Abstract
The anaerobic digestion (AD) of organic waste for biogas production has received much attention in recent years due to the increasing need for renewable energy and environmentally friendly waste management systems. Identification of the microbial community involved in AD aids in better understanding and optimising of the process. The choice of DNA extraction method is an integral step in any molecular biodiversity study. In the present study, potential biases introduced by DNA extraction methods were examined by comparing quality, quantity and representability of DNA extracted from AD samples using various extraction methods. In spite of the non-kit based method (cetyltrimethylammonium bromide) yielding the largest quantity of DNA (approximately 44 µg DNA per gram dry weight), the extracted DNA contained PCR inhibitors. Furthermore, the quantity of extracted DNA was not proportional to species diversity. Diversity, determined using denaturing gradient gel electrophoresis (DGGE), was strongly linked to the type of extraction method used. The spin-column filter-based kit that incorporated mechanical and chemical lysis (Macherey-Nagel kit) gave the best results in terms of bacterial and archaeal diversity (Shannon–Wiener indices: average 2.5 and 2.6, respectively). Furthermore, this kit was the most effective at lysing hard-to-lyse bacterial and archaeal cells. The choice of DNA extraction method significantly influences the reliability and comparability of results obtained during AD microbial ecology investigations. Moreover, the careful selection of the DNA extraction method is of particular importance when analysing AD samples since these samples are rich in PCR inhibitors and hard-to-lyse cells such as archaea and gram-positive bacteria.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-1009-x) contains supplementary material, which is available to authorized users.
Keywords: DNA extraction, Bacteria, Archaea, Anaerobic digestion, Diversity
Introduction
Much interest has been directed to the use of anaerobic digestion (AD) for the conversion of organic waste to bioenergy because this process contributes to waste management, renewable energy production and food security. Microorganisms such as bacteria and archaea play a key role in the AD process, hence, it is of great importance that the microbiology of the AD system be explored (Bergmann et al. 2009; Roopnarain and Adeleke 2017). This can be achieved using culture dependent or culture independent techniques. Culture dependent techniques are notorious for the underestimation of microbial diversity in most environments because only a small percentage of the population is culturable (Amann et al. 1995). Currently, culture independent, molecular approaches are frequently exploited for the investigation of community structure and diversity in practically all environments (Theron and Cloete 2000).
The basis of molecular biodiversity analyses is to obtain a representative nucleic acid extract from the entire microbial community that is under investigation. The quality and representability of the nucleic acid extract are directly influenced by the choice of the extraction method that is used (Carrigg et al. 2007). Inefficiencies at various stages in the extraction process could negatively affect the quality of the final extract. Such inefficiencies include incomplete cell lysis, damage of the extracted DNA, DNA sorption to the surface of various particles in the sample, the loss of DNA at different stages in the extraction process and the co-extraction of various enzymatic inhibitors that could interfere with downstream processing of the DNA, e.g. PCR inhibitors (Miller et al. 1999; Claassen et al. 2013). The efficacy of the DNA extraction process is further influenced by the source of the sample. Samples consisting of a complex microbial matrix and large amounts of inhibitors such as activated sludge and soil contribute to the challenges in DNA extraction (Vanysacker et al. 2010).
Commercial DNA extraction kits and laboratory designed protocols are frequently used for the extraction of DNA from environmental samples. Commercial kits are often used because they are designed to optimise DNA yield and ensure the reproducibility of the extraction. These kits are easy to use and require considerably shorter durations for complete extraction in comparison to conventional methods (Herrera and Cockell 2007). Regardless of the type of method used for DNA extraction, one or more of the following processes are incorporated: chemical lysis, physical disruption and/or enzymatic lysis (Miller et al. 1999). These processes should ensure that sufficient amounts of high molecular weight DNA are extracted with minimal inhibitors and the extract should reflect an accurate representation of the total microbial diversity within the sample (Yeates et al. 1998). Furthermore, the method of DNA extraction should be efficient and reproducible. In addition, the method should also be applicable to a wide range of sample types and be cost effective (Fahle and Fischer 2000).
At present, there are no commercial DNA isolation kits that are specifically designed for the extraction of DNA from anaerobic digester samples. Kits that are frequently used for digester samples include soil (Garcia-Peña et al. 2011; Xu et al. 2014) and stool kits (Slana et al. 2011; Kampmann et al. 2012). However, digester samples are generally rich in inhibitors such as humic acids, which are a by-product of the AD process (Bergmann et al. 2009). Furthermore, a wide range of microorganisms such as bacteria and archaea are evident in digester samples. These microorganisms are integral to the AD process, hence, both bacterial and archaeal communities are frequently analysed when conducting AD studies (Ariesyady et al. 2007; Riviere et al. 2009; Sundberg et al. 2013). One of the major factors influencing the choice of the extraction method used for digester samples is the ability to achieve complete cell disruption of both bacterial and archaeal cells in the sample.
In the present study, various methods of DNA extraction, including commercially available kits and laboratory designed protocols, were tested on samples obtained at different stages of the AD process. The study was aimed at determining the suitability of various DNA extraction methods for AD samples and to determine how the method of extraction impacts on the observed diversity of bacteria and archaea (using DGGE analysis). This will enable the proposal of a single extraction method that facilitates DNA extraction from the majority of or all bacteria and archaea involved in the AD process which would aid significantly in AD microbial ecology studies.
Methods
Sample collection and dry weight measurements
Samples were collected from the inlet (fresh feed), digester chamber (partially digested feed) and slurry/outlet (completely digested feed) of a pre-fabricated digester situated in QwaQwa village, Free State province, South Africa (latitude − 28,566867°S, longitude 28,678145°E). The digester has been working since August 2012 and is fed continuously with a mixture of cow dung and water. The samples were collected in sterile plastic bags and transported on ice, in a cooler bag, to the Agricultural Research Council—Institute for Soil, Climate and Water microbiology laboratory in Pretoria (Gauteng province). Upon arrival, the samples were mixed, aliquoted into sterile centrifuge tubes (50 ml Falcon® tubes) and stored at − 20 °C until further analysis.
To determine dry weight, frozen samples were thawed at 4 °C and pre-weighed aliquots of the samples were incubated at 105 °C for 24 h. The dried samples were weighed and standard curves were constructed showing the correlation between wet and dry weight measurements (Fig. A1). These standard curves were used to determine dry weight from wet weight measurements.
DNA extraction
Samples were thawed at 4 °C and centrifuged at 16,000×g for 10 min for the collection of solids and microorganisms. The supernatant was discarded and the pellet was used as the substrate for various DNA extraction methods. Eight methods of DNA extraction were tested in this study (Table 1): one laboratory-based extraction method (CTAB method) and seven commercially available kits were evaluated. The selection of commercial kits used was based on popularity, cost, availability, novelty, variations in methods of cell lysis and variations in the format of DNA purification. All the methods listed in Table 1 were used to extract DNA from samples obtained from the inlet, digester chamber and the outlet of a working digester. The extractions were conducted in triplicate for each sample to determine the reproducibility of the various methods.
Table 1.
Methods of DNA extraction evaluated in the study
| Kit/extraction method name | Abbreviation | Recommended source material | Method of substrate homogenisation and cell lysis | Format of DNA purification | Approximate duration of extraction process (9 samples) (h) | Weight of starting material (mg) |
|---|---|---|---|---|---|---|
| ZR Soil Microbe DNA MiniPrep—Zymo Research | ZR | Soil samples | Bead beating and cell lysis buffer | Spin column filter based | 4 | 150 |
| QIAamp Fast DNA Stool Mini Kit—QIAGEN | QIA | Stool samples | Cell lysis buffer and heat | Spin column filter based | 3.5 | 220 |
| NucleoSpin Soil Kit—Macherey-Nagel | MN | Soil, sludge and sediment samples | Bead beating and cell lysis buffer | Spin column filter based | 4.5 | 500 |
| MagMAX Total Nucleic Acid Isolation Kit—Manual—Thermo Fisher Scientific | Mag-Man | Broad range—biological and environmental samples | Bead beating and cell lysis buffer | Paramagnetic bead based-manual | 5 | 300 |
| MagMAX Total Nucleic Acid Isolation Kit—Automated—Thermo Fisher Scientific | Mag-Aut | Broad range—biological and environmental samples | Bead beating and cell lysis buffer | Paramagnetic bead based-automated | 2.5 | 300 |
| Powersoil DNA Isolation Kit—MO BIO laboratories | PS | Soil, compost, sediment and manure | Bead beating and cell lysis buffer | Spin column filter based | 3.5 | 250 |
| Meta-G-Nome DNA Isolation Kit—Epicentre | EPI | Water or soil samples | Cell lysis buffer | Solution based | 11 | 1000 |
| Cetyltrimethylammonium bromide based extraction (Minas et al. 2011) | CTAB | Rumen fluid, plant and bacterial pure cultures | Bead beating and cell lysis buffer | Solution based | 26 | 250 |
For all kits tested, the amount of starting material was determined by the protocols available in the kit. The maximum quantity was used in each instance (e.g. if the kit required 0.5–1 g, 1 g of the sample was used as the starting material; see Table 1). The kit extractions were conducted as per manufacturer instructions with minor amendments. For instance, for the ZR kit, a standard benchtop vortex (MX-S; Dragonlab) was used instead of a bead beater. For the QIA kit, a lysis temperature of 95 °C was used instead of 70 °C. For the MN kit, buffer SL1 was used instead of buffer solution SL2, 75 µl Enhancer SX was used instead of 150 and 50 µl elution buffer was used. For the EPI kit, sterile cheesecloth was used instead of miracloth. The CTAB extraction was conducted as described in Minas et al. (2011) with minor deviations. The standard method in 2.0 ml microcentrifuge tubes was used. For the CTAB method, DNA was extracted from 250 mg samples. Cells were lysed by vortexing the material in microcentrifuge tubes containing 2 mm glass beads with 900 µl CTAB lysis buffer.
All the extraction methods tested were direct methods (i.e. cells were lysed directly within the sample) with the exception of the EPI kit which was an indirect method of DNA extraction (i.e. cells are removed from the samples prior to cell lysis and DNA extraction) (Delmont et al. 2011).
Quality and quantity of extracted DNA
DNA yield was measured using two methods: Nanodrop (Nanodrop One, Thermoscientific, USA) and the Qubit fluorometer (Invitrogen, USA, using the Qubit® dsDNA HS Assay Kit). DNA yield measurements were normalised based on the dry weight of the respective samples. DNA purity was determined spectrophotometrically using a Nanodrop (Nanodrop One, Thermoscientific, USA). Protein contamination was measured using the ratio of absorbances at 260 and 280 nm. A ratio between 1.8 and 2.2 was indicative of no protein contamination (Weiss et al. 2007). The ratio of absorbances 260 and 230 nm was used to determine contamination by aromatic compounds, phenols and carbohydrates (Roh et al. 2006). Ratios between 1.5 and 1.8 were taken as an indication of DNA without aromatic compound contamination (Weiss et al. 2007). The integrity of the DNA extracts was evaluated by gel electrophoresis on a 1% agarose gel (w/v) stained with ethidium bromide and run in 1 × TAE buffer at 100 V.
Two-way ANOVA was used to compare the mean differences between the DNA yield and purity measurements obtained using the various methods of DNA extraction. Student’s t LSD (Least significant difference) was calculated at a 5% significance level to compare means of significant source effects. The above analysis was performed using Genstat Release 18.
PCR
PCR amplification of the 16S rDNA with the universal bacterial primer set 341F-GC and 907R (Table 2) was carried out (Muyzer et al. 1993). Methanogenic archaeal DNA was first amplified using the primer set for methanogenic archaea i.e. 0357F and 0915aR. The resulting PCR products were re-amplified using primer set 0357F-GC and 0691R (Table 2; Ikenaga et al. 2004; Watanabe et al. 2004).
Table 2.
Primers used in the study
| Primer | Sequence (5′-3′) | Annealing temp (°C) |
|---|---|---|
| 341F-GC | CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGGCCTAC GGGAGGCAGCAG |
65–55a |
| 907R | CCGTCAATTCCTTTGAGTTT | 65–55a |
| 0357F | CCCTACGGGGCGCAGCAG | 69 |
| 0915aR | GTGCTCCCCCGCCAATTCCT | 69 |
| 0357F-GC | CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCCTAC GGGGCGCAGCAG |
57 |
| 0691R | GGATTACARGATTTCAC | 57 |
a These primers were used in a touchdown protocol where the annealing temperature decreased from 65 to 55 °C in 20 cycles
DGGE
Denaturing gradient gel electrophoresis (DGGE) was used to establish microbial community profiles of the DNA samples obtained using the various extraction methods. For DGGE analysis, triplicate DNA extracts from each kit and respective samples (inlet, digester or slurry) were pooled. DGGE was performed as described by Muyzer et al. (1993) with slight variations. The DCode Universal Mutation Detection System (Bio-Rad Laboratories, Hercules, CA) was used. Amplicon separation proceeded on an 8% (wt/vol) polyacrylamide gel (40% acrylamide/bis solution, 37.5:1) using denaturing gradient ranges of 40–60% for bacterial samples and 25–60% for archaeal samples. The 100% denaturant consisted of 40% formamide and 7M urea. Glycerol (2%) was added to the gel to increase gel flexibility. To enable gel comparisons, a mixture of 5 μl of PCR amplicons from four pure isolates was used as a marker. Electrophoresis was performed at 200 V for 15 min, then at 100 V for 16 h. The 0.5 × TAE buffer was maintained at 60 °C throughout the run. Gels were stained with GelRed and photographed using a UV transilluminator (GelDoc XR; Bio-Rad Laboratories, Hercules, CA). Bands were excised from the gels and DNA was eluted overnight in 10 µl sterile distilled water. Bacterial DNA were re-amplified with the primer pair 341F and 907R and archaeal DNA with 0357F and 0691R. The resulting PCR amplicons were sequenced. Sequences were inspected and edited using BioEdit sequence alignment editor (http://www.mbio.ncsu.edu/bioedit/bioedit) and identified using NCBI Blast and EzTaxon (http://www.ezbiocloud.net/eztaxon).
Images of the DGGE gels were analysed using the Image Lab software (Bio-Rad). Each DGGE gel is composed of numerous lanes that were loaded with individual samples. These samples were separated into several bands of varying intensities. The software detects each band and calculates the relative contribution of the individual band to the overall signal in the lane of interest. The resultant data were used to construct Lorenz distribution curves as previously described (Mertens et al. 2005). Furthermore, two widely used diversity indices, viz. Shannon–Wiener (H’) and Simpsons indices (D) were calculated using the following formulae:
where n is the total number of bands in the lane/community and p i is the relative abundance/intensity of the ith band in the lane/community (Magurran 1988). To ensure that the Simpsons index increases with increasing diversity, 1/D was used instead of the original formulation.
Results and discussion
DNA yield and purity
Maximal DNA yield and purity is important when selecting a DNA isolation procedure. Differences in DNA yield from the same sample, using different DNA extraction methods, could indicate variations in efficacy of cellular lysis. This could imply that only the cells most sensitive to the lytic protocol have been lysed which skews community analysis data. Purity of the extract is important for downstream molecular techniques required for community analysis such as PCR (Krsek and Wellington 1999).
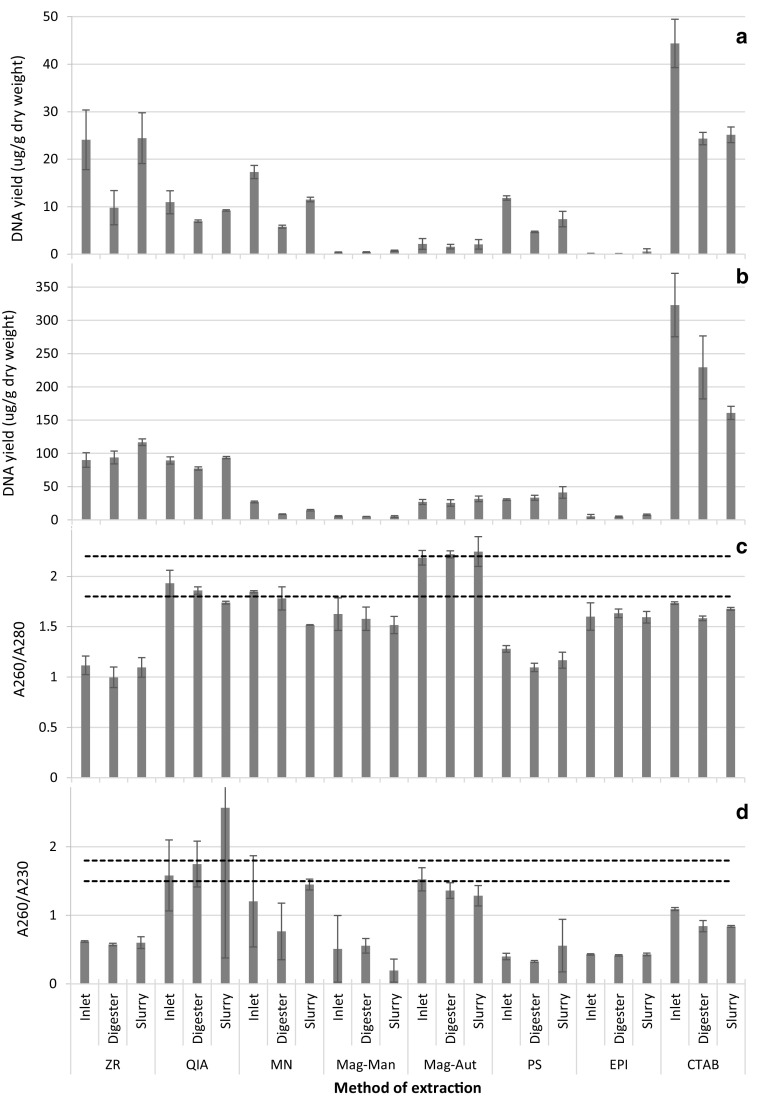
All DNA extraction methods that were tested were successful in extracting DNA from samples obtained from various stages in the AD process. However, the DNA yield significantly varied between extraction methods (P < 0.05) (Fig. 1a, b). For most extraction methods tested, a negative correlation existed between the DNA yield and the weight of the starting material used. This corroborated the results obtained by Ariefdjohan et al. (2010) when extracting DNA from human faecal samples. Elevated DNA yield obtained with smaller sample weights may be attributed to increased contact between the sample, lysis buffer and beads. The CTAB method resulted in the largest DNA yield and the EPI kit resulted in the smallest DNA yield (Fig. 1a, b). The low DNA yield obtained using the EPI kit is justifiable because this kit is an indirect method of DNA extraction. Similar results have been reported in previous studies where it was concluded that DNA yield is greater with direct extractions in comparison to indirect extractions (Leff et al. 1995; Delmont et al. 2011).
Fig. 1.
Yield and quality of DNA extracted from samples obtained from the inlet, digester and slurry using various methods of DNA extraction. a DNA quantified with Qubit. b DNA quantified with NanoDrop. c DNA quality determined by A260/A280 ratio. d DNA quality determined by A260/A230 ratio. Area between perforated lines in c and d is indicative of pure DNA, i.e. DNA with no protein contamination in c and DNA with no contamination by aromatic compounds, phenols and carbohydrates in d. Error bars represent standard deviation (n = 3)
The minute DNA yield obtained in this study when using the EPI kit could also be a function of the limitations of the kit when using cow dung samples at various stages of the AD process. Even though the recommended source material included soil (Table 1), which is quite granular, significant clogging of the filters was observed for all samples used. This could have resulted in loss of DNA because of potential exclusion of certain microbes prior to the second stage of the extraction process (i.e. DNA extraction from filters containing cells). Furthermore, the clogging of filters contributed to the extended duration of the extraction process (Table 1). Clogging of the filters might have been avoided if Miracloth was used for the efficient removal of large particulates in the initial filtration as opposed to cheesecloth. Unlike with cheesecloth, Miracloth has uniform pore sizes, therefore, enabling adequate filtration (Endres et al. 2003). In the present study, sterile cheesecloth was used since it was mentioned as an option in the EPI protocol.
Clogging of filters was also experienced when using the ZR kit, which utilises solid phase nucleic acid extraction. This method of DNA extraction consists of four key steps: cellular lysis, adsorption of nucleic acids, washing and final elution of pure DNA (Kojima and Ozawa 2002; Shaw et al. 2009). Clogging of the filters resulted in reduced adsorption of nucleic acids to the filter material because all of the lysate was unable to pass through. This resulted in the inconsistency of the kit as evidenced by the large standard deviation between DNA yield replicates (Fig. 1a). Reproducibility of DNA extraction is very important when selecting a DNA extraction method (Tan and Yiap 2009). As with the EPI kit, the recommended source material for the ZR kit is soil samples (Table 1). This implies that the kit should be well suited for granular material such as cow dung.
All direct methods of DNA extraction resulted in significantly greater DNA yields in comparison to the paramagnetic bead based extractions (Mag-Man and Mag-Aut; P < 0.05; Fig. 1a). Similar results have been reported elsewhere when comparing DNA yields obtained using magnetic bead extractions versus organic extractions (Kishore et al. 2006; Montpetit et al. 2005). The general consensus was that magnetic based extractions were sub-optimal with low yield and degraded samples as is the case with certain forensic samples (Kishore et al. 2006). However, the samples used in the present study were expected to contain large amounts of intact DNA. The low DNA yield obtained in the present study, when using paramagnetic bead based technology, is corroborated by the study of Brownlow et al. (2012). They reported that the DNA yield obtained using automated paramagnetic technology was significantly lower than that obtained when using automated and manual spin column, silica based technology, regardless of the amount of DNA in the sample material (Brownlow et al. 2012).
The reduced DNA yield when using paramagnetic DNA extraction in the present study may be due to the type of samples used. However, the MagMax kit should efficiently extract DNA from a broad range of sample types including manure (environmental samples; Table 1). Reduced yield may also be due to non-specific adhesion of the DNA to the walls of the extraction tubes. The paramagnetic method resulted in extended periods when the extracted DNA is in direct contact with the tube whereas other methods such as the filter based techniques involve the entrapment of DNA on filters. Furthermore, low DNA yield may also be due to the incomplete release of DNA from the magnetic beads, thus preventing complete elution. This is verified by the present work where manual and automated DNA extraction were tested using a single paramagnetic kit (Mag-Man and Mag-Aut, respectively; Table 1). The DNA yield obtained using the automated system was up to six times greater than that obtained from the same samples using manual extraction (Fig. 1a). For both manual and automated paramagnetic DNA isolation, the agitation of the magnetic beads in specific reagents is required for DNA binding, washing and final elution. This is advantageous as it prevents problems usually associated with other kits, e.g. filter clogging (Fang et al. 2007). However, insufficient agitation may result in reduced DNA yields (Adamowicz et al. 2014). Agitation in the automated system is achieved via the up and down movement of magnetic rods, whereas the manual method (Mag-Man) is agitated by low speed shaking on an orbital shaker. The speed has to be minimal to avoid spillage which could result in cross-contamination of samples in the processing plate. Hence, agitation was limited when conducting manual paramagnetic DNA extraction. Limited agitation probably contributed to incomplete release of DNA from the magnetic beads and the resultant lower DNA yield when using Mag-Man extraction.
The results showed a general trend in the yield of DNA obtained from the various samples. The same samples from the inlet, digester and slurry were used for all extraction methods to enable a clear comparison between kits. The Qubit data showed that the DNA yield, using all methods of extraction, was largest in the inlet, intermediate in the slurry and smallest in the digester (Fig. 1a). However, such clear trends were not evident when using the Nanodrop for DNA quantification (Fig. 1b). Furthermore, the DNA yield was highly overestimated (on average approximately 10 times; Fig. A2) when using the Nanodrop in comparison to the Qubit for quantification (cf. Fig. 1a, b). Similar results were observed in previous studies when comparing data obtained using the Qubit and Nanodrop (Sironen et al. 2008; Guo and Zhang 2013). Qubit quantification is fluorescence based whereas quantification using the Nanodrop is based on UV absorbance. Elevated yield measurements obtained when using the Nanodrop is due to co-extracted impurities in the eluted DNA contributing to the DNA yield measurements (Guo and Zhang 2013). However, the Nanodrop measurements were overestimated to a larger degree in the digester samples than in the inlet and slurry samples with majority of the extraction methods (Fig. A2). This indirectly implies that the digester extracts contain more impurities, which is not surprising considering that the AD process results in the production of substances such as humic acids. This further highlights the need for DNA extraction methods that suit each stage of the process since impurities and inhibitors are present in varying amounts in the inlet, digester chamber and slurry.
Such co-extracted impurities are undesirable because they may negatively influence downstream applications such as PCR. The Nanodrop is advantageous in that it may be used to determine the approximate level of DNA contamination. This is achieved via the analysis of ratios of absorbances at 260/280 nm and at 260/230 nm which represent protein and aromatic compound (e.g. humic acid) contamination, respectively (Roh et al. 2006; Weiss et al. 2007). Protein contamination was mostly evident in DNA samples extracted with the ZR and PS kits, whereas aromatic compound contamination was evident in all samples with the possible exception of the QIA and Mag-Aut kits (Fig. 1c, d). Protein and aromatic compound contamination did not affect PCR for all DNA extracts with the exception of the CTAB method (Fig. A3). Furthermore, upon visual inspection the only DNA extract that was not clear was the one that did not amplify using PCR, i.e. the CTAB extract. However, PCR was successful upon dilution of the CTAB extract. Dilution resulted in the reduction in the concentration of contaminants and DNA. However, the dilution of the DNA did not negatively affect the PCR process because large amounts of DNA were obtained using the CTAB method (Fig. 1a).
DNA integrity
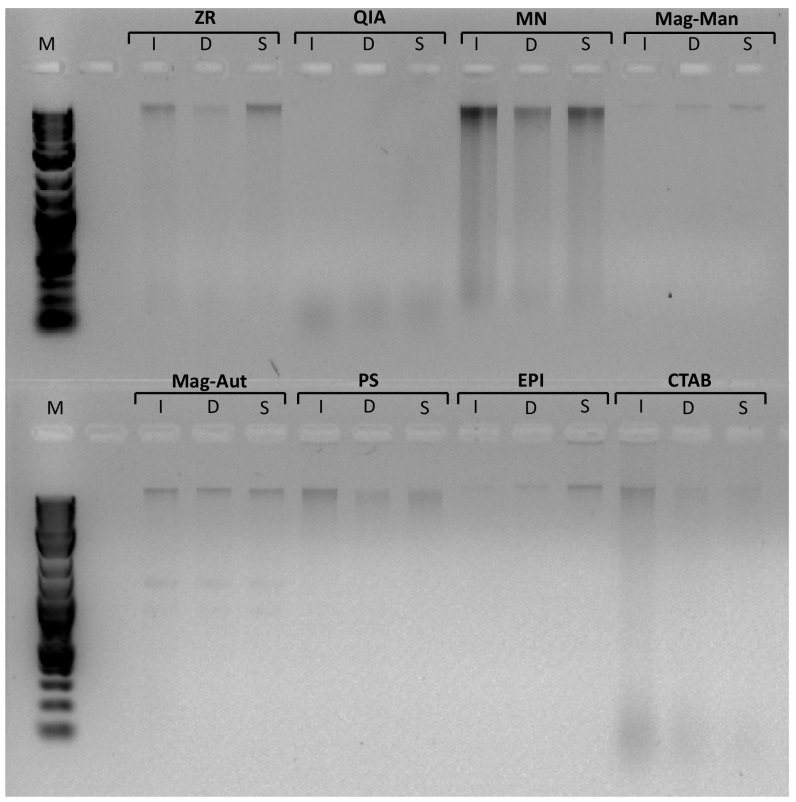
Cell lysis is the first and fundamental step of DNA extraction methods. For all extraction methods tested, the cells were lysed using mechanical, chemical, heat or combinations of various methods (Table 1). Mechanical methods of DNA extraction such as bead beating have been attempted to increase DNA yield via the improved lysis of bacterial cells. However, mechanical lysis may also result in the shearing/fragmenting of genomic DNA (Wintzingerode et al. 1997).
In the present study, the indirect method of DNA extraction (EPI) resulted in minimal or no DNA shear (Fig. 2). These results corroborate the observations of Roh et al. (2006) who showed that DNA shear was more prevalent when using direct extractions as opposed to indirect extractions. Only one direct DNA extraction method resulted in limited/no DNA shear in the present study, namely the paramagnetic extraction method (Mag-Man and Mag-Aut) (Fig. 2). All other direct methods, including mechanical, heat and chemical-based methods, resulted in a significant amount of DNA shear (as indicated by the smear on the gel in Fig. 2). The integrity of the DNA was lowest in the QIA samples where all the extracted DNA was fragmented into between 0.1 and 0.3 kb fragments (the only method with no high molecular weight fragments at all). Interestingly, no bead beating was conducted when using the QIA kit. Cells were lysed using chemical means and heat (Table 1). Due to the proprietary nature of the reagents of the kit, we are unable to deduce the reason for the elevated level of DNA shear, but other investigators have also noted that slow cell disruption such as the addition of lysis solution with no additional physical disruption may lead to DNA degradation (Chaudhary et al. 2011). The integrity of the DNA obtained using the CTAB method was also low. Unlike with the QIA kit, the CTAB method yielded a portion of high molecular weight DNA but the DNA extract was highly fragmented with fragments that were even smaller than 0.1 kb in size (Fig. 2). Fragmented nucleic acids are not ideal because they may contribute to the formation of chimeric PCR products and are also sources of artefacts in PCR amplification or reverse transcription (Liesack et al. 1991; Wintzingerode et al. 1997).
Fig. 2.
DNA isolated from inlet, digester and slurry using various DNA extraction methods. M DNA ladder, I inlet, D digester, S slurry, ZR ZR soil microbe DNA MiniPrep—Zymo research, QIA QIAamp Fast DNA Stool Mini Kit—QIAGEN, MN NucleoSpin Soil Kit—Macherey-Nagel, Mag-Man MagMAX Total Nucleic Acid Isolation Kit—Manual—Thermo Fisher Scientific, Mag-Aut MagMAX Total Nucleic Acid Isolation Kit—Automated—Thermo Fisher Scientific, PS Powersoil DNA Isolation Kit—MO BIO Laboratories, EPI Meta-G-Nome DNA Isolation Kit—Epicentre, CTAB CTAB based extraction
Species richness
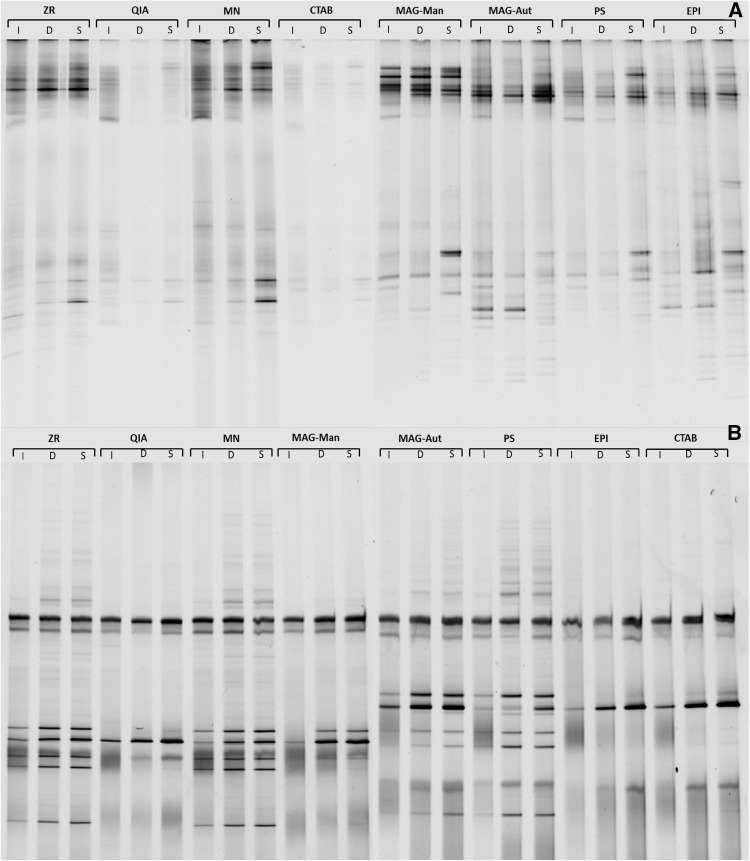
Theoretically, all DNA extracts from the same source, i.e. inlet, digester chamber or slurry, should have identical species composition and abundance/spread because the respective samples originated from the same location. However, visual inspection of the bacterial and archaeal DGGE profiles shows that this was not evident. This was further reiterated by the wide range of bacterial and archaeal diversity indices (Table 3) and the extensive variation of evenness (Fig. A4) evidenced in the microbial communities obtained using the various extraction methods. Some methods of extraction resulted in vastly different or wider community profiles than others (Fig. 3). The QIA and CTAB methods of extraction were the least effective at extracting DNA from bacterial species (as demonstrated by the limited banding pattern in Fig. 3a) whereas the QIA, MAG-Man, EPI and CTAB extraction methods did not successfully extract DNA from all archaeal species (Fig. 3b).
Table 3.
Bacterial and archaeal diversity indices obtained when extracting DNA using various extraction methods
| KIT | Bacteria | Archaea | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inlet | Digester | Slurry | Inlet | Digester | Slurry | |||||||
| H′ | 1/D | H′ | 1/D | H′ | 1/D | H′ | 1/D | H′ | 1/D | H′ | 1/D | |
| ZR | 2.3 | 7.6 | 2.1 | 5.6 | 2.2 | 6.6 | 2.4 | 9.1 | 2.6 | 10.5 | 2.6 | 11.3 |
| QIA | 2.5 | 10.3 | 1.4 | 3.7 | 2.0 | 6.2 | 1.8 | 5.0 | 2.0 | 6.4 | 1.9 | 5.4 |
| MN | 2.7 | 11.1 | 2.4 | 7.9 | 2.4 | 8.6 | 2.4 | 9.0 | 2.6 | 12.1 | 2.8 | 13.0 |
| Mag-Man | 2.1 | 6.1 | 2.3 | 7.3 | 2.2 | 6.7 | 1.7 | 6.0 | 2.2 | 7.6 | 2.1 | 5.9 |
| Mag-Aut | 2.6 | 10.1 | 2.2 | 6.1 | 2.3 | 7.1 | 2.2 | 8.1 | 2.0 | 6.2 | 2.3 | 7.6 |
| PS | 2.5 | 10.8 | 2.3 | 7.7 | 2.4 | 8.2 | 1.6 | 3.6 | 2.6 | 10.3 | 2.6 | 10.6 |
| EPI | 2.6 | 10.3 | 2.7 | 10.2 | 2.6 | 9.3 | 1.7 | 4.4 | 2.1 | 7.3 | 2.1 | 6.8 |
| CTAB | 2.5 | 11.0 | 1.8 | 5.4 | 2.0 | 6.1 | 2.2 | 7.1 | 1.9 | 4.9 | 2.1 | 6.4 |
Fig. 3.
DGGE banding patterns of a bacterial and b archaeal DNA isolated from samples obtained from the inlet, digester and slurry using various DNA extraction methods. I Inlet, D digester, S slurry, ZR ZR Soil Microbe DNA MiniPrep—Zymo research, QIA QIAamp Fast DNA Stool Mini Kit—QIAGEN, MN NucleoSpin Soil Kit—Macherey-Nagel, Mag-Man MagMAX Total Nucleic Acid Isolation Kit—Manual—Thermo Fisher Scientific, Mag-Aut MagMAX Total Nucleic Acid Isolation Kit—Automated—Thermo Fisher Scientific, PS Powersoil DNA Isolation Kit—MO BIO Laboratories, EPI Meta-G-Nome DNA Isolation Kit—Epicentre, CTAB CTAB based extraction
The present study proves that some methods of extraction work optimally for bacterial DNA extraction from AD samples but are not as efficient for archaeal DNA extraction (e.g. EPI; Table 3) and vice versa (e.g. ZR; Table 3). Whilst other methods of extraction worked poorly for both bacterial and archaeal DNA extraction (e.g. QIA and CTAB; Fig. 3). Extraction methods also varied in efficacy based on the stage of AD at which the samples were collected. The highest Shannon Weiner (H′) and inverse Simpsons indices (1/D) were observed when using the EPI kit for bacterial DNA extraction from digester and slurry samples. However, the EPI kit was not as efficient when extracting bacterial DNA from the inlet sample (Table 3). Inlet, digester and slurry samples vary in the degree of digestion where the inlet samples are the least digested (most granular) and the slurry samples are the most digested (least granular). Granular samples need to be homogenised sufficiently to remove all adhering cells that may be hidden in various crevices of the sample, thus enabling complete downstream DNA extraction, whilst less granular samples require a lower degree of homogenisation for sufficient cell removal and subsequent cell lysis. The EPI kit incorporated a solvent-based method of substrate homogenisation (Table 1). This method worked optimally for the digester and slurry samples but did not offer adequate homogenisation of the more granular inlet samples. The highest H’ and 1/D indices were observed when using the MN kit for inlet, bacterial DNA extraction (Table 3). The MN kit incorporated bead beating and cell lysis buffer for sample homogenisation and cell lysis. Bead beating enables the even infiltration of the lysis buffer to the entire sample, regardless of its granularity/consistency, while chemical lysis methods alone may contribute to biases in extraction from granular substrates because the spatial access to all target organisms may be limited (Salonen et al. 2010).
Unlike with the bacterial DNA extracts, the EPI kit yielded poor banding patterns (Fig. 3) and H’ and 1/D indices (Table 3) for archaeal extracts from all sources (I, D and S). This may be attributed to the differences between the cell surface structure of archaeal and bacterial cells. Bacteria are covered by a peptidoglycan layer whereas the archaeal surface structure is divided into various groups ranging from S-layers to methanochondroitin (König 1988; Kubota et al. 2008). Based on the surface covering, some archaeal cells may be more resistant to lysis than others. The EPI kit incorporated proteinase K and a lysozyme solution for cell lysis. These reagents worked optimally for bacterial DNA extraction but showed limited efficacy on hard to lyse archaeal cells (Fig. 3; Table 3). Furthermore, the lack of a mechanical lysis stage may have also contributed to the limited observed archaeal diversity. This is corroborated by a study conducted by Salonen et al. (2010) where they deduced that mechanical cell disruption was more effective than enzymatic means for the extraction of archaeal DNA. In the present study, the MN extraction method resulted in the highest H’ and 1/D indices for archaeal extracts (Table 3). The MN kit incorporates mechanical and chemical lysis. Furthermore, a proprietary ‘enhancer solution’ is also included in the kit. This solution, in combination with the lysis buffer, ensures that the highest possible DNA yield is obtained from the sample.
A better understanding of the link between cellular surface structure and lytic requirement is necessary to enable optimisation of methods of DNA extraction from bacteria and archaea. Hence, bands were isolated from the DGGE gels and sequenced. For the bacterial DGGE profiles, bands that were not well represented in all lanes were excised. These bands represent hard to lyse bacteria. Interestingly, all of the excised bands were gram-positive bacteria (Table A1). Gram-positive bacteria are harder to lyse than gram-negative bacteria due to varying cell structures. Gram-positive and gram-negative bacteria have cell walls containing peptidoglycan but gram-positive cells differ in that the thickness, quantity, length distribution and degree of crosslinking of the peptidoglycan is more extensive than in gram-negative cells (Fig. A5) (Cabeen and Jacobs-Wagner 2005; Mahalanabis et al. 2009). The ideal method of DNA extraction should ensure that all cells are equally lysed, including hard to lyse gram-positive and archaeal cells. Some extraction methods such as the MN kit and the Mag-Aut kit were more successful at extracting DNA from gram-positive bacteria than others (e.g. QIA, CTAB; Table A.1). These MN and Mag-Aut extraction methods incorporated bead beating in the lytic protocol which implies that mechanical methods of cell lysis are effective for some hard to lyse gram-positive bacterial cells.
For the archaeal DGGE profiles, bands were randomly excised. These bands represent a variety of archaeal species, i.e. both hard and easy to lyse cells. Archaea possess cell walls of various types including protein surface layers (S-layer), pseudomurein, methanochondroitin, sheath layers and combinations of the various polymers and the S-layer (Albers and Meyer 2011; Fig. A5). The unusual cell wall structure renders certain archaeal cells resistant to lytic protocols that work well for bacterial cells (Jarrell et al. 1992). This was evident in the present study (see EPI kit in Fig. 2). Interestingly, the sequence data shows that species possessing an S-layer were not lysed using all methods of extraction whereas the species lacking the S-layer were lysed using all extraction methods, albeit at varying levels (Table A1). Methods that effectively lysed most S-layer containing cells included the ZR, MN, PS and Mag-Aut methods. The Mag-Man method also successfully lysed some S-layer containing cells, although to a lower degree than the Mag-Aut method (Table A1). This may be as a consequence of the rapid agitation achieved by automated DNA extraction in comparison to manual extraction. The ZR, MN and PS methods of DNA extraction are spin column filter based methods that incorporate bead beating and cell lysis buffer for cell lysis (Table 1), whereas the Mag-Man and Mag-Aut methods are paramagnetic bead based methods that also incorporate bead beating and cell lysis buffer (Table 1). The common variable that may have resulted in adequate archaeal cell lysis may have been the incorporation of bead beating and lysis buffer. The CTAB method also incorporated bead beating in the lysis step, however, the CTAB and EPI methods are solution based (Table 1). Both solution based methods yielded poor results for archaeal DNA extraction from cells surrounded by an S-layer.
Bacteria and archaea play integral roles in the anaerobic digestion process. Hence, both are frequently explored when conducting microbial community analyses of biogas reactors (Ariesyady et al. 2007; Riviere et al. 2009; Sundberg et al. 2013). Ideally, a single DNA extraction method should be used for DNA sequestration from both bacteria and archaea since this will minimise costs and time. Of the extraction methods tested the QIA kit generally performed the poorest in terms of both bacterial and archaeal species diversity analyses (Table 3). This may be a function of the reduced integrity of the initial DNA extract (Fig. 2) and the reliance on chemical and heat lysis instead of mechanical lysis when using the QIA kit, preventing DNA extraction from hard to lyse cells (Tables 1, 2). The limited diversity obtained when using the QIA kit in the present study is consistent with what has been previously reported by Claassen et al. (2013) and Ariefdjohan et al. (2010). The extraction method that yielded optimal results (H’ and 1/D) for both bacterial and archaeal community profiling was the MN kit (Table 3).
Conclusions
It may be concluded that all DNA extraction methods tested were successful at extracting DNA from AD samples. However, the yield of extracted DNA varied between methods. The microbial diversity was significantly influenced by the choice of DNA extraction method used. However, there was no correlation between DNA yield and diversity. Maximal species diversity and richness in all samples (inlet, digester and slurry) was achieved when using a spin-column filter-based kit that incorporated mechanical and chemical lysis (MN kit). This study proves that it is important to take the method of extraction into consideration when comparing microbial communities obtained by different researchers and laboratories.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Biogas research at ARC-ISCW, South Africa is supported by the National Research Foundation (NRF; Grant Number 96735), the South Africa—Norway Research Co-operation (SANCOOP; Grant Number RCN 234203) and the Water Research Commission (WRC; Grant Number K5/2543). Opinions expressed and conclusions reached are those of the authors and not necessarily endorsed by the NRF, SANCOOP or WRC.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-1009-x) contains supplementary material, which is available to authorized users.
Contributor Information
Ashira Roopnarain, Email: RoopnarainA@arc.agric.za.
Mashudu Mukhuba, Email: MukhubaM@arc.agric.za.
Rasheed Adeleke, Phone: +27 123102519, Email: AdelekeR@arc.agric.za.
Mokhele Moeletsi, Email: MoeletsiM@arc.agric.za.
References
- Adamowicz MS, Stasulli DM, Sobestanovich EM, Bille TW. Evaluation of methods to improve the extraction and recovery of DNA from cotton swabs for forensic analysis. PLoS ONE. 2014;9(12):e116351. doi: 10.1371/journal.pone.0116351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers SV, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9(6):414–426. doi: 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59(1):143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariefdjohan MW, Savaiano DA, Nakatsu CH. Comparison of DNA extraction kits for PCR-DGGE analysis of human intestinal microbial communities from fecal specimens. J Nutr. 2010;9(1):1. doi: 10.1186/1475-2891-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariesyady HD, Ito T, Okabe S. Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester. Water Res. 2007;41(7):1554–1568. doi: 10.1016/j.watres.2006.12.036. [DOI] [PubMed] [Google Scholar]
- Bergmann I, Mundt K, Sontag M, Baumstark I, Nettmann E, Klocke M. Influence of DNA isolation on Q-PCR-based quantification of methanogenic Archaea in biogas fermenters. Syst Appl Microbiol. 2009;33:78–84. doi: 10.1016/j.syapm.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Brownlow RJ, Dagnall KE, Ames CE. A comparison of DNA collection and retrieval from two swab types (cotton and nylon flocked swab) when processed using three QIAGEN extraction methods. J Forensic Sci. 2012;57(3):713–717. doi: 10.1111/j.1556-4029.2011.02022.x. [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. Bacterial cell shape. Nat Rev Microbiol. 2005;3(8):601–610. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- Carrigg C, Rice O, Kavanagh S, Collins G, O’Flaherty V. DNA extraction method affects microbial community profiles from soils and sediment. Appl Microbiol Biotechnol. 2007;77(4):955–964. doi: 10.1007/s00253-007-1219-y. [DOI] [PubMed] [Google Scholar]
- Chaudhary PP, Sirohi SK, Kumar S. Improved extraction of quality DNA from methanogenic archaea present in rumen liquor for PCR application. Asian J Anim Sci. 2011;5(3):166–174. doi: 10.3923/ajas.2011.166.174. [DOI] [Google Scholar]
- Claassen S, du Toit E, Kaba M, Moodley C, Zar HJ, Nicol MP. A comparison of the efficiency of five different commercial DNA extraction kits for extraction of DNA from faecal samples. J Microbiol Methods. 2013;94(2):103–110. doi: 10.1016/j.mimet.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmont TO, Robe P, Clark I, Simonet P, Vogel TM. Metagenomic comparison of direct and indirect soil DNA extraction approaches. J Microbiol Methods. 2011;86(3):397–400. doi: 10.1016/j.mimet.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Endres HN, Johnson JA, Ross CA, Welp JK, Etzel MR. Evaluation of an ion-exchange membrane for the purification of plasmid DNA. Biotechnol Appl Biochem. 2003;37(3):259–266. doi: 10.1042/BA20030025. [DOI] [PubMed] [Google Scholar]
- Fahle GA, Fischer SH. Comparison of six commercial DNA extraction kits for recovery of cytomegalovirus DNA from spiked human specimens. J Clin Microbiol. 2000;38(10):3860–3863. doi: 10.1128/jcm.38.10.3860-3863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Willis RC, Burrell A, Evans K, Hoang Q, Xu W, Bounpheng M. Automation of nucleic acid isolation on KingFisher magnetic particle processors. J Lab Autom. 2007;12(4):195–201. doi: 10.1016/j.jala.2007.05.001. [DOI] [Google Scholar]
- Garcia-Peña EI, Parameswaran P, Kang DW, Canul-Chan M, Krajmalnik-Brown R. Anaerobic digestion and co-digestion processes of vegetable and fruit residues: process and microbial ecology. Bioresour Technol. 2011;102(20):9447–9455. doi: 10.1016/j.biortech.2011.07.068. [DOI] [PubMed] [Google Scholar]
- Guo F, Zhang T. Biases during DNA extraction of activated sludge samples revealed by high throughput sequencing. Appl Environ Microbiol. 2013;97(10):4607–4616. doi: 10.1007/s00253-012-4244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera A, Cockell CS. Exploring microbial diversity in volcanic environments: a review of methods in DNA extraction. J Microbiol Methods. 2007;70(1):1–12. doi: 10.1016/j.mimet.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Ikenaga M, Asakawa S, Muraoka Y, Kimura M. Methanogenic archaeal communities in rice roots grown in flooded soil pots: estimation by PCR-DGGE and sequence analyses. Soil Sci Plant Nutr. 2004;50(5):701–711. doi: 10.1080/00380768.2004.10408526. [DOI] [Google Scholar]
- Jarrell KF, Faguy D, Hebert AM, Kalmokoff ML. A general method of isolating high molecular weight DNA from methanogenic archaea (archaebacteria) Can J Microbiol. 1992;38(1):65–68. doi: 10.1139/m92-010. [DOI] [PubMed] [Google Scholar]
- Kampmann K, Ratering S, Kramer I, Schmidt M, Zerr W, Schnell S. Unexpected stability of Bacteroidetes and Firmicutes communities in laboratory biogas reactors fed with different defined substrates. Appl Environ Microbiol. 2012;78(7):2106–2119. doi: 10.1128/AEM.06394-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore R, Reef Hardy W, Anderson VJ, Sanchez NA, Buoncristiani MR. Optimization of DNA extraction from low-yield and degraded samples using the BioRobot® EZ1 and BioRobot® M48. J Forensic Sci. 2006;51(5):1055–1061. doi: 10.1111/j.1556-4029.2006.00204.x. [DOI] [PubMed] [Google Scholar]
- Kojima K, Ozawa S (2002) US Patent No 20,020,192,667. Washington, DC, US Patent and Trademark Office
- König H. Archaeobacterial cell envelopes. Can J Microbiol. 1988;34(4):395–406. doi: 10.1139/m88-071. [DOI] [Google Scholar]
- Krsek M, Wellington EMH. Comparison of different methods for the isolation and purification of total community DNA from soil. J Microbiol Methods. 1999;39(1):1–16. doi: 10.1016/S0167-7012(99)00093-7. [DOI] [PubMed] [Google Scholar]
- Kubota K, Imachi H, Kawakami S, Nakamura K, Harada H, Ohashi A. Evaluation of enzymatic cell treatments for application of CARD-FISH to methanogens. J Microbiol Methods. 2008;72(1):54–59. doi: 10.1016/j.mimet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Leff LG, Dana JR, McArthur JV, Shimkets LJ. Comparison of methods of DNA extraction from stream sediments. Appl Environ Microbiol. 1995;61(3):1141–1143. doi: 10.1128/aem.61.3.1141-1143.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesack W, Weyland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb Ecol. 1991;21(1):191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- Magurran AE. Ecological diversity and its measurement. Princeton: Princeton University Press; 1988. [Google Scholar]
- Mahalanabis M, Al-Muayad H, Kulinski MD, Altman D, Klapperich CM. Cell lysis and DNA extraction of gram-positive and gram-negative bacteria from whole blood in a disposable microfluidic chip. Lab Chip. 2009;9(19):2811–2817. doi: 10.1039/b905065p. [DOI] [PubMed] [Google Scholar]
- Mertens B, Boon N, Verstraete W. Stereospecific effect of hexachloro- cyclohexane on activity and structure of soil methanotrophic communities. Environ Microbiol. 2005;7:660–669. doi: 10.1111/j.1462-2920.2005.00735.x. [DOI] [PubMed] [Google Scholar]
- Miller DN, Bryant JE, Madsen EL, Ghiorse WC. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol. 1999;65(11):4715–4724. doi: 10.1128/aem.65.11.4715-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minas K, McEwan NR, Newbold CJ, Scott KP. Optimization of a high-throughput CTAB-based protocol for the extraction of qPCR-grade DNA from rumen fluid, plant and bacterial pure cultures. FEMS Microbiol Lett. 2011;325(2):162–169. doi: 10.1111/j.1574-6968.2011.02424.x. [DOI] [PubMed] [Google Scholar]
- Montpetit SA, Fitch IT, O’Donnell PT. A simple automated instrument for DNA extraction in forensic casework. J Forensic Sci. 2005;50(3):555–563. doi: 10.1520/JFS2004181. [DOI] [PubMed] [Google Scholar]
- Muyzer G, De Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59(3):695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S, Weissenbach J, Sghir A. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 2009;3(6):700–714. doi: 10.1038/ismej.2009.2. [DOI] [PubMed] [Google Scholar]
- Roh C, Villatte F, Kim BG, Schmid RD. Comparative study of methods for extraction and purification of environmental DNA from soil and sludge samples. Appl Biochem Biotechnol. 2006;134:97–112. doi: 10.1385/ABAB:134:2:97. [DOI] [PubMed] [Google Scholar]
- Roopnarain A, Adeleke R. Current status, hurdles and future prospects of biogas digestion technology in Africa. Renew Sustain Energy Rev. 2017;67:1162–1179. doi: 10.1016/j.rser.2016.09.087. [DOI] [Google Scholar]
- Salonen A, Nikkilä J, Jalanka-Tuovinen J, Immonen O, Rajilić-Stojanović M, Kekkonen RA, de Vos WM. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81(2):127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Shaw KJ, Thain L, Docker PT, Dyer CE, Greenman J, Greenway GM, Haswell SJ. The use of carrier RNA to enhance DNA extraction from microfluidic-based silica monoliths. Anal Chim Acta. 2009;652(1):231–233. doi: 10.1016/j.aca.2009.03.038. [DOI] [PubMed] [Google Scholar]
- Sironen A, Uimari P, Vilkki J. Comparison of different DNA extraction methods from hair root follicles to genotype Finnish Landrace boars with the Illumina PorcineSNP60 BeadChip. Agric Food Sci. 2008;20(2):143–150. doi: 10.2137/145960611797215709. [DOI] [Google Scholar]
- Slana I, Pribylova R, Kralova A, Pavlik I. Persistence of Mycobacterium avium subsp. paratuberculosis at a farm-scale biogas plant supplied with manure from paratuberculosis-affected dairy cattle. Appl Environ Microbiol. 2011;77(9):3115–3119. doi: 10.1128/AEM.02407-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, Karlsson A. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol. 2013;85(3):612–626. doi: 10.1111/1574-6941.12148. [DOI] [PubMed] [Google Scholar]
- Tan SC, Yiap BC. DNA, RNA, and protein extraction: the past and the present. Biomed Res Int. 2009 doi: 10.1155/2009/574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theron J, Cloete TE. Molecular techniques for determining microbial diversity and community structure in natural environments. Crit Rev Microbiol. 2000;26(1):37–57. doi: 10.1080/10408410091154174. [DOI] [PubMed] [Google Scholar]
- Vanysacker L, Declerck SA, Hellemans B, De Meester L, Vankelecom I, Declerck P. Bacterial community analysis of activated sludge: an evaluation of four commonly used DNA extraction methods. Appl Microbiol Biotechnol. 2010;88(1):299–307. doi: 10.1007/s00253-010-2770-5. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Asakawa S, Nakamura A, Nagaoka K, Kimura M. DGGE method for analyzing 16S rDNA of methanogenic archaeal community in paddy field soil. FEMS Microbiol Lett. 2004;232(2):153–163. doi: 10.1016/S0378-1097(04)00045-X. [DOI] [PubMed] [Google Scholar]
- Weiss A, Jerome V, Freitag R. Comparison of strategies for the isolation of PCR-compatible, genomic DNA from a municipal biogas plants. J Chromatogr B. 2007;853:190–197. doi: 10.1016/j.jchromb.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Wintzingerode FV, Göbel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21(3):213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- Xu Z, Zhao M, Miao H, Huang Z, Gao S, Ruan W. In situ volatile fatty acids influence biogas generation from kitchen wastes by anaerobic digestion. Bioresour Technol. 2014;163:186–192. doi: 10.1016/j.biortech.2014.04.037. [DOI] [PubMed] [Google Scholar]
- Yeates C, Gillings MR, Davison AD, Altavilla N, Veal DA. Methods for microbial DNA extraction from soil for PCR amplification. Biol Proced Online. 1998;1(1):40–47. doi: 10.1251/bpo6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.