Abstract
Parkinson’s disease (PD) is a neurodegenerative movement disorder characterized by the progressive loss of dopaminergic (DA) neurons. Most PD cases are sporadic; however, rare familial forms have been identified. Autosomal recessive PD (ARPD) results from mutations in Parkin, PINK1, DJ-1, and ATP13A2, while rare, atypical juvenile ARPD result from mutations in FBXO7, DNAJC6, SYNJ1, and PLA2G6. Studying these genes and their function has revealed mitochondrial quality control, protein degradation processes, and oxidative stress responses as common pathways underlying PD pathogenesis. Understanding how aberrancy in these common processes leads to neurodegeneration has provided the field with numerous targets that may be therapeutically relevant to the development of disease-modifying treatments.
Keywords: Autosomal recessive Parkinson’s disease, mitochondria, oxidative stress, protein degradation, Parkin, PINK1
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disorder, affecting more than 4 million people worldwide (Calabrese et al., 2007). Clinical manifestations of PD include bradykinesia, postural instability, resting tremor, and rigidity as well as increased susceptibility to memory impairment and behavioral alterations (Savitt et al., 2006). Pathologically, the disease is characterized by the degeneration of dopamine (DA) neurons in the substantia nigra pars compacta (SNpc) and the presence of cytoplasmic aggregates called Lewy Bodies. Though the clinical hallmarks and macro-level pathology of PD have been extensively characterized, understanding the exact molecular basis underlying neurodegeneration remains unclear.
Traditionally, PD has been classified as a non-genetic disease. Greater than 90% of cases are sporadic; however, rare familial forms have been identified. Autosomal recessive PD (ARPD) results from mutations in parkin, PINK1, DJ-1, ATP13A2, FBXO7, DNAJC6, SYNJ1, and PLA2G6. Studying these genes and the role they play in the disease progression has helped to elucidate common pathways in PD pathogenesis. These pathways include mitochondrial quality control, protein degradation, and oxidative stress response, all which result in dopaminergic cell death. Despite these findings, the gold standard treatment for PD patients continues to be DA replacement therapy. This therapy is symptomatically based, addressing primarily motor function, and does not alter disease progression. As a result, there is a dire need for disease-modifying therapies that halt neurodegeneration. In this review, we will discuss the genes underlying ARPD, the common pathways they are involved in, and potential strategies to therapeutically target ARPD genes and the pathways they regulate.
Parkin
Mutations in parkin were first identified in Japanese families exhibiting juvenile PD (Kitada et al., 1998; Periquet, 2003). The gene was mapped to an area on chromosome 6, with deletions of exons 3–7 associated with ARPD. Further studies have identified greater than 120 various mutations including exon rearrangements, indels, and point mutations, which result in decreased parkin function. Mutations in parkin are the second most frequent form of familial PD and account for the majority of ARPD cases including both autosomal recessive juvenile PD (ARJPD) and late onset PD. In fact, nearly 50% of ARJPD cases after age 25 and up to 7% of ARJPD cases between 30 and 45 years of age carry mutations in parkin. Additionally, parkin mutations account for approximately 8.6% of early-onset PD cases (Lill, 2016). Patients with parkin mutations are typically L-DOPA responsive and maintain a slow disease course with prominent dystonia (Bonifati, 2014).
Parkin spans 1.3 Mb of DNA and is translated into a 465 amino acid E3 ubiquitin ligase with an N-terminal ubiquitin like (Ubl) domain, two RING domains, and a unique parkin domain, dubbed RING0 (Trempe and Fon, 2013). Unlike a RING-type E ligase, which catalyzes the transfer of E2-bound ubiquitin directly to its substrate, parkin uses a HECT-type E3 ligase reaction (Zhang et al., 2016). In this reaction, E2-Ub binds to parkin, whereupon a thioester bond between ubiquitin and a cysteine side chain in RING2 forms, which is then followed by ubiquitin transfer to the substrate. Parkin is capable of facilitating mono- and poly-ubiquitination to lysine 29, 48, or 63 and resides in an autoinhibited state. Parkin has been implicated in mitochondrial homeostasis, the ubiquitin-proteasome pathway, oxidative stress response, and in cell death pathways.
PINK1
In 2004, scientists connected mutations in PTEN-induced putative kinase 1 (PINK1) to PD through linkage analysis of consanguineous families (Valente, 2004). The gene was mapped to chromosome 1 on the PARK6 locus. Since its initial discovery, more than 70 mutations of PINK1 have been discovered, the majority of which lead to its loss of function. Mutations in PINK1 account for approximately 3.7% of early onset cases and are the second most common cause of ARPD (Lill, 2016). Post mortem analysis of a single patient with PINK1-mediated PD reveal LB pathology, though data from this patient is currently the only data available (Bonifati, 2014). Clinically these patients exhibit a slow disease course, are responsive to L-DOPA, and have a higher reporting of psychiatric issues.
PINK1 encodes a ubiquitously expressed 581-amino acid protein with an N-terminal mitochondrial targeting sequence, a serine/threonine kinase domain, and a C-terminal regulatory domain (Trempe and Fon, 2013). Several studies have found that PINK1 localizes to both the mitochondria and the cytosol, with reports of both full length and shortened “mature” forms in each location (Lin and Kang, 2010). When localized to the mitochondria, PINK1 tethers to the outer mitochondrial membrane (OMM) with both its kinase domain and C-terminus facing the cytosol, suggesting that it acts on cytosolic substrates (Zhou et al., 2008). PINK1 is involved in mitochondrial quality control, stress pathways, and metabolism.
DJ-1
Mutations in DJ-1 (PARK7) were first identified in consanguineous Dutch and Italian families that displayed early onset PD (Bonifati et al., 2003). Initial studies found that mutations were caused by a large deletion in the DJ-1 coding region and by missense mutations affecting a highly conserved cysteine residue in exon 7. Since then, a variety of mutations that result in its loss of function or mislocalization. These mutations account for less than 1% of familial PD cases and approximately 0.4% of early-onset cases (Lill, 2016). While the pathology of DJ-1 patients is not known, patients with DJ-1 mutations have a clinical phenotype very similar to parkin- and PINK-1-mediated PD.
DJ-1 is a 24kb gene that encodes for a ubiquitously expressed, 189-amino acid protein (Moore et al., 2006; Moore et al., 2005; Trempe and Fon, 2013). DJ-1 has been shown to function as a dimer in solution and contains an essential cysteine residue within its active site that functions as an oxidative sensor. DJ-1 has been implicated in a number of pathways including transcriptional regulation, mitochondrial quality control, and the oxidative stress response.
ATP13A2
Mutations in ATP13A2 (PARK9) were first discovered in consanguineous families that exhibited a rare form of juvenile-onset PD known as Kufor-Rakeb syndrome (Ramirez et al., 2006). These studies found that ATP13A2 deficiencies were caused by a single nucleotide deletion, resulting in a frameshift, or a single nucleotide substitution. Further studies have found various mutations that result in ATP13A2 loss of function. The pathology in patients with ATP13A2 mutations remains unknown (Bonifati, 2014). The clinical phenotype of DJ-1 mediated PD includes pyramidal signs, poor L-DOPA response, dystonia, supranuclear palsy, and dystonia.
ATP13A2 encodes for a 5 P-Type ATPase located on lysosomal and late-endosomal membranes (van Veen et al., 2014). Studies suggest that endolysomal protein ATPase13A2 is involved in mitochondrial homeostasis, protein degradation pathways, and oxidative stress, potentially through the regulation of mono- and divalent cations, such as Zn2+ and H+ (Kett and Dauer, 2016).
Other Rare ARPD Genes
Mutations in FBXO7, DNAJC6, SYNJ1, and PLA2G6 have been identified in exceedingly rare, atypical forms of juvenile ARPD (Bonifati, 2014). Due to rarity and lack of patient tissue samples, data and mechanistic details underpinning pathology caused by these mutations are limited. As a result, we will provide a brief discussion of these genes and the pathways they are implicated in; however, our review will be focused primarily on parkin, PINK1, DJ-1 and ATP13A2 pathology and therapeutically targeting said pathways.
Mutations in FBXO7 were first identified in a consanguineous Iranian family with pyramidal disease(Di Fonzo et al., 2009). These patients and others with pathogenic FBXO7 mutations exhibit pyramidal symptoms and variable L-DOPA responsiveness. FBXO7 is a member of the Skp1-Cullin-F-box-type E3 ubiquitin ligase and has been implicated in both parkin-dependent and –independent proteasome-mediated degradation and mitophagy (Zhou et al., 2016). Exome sequencing and homozygosity mapping of several consanguineous families revealed DNAJC6 and SYNJ1 as causative ARPD genes (Bonifati, 2014). (Bonifati, 2014). The clinical phenotype of DNAJC6-mediated PD includes a rapid disease course with poor response to L-DOPA, pyramidal signs, seizures, mental retardation, and dystonia (Olgiati et al., 2016). Patients with pathogenic SYNJ1 mutations also present with a rapid disease course; however, progression stabilizes in later stages of the disease. These patients have variable symptoms including seizures, cognitive impairment, and developmental issues (Drouet, 2014). DNAJC6 encodes Auxilin, a member of the Hsp40 chaperone family, while SYNJ1 encodes synaptojanin-1, a phosphoinositide phosphatase expressed in nerve terminals. These proteins are thought to work together in the recycling of synaptic vesicles, perhaps in a parkin-dependent manner. Mutations in PLA2G6 were initially associated with brain iron accumulation neurodegeneration and were later linked to ARPD. The clinical phenotype associated with PLA2G6 mutations include good L-DOPA response, dystonia, pyramidal signs, and cognitive/psychiatric issues (Miki et al., 2017). PLA2G6 encodes the calcium-independent phospholipase A2, group VI. Recent evidence suggests that PLA2G6 deficiency is linked to aberrant ER Ca2+ signaling, leading to autophagy impairment, oxidative stress, and potentially aberration in other common ARPD-related pathways (Zhou et al., 2016).
Common Pathways in PD Pathogenesis
1. Mitochondrial Quality Control
Numerous studies have implicated aberrant mitochondrial quality control, a highly dynamic system regulating mitochondrial health, as a primary underlying cause of neurodegeneration in PD (Pickrell and Youle, 2015; Scarffe et al., 2014). Pathways in this system include fission/fusion, mitochondrial transport, mitophagy, and mitochondrial biogenesis. Though parkin and PINK1 have been intimately linked to mitochondrial quality control, evidence suggests that DJ-1, ATP13A2, and FBXO7 may also be involved. Whether these genes play more prominent roles in fission/fusion, transport, mitophagy, or biogenesis in PD progression remains unknown, though elucidating the prominent role is imperative in order to find the most efficient therapeutics.
i. Mitochondrial Fission and Fusion
The balance between fission and fusion is critical in maintaining healthy pools of mitochondria (Scarffe et al., 2014). Studies in Drosophila have found that parkin or PINK1 knockout (KO) causes aggregation of swollen mitochondria in both flight muscles and DA neurons, a sign of anti-fission or pro-fusion (Chen and Chan, 2009). This role is further strengthened by evidence that if fission is downregulated or fusion is increased, the phenotype is enhanced, while inhibiting fusion or upregulating fission results in wildtype rescue. Recently, PINK1 was found to dislodge Protein Kinase A (PKA) from its anchoring protein, allowing PKA to activate DRP1, a mediator of mitochondrial fission (Pryde et al., 2016). This would explain the observation of increased fusion in flies with PINK1 deficiencies; however, further validation by other groups is required to verify this interaction. Contrary to Drosophila data, loss of PINK1 in mammalian cells results in increased mitochondrial fragmentation, indicating that fusion is inhibited or fission is promoted (Scarffe et al., 2014). Supporting evidence shows that overexpression of PINK1 and parkin produces elongated, interconnected mitochondria, a sign of overactive fusion. Fibroblasts from patients with mutant PINK1-mediated PD also show increased mitochondrial fragmentation. A comprehensive screen for PINK1 and parkin substrates has revealed Mitofusin 1 and 2 (Mfn 1/2), GTPases responsible for fusing the OMM, as common substrates, suggesting a direct connection to mitochondrial fusion machinery (Ziviani et al., 2010). However, overexpression of parkin or PINK1 in primary rat DA neuron culture demonstrated increased mitochondrial fragmentation, indicating a role in mitochondrial fission (Yu et al., 2011). Discrepancies across models imply that factors such as tissue specific protein expression, cellular bioenergetic demands, temporal effects, overexpression artifacts, and compensatory pathways may come into play. Taken together, this indicates that further research evaluating the precise mechanisms by which parkin and PINK1 regulate fission and fusion is required.
DJ-1 and ATP13A2 have also been linked to mitochondrial fission and fusion, though their respective roles appear to be independent of parkin and PINK1. Studies in DJ-1 KO mice and in fibroblasts from patients with DJ-1 mutations reveal reduced mitochondrial branching, abnormal morphology, and diminished network connectivity between mitochondria (Krebiehl et al., 2010). DJ-1 deficient cell lines also display increased fragmentation, implying that DJ-1 may be involved in mitochondrial fusion. However, levels of fission and fusion regulators Drp1, Fis1, Mfn2, and OPA1 remain unchanged. This suggests that DJ-1 may regulate fission and fusion balance by a yet unknown mechanism. Alternatively, deficiency in DJ-1 may non-specifically perturb mitochondrial fission and fusion. Studies evaluating ATP13A2 loss of function have observed increased mitochondrial fragmentation in patient derived fibroblasts and in ATP13A2 KO cell lines (Grünewald et al., 2012; Park et al., 2014). Data suggests that fragmentation occurs as a result of Zn2+ dysregulation. The loss of Zn2+ buffering capacity results in prolonged upregulation of cytosolic Zn2+ levels, which in turn induces cellular stress and mitochondrial dysfunction, ultimately leading to an increase in mitochondrial fission. The direct interaction, if any, between ATP13A2 and mitochondrial fission and fusion machinery remains unknown.
ii. Mitochondrial Transport
Mitochondrial transport along axons is essential in maintaining neuronal homeostasis and in keeping up with cellular bioenergetic demands. Interestingly, Mfn 2 has been shown to be necessary for mitochondrial trafficking through its interaction with Miro, an OMM protein involved in mitochondrial transport, and the adaptor protein Milton (Misko et al., 2010). PINK1 has been shown to form a complex with Miro and Milton, whereupon they interact with a kinesin motor protein 5 (KIF5) to begin transport (Liu et al., 2012; Wang et al., 2011b). Furthermore, Miro is phosphorylated by PINK1 and ubiquitinated by parkin, leading to its degradation and halting mitochondrial transport. This allows for enhanced clearance of damaged mitochondria. Therefore, Miro levels, and potentially Mfn2 levels, appear to be regulated in a parkin/PINK1-dependent manner, demonstrating parkin/PINK1 involvement in mitochondrial trafficking.
iii. Mitophagy
Mitophagy is an essential process by which the cell degrades damaged mitochondria, allowing it to maintain a predominantly healthy mitochondria population to support its bioenergetic needs. Mitophagy is a sequential process that includes the recognition of damaged mitochondria, recruitment of autophagy machinery to the mitochondria, formation of the autophagosome, and fusion of the autophagosome with the lysosome for lysosomal degradation (Pickrell and Youle, 2015; Scarffe et al., 2014). Data from the field has linked the majority of ARPD genes to mitophagy and demonstrates that parkin, PINK1, and FBXO7 directly interact with each other in this process. Parkin was initially connected to mitophagy in 2008 when an important study found that chemically-induced mitochondrial depolarization recruits parkin to mitochondria in order to facilitate their degradation (Narendra et al., 2008). Since then, additional studies have broadened ARPD gene involvement in parkin-mediated mitophagy to include PINK1 and FBXO7. These studies reveal that under steady state conditions, PINK1 is targeted to the mitochondria via its mitochondria targeting sequence (MTS) and tethers to the OMM. PINK1 is imported through the TOM20 complex into the TIM complex, where its MTS is removed by mitochondrial processing peptidase (MPP) (Misko et al., 2010). Presenilin-associated rhomboid-like protein (PARL) then cleaves PINK1 within its transmembrane domain, producing a N-terminal truncated version of PINK1 that is released into the cytosol and rapidly degraded by the proteasome system (Pickrell and Youle, 2015). When mitochondrial depolarization occurs importation into the mitochondria is inhibited; therefore, PINK1 binds to TOM20 and accumulates on the OMM. Simultaneously, cytosolic FBXO7 also localizes to the OMM in depolarization conditions in a PINK1-dependent manner (Burchell et al., 2013). Here, PINK1 and FBXO7 work together in recruiting parkin to the mitochondrial membrane. Though the underlying mechanism of PINK1/FBXO7-mediated parkin recruitment is not fully understood, FBXO7 has been shown to interact with both parkin and PINK1, suggesting that it is an essential adaptor protein between the two (Scarffe et al., 2014). In addition to FBXO7 interaction, some groups postulate that PINK1-phosphorylated proteins such as Mfn, Miro, or VDACs on the OMM may act as receptors that aid in parkin docking.
Once parkin is recruited to the OMM, PINK1 phosphorylates both parkin and ubiquitin at Ser65, fully activating parkin. Parkin then ubiquitinates OMM proteins VDAC1, Miro, and Mfn1/2, though these proteins may not be required for the recruitment of autophagy machinery (Narendra et al., 2008). Interestingly, FBXO7 has been shown to be required for efficient Mfn1 ubiquitination and subsequent recruitment of autophagy machinery protein p62, suggesting that its role as an adaptor protein extends beyond parkin recruitment and may aid parkin in initiating mitophagy (Burchell et al., 2013). While it is unknown what exactly initiates mitophagy, studies have found that parkin interacts with autophagy proteins p62/SQSTM1, HDAC6, Beclin, and Ambra1; however, evidence suggests that perhaps p62 and VDAC1 are not essential for initiation (Scarffe et al., 2014).
It should be noted that FBXO7 and PINK1 may also have a role in mitophagy in a parkin-independent manner. Evidence for such roles stem from Drosophila studies in which FBXO7 overexpression rescued mitochondria dysfunction in parkin KO flies but not in PINK1 deficient flies (Burchell et al., 2013). Taken together with its role in Mfn1 ubiquitination, this implies that FBXO7 may interact with other proteins to initiate PINK1-dependent mitophagy, and that targeting FBXO7 expression could potentially circumvent parkin deficiencies. Additionally, a recent study found that PINK1 will recruit synphilin-1 to the mitochondria, thereby inducing mitochondrial depolarization (Szargel et al., 2016). After depolarization, Synphilin-1 was shown to recruit SIAH-1, an E3 ubiquitin ligase, to the mitochondria where it initiates mitophagy. Strikingly, PINK1 disease mutants fail to initiate PINK1-synphilin-SIAH-1 mitophagy, in the presence or absence of parkin. These findings offer alternative pathways that promote mitophagy; however, further validation in PD disease models is needed.
Although parkin/PINK1-dependent mitophagy has been consistently observed in mammalian cells overexpressing parkin, reproducible translocation of endogenous parkin in neurons remains elusive (Grenier et al., 2013). The bioenergetics of neurons, cell culture conditions, and cell types may contribute to variability between studies. Additionally, chemical depolarizing agents induce rapid depolarization of nearly all mitochondria, a phenotype not physiologically relevant to PD pathogenesis. Though studies have failed to see abnormal aggregation of damaged mitochondria in PINK1 KO and parkin KO mice, more recent work using microfluidics and high-resolution microscopy has verified parkin-dependent mitophagy in primary neuronal culture (Ashrafi et al., 2014).
Direct evidence connecting DJ-1 to mitophagy is lacking; however, several studies have shown that DJ-1 localizes to the mitochondria during oxidative stress and that loss of DJ-1 function results in reduced mitochondrial membrane potential (Krebiehl et al., 2010; Thomas et al., 2010). Furthermore, one study found that DJ-1 deficiency resulted in an accumulation of autophagic markers surrounding mitochondria, indicative of impaired mitochondrial clearance (Thomas et al., 2010). Contrarily, some studies have found that DJ-1 loss actually increases basal autophagy (Ren et al., 2010). This suggests that DJ-1 may interact with PINK1, parkin, or FBXO7 in inducing mitophagy in a yet-to-be elucidated mechanism. Recent data demonstrates that DJ-1 binds to Foxo3a transcription factor, which then binds to the promoter of PINK1, resulting in an upregulation of PINK1 transcription (Requejo-Aguilar et al., 2015). Whether this transcriptional regulation provides a protective role in upregulating mitophagy machinery is unclear. Mitochondrial defects related to DJ-1 deficiency appear to be rescued by parkin and PINK1 overexpression, suggesting the DJ-1 may act downstream or in a parallel pathway (Thomas et al., 2010). To date, the exact role of DJ-1 in parkin/PINK1/FBXO7-dependent and -independent mitophagy remains unknown.
An important step in mitophagy is the fusion of the autophagosome with the lysosome, followed by lysosomal degradation of the damaged mitochondria (Scarffe et al., 2014). Although the role of ATP13A2 has not been extensively characterized, studies have demonstrated that ATP13A2 colocalizes with structural lysosomal protein LAMP1/2a and lysotracker in homeostatic conditions, suggesting a role in lysosomal maintenance (Gusdon et al., 2012). A recent study found that ATP13A2 colocalizes with LC3, an autophagosome and late endosomal marker, perhaps specific to the multivesicular body (MVB) (Kett and Dauer, 2016). This suggests that ATP13A2 may be important in fusion of the autophagosome or vesicles from the MVB with the lysosome. ATP13A2 deficient models also exhibit lysosomal dysregulation and impaired mitochondrial clearance as assessed by reduced autophagic flux and increased mitochondrial mass. Additionally, studies have observed lysosomal accumulation of subunit c of mitochondrial ATP synthase and cathepsin D deficiency, indicating deficient lysosomal processing of damaged mitochondria (Kett et al., 2015).
iv. Mitochondrial Biogenesis
Another important arm of mitochondrial quality control is mitochondrial biogenesis, a critical pathway by which the cell is able to replace damaged mitochondria after they have been degraded. A link between parkin/PINK1 and mitochondrial biogenesis was first discovered in 2006, when parkin and PINK1 were found to increase mtDNA replication and to enhance mitochondrial transcription by associating with mitochondrial transcription factor A (TFAM) (Kuroda, 2006). Parkin was also shown to immunoprecipitate with mtDNA, stimulate mtDNA repair, and protect mtDNA against oxidative damage (Rothfuss et al., 2009). In one study, loss of PINK1 function resulted in a decrease in mtDNA levels and synthesis, a reduction in ATP synthesis, and a decline in complex IV activity (Gegg et al., 2009). Further studies using patient samples (Müftüoglu et al., 2003) and parkin and PINK1 KO mice (Gautier et al., 2008) have shown significant deficits in Complex I (COXI) levels, a key component of the ETC, which may suggest that DA neurons are not able to replace mitochondria after turnover. Recently, the discovery of a zinc finger transcriptional repressor called parkin interacting substrate (PARIS) have directly linked parkin to biogenesis (Shin et al., 2011). Under normal physiological conditions, PARIS was shown to be ubiquitinated by parkin and degraded by the proteasome. Loss of parkin function results in the accumulation of PARIS, which in turn suppresses peroxisome proliferator-activated receptor gamma coactivator 1α (PGC-1α), a coregulator of mitochondrial biogenesis. This was validated by analysis of postmortem SNpc tissue of PD patients, in which DA neurons displayed a reduction in PGC-1α levels as well as downstream effectors such as nuclear encoded components of the ETC (Shin et al., 2011). Recent studies indicate that PARIS is phosphorylated by PINK1 to regulates PARIS ubiquitination and clearance by parkin where it controls PGC-1α levels indicating that PARIS provides a link between parkin and PINK1 in the regulation of mitochondrial biogenesis (Lee et al., 2017).
2. Protein degradation pathways
Protein degradation pathways, including the ubiquitin proteasome system (UPS) and autophagy-lysosomal degradation, are essential in clearance of misfolded proteins and recycling of cellular materials. As a ubiquitously expressed E3 ligase, parkin has been shown to be involved in the UPS, and studies in parkin overexpression have confirmed enhanced proteasome activity (Khandelwal, 2010). Screening for parkin substrates has identified hundreds of proteins, including proteins involved in the epidermal growth factor pathway, Wnt signaling, inflammation, mitochondrial homeostasis, Lewy body formation, and apoptosis, suggesting broad involvement of parkin ubiquitination (Charan and LaVoie, 2015; von Coelln et al., 2004). Lack of parkin activity would suggest an accumulation of its substrates, leading to aberrant cell signaling and neurotoxicity. This was confirmed in several studies, including in adult conditional parkin KO mice as they display the accumulation of PARIS and Aminoacyl-tRNA synthetase complex interacting multifunctional protein-2 (AIMP2) and subsequent dopaminergic neurotoxicity (Lee et al., 2013; Shin et al., 2011). Interestingly, studies in germline parkin KO mice fail to show accumulation of parkin specific substrates, implying the presence of alternative regulatory mechanisms or a developmental compensatory pathway (Charan et al., 2014). This may also be due to the restricted number of authentic parkin substrates that the study tested for. Taken together, this implies that there is an intimate relationship between parkin and the UPS in inducing ARPD.
The role of ATP13A2 in lysosomal and late endosomal maintenance may extend far beyond mitophagy. Because of its apparent role in regulating lysosomes, it is easy to infer that ATP13A2 could have critical roles in macro-, micro-, and chaperone-mediated autophagy. This claim is supported by data from ATP13A2 deficient mice, as they display decreased lysosomal proteolytic processing and aberrant lysosomal storage phenotypes (Gusdon et al., 2012; Kett et al., 2015). Interestingly, ATP13A2 has been found to colocalize with α-synuclein aggregates in endosomes, lysosomes, and Lewy bodies (Kett et al., 2015). Furthermore, ATP13A2 deficiency has been shown to promote α-synuclein aggregation in MVBs, suggesting that it plays a role in externalizing α-synuclein and other misfolded proteins. Recent evidence suggests that ATP13A2 may mediate autophagy through the transcriptional and post-translational regulation of SYT11, a putative PD risk locus thought to be involved in lysosome function (Bento et al., 2016). This study demonstrates that ATP13A2 deficiency results in reduced SYT11 expression, lysosomal impairment, and diminished clearance of autophagosomes. Overexpression of SYT11 was found to rescue autophagy dysfunction induced by ATP13A2 knockdown, implying that ATP13A2-mediated autophagy may be dependent on SYT11 function, though validation by other groups is required to confirm this mechanism. These data suggest an intimate role of ATP13A2 in lysosomal regulation, though further evidence is needed to elucidate the mechanism by which loss of ATP13A2 induces autophagic dysfunction, resulting in neurodegeneration.
3. Oxidative stress and cell death pathways
Neurons are post-mitotic, high-energy consuming cells and are particularly susceptible to oxidative stress; therefore, it is no surprise that oxidative stress, stress-induced apoptosis, and cell death pathways have been repeatedly implicated in PD pathogenesis. ARPD genes have demonstrated varying degrees of antioxidant properties and involvement in preventing dopaminergic cell death. Whether ARPD genes indirectly or directly interact with each other in these responses, however, remains unknown.
i. Parkin involvement in oxidative stress and cell death pathways
Parkin’s neuroprotective role has been extensively observed in a variety of oxidative stress conditions, including mitochondrial, ER, proteolytic, and dopamine stress (Charan and LaVoie, 2015). Though parkin’s primary function in mediating oxidative stress appears to be in regulating mitophagy, it has been implicated in other pro-survival roles. For example, in oxidative conditions parkin will polyubiquitinate NEMO, allowing for the nuclear translocation of NF-κB and subsequent upregulation of prosurvival factors (Müller-Rischart et al., 2013). These prosurvival factors, including mitochondrial fusion regulator OPA1, prevent cytochrome c release, maintain mitochondrial integrity, and inhibit stress-induced cell death. Additionally, several studies suggest that parkin may interact with pro-apoptotic Bax and anti-apoptotic Bcl-2; however, the precise mechanism by which parkin interacts with Bcl-2 family proteins to regulate cell death remains unclear (Charan and LaVoie, 2015).
A more concrete role of parkin in mediating cell death pathways and promoting cell survival is through its function in the UPS. As discussed previously, parkin has been shown to polyubiquitinate AIMP2, a protein found in Lewy bodies of PD patients, leading to its degradation by the proteasome (Lee et al., 2013). Loss of parkin leads to the accumulation of AIMP2, resulting in its nuclear translocation and subsequent activation of poly ADP-ribose (PAR) polymerase 1 (PARP1). Overactivation of PARP1 leads to increased PAR levels, which act as a signal from the nucleus to the mitochondria where PAR binds to Apoptosis Inducing Factor-1 (AIF), resulting in its translocation from the mitochondria to the nucleus. AIF interacts with macrophage migration inhibitory factor (MIF) where it recruits MIF to the nuclease (Wang et al., 2016). Once inside the nucleus MIF’s nuclease activity leads to extensive DNA fragmentation and caspase-independent cell death (Wang et al., 2016). These data establish a direct link between parkin and the cell death pathway called parthanatos, leading to DA neurodegeneration.
ii. PINK1-mediated oxidative-stress and anti-apoptotic response
Studies have found that PINK1 deficiency leads to increased levels of mitochondrial-produced reactive oxygen species (ROS) and enhanced susceptibility to mitochondrial stress-induced apoptosis (Wang et al., 2011a). Though mitochondrial membrane depolarization is known to stabilize PINK1 on the OMM and recruit parkin in mitophagy, loss of PINK1 itself has also been shown to induce mitochondrial depolarization. This implies a mitophagy-independent pathway in which PINK1 helps to maintain homeostatic mitochondrial membrane potential, thus preventing the release of ROS and cytochrome c. This pathway may be through the mitochondrial N+/Ca2+ exchanger, as studies have demonstrated that PINK1 interacts with this transporter to regulate calcium efflux from the mitochondria, and aberrant mitochondrial calcium signaling has been shown to induce cellular stress and death (Kostic et al., 2015). Additionally, PINK1 has been found to interact with a number substrates including TNF receptor associated protein 1 (TRAP1) and aconitase. Studies demonstrate that PINK1 phosphorylates TRAP1, perhaps selectively in low mitochondrial stress conditions, thereby preventing oxidative-stressed induced apoptosis by inducing pro-survival signals (Zhang et al., 2013). Aconitase, an intermediate in the TCA cycle, is reduced in PINK1 deficient cells and inactivated in DA-induced oxidative stress (Esposito et al., 2013). Loss of aconitase function results in increased ROS production, increased OMM permeability, and cytochrome c-induced cell death. Taken together, these data suggest a role for PINK1 activity in regulating the cell’s stress response.
iii. DJ-1-mediated oxidative-stress and anti-apoptotic response
Since its identification, DJ-1 has been touted as an endogenous redox sensor that responds to oxidative stress. Evidence for this role stems from DJ-1 deficient models, as they show increased susceptibility to oxidative stress, and the neuroprotective ability of DJ-1 in the presence of ROS, which prevents the cell from undergoing stress-induced apoptosis (Shadrach et al., 2013). The enzyme has been reported to work through a variety of mechanisms, including self-oxidation, transcriptional regulation, and regulation of anti-oxidative stress pathways. DJ-1 has a well-documented, highly conserved cysteine residue, Cys106, that oxidizes in the presence of ROS, suggesting that DJ-1 may have a prominent role in ROS scavenging (Blackinton et al., 2009). Studies in DJ-1 knockout mice indicate that DJ-1 functions in vivo as an atypical peroxiredoxin-like peroxidase to scavenge mitochondrial hydrogen peroxidase (Andres-Mateos et al., 2007). Additionally, it has been widely observed that DJ-1 translocates to the mitochondria during oxidative stress. Studies have found that translocation results in its binding to COXI, the main electron transport subunit responsible for ROS leakage (Hayashi et al., 2009). This implies that the cysteine residues of DJ-1 may sop up ROS released through COXI, thus stabilizing the mitochondria and preventing ROS-induced cell stress.
DJ-1 also regulates factors in stress-induced cell death. For example, Drosophila studies have found that DJ-1 suppresses PTEN activity, thereby promoting cell growth and promoting cellular defense against ROS through PI3K/Akt signaling (Chan and Chan, 2015). In vitro data demonstrates a direct interaction between DJ-1 and death domain-associated protein (Daxx) in the nucleus. Studies evaluating MPTP-treated mice have complemented in vitro evidence, demonstrating a significant reduction in nuclear levels of DJ-1, thus preventing the association of DJ-1 with Daxx (Karunakaran et al., 2007). This in turn allows the translocation of Daxx to the cytosol where it interacts with apoptosis signal-regulating kinase 1 (ASK1) to induce cell death. However, further evidence using in vivo models, such as DJ-1 KO mice, is needed to solidify a direct interaction between DJ-1 and Daxx-induced cell death.
iv. ATP13A2 in mediating oxidative stress
ATP13A2 expression appears to be upregulated in oxidative stress conditions, suggesting it plays an important role in mediating cellular stress (Xu et al., 2012). Many studies have implicated Mn2+, Mg2+, H+, and Zn2+ as cations transported by ATP13A2, though extensive data and exact mechanisms remain unknown (van Veen et al., 2014). Dysregulation of cation homeostasis as a result of ATP13A2 loss can have profound issues, including heightened cellular stress leading to programmed cell death. For example, ATP13A2 loss reduces Zn2+ buffering capacity and OMM potential, resulting in the release of mitochondrial ROS and a stress-induced response. These data imply that ATP13A2 mediates oxidative stress through cation regulation and mitochondrial maintenance.
Therapeutics
There are a wide variety of therapeutic strategies to target pathways regulated by ARPD genes. In this section, we will discuss strategies such as increasing parkin activity and enhancing or circumventing pathways that are abnormal in ARPD. Though this is by no means an exhaustive list, we hope to provide an overview of the most relevant potential targets that have been elucidated by understanding the function of ARPD genes and their roles in maintaining homeostasis. These targets may be crucial in the development of disease-modifying therapies for PD.
1. Targeting Parkin Function
Parkin overexpression has been shown to be neuroprotective under a variety of stressors and to ameliorate mitochondrial health in both in vitro and in vivo models. As a result, therapies that enhance parkin function are incredibly desirable. One way to increase parkin function is to promote its activation. Studies investigating parkin activation have found that phosphorylation of Ser65 is required for activation and that phosphoubiquitin serves as an allosteric enhancer of parkin function (Pickrell and Youle, 2015). Parkin’s crystal structure revealed that phosphoubiquitin binding to parkin results in a conformational change of the RING1 domain, which releases the Ubl domain from parkin’s core to yield a partially-active Parkin. Release of Ubl allows for PINK1 phosphorylation of Ubl, causing parkin to open and become fully active. As a result, screening for small allosteric modulators that induce conformational changes to expose the catalytic site may be useful in enhancing parkin activation. Other serines on parkin have also been identified as phosphorylation sites, suggesting alternative parkin regulation and activation. Elucidating the function of alternative phosphorylation sites and their role in parkin’s activation may lead to promising therapeutic targets.
Because PINK1 plays a critical role in activating parkin through direct phosphorylation and in phosphorylating ubiquitin, the allosteric parkin activator, therapies that increase PINK1 activity are highly attractive. Developing therapies that target PINK1 are limited, however, because allosteric regulatory sites of PINK1 have not been identified. Recently, a group found that Kinetin triphosphate (KTP), an ATP analog, ameliorates kinase activity in mutant PINK1 to nearly a full rescue to wild-type function (Hertz et al., 2013). KTP may prove to be an ideal therapeutic, as initial in vitro studies demonstrate that the KTP precursor, kinetin, crosses the plasma membrane and is converted into KTP by endogenous enzymes. Because phospho-ubiquitin plays a vital role in parkin activation, screening for complementary endogenous kinases that can produce phospho-ubiquitin to activate parkin may also have therapeutic merit. Alternatively, identifying phosphatases that dephosphorylate PINK1 substrates, such as parkin, and screening for molecules that inhibit said phosphatases may be also therapeutically beneficial.
Another strategy to improve parkin function is to regulate its post-translational modifications. Many post-translational modifications, such as s-nitrosylation, oxidation, phosphorylation, and dopamine conjugation, reduce the stability and solubility of parkin, resulting in diminished activity (Charan and LaVoie, 2015). Melatonin, taurine, and vitamin have been shown to protect against oxidative stress and may protect parkin against some of these detrimental modifications. Studies investigating post-translational modifications of parkin have found that c-Abl, a tyrosine kinase activated in brain tissue of PD patients and in MPTP-treated mice, is responsible for phosphorylating parkin and inhibiting its E3 ligase function (Imam et al., 2013; Ko et al., 2010). Furthermore, data has demonstrated that administration of the c-Abl inhibitors is neuroprotective against MPTP-treatment in mice, perhaps by preventing parkin phosphorylation or preventing the accumulation parkin substrates, such as PARIS (Imam et al., 2013; Karuppagounder et al., 2014). Other studies have shown that sulfhydration increases parkin function and that PD patients display striking reductions in levels of parkin sulfhydration (Vandiver et al., 2013). Researchers have found that hydrogen sulfide (H2S) protects against neurodegeneration in rodent models, suggesting that therapies employing H2S donors that selectively sulfhydrate parkin may be beneficial in enhancing parkin function. Understanding the effects of post-translational modifications of parkin and the enzymes that conduct these modifications may be essential to parkin’s pharmacological manipulation.
Parkin activity is, in part, related to its ability to translocate to damaged mitochondria, thus allowing it to become activated by PINK1. Several studies have identified SMURF1, HK1, HK2, FBXW7, IF1, HSPA1L, and SREBF1 as key modifiers that promote translocation (Pickrell and Youle, 2015). Others, such as BAG4 and SIAH3, have been identified as inhibitors of parkin translocation by directly interacting with parkin or inhibiting the accumulation of PINK1 on depolarized OMM, respectively. Understanding the unique role of each modifier and how they work together to regulate parkin translocation is critical, as increasing proteins that promote translocation and/or inhibiting those that suppress translocation may be therapeutically relevant to enhancing parkin’s activity in the setting of mitochondrial depolarization.
2. Exploiting Parallel Pathways to Parkin
A compelling issue in the PD field is that germline parkin KO mice fail to show neurodegeneration, motor deficits, or an accumulation of parkin substrates (Charan et al., 2014; Charan and LaVoie, 2015). The lack of a Parkinsonian phenotype suggests the presence of a developmental or compensatory pathway. This would make sense because ubiquitination is a sequential process involving E1 (ubiquitin activating enzymes), E2 (ubiquitin conjugating enzymes), and E3 (ubiquitin ligases), implicating a large network of protein involvement. For example, a bioinformatics study has identified approximately 617 human E3 ligases and substrate recognition subunits of E3 complexes, most with unknown function (Li et al., 2008). Screening through proteins involved in the ubiquitin-proteasome pathway may elucidate therapeutic targets involved in mitophagy and protein degradation pathways that work parallel to parkin, potentially identifying a therapeutic target to circumvent parkin deficiencies.
3. Targeting Mitochondrial Dysfunction
Abundant evidence suggests that defects in ARPD genes result in aberrant mitochondrial quality control, leading to mitochondrial dysfunction and subsequent neurodegeneration. Therefore, one could speculate that targeting mitochondrial dysfunction using mitochondrial enhancing drugs could be useful in generating disease-modifying therapies. Despite promising preclinical data, human clinical trials supplementing with mitochondrial enhancing drugs, including coenzyme Q10 and creatine have consistently failed to display significant neuroprotection or alter disease progression (Chaturvedi and Beal, 2013). As a result, targeting upstream pathways, such as mitophagy or mitochondrial biogenesis, may be more efficacious.
4. Targeting Mitophagy
Recent evidence suggests that deubiquitinases (DUBs) are essential in regulating parkin/PINK1 mediated mitophagy (Pickrell and Youle, 2015). USP30, a DUB that localizes to the mitochondria, is responsible for deubiquitinating OMM proteins such as TOM20 and MIRO1, key parkin substrates that promote mitophagy, and in delaying parkin recruitment (Cunningham et al., 2015; Wang et al., 2015). siRNA knockdown of USP30 in mutant parkin SHSY5Y lines rescued defects in mitophagy. Additionally, USP30 knockdown in parkin or PINK1 deficient flies and parkin or PINK1 deficient neurons rescued mitophagy deficits. Another DUB, USP15, similarly antagonizes parkin-mediated mitophagy by deubiquitinating mitochondrial parkin substrates (Cornelissen et al., 2014). Furthermore, knockdown of USP15 Drosophila homolog, DUB CG834, significantly improved mitochondrial deficits in parkin RNAi flies. Newly identified USP35 has also been shown to oppose parkin, perhaps by regulating Mfn2 levels, though its mechanism of action is not entirely understood (Wang et al., 2015). USP8 (Durcan et al., 2014) and Ataxin-3 (Durcan et al., 2011) directly remove ubiquitin moieties from parkin, inhibiting mitophagy by limiting parkin recruitment. USP8 removes K6-linked ubiquitin, which is thought to stabilize parkin (Durcan et al., 2014). Ataxin-3 prevents parkin ubiquitination by stabilizing parkin binding with E2-Ub and inhibiting the transfer of Ub onto parkin (Durcan et al., 2011). As a result, screening for compounds that can selectively inhibit these DUBs are desirable to circumvent parkin deficiencies and promote mitophagy.
The recruitment of autophagy adaptors is essential to the initiation of mitophagy (Scarffe et al., 2014). Researchers have recently discovered that PINK1 generated phospho-ubiquitin is critical for recruiting receptors OPTN and NDP52 and initiates both parkin-dependent and -independent mitophagy (Heo et al., 2015). Understanding the precise roles of these receptors and screening for compounds and enzymes that can promote receptor recruitment to the mitochondria may provide an ideal therapeutic target, especially in patients with parkin deficiencies. Additionally, parkin interactors, such as Ambra1 and Beclin-1, have been strongly implicated in carrying out efficient mitophagy. Several studies have suggested a variety of roles for Ambra1 including: (1) interaction with parkin during mitochondrial depolarization, whereupon Ambra1 activates class III PI3K (Van Humbeeck et al., 2011); (2) Ambra1 mediated mitochondrial depolarization and subsequent ubiquitination by parkin; (3) induction of parkin-independent mitophagy; and (4) binding of LC3 to complete autophagophore formation (Strappazzon et al., 2015). Beclin-1 has been shown to bind to both PINK1 and parkin to promote mitophagy and parkin translocation (Choubey et al., 2014). Because of their importance in mitophagy initiation, inducing Ambra1 and Beclin-1 translocation to the mitochondria and/or enhancing their function has the potential to enhance parkin-dependent and -independent mitophagy, thereby enabling the clearance of damaged mitochondria.
5. Enhancing Mitochondrial Biogenesis
Recent data suggest an increasingly crucial involvement of parkin in mitochondrial biogenesis. As such, finding drugs that can enhance mitochondrial biogenesis or inhibit repressors of biogenesis are extremely appealing. Studies have found that knockdown of PARIS in adult conditional parkin KO mice rescues mitochondrial defects and neurodegeneration (Shin et al., 2011). PARIS seems to be particularly important in the PINK1 and parkin pathway of neurodegeneration in PD since a reduction in PARIS levels rescues the neurodegeneration in adult conditional PINK1 knockdown mice (Lee et al., 2017). Moreover, defects in mitochondrial biogenesis drive the loss of DA neurons since adult conditional parkin KO mice have primary defects in mitochondrial biogenesis in the absence of measurable defects in mitophagy, consistent with rescue of degeneration by reducing PARIS levels (Stevens et al., 2015). Because PARIS is one of the only known transcriptional suppressors of the key mitochondrial biogenesis regulator PGC-1α, it is an attractive therapeutic target. Screening for small molecules that (1) inhibit PARIS from binding to the PGC-1α promoter, (2) block the nuclear localization signal on PARIS, or (3) sequester PARIS and promote its degradation by the proteasome could potentially modify PD progression in patients.
Recent evidence has demonstrated that adult conditional PGC-1α KO mice display a significant loss of DA neurons in the SNpc (Jiang et al., 2016). Taken together with PGC-1α overexpression data, which has been shown to rescue mitochondrial biogenesis defects and accompanying neurodegeneration, it stands to reason that finding compounds which upregulate PGC-1α expression may be therapeutically beneficial. Many studies have analyzed the effects of flavonoids, plant-based compounds, in activating PGC-1α expression. For example, Quercetin, a common dietary bioflavanoid, has been studied in ischemic rodent models and has been found to activate CREB, leading to transcriptional activation of PGC-1α and enhanced mitochondrial biogenesis (Li et al., 2016). This therapy is attractive, as flavonoids can be obtained through diet and have limited side effects. However, to our knowledge, no studies investigating the effects of Quercetin administration have been performed in PD models. Recently, necidin, a MAGE family protein, was found to posttranslationally upregulate PGC-1α, thus enhancing mitochondrial biogenesis. This study found demonstrated that necidin worked by inhibiting PGC-1α degradation by the UPS, thereby stabilizing endogenous PGC-1α and preventing MPTP-induced neurodegeneration (Hasegawa et al., 2016). This suggests that identifying endogenous proteins that prolong the half-life of PGC-1α may be therapeutically beneficial in promoting mitochondria biogenesis. Alternatively, screening for molecules that prevent PGC-1α degradation by the proteasome or that inherently stabilize PGC-1α could potentially modify disease progression in PD patients.
There are data that suggest that prolonged overexpression of PGC-1α sensitizes neurons to MPTP damage, resulting in neuronal death (Hasegawa et al., 2016). These data suggest that therapies targeting overexpression should aim to elevate and maintain PGC-1α at physiologically relevant levels. It should be also noted that PGC-1α is involved in several pathways outside of mitochondrial biogenesis; as a result, targeting PGC-1α to selectively enhance mitochondrial biogenesis is a large hurdle that has yet to be tackled.
6. Targeting oxidative stress
Given the link between the function of ARPD genes, mitochondrial dysfunction, and ROS production in PD, strategies that target the oxidative stress response may be beneficial. Strategies include targeting endogenous antioxidant enzymes and non-enzymatic antioxidant molecules. There are several endogenous antioxidant enzymes, including peroxidases, SOD, catalase, and thioredoxin, and non-enzymatic antioxidant molecules, such as GSH and uric acid (Chan and Chan, 2015). Screening for compounds that enhance endogenous antioxidant enzymatic activity or increase the production of endogenous antioxidants have been found to be neuroprotective in laboratory settings and have the potential to be therapeutically beneficial in treating PD.
Another strategy is the supplementation of exogenous antioxidants, such as vitamin E and CoQ10 (Kim et al., 2015). Many exogenous antioxidants have demonstrated promising neuroprotective effects in vivo; however, most antioxidant-based human clinical trials have failed to show amelioration of PD symptoms or modification of disease progression. Dosing, timing, and stage of the disease may be plausible factors; however, the more likely explanation is that oxidative stress is not the primary cause of neurodegeneration. As a result, primarily targeting oxidative stress, as opposed to targeting the cause of stress and ROS production, is probably not sufficient in altering PD progression. Consequently, administering antioxidant cocktails in combination with previously discussed therapeutic strategies may be more efficacious in halting neurodegeneration.
7. Preventing Dopaminergic Cell Death
Dysfunctional mitochondrial quality control, abnormal protein degradation pathways, and oxidative stress all culminate in the death of DA neurons. Therefore, inhibiting cell death pathways or enhancing pro-survival responses may be a viable therapeutic strategy. Initiator and effector caspases are activated in stressed-induced apoptotic pathways, and many studies have investigated the inhibition of caspase activity in disease. For example, caspase inhibitors, such as the caspase-1 inhibitor Pralnacasan, have been shown to markedly reduce brain damage in ischemic rats (Ross et al., 2007); however, to our knowledge caspase-inhibitor efficacy in PD models has not yet been tested.
Because of parkin’s role in regulating AIMP2-induced parthanatos, screening for AIMP2 and PARP1inhibitors may aid in halting neurodegeneration. Since AIMP2 localizes to the nucleus to interact with PARP1, blocking its nuclear localization sequence may suppress its ability to induce parthanatos (Lee et al., 2013). PARP1 inhibitors have been shown to be effective in human cancer clinical trials and in preclinical models of PD; however human clinical testing of PARP1 inhibitor treatment in the context of PD has not yet been published. This may be in part due to concerns in prolonged PARP1 inhibition, as PARP1 plays an essential role in mediating the DNA-damage response. Alternatively, identifying and targeting specific substrates of PARP1 that induce neurodegeneration may be safer as it is more selective. It should be noted that neuronal death is late in disease progression; therefore, one can imagine that inhibiting cell death may not be a permanent solution, as the underlying cause is not treated. However, inhibiting cell death in combination with previously mentioned therapeutic strategies may be therapeutically relevant.
Concluding Remarks
To date, there are no effective disease-modifying therapies for PD. Lack of treatment efficacy may be due to our restricted knowledge of molecular mechanisms involved, the complexity of PD pathogenesis, and perhaps the predominant use of toxin based models in drug testing. The identification of genes causing ARPD has provided etiologically based PD models and has illuminated common pathways involved in ARPD pathology, highlighting potential therapeutic targets that may halt disease progression. parkin, PINK1, DJ-1, ATP13A2, and FBXO7 have all been linked to mitochondrial quality control, indicating that mitochondrial dysfunction may be a primary cause of DA neurodegeneration. As a result, pharmacologically manipulating pathways such as mitophagy and mitochondrial biogenesis, perhaps in combination with enhancing mitochondrial health, may prove to be efficacious.
Oxidative stress and stress-induced cell death appear to be regulated by ARPD genes. Studies have found that upregulating endogenous antioxidants or supplementing with exogenous antioxidants dramatically decreases neuronal death. These studies, however, have repetitively failed to show neuroprotective benefits in clinic, implying that oxidative stress and apoptosis are end results of more upstream issues. Additional research investigating oxidative stress in genetic ARPD models may provide better pathological and therapeutic relevance. Furthermore, using antioxidants and/or cell death inhibitors in combination with drugs that selectively target upstream pathways, such as mitochondrial quality control, may be more therapeutically efficacious.
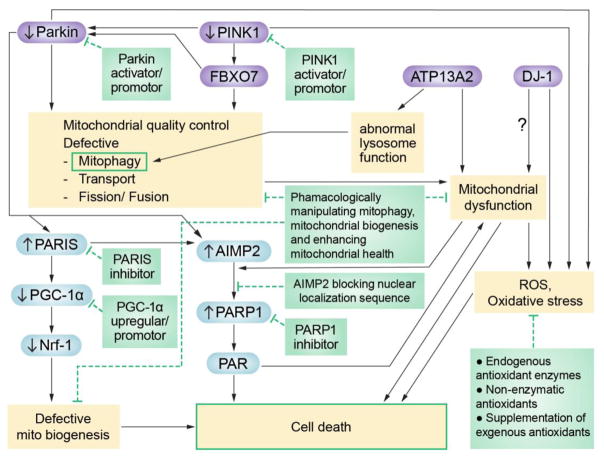
Figure 1. Schematic of common pathways regulated by ARPD genes and their potential therapeutic targets.
Mutations in each gene (in purple) lead to the dysregulation of shared pathways and cellular functions (in yellow) as well as specific transcription factors and proteins (in blue). Shaded green boxes offer potential therapeutic strategies to inhibit specific targets that lead to disease progression.
Table 1.
Overview of verified ARPD genes, their disease prevalence, and pathological/clinical phenotypes.
| Gene | Disease Prevalence | Pathological Phenotype | Clinical Phenotype | Reference |
|---|---|---|---|---|
| Parkin | ~50% of ARPD cases; 8.6% of early-onset PD | LB absent in most cases | Early-onset, good L- DOPA responsiveness, slow disease course, dystonia, variable psychiatric issues | (Bonifati, 2014; Lill, 2016) |
| PINK1 | 3.7% of early-onset PD | LB present in the only post mortem brain autopsied | Early-onset, L-DOPA responsive slow disease course | (Bonifati, 2014; Lill, 2016) |
| DJ-1 | 0.4% of early- onset PD | unknown | Early-onset, L-DOPA responsive slow disease course | (Bonifati, 2014; Lill, 2016) |
| FBXO7 | unknown | unknown | Juvenile-onset, atypical PD characterized by pyramidal symptoms and varying L-DOPA responsiveness | (Hernandez et al., 2016) |
| ATP13A2 | unknown | unknown | Juvenile-onset, atypical PD known as Kufor– Rakeb syndrome. characterized by pyramidal symptoms, diminished L-DOPA responsiveness, dystonia, dementia, and supranuclear palsy | (Bonifati, 2014) |
| DNAJC6 | unknown | unknown | Juvenile-onset, atypical PD characterized by poor LDOPA responsiveness, mental retardation, seizures, pyramidal signs, dystonia, and rapid disease course | (Olgiati et al., 2016) |
| SYNJ1 | unknown | unknown | Juvenile-onset, atypical PD characterized by poor LDOPA responsiveness, rapid progression in initial stages followed by stabilization in later stages, and variable atypical signs such as seizures, cognitive impairment, developmental issues | (Drouet, 2014) |
| PLA2G6 | unknown | unknown | Juvenile-onset, atypical PD characterized by L- DOPA responsiveness, pyramidal symptoms, dystonia, and cognitive/psychiatric issues | (Miki et al., 2017) |
Table 2.
Overview of verified ARPD genes and key findings that have been critical in understanding their function as well as the pathways they regulate.
| ARPD Genes | Key Findings |
|---|---|
| parkin (PARK 2) | RING-Type E3 ligase. Remains in an autoinhibited state until activated. Is recruited to OMM during mitochondrial damage where it ubiquitinates OMM proteins and autophagy adaptors involved in mitophagy. Ubiquitinates substrates such as PARIS, Miro, and AIMP2, key factors involved in mitochondrial biogenesis, mitochondrial transport, and parthanatos, respectively. |
| PINK1 (PARK 6) | Serine/threonine kinase. Localizes to the OMM in depolarization conditions where it phosphorylates a number of substrates including parkin, ubiquitin, and autophagy adaptors involved in mitophagy. Phosphorylates Miro, a key protein in mitochondrial trafficking. It also phosphorylates PARIS priming it for ubiquitination by parkin. May aid in mitigating oxidative stress by regulating calcium efflux from the mitochondria. |
| DJ-1 (PARK 7) | Endogenous redox sensor that self-oxidizes in oxidative stress conditions. |
| ATP13A2 (PARK 9) | 5 P-Type ATPase. Localizes to lysosomal and late-endosomal membranes. Mutations in ATP13A2 cause lysosomal dysregulation as well as reduced autophagic flux and mitochondrial clearance. Thought to be involved in the oxidative stress response through the regulation of cations, such as Zn2+. |
| FBXO7 (PARK 15)* | Member of the Skp1-Cullin-F-Box type E3 ligase. Works with PINK1 to recruit parkin to OMM in mitochondrial depolarization conditions. Is required for Mfn1 ubiquitination and subsequent recruitment of autophagy machinery to the mitochondria. |
| DNJC6 (PARK 19)* | Encodes Auxilin, a member of the Hsp40 chaperone family. Works in tandem with SYNJ1to recycle synaptic vesicles. |
| SYNJ1 (PARK 20)* | Encodes synaptojanin-1, a phosphoinositide phosphatase. Works in tandem with DNAJC6 to recycle synaptic vesicles. |
| PLA2G6 (PARK 14)* | Calcium-independent phospholipase A2, group VI. Mutations in PLA2G6 cause aberrant ER calcium signaling which may impede mitophagy and the oxidative stress response. |
Mutations in these genes cause exceedingly rare, atypical forms of ARPD.
Acknowledgments
Supported by NIH/NINDS grant P50 NS38377 and the JPB Foundation. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases. X.B., V.L.D. and T.M.D. acknowledge the joint participation by the Adrienne Helis Malvin Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with the Johns Hopkins Hospital and the Johns Hopkins University School of Medicine and the Foundation’s Parkinson’s Disease Program M-2014.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Andres-Mateos E, Perier C, Zhang L, Blanchard-Fillion B, Greco TM, Thomas B, Ko HS, Sasaki M, Ischiropoulos H, Przedborski S, Dawson TM, Dawson VL. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc Natl Acad Sci U S A. 2007;104:14807–14812. doi: 10.1073/pnas.0703219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi G, Schlehe JS, LaVoie MJ, Schwarz TL. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. The Journal of Cell Biology. 2014;206:655–670. doi: 10.1083/jcb.201401070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento CF, Ashkenazi A, Jimenez-Sanchez M, Rubinsztein DC. The Parkinson’s disease-associated genes ATP13A2 and SYT11 regulate autophagy via a common pathway. Nature Communications. 2016;7:11803. doi: 10.1038/ncomms11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackinton J, Lakshminarasimhan M, Thomas KJ, Ahmad R, Greggio E, Raza AS, Cookson MR, Wilson MA. Formation of a stabilized Cysteine Sulfinic acid is critical for the Mitochondrial function of the Parkinsonism protein DJ-1. Journal of Biological Chemistry. 2009;284:6476–6485. doi: 10.1074/jbc.M806599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifati V. Genetics of Parkinson’s disease – state of the art, 2013. Parkinsonism & Related Disorders. 2014;20:S23–S28. doi: 10.1016/S1353-8020(13)70009-9. [DOI] [PubMed] [Google Scholar]
- Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- Burchell VS, Nelson DE, Sanchez-Martinez A, Delgado-Camprubi M, Ivatt RM, Pogson JH, Randle SJ, Wray S, Lewis PA, Houlden H, Abramov AY, Hardy J, Wood NW, Whitworth AJ, Laman H, Plun-Favreau H. The Parkinson’s disease–linked proteins Fbxo7 and Parkin interact to mediate mitophagy. Nature Neuroscience. 2013;16:1257–1265. doi: 10.1038/nn.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese VP, Dorsey ER, Constantinescu R, Thompson JP, Biglan KM, Holloway RG, Kieburtz K, Marshall FJ, Ravina BM, Schifitto G, Siderowf A, Tanner CM. Projected Number Of People With Parkinson Disease In The Most Populous Nations, 2005 Through 2030. Neurology. 2007;69:223–224. doi: 10.1212/01.wnl.0000247740.47667.03. [DOI] [PubMed] [Google Scholar]
- Chan JYH, Chan SHH. Activation of endogenous antioxidants as a common therapeutic strategy against cancer, neurodegeneration and cardiovascular diseases: A lesson learnt from DJ-1. Pharmacology & Therapeutics. 2015;156:69–74. doi: 10.1016/j.pharmthera.2015.09.005. [DOI] [PubMed] [Google Scholar]
- Charan RA, Johnson BN, Zaganelli S, Nardozzi JD, LaVoie MJ. Inhibition of apoptotic Bax translocation to the mitochondria is a central function of parkin. Cell Death and Disease. 2014;5:e1313. doi: 10.1038/cddis.2014.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charan RA, LaVoie MJ. Pathologic and therapeutic implications for the cell biology of parkin. Molecular and Cellular Neuroscience. 2015;66:62–71. doi: 10.1016/j.mcn.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi RK, Beal MF. Mitochondria targeted therapeutic approaches in Parkinson’s and Huntington’s diseases. Molecular and Cellular Neuroscience. 2013;55:101–114. doi: 10.1016/j.mcn.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey V, Cagalinec M, Liiv J, Safiulina D, Hickey MA, Kuum M, Liiv M, Anwar T, Eskelinen EL, Kaasik A. BECN1 is involved in the initiation of mitophagy. Autophagy. 2014;10:1105–1119. doi: 10.4161/auto.28615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen T, Haddad D, Wauters F, Van Humbeeck C, Mandemakers W, Koentjoro B, Sue C, Gevaert K, De Strooper B, Verstreken P, Vandenberghe W. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Human Molecular Genetics. 2014;23:5227–5242. doi: 10.1093/hmg/ddu244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CN, Baughman JM, Phu L, Tea JS, Yu C, Coons M, Kirkpatrick DS, Bingol B, Corn JE. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nature Cell Biology. 2015;17:160–169. doi: 10.1038/ncb3097. [DOI] [PubMed] [Google Scholar]
- Di Fonzo A, Dekker MC, Montagna P, Baruzzi A, Yonova EH, Correia Guedes L, Szczerbinska A, Zhao T, Dubbel-Hulsman LO, Wouters CH, de Graaff E, Oyen WJ, Simons EJ, Breedveld GJ, Oostra BA, Horstink MW, Bonifati V. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology. 2009;72:240–245. doi: 10.1212/01.wnl.0000338144.10967.2b. [DOI] [PubMed] [Google Scholar]
- Drouet VaSL. Synaptojanin 1 Mutation in Parkinson’s Disease Brings Further Insight into the Neuropathological Mechanisms. BioMed Research International. 2014 doi: 10.1155/2014/289728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcan TM, Kontogiannea M, Bedard N, Wing SS, Fon EA. Ataxin-3 Deubiquitination is coupled to Parkin Ubiquitination via E2 Ubiquitin-conjugating enzyme. Journal of Biological Chemistry. 2011;287:531–541. doi: 10.1074/jbc.M111.288449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcan TM, Tang MY, Perusse JR, Dashti EA, Aguileta MA, McLelland GL, Gros P, Shaler TA, Faubert D, Coulombe B, Fon EA. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. The EMBO Journal. 2014;33:2473–2491. doi: 10.15252/embj.201489729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Vos M, Vilain S, Swerts J, De Sousa Valadas J, Van Meensel S, Schaap O, Verstreken P. Aconitase causes iron toxicity in Drosophila pink1 Mutants. PLoS Genetics. 2013;9:e1003478. doi: 10.1371/journal.pgen.1003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proceedings of the National Academy of Sciences. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg ME, Cooper JM, Schapira AHV, Taanman JW. Silencing of PINK1 expression affects Mitochondrial DNA and Oxidative Phosphorylation in DOPAMINERGIC cells. PLoS ONE. 2009;4:e4756. doi: 10.1371/journal.pone.0004756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier K, McLelland G-L, Fon EA. Parkin- and PINK1-Dependent Mitophagy in Neurons: Will the real pathway please stand up? Frontiers in Neurology. 2013:4. doi: 10.3389/fneur.2013.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald A, Arns B, Seibler P, Rakovic A, Münchau A, Ramirez A, Sue CM, Klein C. ATP13A2 mutations impair mitochondrial function in fibroblasts from patients with Kufor-Rakeb syndrome. Neurobiology of Aging. 2012;33:1843.e1841–1843.e1847. doi: 10.1016/j.neurobiolaging.2011.12.035. [DOI] [PubMed] [Google Scholar]
- Gusdon AM, Zhu J, Van Houten B, Chu CT. ATP13A2 regulates mitochondrial bioenergetics through macroautophagy. Neurobiology of Disease. 2012;45:962–972. doi: 10.1016/j.nbd.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Yasuda T, Shiraishi C, Fujiwara K, Przedborski S, Mochizuki H, Yoshikawa K. Promotion of mitochondrial biogenesis by necdin protects neurons against mitochondrial insults. Nature Communications. 2016;7:10943. doi: 10.1038/ncomms10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Ishimori C, Takahashi-Niki K, Taira T, Kim Y-c, Maita H, Maita C, Ariga H, Iguchi-Ariga SMM. DJ-1 binds to mitochondrial complex I and maintains its activity. Biochemical and Biophysical Research Communications. 2009;390:667–672. doi: 10.1016/j.bbrc.2009.10.025. [DOI] [PubMed] [Google Scholar]
- Heo JM, Ordureau A, Paulo Joao A, Rinehart J, Harper JW. The PINK1-PARKIN Mitochondrial Ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote Mitophagy. Molecular Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Reed X, Singleton AB. Genetics in Parkinson disease: Mendelian versus non-Mendelian inheritance. J Neurochem. 2016;139(Suppl 1):59–74. doi: 10.1111/jnc.13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz Nicholas T, Berthet A, Sos Martin L, Thorn Kurt S, Burlingame Al L, Nakamura K, Shokat Kevan M. A neo-substrate that amplifies catalytic activity of Parkinson’s-disease-related Kinase PINK1. Cell. 2013;154:737–747. doi: 10.1016/j.cell.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam SZ, Trickler W, Kimura S, Binienda ZK, Paule MG, Slikker W, Li S, Clark RA, Ali SF. Neuroprotective efficacy of a new brain-penetrating C-Abl inhibitor in a Murine Parkinson’s disease model. PLoS ONE. 2013;8:e65129. doi: 10.1371/journal.pone.0065129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Kang SU, Zhang S, Karuppagounder S, Xu J, Lee YK, Kang BG, Zhang J, Pletnikova O, Troncoso JC, Pirooznia S, Andrabi SA, Dawson VL, Dawson TM. Adult conditional knockout of PGC-1 leads to loss of Dopamine Neurons. eNeuro. 2016:3. doi: 10.1523/ENEURO.0183-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunakaran S, Diwakar L, Saeed U, Agarwal V, Ramakrishnan S, Iyengar S, Ravindranath V. Activation of apoptosis signal regulating kinase 1 (ASK1) and translocation of death-associated protein, Daxx, in substantia nigra pars compacta in a mouse model of Parkinson’s disease: Protection by -lipoic acid. The FASEB Journal. 2007;21:2226–2236. doi: 10.1096/fj.06-7580com. [DOI] [PubMed] [Google Scholar]
- Karuppagounder SS, Brahmachari S, Lee Y, Dawson VL, Dawson TM, Ko HS. The c-Abl inhibitor, nilotinib, protects dopaminergic neurons in a preclinical animal model of Parkinson’s disease. Sci Rep. 2014;4:4874. doi: 10.1038/srep04874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kett LR, Dauer WT. Endolysosomal dysfunction in Parkinson’s disease: Recent developments and future challenges. Mov Disord. 2016;31:1433–1443. doi: 10.1002/mds.26797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kett LR, Stiller B, Bernath MM, Tasset I, Blesa J, Jackson-Lewis V, Chan RB, Zhou B, Di Paolo G, Przedborski S, Cuervo AM, Dauer WT. -Synuclein-Independent Histopathological and motor deficits in mice lacking the Endolysosomal Parkinsonism protein Atp13a2. Journal of Neuroscience. 2015;35:5724–5742. doi: 10.1523/JNEUROSCI.0632-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal P. The relationship between parkin and protein aggregation in neurodegenerative diseases. Frontiers in Psychiatry. 2010:1. doi: 10.3389/fpsyt.2010.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GH, Kim JE, Rhie SJ, Yoon S. The Role of Oxidative Stress in Neurodegenerative Diseases. Exp Neurobiol. 2015;24:325–340. doi: 10.5607/en.2015.24.4.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Ko HS, Lee Y, Shin JH, Karuppagounder SS, Gadad BS, Koleske AJ, Pletnikova O, Troncoso JC, Dawson VL, Dawson TM. Phosphorylation by the c-Abl protein tyrosine kinase inhibits parkin’s ubiquitination and protective function. Proc Natl Acad Sci U S A. 2010;107:16691–16696. doi: 10.1073/pnas.1006083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic M, Ludtmann Marthe HR, Bading H, Hershfinkel M, Steer E, Chu Charleen T, Abramov Andrey Y, Sekler I. PKA Phosphorylation of NCLX reverses Mitochondrial calcium overload and Depolarization, promoting survival of PINK1-Deficient Dopaminergic Neurons. Cell Reports. 2015;13:376–386. doi: 10.1016/j.celrep.2015.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebiehl G, Ruckerbauer S, Burbulla LF, Kieper N, Maurer B, Waak J, Wolburg H, Gizatullina Z, Gellerich FN, Woitalla D, Riess O, Kahle PJ, Proikas-Cezanne T, Krüger R. Reduced Basal Autophagy and impaired Mitochondrial dynamics due to loss of Parkinson’s disease-associated protein DJ-1. PLoS ONE. 2010;5:e9367. doi: 10.1371/journal.pone.0009367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda Y. Parkin enhances mitochondrial biogenesis in proliferating cells. Human Molecular Genetics. 2006;15:883–895. doi: 10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]
- Lee Y, Karuppagounder SS, Shin JH, Lee YI, Ko HS, Swing D, Jiang H, Kang SU, Lee BD, Kang HC, Kim D, Tessarollo L, Dawson VL, Dawson TM. Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss. Nat Neurosci. 2013;16:1392–1400. doi: 10.1038/nn.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Stevens DA, Kang S-U, Jiang H, Lee Y-I, Ko HS, Scarffe LA, Umanah GE, Kang H, Ham S, Kam T-I, Allen K, Brahmachari S, Kim JW, Neifert S, Yun SP, Fiesel FC, Springer W, Dawson VL, Shin J-H, Dawson TM. PINK1 primes parkin-mediated ubiquitination of PARIS in dopaminergic neuronal survival. Cell Reports. 2017 doi: 10.1016/j.celrep.2016.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CAP. Genome-wide and functional Annotation of human E3 Ubiquitin Ligases identifies MULAN, a Mitochondrial E3 that regulates the Organelle’s dynamics and signaling. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang H, Gao Y, Li L, Tang C, Wen G, Zhou Y, Zhou M, Mao L, Fan Y. Protective effects of Quercetin on Mitochondrial Biogenesis in experimental traumatic brain injury via the Nrf2 signaling pathway. PLOS ONE. 2016;11:e0164237. doi: 10.1371/journal.pone.0164237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill CM. Genetics of Parkinson’s disease. Mol Cell Probes. 2016;30:386–396. doi: 10.1016/j.mcp.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Lin W, Kang U. Structural determinants of PINK1 topology and dual subcellular distribution. BMC Cell Biology. 2010;11:90. doi: 10.1186/1471-2121-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Sawada T, Lee S, Yu W, Silverio G, Alapatt P, Millan I, Shen A, Saxton W, Kanao T, Takahashi R, Hattori N, Imai Y, Lu B. Parkinson’s Disease–Associated Kinase PINK1 regulates Miro protein level and Axonal transport of Mitochondria. PLoS Genetics. 2012;8:e1002537. doi: 10.1371/journal.pgen.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Tanji K, Mori F, Kakita A, Takahashi H, Wakabayashi K. PLA2G6 accumulates in Lewy bodies in PARK14 and idiopathic Parkinson’s disease. Neurosci Lett. 2017;645:40–45. doi: 10.1016/j.neulet.2017.02.027. [DOI] [PubMed] [Google Scholar]
- Misko A, Jiang S, Wegorzewska I, Milbrandt J, Baloh RH. Mitofusin 2 is necessary for transport of Axonal Mitochondria and interacts with the Miro/Milton Complex. Journal of Neuroscience. 2010;30:4232–4240. doi: 10.1523/JNEUROSCI.6248-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DJ, Dawson VL, Dawson TM. Lessons from Drosophila models of DJ-1 deficiency. Sci Aging Knowledge Environ. 2006;2006:pe2. doi: 10.1126/sageke.2006.2.pe2. [DOI] [PubMed] [Google Scholar]
- Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson’s disease. Annu Rev Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- Müftüoglu M, Elibol B, Dalm zrak O, Ercan A, Kulaksız G, Ögüs H, Dalkara T, Özer N. Mitochondrial complex I and IV activities in leukocytes from patients with parkin mutations. Movement Disorders. 2003;19:544–548. doi: 10.1002/mds.10695. [DOI] [PubMed] [Google Scholar]
- Müller-Rischart Anne K, Pilsl A, Beaudette P, Patra M, Hadian K, Funke M, Peis R, Deinlein A, Schweimer C, Kuhn PH, Lichtenthaler Stefan F, Motori E, Hrelia S, Wurst W, Trümbach D, Langer T, Krappmann D, Dittmar G, Tatzelt J, Winklhofer Konstanze F. The E3 Ligase Parkin maintains Mitochondrial integrity by increasing linear Ubiquitination of NEMO. Molecular Cell. 2013;49:908–921. doi: 10.1016/j.molcel.2013.01.036. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of Cell Biology. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olgiati S, Quadri M, Fang M, Rood JP, Saute JA, Chien HF, Bouwkamp CG, Graafland J, Minneboo M, Breedveld GJ, Zhang J, Verheijen FW, Boon AJ, Kievit AJ, Jardim LB, Mandemakers W, Barbosa ER, Rieder CR, Leenders KL, Wang J, Bonifati V International Parkinsonism Genetics N. DNAJC6 Mutations Associated With Early-Onset Parkinson’s Disease. Ann Neurol. 2016;79:244–256. doi: 10.1002/ana.24553. [DOI] [PubMed] [Google Scholar]
- Park JS, Koentjoro B, Veivers D, Mackay-Sim A, Sue CM. Parkinson’s disease-associated human ATP13A2 (PARK9) deficiency causes zinc dyshomeostasis and mitochondrial dysfunction. Human Molecular Genetics. 2014;23:2802–2815. doi: 10.1093/hmg/ddt623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periquet M. Parkin mutations are frequent in patients with isolated early-onset parkinsonism. Brain. 2003;126:1271–1278. doi: 10.1093/brain/awg136. [DOI] [PubMed] [Google Scholar]
- Pickrell Alicia M, Youle Richard J. The roles of PINK1, Parkin, and Mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde KR, Smith HL, Chau KY, Schapira AHV. PINK1 disables the anti-fission machinery to segregate damaged mitochondria for mitophagy. The Journal of Cell Biology. 2016;213:163–171. doi: 10.1083/jcb.201509003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez A, Heimbach A, Grundemann J, Stiller B, Hampshire D, Cid LP, Goebel I, Mubaidin AF, Wriekat AL, Roeper J, Al-Din A, Hillmer AM, Karsak M, Liss B, Woods CG, Behrens MI, Kubisch C. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- Ren H, Fu K, Mu C, Li B, Wang D, Wang G. DJ-1, a cancer and Parkinson’s disease associated protein, regulates autophagy through JNK pathway in cancer cells. Cancer Letters. 2010;297:101–108. doi: 10.1016/j.canlet.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Requejo-Aguilar R, Lopez-Fabuel I, Jimenez-Blasco D, Fernandez E, Almeida A, Bolaños Juan P. DJ1 represses glycolysis and cell proliferation by transcriptionally up-regulating pink1. Biochemical Journal. 2015;467:303–310. doi: 10.1042/BJ20141025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Brough D, Gibson R, Loddick S, Rothwell N. A selective, non-peptide caspase-1 inhibitor, VRT-018858, markedly reduces brain damage induced by transient ischemia in the rat. Neuropharmacology. 2007;53:638–642. doi: 10.1016/j.neuropharm.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Rothfuss O, Fischer H, Hasegawa T, Maisel M, Leitner P, Miesel F, Sharma M, Bornemann A, Berg D, Gasser T, Patenge N. Parkin protects mitochondrial genome integrity and supports mitochondrial DNA repair. Human Molecular Genetics. 2009;18:3832–3850. doi: 10.1093/hmg/ddp327. [DOI] [PubMed] [Google Scholar]
- Savitt JM, Dawson VL, Dawson TM. Diagnosis and treatment of Parkinson disease: molecules to medicine. J Clin Invest. 2006;116:1744–1754. doi: 10.1172/JCI29178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarffe LA, Stevens DA, Dawson VL, Dawson TM. Parkin and PINK1: Much more than mitophagy. Trends in Neurosciences. 2014;37:315–324. doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadrach KG, Rayborn ME, Hollyfield JG, Bonilha VL. DJ-1-Dependent regulation of Oxidative stress in the retinal pigment Epithelium (RPE) PLoS ONE. 2013;8:e67983. doi: 10.1371/journal.pone.0067983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Ko Han S, Kang H, Lee Y, Pletinkova O, Troconso Juan C, Dawson Valina L, Dawson Ted M. PARIS (ZNF746) repression of PGC-1α contributes to Neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens DA, Lee Y, Kang HC, Lee BD, Lee YI, Bower A, Jiang H, Kang SU, Andrabi SA, Dawson VL, Shin JH, Dawson TM. Parkin loss leads to PARIS-dependent declines in mitochondrial mass and respiration. Proc Natl Acad Sci U S A. 2015;112:11696–11701. doi: 10.1073/pnas.1500624112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strappazzon F, Nazio F, Corrado M, Cianfanelli V, Romagnoli A, Fimia GM, Campello S, Nardacci R, Piacentini M, Campanella M, Cecconi F. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015;22:419–432. doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szargel R, Shani V, Abd Elghani F, Mekies LN, Liani E, Rott R, Engelender S. The PINK1, synphilin-1 and SIAH-1 complex constitutes a novel mitophagy pathway. Hum Mol Genet. 2016;25:3476–3490. doi: 10.1093/hmg/ddw189. [DOI] [PubMed] [Google Scholar]
- Thomas KJ, McCoy MK, Blackinton J, Beilina A, van der Brug M, Sandebring A, Miller D, Maric D, Cedazo-Minguez A, Cookson MR. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Human Molecular Genetics. 2010;20:40–50. doi: 10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe J-F, Fon EA. Structure and function of Parkin, PINK1, and DJ-1, the Three Musketeers of Neuroprotection. Frontiers in Neurology. 2013:4. doi: 10.3389/fneur.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Van Humbeeck C, Cornelissen T, Hofkens H, Mandemakers W, Gevaert K, De Strooper B, Vandenberghe W. Parkin interacts with Ambra1 to induce Mitophagy. Journal of Neuroscience. 2011;31:10249–10261. doi: 10.1523/JNEUROSCI.1917-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen S, Sørensen DM, Holemans T, Holen HW, Palmgren MG, Vangheluwe P. Cellular function and pathological role of ATP13A2 and related p-type transport ATPases in Parkinson’s disease and other neurological disorders. Frontiers in Molecular Neuroscience. 2014:7. doi: 10.3389/fnmol.2014.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandiver MS, Paul BD, Xu R, Karuppagounder S, Rao F, Snowman AM, Seok Ko H, Il Lee Y, Dawson VL, Dawson TM, Sen N, Snyder SH. Sulfhydration mediates neuroprotective actions of parkin. Nature Communications. 2013;4:1626. doi: 10.1038/ncomms2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Coelln R, Dawson VL, Dawson TM. Parkin-associated Parkinson’s disease. Cell Tissue Res. 2004;318:175–184. doi: 10.1007/s00441-004-0924-4. [DOI] [PubMed] [Google Scholar]
- Wang HL, Chou AH, Wu AS, Chen SY, Weng YH, Kao YC, Yeh TH, Chu PJ, Lu CS. PARK6 PINK1 mutants are defective in maintaining mitochondrial membrane potential and inhibiting ROS formation of substantia nigra dopaminergic neurons. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2011a;1812:674–684. doi: 10.1016/j.bbadis.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011b;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, An R, Umanah GK, Park H, Nambiar K, Eacker SM, Kim B, Bao L, Harraz MM, Chang C, Chen R, Wang JE, Kam TI, Jeong JS, Xie Z, Neifert S, Qian J, Andrabi SA, Blackshaw S, Zhu H, Song H, Ming GL, Dawson VL, Dawson TM. A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science. 2016:354. doi: 10.1126/science.aad6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Serricchio M, Jauregui M, Shanbhag R, Stoltz T, Di Paolo CT, Kim PK, McQuibban GA. Deubiquitinating enzymes regulate PARK2-mediated mitophagy. Autophagy. 2015;11:595–606. doi: 10.1080/15548627.2015.1034408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Guo H, Zhang X, Tang B, Cai F, Zhou W, Song W. Hypoxia regulation of ATP13A2 (PARK9) gene transcription. Journal of Neurochemistry. 2012;122:251–259. doi: 10.1111/j.1471-4159.2012.07676.x. [DOI] [PubMed] [Google Scholar]
- Yu W, Sun Y, Guo S, Lu B. The PINK1/Parkin pathway regulates mitochondrial dynamics and function in mammalian hippocampal and dopaminergic neurons. Human Molecular Genetics. 2011;20:3227–3240. doi: 10.1093/hmg/ddr235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C-W, Hang L, Yao T-P, Lim K-L. Parkin regulation and Neurodegenerative disorders. Frontiers in Aging Neuroscience. 2016:7. doi: 10.3389/fnagi.2015.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Karsten P, Hamm S, Pogson JH, Muller-Rischart AK, Exner N, Haass C, Whitworth AJ, Winklhofer KF, Schulz JB, Voigt A. TRAP1 rescues PINK1 loss-of-function phenotypes. Human Molecular Genetics. 2013;22:2829–2841. doi: 10.1093/hmg/ddt132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, Dauer W, Schon EA, Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proceedings of the National Academy of Sciences. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZD, Sathiyamoorthy S, Angeles DC, Tan EK. Linking f-box protein 7 and parkin to neuronal degeneration in Parkinson’s disease (PD) Molecular Brain. 2016:9. doi: 10.1186/s13041-016-0218-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ. Drosophila Parkin requires PINK1 for mitochondrial translocation and ubiquitinates Mitofusin. Proceedings of the National Academy of Sciences. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]