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.