Abstract
Purpose
Accumulating evidence has shown that dysregulation of microRNA-191 (miR-191) is closely associated with tumorigenesis and progression in a wide range of cancers. This study aimed to explore the potential role of miR-191 in esophageal squamous cell carcinoma (ESCC).
Materials and Methods
miR-191 expression was assessed in 93 ESCC tissue specimens by real-time polymerase chain reaction, and survival analysis was performed via Kaplan-Meier and Cox regression analyses. 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide, plate colony-forming, BrdU, and Transwell assays were conducted to observe the effect of miR-191 on ESCC proliferation and invasion. Luciferase reporter and western blot assays were taken to identify target genes of miR-191.
Results
miR-191 was overexpressed in 93 cases of ESCC, compared with adjacent normal tissues, and miR-191 expression was significantly related to differentiation, depth of invasion, TNM stage, lymph node metastasis, and distant metastasis of tumor. Kaplan-Meier and Cox regression analyses demonstrated that overexpression of miR-191 was an independent and significant predictor of ESCC prognosis. Both gain-of-function and loss-of-function experiments showed that miR-191 promoted ESCC cell proliferation and invasion activities in vitro. Early growth response 1 (EGR1), a tumor suppressor, was predicted as a direct target of miR-191. Luciferase reporter and western blot assays proved that miR-191 reduced EGR1 expression by directly binding its 3' untranslated region. Moreover, EGR1 knockdown by siRNA enhanced ESCC cell growth and invasion.
Conclusion
Our findings provide specific biological roles of miR-191 in ESCC survival and progression. Targeting the novel miR-191/EGR1 axis represents a potential new therapeutic way to block ESCC development.
Keywords: miR-191, esophageal squamous cell carcinoma, early growth response 1, prognosis, proliferation, invasion
INTRODUCTION
Esophageal cancer (EC) is one of the most frequent malignant neoplasms worldwide and ranks as the sixth-leading cause of cancer-related death.1 EC consists of two main subtypes, namely esophageal adenocarcinoma, which is frequently occurring in developed countries, such as North America and Europe, and esophageal squamous cell carcinoma (ESCC), which is more common in developing countries, such as East Asian countries.2,3 ESCC remains the main histological subtype, accounting for more than 90% of all ECs, and the 5-year survival rate of patients with ESCC is less than 10%.2,3 Therefore, it is observably necessary to search and explore novel molecular targets for ESCC therapy.
Recently, microRNAs (miRNAs) have attracted more and more attention in various biological processes, including cell proliferation, migration, differentiation, and apoptosis.4,5 It is not surprising that miRNA dysregulation correlates with tumorigenesis in many cancers including ESCC.6 Many studies reported that microRNA-191 (miR-191) can act as a multifunctional miRNA, with important roles in diabetes-type 2, Crohn's, pulmonary hypertension, Alzheimer's, and neoplastic diseases.7,8,9,10,11 It has been reported that miR-191 expression is dysregulated in several cancers and correlated with malignant clinical features and tumor prognosis.12,13,14,15,16,17 On the one hand, miR-191 is overexpressed in several cancers, including breast cancer, colon cancer, and lung cancer, and plays an oncogenic role.12,13,14,18 On the other hand, miR-191 has been found to be down-regulated in several other cancer types, including retinoblastoma, thyroid follicular tumor, and melanoma, suggesting its tumor suppressive role.15,16,17 However, its clinical significance in human ESCC remains unclear.
The aims of the present study were to evaluate the relationships between miR-191 expression level and the clinicopathological characteristics and outcomes of ESCC patients and to explore the functional role of miR-191 in ESCC cell lines. We found that miR-191 overexpression independently affects prognosis in ESCC. miR-191 promotes ESCC cell proliferation and invasion in vitro by directly targeting early growth response 1 (EGR1).
MATERIALS AND METHODS
Patients and specimens
Specimens of 93 ESCCs and 58 adjacent normal tissues were obtained from 93 patients who underwent surgical treatment of ESCC at Affiliated Hospital of Guangdong Medical University between January 2010 and December 2013. All the patients were diagnosed by pathological examination, and none of the patients had received preoperative chemotherapy or radiotherapy. Patient data, including kinds of clinicopathological features, was collected, and follow-up information after surgery was acquired by telephone interview. The study protocol was approved by the Ethics Committee of Guangdong Medical University, and all the patients provided written informed consent (Approval no. GMU2009035).
RNA isolation and quantitative real-time polymerase chain reaction (RT-PCR)
Total RNA from all tissues and cells were isolated using Trizol reagent (Invitrogen, Carlsbad, NM, USA). RT-PCR was carried out applying SYBR Premix Ex Taq II (TaKaRa, Dalian, China) and detected in a LightCycler 480 system (Roche, Basel, Switzerland) according the manufacturers' instructions. All miRNA primers were obtained from RiBoBio Co. (Guangzhou, China). EGR1 primer (TaKaRa) is shown in Supplementary Table 1 (only online). U6 or glyceraldehyde-3-phosphate dehydrogenase was used as an internal control, and the fold change was calculated by 2-ΔΔCt.
Cell lines and transfection
ESCC cell lines EC9706 and TE-1 were purchased from the Chinese Academy of Medical Science (Beijing, China) and maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 µg/mL streptomycin at 37℃ in a humidified incubator with 5% CO2. miR-191 mimic, mimic control, miR-191 inhibitor, and inhibitor control were purchased from RiBoBio. EGR1 siRNA was obtained from Santa Cruz Biotechnology Inc. (sc-29303; Santa Cruz, Dallas, TX, USA). Cell transfection was performed applying Lipofectamine 2000 (Invitrogen).
MTT cell proliferation assay
Cell proliferation was measured by the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay. Briefly, the cells (1×103 cells/well) were seeded into each well of seven 96 well plates. At a different time point after transfection, 20 µL of MTT (5 mg/mL, Sigma, MO, USA) was added. Then, the reaction density was read at 490 nm on a Varioskan Flash Multimode Reader (Thermo Fisher, Waltham, MA, USA).
Plate colony-forming assay
Log phase cells were trypsinized into single cell suspensions and plated in 10-cm plates at indicated cell numbers. The colonies were stained with Giemsa, and the total number of colonies was counted.
BrdU assay
The BrdU assay was performed using a BrdU Cell Proliferation Chemiluminescent Assay Kit (Cell Signaling Technology Inc., Denver, CO, USA) according to the manufacturer's instructions.
Transwell assay
Migration and invasion was detected by Transwell assay. Briefly, 1×105 transfected cells were seeded in FBS-free media into the upper chamber of each transwell, which was pre-coated with Matrigel. Medium with 20% FBS was placed in the lower chamber. 24 hours later, non-invasive cells in the upper chamber were removed with a cotton swab, while the cells on the lower surface were fixed and stained with 0.1% crystal violet and photographed. Pictures of three random fields from triplicate wells were recorded, and the number of cells was counted. For the migration assays, all procedures were the same as in invasion assays, except each chamber without Matrigel coating.
Luciferase reporter assays
Wild-type 3′UTR of EGR1 and mutant constructs, which were produced via mutations in the binding site of miR-191 in EGR1's 3′UTR were cloned downstream of the Luciferase gene in the psiCHECK-2 Luciferase vector. Luciferase reporter vectors and miR-191 mimic were transfected into HEK293T and EC9706 cells with Lipofectamine 2000, and Luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Western blot
Whole-cell lysates were obtained using RIPA buffer (NobleRyder, Beijing, China) according to the manufacturer's instructions. Western blot analysis was conducted as shown previously.19 The primary antibodies used were anti-EGR1 (Abcam, Cambridge, UK) and β-actin (Sigma).
Statistical analysis
All statistical analyses were conducted using SPSS software (version 21.0, IBM Corp., Armonk, NY, USA). χ2 test was used to analyze relationships between miR-191 expression levels and clinicopathological factors. Overall survival curves were analyzed with the Kaplan-Meier method and compared through the log-rank test. On the basis of Cox proportional hazards model, univariate and multivariable survival analyses were conducted. p<0.05 was regarded as statistically significant.
RESULTS
miR-191 is markedly up-regulated in ESCC tissues and correlated with malignant clinicopathological features
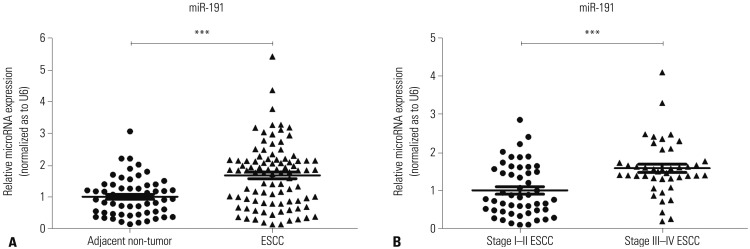
To classify the clinical significance of miR-191 in ESCC, we investigated its expression in 93 cases of ESCC tissues with RT-PCR. The expression level of miR-191 was much higher in ESCC tissues, compared with adjacent normal samples (Fig. 1A). In addition, miR-191 expression in Stage I–II were lower than that in Stage III–IV (Fig. 1B). Next, we investigated the relationships between miR-191 expression and clinicopathological factors of ESCC patients. According to the mean level of miR-191 expression, ESCC patients were divided into two groups, with 41 in the low-expression group and 52 in the high-expression group. As shown in Table 1, miR-191 expression was significantly correlated with ESCC differentiation, depth of invasion, TNM stage, lymph node metastasis, and distant metastasis, but not sex, age, and tumor size.
Fig. 1. miR-191 is overexpressed in ESCC tissues. (A) The relative expression levels of miR-191 in adjacent non-cancerous esophageal and ESCC samples. (B) The relative expression levels of miR-191 in Stage I–II and III–IV ESCC tissues. ***p<0.001. miRNA, microRNA; miR-191, microRNA-191; ESCC, esophageal squamous cell carcinoma.
Table 1. Associations of miR-191 Expression in ESCC Tissues with Demographic and Clinicopathologic Characteristics.
| Category | n=93 | miR-191 expression | p value* | |
|---|---|---|---|---|
| Low (n=41) | High (n=52) | |||
| Gender | 0.976 | |||
| Male | 77 | 34 | 43 | |
| Female | 16 | 7 | 9 | |
| Age | 0.144 | |||
| ≤65 | 51 | 19 | 32 | |
| >65 | 42 | 22 | 20 | |
| Tumor size, cm | 0.697 | |||
| <5 | 52 | 22 | 30 | |
| ≥5 | 41 | 19 | 22 | |
| Differentiation | 0.037 | |||
| Well and moderate | 57 | 30 | 27 | |
| Poorly and not | 36 | 11 | 25 | |
| Depth of invasion | 0.010 | |||
| T1–T2 | 24 | 16 | 8 | |
| T3–T4 | 69 | 25 | 44 | |
| Stages | <0.001 | |||
| I–II | 50 | 32 | 18 | |
| III–IV | 43 | 9 | 34 | |
| Lymph node metastases | <0.001 | |||
| 0 | 45 | 28 | 17 | |
| ≥1 | 48 | 13 | 35 | |
| Metastases to other organs | 0.025 | |||
| Present | 6 | 0 | 6 | |
| Absent | 87 | 41 | 46 | |
miR-191, microRNA-191; ESCC, esophageal squamous cell carcinoma.
*p values were calculated through the χ2 test to analyze the relationship between miR-191 expression and clinicopathologic characteristics.
miR-191 is an independent predictor of ESCC prognosis
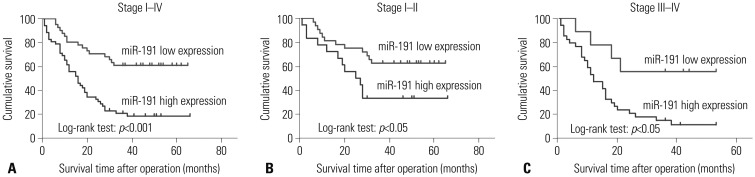
In order to explore the prognostic value of miR-191 in ESCC, Kaplan-Meier analysis and log-rank test were performed to observe relationships with overall survival. Kaplan-Meier survival curves indicated that higher miR-191 expression brought a shorter survival (p<0.001) (Fig. 2A). Higher expression of miR-191 led to a poorer prognosis, compared with low miR-191, with the unadjusted hazard ratio (HR) being 3.449 [95% confidence interval (CI): 1.900–6.260, p<0.001] (Table 2). Furthermore, the difference of overall survival between the two groups seemed to be more distinct in Stage III–IV than Stage I–II (Fig. 2B and C). The factors found to be statistically important in univariatedistant metastasis, and analysis, including depth of invasion, TNM stage, differentiation, lymph node metastasis, distant metastasis, and miR-191 expression, were selected to perform multivariate analysis. Therein, distant metastasis and miR-191 expression were independent prognostic factors for ESCC (Table 2). Taken together, these findings suggested that miR-191 expression is an independent and significant predictor of ESCC prognosis.
Fig. 2. Kaplan-Meier survival curves of ESCC patients with different level of miR-191 expression stratified by the TNM stage. (A) Association of miR-191 expression with overall survival (cumulative survival) in all stages. (B) Association of miR-191 expression with overall survival in Stage I–II. (C) Association of miR-191 expression with overall survival in Stage III–IV. ESCC, esophageal squamous cell carcinoma; miR-191, microRNA-191.
Table 2. Univariate Analysis and Multivariate Analysis of Correlations among Clinicopathological Parameters and Survival Time of Patients with Gastric Cancer.
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI for HR | p | HR | 95% CI for HR | p | |
| Gender | ||||||
| Male vs. female | 0.827 | 0.405–1.685 | 0.600 | |||
| Age | ||||||
| ≤65 vs. >65 | 0.886 | 0.525–1.496 | 0.651 | |||
| Tumor size (cm) | ||||||
| <5 vs. ≥5 | 0.924 | 0.546–1.565 | 0.769 | |||
| Depth of invasion | ||||||
| T1–T2 vs. T3–T4 | 3.417 | 1.543–7.566 | 0.002 | 2.547 | 0.964–6.728 | 0.059 |
| Differentiation | ||||||
| Well and moderate vs. poorly and not | 1.745 | 1.036–2.940 | 0.036 | 1.324 | 0.748–2.343 | 0.335 |
| Stages | ||||||
| I–II vs. III–IV | 2.572 | 1.506–4.393 | 0.001 | 0.937 | 0.159–5.535 | 0.943 |
| Lymph node metastases | ||||||
| 0 vs. ≥1 | 2.182 | 1.269–3.753 | 0.005 | 1.631 | 0.303–8.783 | 0.569 |
| Distant metastasis | ||||||
| Present vs. absent | 4.274 | 1.769–10.325 | 0.001 | 2.593 | 1.038–6.475 | 0.041 |
| miR-191 expression | ||||||
| Low vs. high | 3.449 | 1.900–6.260 | <0.001 | 2.245 | 1.143–4.410 | 0.019 |
miR-191, microRNA-191; CI, confidence interval; HR, hazard ratio.
miR-191 promotes ESCC cell proliferation and invasion in vitro
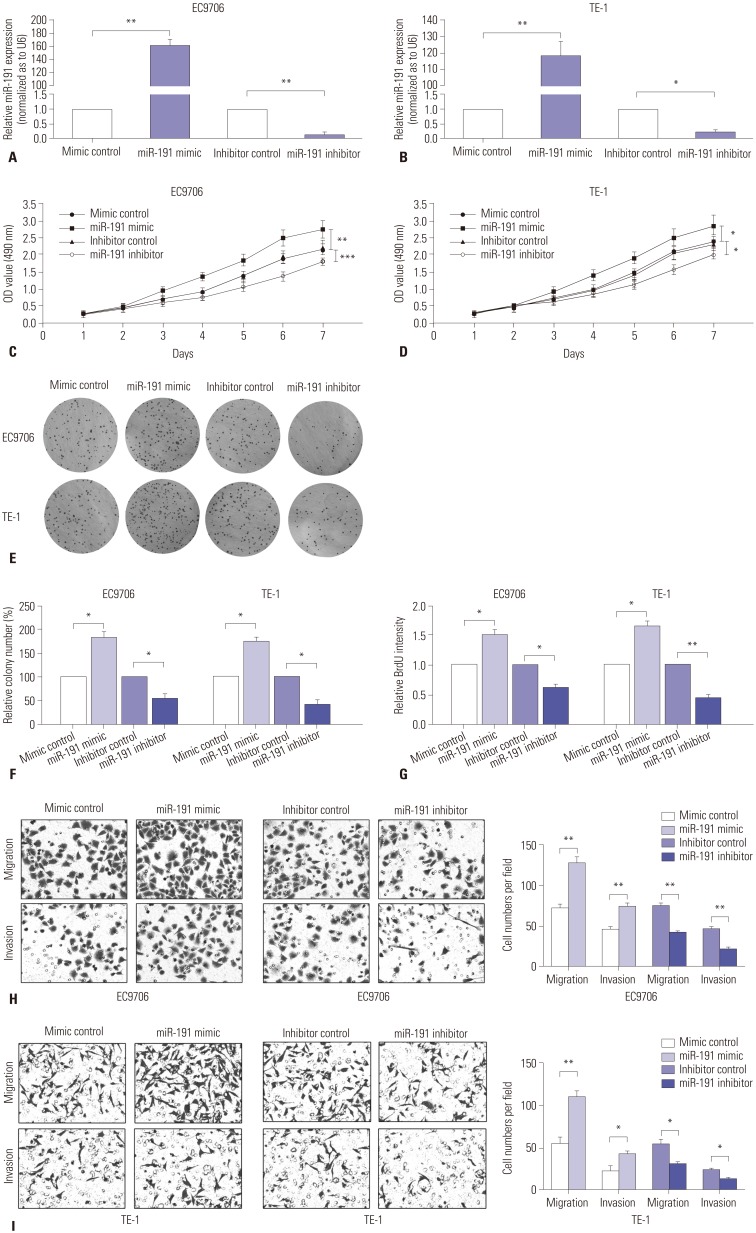
To further characterize the role of miR-191 in ESCC tumorigenesis, miR-191 mimic and inhibitor were transfected into EC9706 or TE-1 cells, respectively, to conduct gain-of-function and loss-of-function assays. RT-PCR results displayed that miR-191 mimic dramatically up-regulated miR-191 expression in EC9706 and TE-1 cells, compared with mimic control, while miR-191 inhibitor significantly reduced miR-191 expression (Fig. 3A and B). MTT assays showed that up-regulation of miR-191 significantly increased EC9706 and TE-1 cell proliferation, while down-regulation of miR-191 decreased cell growth (Fig. 3C and D). A significantly reduced number of cell colonies were observed in plates where EC9706 or TE-1 cells with down-regulation of miR-191 were seeded, and ESCC cells with miR-191 mimic transfection exhibited stronger capacity to form colonies (Fig. 3E and F). As DNA replication is a key step during cell mitosis, we also used BrdU to evaluate cell proliferation. By doing so, we noted that ectopic miR-191 increased BrdU intensity, while miR-191 suppression decreased BrdU intensity in ESCC cells (Fig. 3G). Next, the function of miR-191 in ESCC cell migration and invasion was observed. We found that up-regulation of miR-191 in EC9706 and TE-1 cells significantly promoted the migration of cells in Transwell assays without Matrigel and increased the invasion in Transwell assays with Matrigel (Fig. 3H). In contrast, down-regulation of miR-191 reduced ESCC cell migration and invasion (Fig. 3I). Collectively, these observations suggested that miR-191 acts as an oncogene in ESCC progression.
Fig. 3. miR-191 promotes ESCC cell proliferation and invasion in vitro. (A) RT-PCR analysis of miR-191 expression in EC9706 cells transfected with miR-191 mimic or inhibitor and corresponding control. (B) RT-PCR analysis of miR-191 expression in TE-1 cells transfected with miR-191 mimic or inhibitor and corresponding control. (C) MTT assay of EC9706 cells transfected with miR-191 mimic or inhibitor and corresponding control. (D) MTT assay of TE-1 cells transfected with miR-191 mimic or inhibitor and corresponding control. (E) The colony formation assays in EC9706 and TE-1 cells transfected with miR-191 mimic or inhibitor and corresponding control. Colonies were evaluated and values were reported as the ratio. (G) The BrdU assays from EC9706 and TE-1 cells transfected with miR-191 mimic or inhibitor and corresponding control. *p<0.05, **p<0.01, ***p<0.001 miR-191, microRNA-191; ESCC, esophageal squamous cell carcinoma; RT-PCR, real-time polymerase chain reaction; MTT, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide; OD, optical density. (H) Transwell assays of EC9706 cells transfected with miR-191 mimic or inhibitor and corresponding control. Left panel: representative images. Right panel: quantification of 10 randomly selected fields. (I) Transwell assays of TE-1 cells transfected with miR-191 mimic or inhibitor and corresponding control. *p<0.05, **p<0.01. miR-191, microRNA-191; ESCC, esophageal squamous cell carcinoma.
miR-191 regulates EGR1 expression by directly targeting its 3′UTR
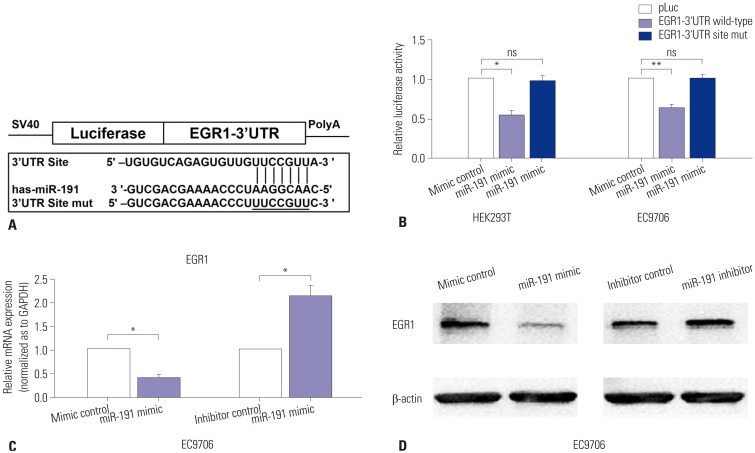
We next investigated the specific mechanism of miR-191 in ESCC proliferation and metastasis, and bioinformatic methods were performed to identify the downstream genes of miR-191. According to the results from three different bioinformatic programs (miRanda, Memorial Sloan-Kettering Cancer Center, New York, NY, USA; Targetscan, Whitehead Institute for Biomedical Research, Cambridge, MA, USA; and Pictar, Rajewsky Lab, New York, NY, USA), EGR1, a tumor suppressor, was predicted as a potential target gene of miR-191 (Fig. 4A). In order to validate whether miR-191 directly bound 3′UTR of EGR1 to suppress its expression, reporter vectors containing wildtype or mutant EGR1 3′UTR binding sites were transfected into HEK293T and EC9706 cells, respectively. Luciferase reporter assays indicated that miR-191 decreased the Luciferase activity in the vector containing wild-type EGR1 3′UTR, but not the one containing mutant site, suggesting a direct interaction of miR-191 and EGR1 3′UTR (Fig. 4B). In addition, both RT-PCR and western blot assays showed that overexpression of miR-191 reduced EGR1 expression in EC9706 cells, while down-regulation of miR-191 increased EGR1 expression (Fig. 4C and D). Taken together, these results indicated that miR-191 might suppress EGRI expression by directly targeting its 3′UTR to enhance ESCC progression.
Fig. 4. miR-191 down-regulates EGR1 expression by directly targeting its 3′UTR. (A) Diagram of EGR1 3′UTR-containing reporter construct. Mutations were generated at the predicted miR-191 binding site located in the EGR1 3′UTR. (B) Relative Luciferase activity after the wild type or mutant reporter plasmids were co-transfected with miR-191 mimic or mimic control in HEK293T and EC9706 cells. (C) RT-PCR analysis of EGR1 mRNA expression in EC9706 cells transfected with miR-191 mimic or inhibitor and corresponding control. (D) Western blot analysis of EGR1 protein expression in EC9706 cells transfected with miR-191 mimic or inhibitor and corresponding control. *p<0.05, **p<0.01. ns, no significance; EGR1, early growth response 1; miR-191, microRNA-191; mRNA, messenger RNA.
Down-regulation of EGR1 expression promotes ESCC cell growth and invasion
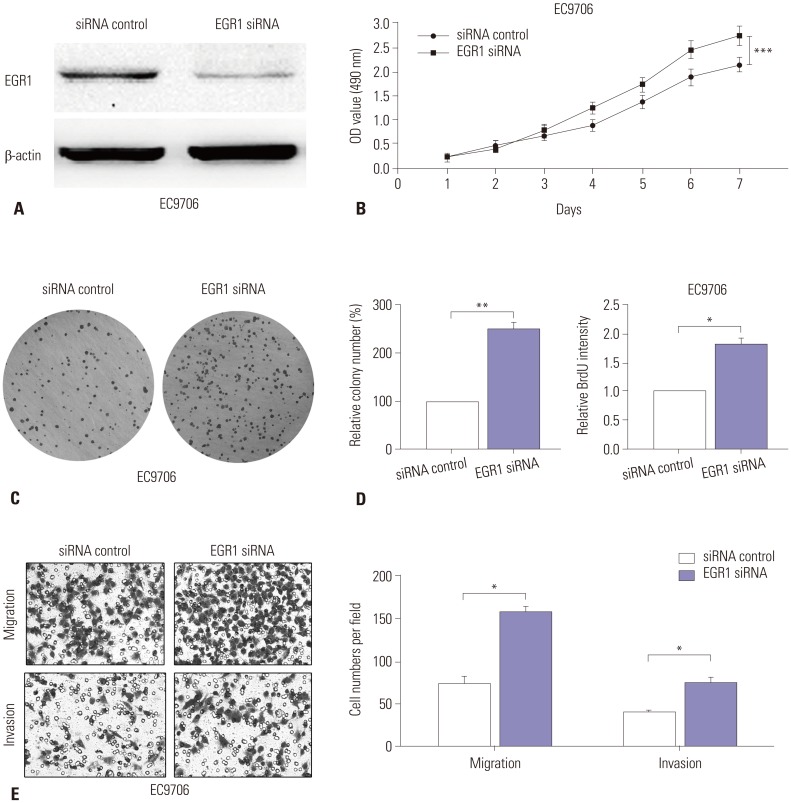
In order to confirm whether miR-191 promotes ESCC progression via EGR1, we next examined the exact function of EGR1 during ESCC cell proliferation and invasion. EGR1 siRNA was transfected into EC9706 cells, and western blot assay confirmed that it could significantly reduce EGR1 expression (Fig. 5A). All of the MTT, plate clone formation, and BrdU assays indicated that down-regulation of EGR1 accelerated EC9706 cell proliferation and growth (Fig. 5B, C, and D). Moreover, Transwell assay showed that EGR1 knockdown increased cell migration and invasion in vitro (Fig. 5E). Collectively, these observations indicated that down-regulation of EGR1 promotes ESCC proliferation and invasion.
Fig. 5. Down-regulation of EGR1 promotes ESCC cell proliferation and invasion. (A) Western blot analysis of EGR1 expression in EC9706 cells transfected with EGR1 siRNA or corresponding control. β-actin was used as an internal control. (B) MTT assay of EC9706 cells transfected with EGR1 siRNA or control. (C) The colony formation assays in EC9706 transfected with EGR1 siRNA or control. Colonies were evaluated and values were reported as the ratio. (D) The BrdU assays from EC9706 transfected with EGR1 siRNA or control. (E) Transwell assays of EC9706 cells transfected with EGR1 siRNA or control. Left panel: representative images. Right panel: quantification of 10 randomly selected fields. *p<0.05, **p<0.01, ***p<0.001. EGR1, early growth response 1; ESCC, esophageal squamous cell carcinoma; MTT, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide; OD, optical density.
DISCUSSION
Accumulating evidence has shown that miRNAs can act either as oncogenes or as tumor suppressors in ESCC and that measurement of miRNA expression in malignancies may have diagnostic and prognostic implications.20 Their very small size, in principle, makes them less prone to degradation processes, unlike messenger RNAs, which were previously proposed as molecular markers. The dysregulated expression of several miRNAs, including miR-21, miR-126, miR-138, miR-183, miR-200b, miR-375, and miR-508, has been associated with ESCC in recent literature.20,21,22,23,24,25,26,27 However, there is still a dearth of molecular biomarkers suitable for clinical application in this disease.
MiR-191, together with miR-425, belongs to the miR-191/425 cluster, which is highly conserved in several metazoan species (miR-425 in 26 species, miR-191 in 30 species), suggesting it to be an important player in higher eukaryotes.7 The aberrant expression of miR-191 has been observed in more than 20 different cancers. However, it remains controversial as to whether miR-191 acts as an oncogene or tumor suppressor in carcinogenesis. MiR-191 functions as an oncomiRNA in hepatocellular carcinoma, and its up-regulation enhanced cell growth and reduced apoptosis.28 It was also reported that miR-191 acts as an estrogen inducible oncogene and promotes breast cancer cell proliferation and invasion via targeting special AT-rich sequence-binding protein 1 (SATB1).12 Zhang, et al.13 showed that miR-191 promotes colorectal cancer carcinogenesis via targeting CCAAT/enhancer-binding protein β (C/EBPβ) and adjusting related signaling pathways. On the contrary, Di Leva, et al.29 showed that the overexpression of miR-191 suppresses cell growth and blocks carcinogenesis in aggressive breast cancer cells. Furthermore, miR-191 decreases cell proliferation and invasion by binding 3′UTR of CDK6 in thyroid cancer.16 However, to our knowledge, the clinical significance of miR-191 in human ESCC remains unclear.
Herein, we revealed that miR-191 was frequently overexpressed in ESCC tissues and significantly related to advanced clinical stage, metastasis, and poor survival rate of ESCC. Moreover, multivariate Cox regression analysis indicated that up-regulation of miR-191 was an independent prognostic factor for poor survival of patients with ESCC. Previous studies have demonstrated that measurement of miR-191 expression level may represent as a novel approach of diagnosis and prognosis in cancers.20 It was reported that miR-191 expression is a novel independent prognostic factor of patients with colorectal cancer.30 Peng, et al.18 revealed the oncogenic role of miR-191 in gastric carcinogenesis and indicated the potential use of serum miR-191 as a novel and stable biomarker for diagnosis. The present study further confirmed that miR-191 could be a novel potential prognosis biomarker in ESCC.
In the present study, we also showed that miR-191 could increase cell proliferation and invasion, implying that miR-191 is an oncomiRNA in ESCC. Further mechanism investigation demonstrated that miR-191 might reduce EGR1 expression by binding its 3′UTR, to be involved in ESCC progression. As a transcription factor, EGR1 is involved in tumorigenesis and development of many cancers, and plays a tumor suppressive role.31,32,33,34,35 It was demonstrated that decreased EGR1 expression is an important predictor of poor over survival in non-small cell lung cancer.31 Clinically, loss of EGR1 results in enhancive tumor transformation, followed by patient morbidity and mortality.31,32,33 Moreover, EGR1 is down-regulated in ESCC tissues and plays a significant role in ESCC prognosis.34 Dong, et al.35 reported that EGR1 regulates cisplatin-induced apoptosis in ESCC and activates apoptosis related pathways. On the basis of these findings, this study further confirmed that EGR1 down-regulation promotes ESCC cell growth and invasion, providing new insight of EGR1 function in ESCC development and progression.
Several limitations need to be addressed in our study. Firstly, the clinical sample size for evaluating prognostic value of miR-191 was slightly inadequate. Secondly, due to the limited information about disease-free survival, we were unable to classify the role of miR-191 in disease-free survival prediction. Th-irdly, all available function studies of miR-191 and EGR1 were conducted in vitro, and further in vivo research remains to be studied.
In summary, overexpression of miR-191 predicted poor prognosis in patients with ESCC and miR-191 promoted ESCC cell proliferation and invasion by targeting EGR1. The results from this study suggest that miR-191 may be an important molecular marker for the prognosis of ESCC. However, more rigorous clinical and basic studies are needed to confirm the present results.
ACKNOWLEDGEMENTS
We thank all the people and patients who participated in this study.
This work was conducted in Guangdong Medical University between January 2010 and December 2016.
Footnotes
The authors have no financial conflicts of interest.
SUPPLEMENTARY MATERIAL
Primers for RT-PCR
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 3.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 4.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Nagpal N, Kulshreshtha R. miR-191: an emerging player in disease biology. Front Genet. 2014;5:99. doi: 10.3389/fgene.2014.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dangwal S, Stratmann B, Bang C, Lorenzen JM, Kumarswamy R, Fiedler J, et al. Impairment of wound healing in patients with type 2 diabetes mellitus influences circulating microRNA patterns via inflammatory cytokines. Arterioscler Thromb Vasc Biol. 2015;35:1480–1488. doi: 10.1161/ATVBAHA.114.305048. [DOI] [PubMed] [Google Scholar]
- 9.Tan L, Yu JT, Tan MS, Liu QY, Wang HF, Zhang W, et al. Genomewide serum microRNA expression profiling identifies serum biomarkers for Alzheimer's disease. J Alzheimers Dis. 2014;40:1017–1027. doi: 10.3233/JAD-132144. [DOI] [PubMed] [Google Scholar]
- 10.Wei C, Henderson H, Spradley C, Li L, Kim IK, Kumar S et al. Circulating miRNAs as potential marker for pulmonary hypertension. PLoS One. 2013 May 23; doi: 10.1371/journal.pone.0064396. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paraskevi A, Theodoropoulos G, Papaconstantinou I, Mantzaris G, Nikiteas N, Gazouli M. Circulating MicroRNA in inflammatory bowel disease. J Crohns Colitis. 2012;6:900–904. doi: 10.1016/j.crohns.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Nagpal N, Ahmad HM, Molparia B, Kulshreshtha R. MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis. 2013;34:1889–1899. doi: 10.1093/carcin/bgt107. [DOI] [PubMed] [Google Scholar]
- 13.Zhang XF, Li KK, Gao L, Li SZ, Chen K, Zhang JB, et al. miR-191 promotes tumorigenesis of human colorectal cancer through targeting C/EBPβ. Oncotarget. 2015;6:4144–4158. doi: 10.18632/oncotarget.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu W, Ji J, Xu Y, Liu Y, Shi L, Liu Y, et al. MicroRNA-191, by promoting the EMT and increasing CSC-like properties, is involved in neoplastic and metastatic properties of transformed human bronchial epithelial cells. Mol Carcinog. 2015;54(Suppl 1):E148–E161. doi: 10.1002/mc.22221. [DOI] [PubMed] [Google Scholar]
- 15.McEvoy J, Ulyanov A, Brennan R, Wu G, Pounds S, Zhang J, et al. Analysis of MDM2 and MDM4 single nucleotide polymorphisms, mRNA splicing and protein expression in retinoblastoma. PLoS One. 2012;7:e42739. doi: 10.1371/journal.pone.0042739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colamaio M, Borbone E, Russo L, Bianco M, Federico A, Califano D, et al. miR-191 down-regulation plays a role in thyroid follicular tumors through CDK6 targeting. J Clin Endocrinol Metab. 2011;96:E1915–E1924. doi: 10.1210/jc.2011-0408. [DOI] [PubMed] [Google Scholar]
- 17.Xiao D, Barry S, Kmetz D, Egger M, Pan J, Rai SN, et al. Melanoma cell-derived exosomes promote epithelial-mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett. 2016;376:318–327. doi: 10.1016/j.canlet.2016.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng WZ, Ma R, Wang F, Yu J, Liu ZB. Role of miR-191/425 cluster in tumorigenesis and diagnosis of gastric cancer. Int J Mol Sci. 2014;15:4031–4048. doi: 10.3390/ijms15034031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan L, Zhang Y, Xia J, Liu B, Zhang Q, Liu J, et al. Resveratrol induces cell cycle arrest via a p53-independent pathway in A549 cells. Mol Med Rep. 2015;11:2459–2464. doi: 10.3892/mmr.2014.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harada K, Baba Y, Ishimoto T, Shigaki H, Kosumi K, Yoshida N, et al. The role of microRNA in esophageal squamous cell carcinoma. J Gastroenterol. 2016;51:520–530. doi: 10.1007/s00535-016-1161-9. [DOI] [PubMed] [Google Scholar]
- 21.Komatsu S, Ichikawa D, Kawaguchi T, Miyamae M, Okajima W, Ohashi T, et al. Circulating miR-21 as an independent predictive biomarker for chemoresistance in esophageal squamous cell carcinoma. Am J Cancer Res. 2016;6:1511–1523. [PMC free article] [PubMed] [Google Scholar]
- 22.Liu R, Gu J, Jiang P, Zheng Y, Liu X, Jiang X, et al. DNMT1-microRNA126 epigenetic circuit contributes to esophageal squamous cell carcinoma growth via ADAM9-EGFR-AKT signaling. Clin Cancer Res. 2015;21:854–863. doi: 10.1158/1078-0432.CCR-14-1740. [DOI] [PubMed] [Google Scholar]
- 23.Gong H, Song L, Lin C, Liu A, Lin X, Wu J, et al. Downregulation of miR-138 sustains NF-κB activation and promotes lipid raft formation in esophageal squamous cell carcinoma. Clin Cancer Res. 2013;19:1083–1093. doi: 10.1158/1078-0432.CCR-12-3169. [DOI] [PubMed] [Google Scholar]
- 24.Yang M, Liu R, Li X, Liao J, Pu Y, Pan E, et al. miRNA-183 suppresses apoptosis and promotes proliferation in esophageal cancer by targeting PDCD4. Mol Cells. 2014;37:873–880. doi: 10.14348/molcells.2014.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang HF, Alshareef A, Wu C, Jiao JW, Sorensen PH, Lai R, et al. miR-200b induces cell cycle arrest and represses cell growth in esophageal squamous cell carcinoma. Carcinogenesis. 2016;37:858–869. doi: 10.1093/carcin/bgw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osako Y, Seki N, Kita Y, Yonemori K, Koshizuka K, Kurozumi A, et al. Regulation of MMP13 by antitumor microRNA-375 markedly inhibits cancer cell migration and invasion in esophageal squamous cell carcinoma. Int J Oncol. 2016;49:2255–2264. doi: 10.3892/ijo.2016.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin C, Liu A, Zhu J, Zhang X, Wu G, Ren P, et al. miR-508 sustains phosphoinositide signalling and promotes aggressive phenotype of oesophageal squamous cell carcinoma. Nat Commun. 2014;5:4620. doi: 10.1038/ncomms5620. [DOI] [PubMed] [Google Scholar]
- 28.Elyakim E, Sitbon E, Faerman A, Tabak S, Montia E, Belanis L, et al. hsa-miR-191 is a candidate oncogene target for hepatocellular carcinoma therapy. Cancer Res. 2010;70:8077–8087. doi: 10.1158/0008-5472.CAN-10-1313. [DOI] [PubMed] [Google Scholar]
- 29.Di Leva G, Piovan C, Gasparini P, Ngankeu A, Taccioli C, Briskin D, et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 2013;9:e1003311. doi: 10.1371/journal.pgen.1003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin S, Zhu Y, Ai F, Li Y, Bai B, Yao W, et al. MicroRNA-191 correlates with poor prognosis of colorectal carcinoma and plays multiple roles by targeting tissue inhibitor of metalloprotease 3. Neoplasma. 2014;61:27–34. [PubMed] [Google Scholar]
- 31.Ferraro B, Bepler G, Sharma S, Cantor A, Haura EB. EGR1 predicts PTEN and survival in patients with non-small-cell lung cancer. J Clin Oncol. 2005;23:1921–1926. doi: 10.1200/JCO.2005.08.127. [DOI] [PubMed] [Google Scholar]
- 32.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70:9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 33.Liu C, Adamson E, Mercola D. Transcription factor EGR-1 suppresses the growth and transformation of human HT-1080 fibrosarcoma cells by induction of transforming growth factor beta 1. Proc Natl Acad Sci U S A. 1996;93:11831–11836. doi: 10.1073/pnas.93.21.11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu MY, Wu XY, Li QS, Zheng RM. Expression of Egr-1 gene and its correlation with the oncogene proteins in non-irradiated and irradiated esophageal squamous cell carcinoma. Dis Esophagus. 2006;19:267–272. doi: 10.1111/j.1442-2050.2006.00575.x. [DOI] [PubMed] [Google Scholar]
- 35.Dong Q, Zhang J, Hendricks DT, Zhao X. GROβ and its downstream effector EGR1 regulate cisplatin-induced apoptosis in WHCO1 cells. Oncol Rep. 2011;25:1031–1037. doi: 10.3892/or.2011.1163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers for RT-PCR