Abstract
Immunotherapy has become a key element in the treatment of several tumors, such as lung carcinoma and melanoma. Immunotherapy, unlike classical chemotherapy and targeted drugs, may yield long-term survival, even in patients who stop treatment due to toxicity. This fact has generated considerable excitement and a real shift in the paradigm of cancer treatment. However, only a small subset of patients benefit from immunotherapy. Survival curves show that most patients have progression of the disease in the first months after starting immunotherapy, followed by a slower decrease over the first 3 years, until curves reach a plateau. This early progression suggests the presence of mechanisms for primary resistance. In addition, some patients have tumor relapse after years of response, suggesting that there is also acquired resistance in a small subset of patients. Resistance mechanisms are now being elucidated. PD-L1 expression in tumor and immune cells correlates with higher chances of response, but melanoma patients with PD-L1 negative tumors can also respond. Several studies have demonstrated an increased probability of clinical benefit when tumors are infiltrated by CD8 T cells, have a high mutation burden or have an interferon gamma signature. But none of these factors has been implemented in the clinical practice, since more studies confirming their value are needed, as well as the development of standardized techniques.
Keywords: CTLA-4, melanoma, PD-1, predictive factors
Introduction
Immune check points antibodies, as anti PD-1/PD-L1 antibodies or anti CTLA-4 antibody, may yield long-term survival in cancer patients, but activity is seen only in 20–40% of patients. Recent studies indicate a number of mechanisms involved in resistance. Basically, the tumor will not respond if there are no antigens or, in the presence of antigens, the antigen-presentation machinery is altered.
Some common genetic alterations in melanoma produce immune evasion. The MAPK pathway activation is involved in the production of vascular-endothelial growth factor and interleukin-8, which impair T-cell recruitment and function (1). PTEN loss correlates with decreased expression of interferon-gamma and T-cell infiltration, and enhances PI3K signalling, which results in resistance to anti-PD1 therapy (2). Increased expression of beta-catenin in related genes, such as WNT, also leads to T-cell exclusion from the tumor microenvironment (3).
A second reason for immunotherapy resistance is the constitutively expression of PD-L1 in tumor cells. When PD-L1 binds to PD-1 on T cells, there is an inhibitory signal that inhibits T-cell response. Constitutively, expression of PD-L1 can be seen as a result of chromosome 9 amplification in a locus that includes PD-L1, PD-L2 and the interferon gamma receptor signalling molecular JAK2 (4), PTEN deletion, PI3K or AKT or EGFR mutations (5), or MYC overexpression (6).
Alterations in the interferon gamma pathway relate to immunotherapy resistance. Interferon gamma coming from an activated T-cell increases the expression of MHC molecules (involved in antigen presentation) and recruits other immune cells. However, some tumor cells downregulate or mutate molecules involved in this pathway, such as receptor associated Janus kinase 1 (JAK1) and receptor associated Janus kinase 2 (JAK2), leading to loss of the anti-tumor effect of interferon-gamma (7) in patients that progressed after a previous response to pembrolizumab (8). Also, other genes involved in mesenchymal transformation and wound healing, appear to confer resistance to anti-PD1 therapy, although the precise mechanism remains unknown (9). Other molecules involved in T-cell inactivation are also being studied: indoleamine-dioxygenase (IDO), carcinoembryonic antigen cell adhesion molecule 1, TIM-3, LAG-3, VISTA, TIGIT and transforming growth factor beta.
Defining predictive factors of response or resistance will help to select the best treatment for all patients. In this review, we try to summarize the state of the art about the development of predictive factors, as well as the most recent data on the predictive factor search.
Clinical factors and peripheral blood parameters
Even when clinical factors and peripheral blood markers are far from accurate, clinicians can use them in the daily practice. On the contrary, analysis of molecular factors requires sophisticated laboratory equipment that is not easily attainable. An elevated LDH level is an indicator of tumor burden that is usually associated with lower response rates to immunotherapy in clinical trials. For instance, in the second line setting, the overall response rate for pembrolizumab is lower for patients with high LDH levels (26%) than for the general population (34%) (10), as well as in the first line setting, pembrolizumab yields a response rate of 40% when LDH is in the normal range, 34% when LDH is up to 2 times the upper normal limit, and 8% when LDH is elevated over 2 times the upper normal limit (11).
Pretreatment leucocyte count also correlates with response to immunotherapy. For instance, benefit from ipilimumab has been seen in patients with low neutrophil count, low neutrophil to lymphocyte ratio, low frequency of myeloid-derived suppressor cells and high eosinophil count (12,13). A retrospective analysis combined different analytical and clinical parameters to predict response to pembrolizumab: low eosinophils and neutrophils counts, high LDH level and visceral metastases beyond the lung were identified as adverse factors (14).
These clinical and peripheral blood markers per se do not directly justify the lack of response, but probably reflect a long time of tumor evolution or a high aggressiveness that also let to the acquisition of molecular mechanisms of resistance.
BRAF status
BRAF melanomas are regarded as more aggressive than wild-type counterparts, but the value of this parameter as a predictive marker for immunotherapy remains controversial. A retrospective analysis of pembrolizumab showed a response rate of 26% in wild-type versus 12% in mutated melanomas in second line (15). Differences narrowed at the first line setting with response rates of 38% for BRAF non-mutant versus 32% in BRAF mutant melanomas. A phase III study of nivolumab showed the opposite result, 2-year survival rate was superior in patients with BRAF-mutant disease (57% vs. 62%) (16). These results suggest that BRAF status does not play a key role in resistance to immunotherapy.
PD-L1 expression
PD-L1 is the ligand of the check point inhibitor PD-1 on immune cells. When PD-1 binds to its ligand PD-L1, an inhibitory signal is sent to the nucleus inhibiting the immune response. PD-L1 can be expressed on tumor cells and it can be induced constitutively by intrinsic alteration that drives tumor development, as well as by mechanisms of adaptative resistance mediated by interferon gamma (17). High number of studies has a try to investigate the clinical value of measuring PD-L1 expression on tumor biopsies, using immunohistochemestry. Although measuring PD-L1 expression makes sense as a predictive marker for anti-PD1 antibodies, its predictive value is not clear, mainly in melanoma. Some anti PD-1 drugs have been approved only for PD-1 positive tumors, as pembrolizumab for non-small cell lung cancer (NSCLC) in first line setting (18), but in melanoma some patients with negative expression can obtain responses and become long-term survivors (19). For this reason, low PD-L1 expression does not preclude treatment with anti-PD1 antibodies in melanoma.
The detection of PD-L1 has not been standardised yet, with every company in the field developing its own immunohistochemistry antibody and method (19,20). As an additional difficulty, the expression of PD-L1 may vary within the tumor and can be induced by previous therapies (21). That being said, patients with low tumor expression of PD-L1 do not respond as well as those with high expression, not only in melanoma, but also across different tumor types (22). In an analysis of melanoma patients treated with pembrolizumab, response rate varied between 57% when melanoma had a high PD-L1 expression, and 8% when there was not PD-L1 expression (23). Other studies using nivolumab, demonstrated no difference between patients PD-L1 positive or negative with a response rate of 57% and 41%, respectively (24). In general, response rate with single anti PD-1 therapy is around 15% when melanoma cells are PD-L1 negative, and 48% when they are PD-L1 positive (Figure 1) (22).
Figure 1.
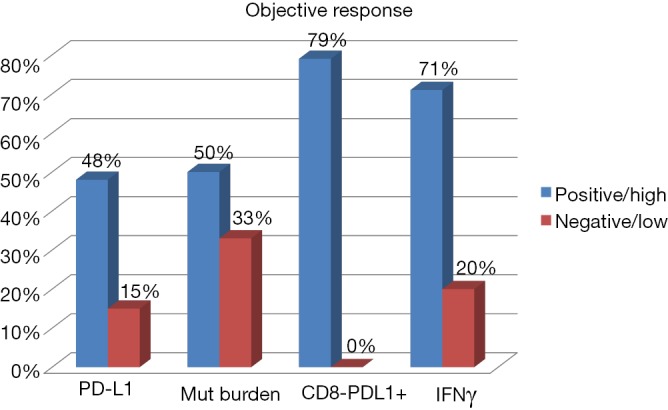
Predictive factors of response to check point inhibitors in melanoma. PD-L1: analysis of PD-L1 expression on tumor cells by immunohistochemestry and correlation with response to anti PD-1 antibodies (22); Mut burden: mutational burden in tumor biopsies and correlation with response to anti CTLA-4 antibodies (25); CD-8/PD-L1+: higher than 20% of lymphocyte tumor infiltration by CD8+ T cells with high expression of PD-1 and CTLA-4 (PD-1hi CTLA-4hi CTL) and correlation with response to anti PD-1 antibodies (23); IFNγ: RNA levels of interferon-gamma low versus high and correlation with response to anti PD-1 antibodies (26).
Data from a phase III study suggested that the combination of nivolumab and ipilimumab may be preferable if PD-L1 expression was low, since patients with negative PD-L1 expression (defined as <5% staining) were those who most benefitted from the combination. But these data were not confirmed with the updated results of the study demonstrating a higher response rate for the combination, regardless of the PD-L1 level (24). An exploratory analysis showed that, in terms of overall survival (OS), those patients with negative PD-L1 (almost two thirds of the population included in the trial), had better OS than those who were PD-L1 positive (HR 0.84 versus 1.05) (27).
These results indicate that PD-L1 expression matters, but cannot be reliably determined, at least with current methods. On the other hand, lack of benefit in some patients with a positive marker suggests that other factors are involved in resistance to anti-PD1 antibodies.
Recently, an interesting report demonstrated that the increase of CD8+ T cells PD1 positive in peripheral blood after 4 weeks of starting the anti PD-1 antibody therapy correlates with clinical benefit in lung cancer patients (28). In melanoma, there are data indicating an increase of CD8 memory T cells into the tumor, and a possible increase in peripheral blood of a subset of dendritic cells, or HLA-DR NK cells, in patients with a response to therapy after anti PD-1 treatment (29).
Mutational load and neoantigens
Mutations have the capacity to generate neoantigens that elicit an immune response. For this reason, mutational load could act as a surrogate marker for immunogenicity and likelihood of response to immune agents. Benefit from anti-CTLA4 antibodies has been reported in patients with melanoma and high mutational load. In two studies based on whole-exome sequencing, a mutational load of more than 100 somatic mutations correlated with increased responses and OS (25,30) (Figure 1). On the other hand, a favourable correlation was also seen for pembrolizumab in patients with either NSCLC (31) or mismatch repair-deficient colorectal cancer (32).
Mutational load is determined by whole genome sequencing, a technique with limited implementation due to cost and informatics requirements. A retrospective study showed that small sets of genes could be used in substitution of whole genome sequencing to predict response to ipilimumab (33).
Tumor burden can be combined with other factors to improve predictive accuracy. For instance, in a study derived from the Cancer Genome Atlas, a combination of high mutational burden and low neoantigen tumor heterogeneity was associated with longer survival in patients with lung cancer, irrespective of treatment (34).
T-cells recognizing neoantigens account for a very small proportion of peripheral T-cells at baseline, although their percentage increases during therapy (31). Assessment of peripheral T-cell receptor repertoire has a potential role as a predictive marker. It has the advantage that peripheral blood is easily available. Most mutations appear to be patient-specific, so mutational markers—either measured in the tumor or in peripheral blood—would require a personalised approach that is far beyond most clinical centers.
Tumor-infiltrating lymphocytes
Lymphocyte infiltration in the tumor results from an immune response, which is thought to improve disease control and might serve as a prognostic biomarker. This factor correlates with survival in melanoma and other tumor types: “inflamed” tumors usually do better than “cold” tumors. Lymphocyte density in tumor biopsies taken after the second dose of ipilimumab has been associated with significant activity of this drug (35). Also in melanoma, patients with higher CD8+ density at the invasive margin and within tumor parenchyma showed higher responses to pembrolizumab (36). However, lymphocyte infiltration also appears when the disease eventually progresses, and there is not a clear cut-off to be used as a clinical marker. A recent study, analysing melanoma biopsies from patients treated with anti PD-1 antibodies, demonstrates a response rate of 79% when pre-treatment tumor biopsies have more than 20% of tumor infiltrating PD-1 high and CTLA-4 high CD8 T cells (PD-1hi CTLA-4hi CTL), versus no responders in patients with fewer than 20% infiltrating PD-1hi CTLA-4hi CTL (37) (Figure 1).
A retrospective analysis of specimens from patients receiving pembrolizumab showed that high CD8 and PD-L1 simultaneous expression was associated with response (36). In this study, infiltrating lymphocytes had a narrow T-cell receptor repertoire when there was a response. Pre-treatment and post-treatment biopsies showed a 10-fold increase in these clones after therapy (36).
After anti PD-1 treatment, the increase of memory CD8 T cells infiltrating the tumors correlates with clinical benefit (29).
Infiltration by other immune cells
Unlike CD8+ T-cells, other immune cells present in the tumour microenvironment may impair the immune response: Tregs, myeloid-derived suppressor cells and M2 macrophages. Myeloid-derived suppressor cells promote angiogenesis and cell invasion and their presence in peripheral blood was associated with decreased efficacy of ipilimumab in a small series of patients (38).
Infiltration by tumor-associated macrophages correlates with poor survival in human cancer and some investigators have suggested that these cells could mediate therapeutic resistance (39).
Although immune cells other than effectors lymphocyte could account for resistance to immunotherapy, their identification is challenging and their clinical value has not been validated.
Gene signatures
Gene signatures allow assessing the expression of hundreds to thousands of genes at a given time. In the case of breast cancer, for instance, they have been introduced as prognostic markers with clinical utility (40). In the field of melanoma, a set of 22 immune-related genes identified patients who had a benefit from ipilimumab (41). With regard to anti-PD1 therapy, another retrospective study performed in patients who had received pembrolizumab described an interferon-gamma score with predictive value (42). The score was identified through a 28-gene signature that included interferon-gamma, granzymes A and B, perforin 1, LAG 3 and indoleamine dioxygenase, among others.
Assessment in serial biopsies throughout the course of treatment is an interesting strategy, because it can better reflect the evolving response of the immune system. Several studies show that some markers offer better predictive information when the biopsy is performed after treatment initiation (9,36,43). Assessment of gene expression in blood samples would facilitate serial sampling, but technical difficulties remain in this field.
Although gene signatures can become easily available for clinical practice, they need validation in the context of immunotherapy. mRNA concentration of PD-L1 and other key regulator molecules are subjected to post-transcriptional regulation, so gene signatures might not properly detect them. This problem could be overcome through the combined use of gene expression and immunohistochemistry. This approach was used in samples from patients with lung cancer receiving the anti PD-L1 antibody, durvalumab, and demonstrated that the response rate was higher, and OS longer, in patients with interferon-gamma positive and PD-L1 positive tumors, whereas response rate was very low if both markers were negative (44). A recent study analysing both melanoma and lung cancer biopsies, demonstrated that patients with high expression of interferon-gamma have a higher chances of response and survival with anti PD-1 antibodies (26). For melanoma patients included in this study response rate was 71% when there was a high expression of interferon-gamma and 20% when levels of interferon-gamma were low (26) (Figure 1).
Conclusions
In summary, predictive factors for immunotherapy are being actively investigated. Table 1 summarizes the main conclusions. Most of the currently available information comes from small retrospective studies. The most consistent results come from the mutational load and assessment of tumor lymphocyte infiltration, although they have not been taken to clinical practice yet. Clinical and peripheral blood markers are more inaccurate, but easily accessible. New molecular markers under study will provide better insight into the mechanisms of resistance that play a role in individual patients and will hopefully be useful to guide clinical decisions.
Table 1. Factors predicting response to anti-PD1 and anti-CTLA4 antibodies.
| Factor | Comments |
|---|---|
| Tumour burden | Clinically available, but inaccurate |
| PD-L1 expression | Easily available (immunohistochemistry); good negative predictive value in lung cancer, but not in melanoma; not standardised |
| Mutational load | Growing evidence, although formal validation required; possibility to combine with other factors to improve accuracy |
| Tumour infiltrating lymphocytes | Not standardised |
| T-cell receptor clonality | Require sophisticated technology, not validated |
| Gene signatures | Possibility to be used in the clinic, not enough validation |
| Molecular factors | Make biological sense, not standardised |
Acknowledgements
None.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Loi S, Dushyanthen S, Beavis PA, et al. RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast cancer: therapeutic cooperation between MEK and PD-1/PD-L1 immune checkpoint inhibitors. Clin Cancer Res 2016;22:1499-509. 10.1158/1078-0432.CCR-15-1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng W, Chen JQ, Liu C, et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov 2016;6:202-16. 10.1158/2159-8290.CD-15-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015;523:231-5. 10.1038/nature14404 [DOI] [PubMed] [Google Scholar]
- 4.Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med 2015;372:311-9. 10.1056/NEJMoa1411087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lastwika KJ, Wilson W, 3rd, Li QK, et al. Control of PD-L1 Expression by Oncogenic Activation of the AKT-mTOR Pathway in Non-Small Cell Lung Cancer. Cancer Res 2016;76:227-38. 10.1158/0008-5472.CAN-14-3362 [DOI] [PubMed] [Google Scholar]
- 6.Casey SC, Tong L, Li Y, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016;352:227-31. 10.1126/science.aac9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benci JL, Xu B, Qiu Y, et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell 2016;167:1540-54. e12. [DOI] [PMC free article] [PubMed]
- 8.Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med 2016;375:819-29. 10.1056/NEJMoa1604958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hugo W, Zaretsky JM, Sun L, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016;165:35-44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribas A, Hamid O, Daud A, et al. Association of Pembrolizumab With Tumor Response and Survival Among Patients With Advanced Melanoma. JAMA 2016;315:1600-9. 10.1001/jama.2016.4059 [DOI] [PubMed] [Google Scholar]
- 11.Blank C, Ribas A, Long GV, et al. Impact of baseline serum lactate dehydrogenase (LDH) concentration on efficacy in the KEYNOTE-006 study of pembrolizumab vs ipilimumab. The Society for Melanoma Research 2016 Congress; Boston, Massachusetts, USA. Available online: https://www.melanomacongress.com/abstracts
- 12.Ferrucci PF, Ascierto PA, Pigozzo J, et al. Baseline neutrophils and derived neutrophil-to-lymphocyte ratio: prognostic relevance in metastatic melanoma patients receiving ipilimumab. Ann Oncol 2016;27:732-8. 10.1093/annonc/mdw016 [DOI] [PubMed] [Google Scholar]
- 13.Martens A, Wistuba-Hamprecht K, Geukes Foppen M, et al. Baseline Peripheral Blood Biomarkers Associated with Clinical Outcome of Advanced Melanoma Patients Treated with Ipilimumab. Clin Cancer Res 2016;22:2908-18. 10.1158/1078-0432.CCR-15-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weide B, Martens A, Hassel JC, et al. Baseline Biomarkers for Outcome of Melanoma Patients Treated with Pembrolizumab. Clin Cancer Res 2016;22:5487-96. 10.1158/1078-0432.CCR-16-0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puzanov I, Ribas A, Daud A, et al. Pembrolizumab for advanced melanoma: effect of BRAFV600 mutation status and prior BRAF inhibitor therapy. 12th International Congress of the Society for Melanoma Research. San Francisco, CA, USA. November 21, 2015. [Google Scholar]
- 16.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Overall survival results from a phase III trial of nivolumab combined with ipilimumab in treatment-naïve patients with advanced melanoma (CheckMate-067): press conference. Proceedings from the 2017 American Association for Cancer Research Annual Meeting. Washington DC, USA. April 2 to 5, 2017. [Google Scholar]
- 17.Brogden KA, Vali S, Abbasi T. PD-L1 is a diverse molecule regulating both tumor-intrinsic signaling and adaptive immunosuppression. Transl Cancer Res 2016;5:S1396-9. 10.21037/tcr.2016.12.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 19.Teixidó C, Gonzalez-Cao M, Karachaliou N, et al. Predictive factors for immunotherapy in melanoma. Ann Transl Med 2015;3:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santarpia M, Gonzalez-Cao M, Viteri S, et al. Programmed cell death protein-1/programmed cell death ligand-1 pathway inhibition and predictive biomarkers: understanding transforming growth factor-beta role. Transl Lung Cancer Res 2015;4:728-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443-54. 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sunshine J, Taube JM. PD-1/PD-L1 inhibitors. Curr Opin Pharmacol 2015;23:32-8. 10.1016/j.coph.2015.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daud AI, Wolchok JD, Robert C, et al. Programmed Death-Ligand 1 Expression and Response to the Anti-Programmed Death 1 Antibody Pembrolizumab in Melanoma. J Clin Oncol 2016;34:4102-9. 10.1200/JCO.2016.67.2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolchok JD, Chiarion-Sileni V, Gonzalez R, et al. Updated results from a phase III trial of nivolumab (NIVO) combined with ipilimumab (IPI) in treatment-naive patients (pts) with advanced melanoma (MEL) (CheckMate 067). J Clin Oncol 2016;34 suppl:abstr 9505.
- 25.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207-11. 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karachaliou N, Crespo G, Aldeguer E, et al. Interferon-gamma (INFG), an important marker of response to immune checkpoint blockade (ICB) in non-small cell lung cancer (NSCLC) and melanoma patients. J Clin Oncol 2017;35:abstr 11504. [DOI] [PMC free article] [PubMed]
- 27.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Overall survival (OS) results from a phase III trial of nivolumab (NIVO) combined with ipilimumab (IPI) in treatment-naïve patients with advanced melanoma (CheckMate 067). AACR Annual Meeting 2017. Washington, DC, USA. April 1-5, 2017. [Google Scholar]
- 28.Kamphorst AO, Pillai RN, Yang S, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci U S A 2017;114:4993-8. 10.1073/pnas.1705327114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ribas A, Shin DS, Zaretsky J, et al. PD-1 Blockade Expands Intratumoral Memory T Cells. Cancer Immunol Res 2016;4:194-203. 10.1158/2326-6066.CIR-15-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189-99. 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roszik J, Haydu LE, Hess KR, et al. Novel algorithmic approach predicts tumor mutation load and correlates with immunotherapy clinical outcomes using a defined gene mutation set. BMC Med 2016;14:168. 10.1186/s12916-016-0705-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463-9. 10.1126/science.aaf1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamid O, Schmidt H, Nissan A, et al. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J Transl Med 2011;9:204. 10.1186/1479-5876-9-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568-71. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daud AI, Loo K, Pauli ML, et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest 2016;126:3447-52. 10.1172/JCI87324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer C, Cagnon L, Costa-Nunes CM, et al. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol Immunother 2014;63:247-57. 10.1007/s00262-013-1508-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams CB, Yeh ES, Soloff AC. Tumor-associated macrophages: unwitting accomplices in breast cancer malignancy. NPJ Breast Cancer 2016;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cardoso F, van’t Veer LJ, Bogaerts J, et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N Engl J Med 2016;375:717-29. 10.1056/NEJMoa1602253 [DOI] [PubMed] [Google Scholar]
- 41.Ji RR, Chasalow SD, Wang L, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother 2012;61:1019-31. 10.1007/s00262-011-1172-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribas A, Robert C, Hodi FS, et al. Association of response to programmed death receptor 1 (PD-1) blockade with pembrolizumab (MK-3475) with an interferon-inflammatory immune gene signature. J Clin Oncol 2015;33 suppl:3001. [Google Scholar]
- 43.Chen PL, Roh W, Reuben A, et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov 2016;6:827-37. 10.1158/2159-8290.CD-15-1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higgs BW, Morehouse C, Streicher K, et al. Relationship of baseline tumoral IFNγ mRNA and PD-L1 protein expression to overall survival in durvalumab-treated NSCLC patients. J Clin Oncol 2016;34 suppl:3036. [Google Scholar]


