Abstract
Cigarette smoking is the leading cause of lung cancer worldwide. In this study, we evaluated the serum autoantibody (AAb) repertoires of non-small cell lung cancer (NSCLC) patients and smokers (SM), leading to the identification of overactivated pathways and hubs involved in the pathogenesis of NSCLC. Surface- and solution-phase biopanning were performed on immunoglobulin G purified from the sera of NSCLC and SM groups. In total, 20 NSCLC- and 12 SM-specific peptides were detected, which were used to generate NSCLC and SM protein datasets. NSCLC- and SM-related proteins were visualized using STRING and Gephi, and their modules were analyzed using Enrichr. By integrating the overrepresented pathways such as pathways in cancer, epithelial growth factor receptor, c-Met, interleukin-4 (IL-4) and IL-6 signaling pathways, along with a set of proteins (e.g. phospholipase D (PLD), IL-4 receptor, IL-17 receptor, laminins, collagens, and mucins) into the PLD pathway and inflammatory cytokines network as the most critical events in both groups, two super networks were made to elucidate new aspects of NSCLC pathogenesis and to determine the influence of cigarette smoking on tumour formation. Taken together, assessment of the AAb repertoires using a systems biology approach can delineate the hidden events involved in various disorders.
Introduction
Lung cancer is recognized as a fatal malignancy, accounting for more than a quarter of all cancer deaths in the United States and 1.6 million deaths every year worldwide1,2. Approximately 85% of all new cases of lung cancer are classified as non-small cell lung cancer (NSCLC), including adenocarcinoma, squamous cell carcinoma and large-cell carcinoma2. Despite an overall increase in the rate of survival for most cancers, 5-year survival for lung cancer has not improved significantly1. On the other hand, smoking is a primary risk factor for lung cancer, and nearly 14–25% of lifetime smokers (SM) are expected to develop lung cancer2. Accordingly, the carcinogenic effects of tobacco smoke through deregulation of various cellular pathways and DNA adduct formation have been extensively studied in the past decades3.
The low success rate of lung cancer treatment originates from delayed diagnosis, which reduces the chance of total tumour excision via surgery and acquired resistance to available medication regimens4,5. Imaging methods are usually applied in the diagnosis of lung cancer. However, the high costs of these methods, radiation exposure, and false positive or negative results, restrict their applications as screening tests2,4. Moreover, resistance to conventional chemotherapeutics, such as platinum agents, antitubulars, topoisomerase II inhibitors, and antimetabolites, is quite common among NSCLC patients who eventually develop resistance to chemotherapeutics5.
Autoantibody (AAb) production, as a reaction of the immune system to tumour antigens, is an inevitable phenomenon in cancers6. Since AAb production occurs during the early stages of disease, it may serve as a functional tool for NSCLC diagnosis4,7. Therefore, detection of AAbs in NSCLC patients has been the subject of a few investigations to determine the AAb repertoires and identify AAbs against particular proteins4,7–14. Nevertheless, for comparison of the AAb repertoires and identification of common autoantigens, limited research has been performed on the sera of NSCLC patients and SM.
In contrast to studies assessing lung cancer via bioinformatics tools15–19, a few studies have focused on smoking with a systems biology approach; for instance, expression-based network analysis and assessment of protein-protein interactions have revealed changes in human blood20, human buccal mucosa21, and mouse lung samples22 due to cigarette exposure. For exhibiting the direct correlation between smoking and lung cancer, Ying Liu et al. analysed three protein expression datasets of lung adenocarcinoma and divided them into two subcategories of SM and non-SM. The results revealed Ras signalling pathway (SP) and proteoglycans in cancer as the key pathways in lung cancer development in SM23. Furthermore, Wang et al. constructed a graphical model to assess gross network alterations during transition from normal to cancer tissue in SM. The findings demonstrated a gain of epithelial growth factor receptor (EGFR) and platelet-derived growth factor receptor A (PDGFRA) modules in addition to a loss of interleukin-6 (IL-6) module in lung cancer24.
In the present study, we carried out biopanning with a phage-displayed random peptide library on immunoglobulin G (IgG) purified from the sera of NSCLC patients and SM to detect peptides based on their AAbs. NSCLC- and SM-specific proteins, predicted from the detected peptides, were investigated with systems biology tools to demonstrate their involvement as hubs in significant pathways driving lung cancer pathogenesis. Furthermore, they were used to construct two super networks for clarifying multidimensional contribution of smoking to NSCLC.
Results
Peptide library enrichment led to the generation of NSCLC and SM protein datasets
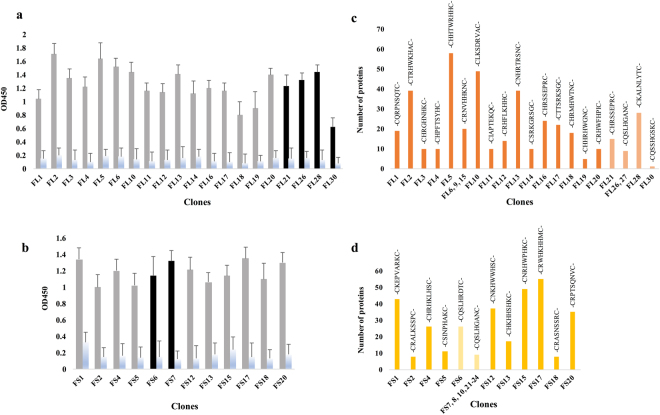
To assess the binding of NSCLC and SM IgG to phage pools after three consecutive rounds of surface- (SU) and solution-phase (SL) biopanning, all the input and output phages were screened via polyclonal phage enzyme-linked immunosorbent assay (ELISA). The highest signals were observed in the third round of SU and SL biopanning (data not shown). Based on monoclonal phage ELISA on random clones obtained from the third round of biopanning, 30 NSCLC (SU, FLU1-20; SL, FLL21-30) and 15 SM (SU, FSU1-5 and FSU11-20; SL, FSL6-10 and FSL21-25) clones were selected, which showed the highest intensities in comparison to the controls (Fig. 1a and b). Among the designated phage clones, three FLL and five FSU clones failed to produce meaningful sequences. In addition, two NSCLC-specific peptides (FLU7 and FLU8) containing sequences with three consecutive histidine residues, and one SM-specific peptide (FSL25) due to its similarity to another peptide sequence in this group, were omitted. Notably, among NSCLC-specific clones, FLU6, FLU9, and FLU15 (-CRNVHHKNC-), FLL22 and FLL29 (-CTRHWPHHC-), in addition to FLL26 and FLL27 (-CQSLHGANC-), showed similar sequences (Fig. 1c). Additionally, clones FSL7, FSL8, FSL10, and FSL21-24 showed an identical amino acid composition (-CQSLHGANC-) to clones FLL26 and FLL27 (Fig. 1d).
Figure 1.
Monoclonal phage ELISA and number of proteins predicted from each selected peptide. (a) Randomly selected NSCLC and (b) SM clones were screened by monoclonal phage ELISA; intensities are reported as mean ± SD. Columns 1–10 (grey) and 11–20 (black) belong to clones from SU and SL biopanning with the highest intensities, respectively, and the blue columns indicate the controls (bovine serum albumin (BSA)). Peptides identified with the purified IgG of (c) NSCLC patients (orange) and (d) SM (yellow) are plotted against the number of human proteins which matched to each sequence after blasting with a cut-off of 18.5 and manual deletion of unrelated entities. Mild orange (c) and (d) mild yellow indicate the peptides obtained from SL biopanning.
Evaluation of the selected peptides in the MimoDB demonstrated that 21 NSCLC- and 13 SM-specific peptides, were true target binders. Blasting peptides FLU22, FLU29, and FSL9 (score ≥18.5) followed by an assessment in the UniProt database and review of the literature, did not present major proteins; accordingly, these peptides were deleted from the final peptide lists. However, blasting the remaining peptide sequences led to the prediction of 531 NSCLC and 346 SM proteins with 50 common proteins (Supplementary dataset). Figure 1c and d show the sequence and quantity of proteins predicted from each peptide. Furthermore, the list of proteins found in more than one clone is presented in Table S1. Peptides FLU19, FLL26, FLL27, and FLL30 accounted for the lowest number of NSCLC-related proteins (<10), while peptides FLU5 and FLU10 represented the highest number of NSCLC-related proteins (>40). Moreover, peptides FSU1, FSU15 and FSU17 were individually related to more than 40 proteins whereas a group of SM-specific peptides (FSU2 and FSU18, and FSL7, FSL8, FSL10, and FSL21-24) accounted for less than 10 proteins in the SM protein dataset. Markedly, retinoic acid-induced protein 1 (RAI1), dermatan-sulfate epimerase (DSE), and protein kinase C delta (PRKCD) were proteins predicted from more than one peptide in both NSCLC and SM groups.
Protein complexes in the NSCLC and SM groups
Gene set enrichment by Consensus PathDB (CPDB) revealed multiple physically associated protein complexes, with at least two members in the NSCLC or SM protein dataset. Proline-rich tyrosine kinase 2 (PYK2)/Src homology region 2-containing protein tyrosine phosphatase 2 (SHP2) complex (two out of two components; 2/2), ADP-ribosylation factor 6 (ARF6)/Guanosine triphosphate (GTP)/Nucleoside diphosphate kinase A (NME1)/T-lymphoma invasion and metastasis-inducing protein 1 (Tiam1) complex (2/3), proto-oncogene tyrosine-protein kinase Src (SRC)-PRKCD-CUB domain-containing protein 1 (CDCP1) complex (2/3), and Janus kinase 1 (JAK1): Interleukin-13 receptor subunit alpha-1 (IL-13RA-1): Interleukin-4 receptor alpha (IL-4RA): Tyrosine kinase 2 (TYK2) complex (2/4) were among the most significant complexes in the NSCLC group (p-value < 0.05). APC-stimulated guanine nucleotide exchange factor 2 (ASEF2)/Adenomatous polyposis coli (APC) complex (2/2), Early elongation complex with hyperphosphorylated RNA polymerase II C-terminal domain (Pol II CTD) (4/4), Positive transcription elongation factor (P-TEFb) complex (2/3), and Tankyrin 1-tankyrin 2-telomeric repeat binding factor (TRF1) complex (2/3) were the most significant complexes in the SM group (p-value < 0.05). Histone deacetylase 2 (HDAC2) and lysine-specific demethylase 1 (LSD1) complexes were common in the datasets. Table S2 shows the overlapped components and p-value of each complex.
Primary classification of all proteins via Pathway Studio®
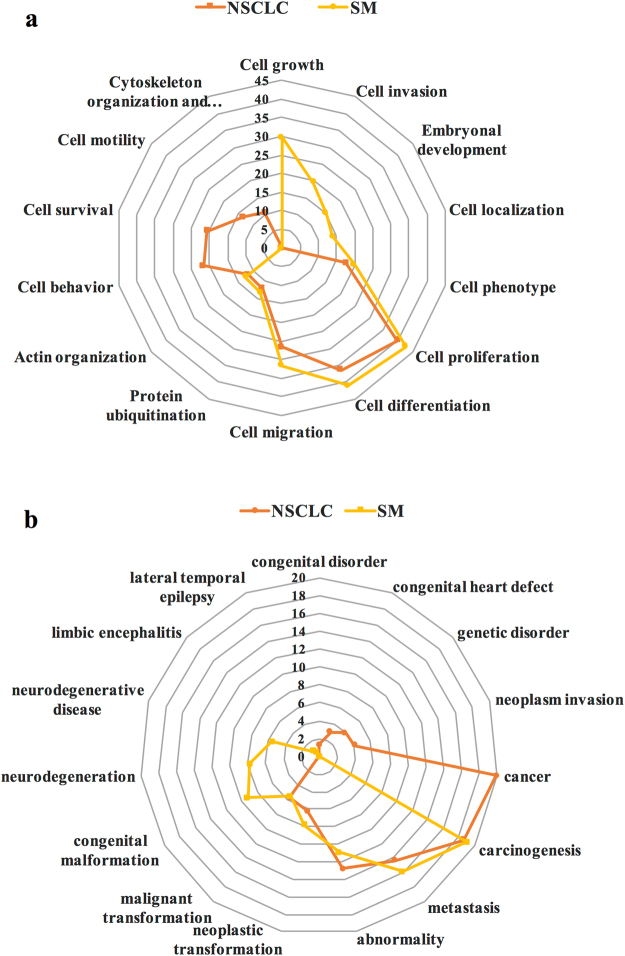
Network analysis of NSCLC and SM protein datasets using Pathway Studio® presented a similar pattern in some cell processes, including proliferation (40%, p-value: 1.7E-13; and 42%, p-value: 7.1E-15, respectively), differentiation (36%, p-value: 3.7E-23; and 41%, p-value: 2.9E-26, respectively), and migration (27%, p-value: 5.0E-19; and 32%, p-value: 2.9E-23, respectively) as the top three processes associated with the highest proteins count (Fig. 2a). Although cell growth seemed to be an important phenomenon in the SM group, lack of cell survival arm was witnessed.
Figure 2.
Top NSCLC- and SM-related cell processes and diseases based on Pathway Studio®. (a) The most significant cell processes. (b) The most significant diseases. Significant cell processes and diseases associated with the NSCLC (orange) and SM (yellow) groups were selected based on the network analysis function in Pathway Studio®. They are reported as the percentage of the number of proteins involved in the cell process or disease, to the total number of proteins in the related dataset (NSCLC or SM; 100 × n involved proteins/n total protein in dataset).
Diseases analysis demonstrated cancer (20%, p-value: 3.1E-6), carcinogenesis (19%, p-value: 2.1E-19), metastasis (14%, p-value: 3.2E-10), abnormality (13%, p-value: 5.6E-10), neoplastic transformation (6%, p-value: 6.8E-7), malignant transformation (6%, p-value: 4.3E-7), and neoplasm invasion (4%, p-value: 4.1E-5), as the most important diseases linked to the NSCLC group (Fig. 2b). Moreover, carcinogenesis (p-value: 1.9E-14) was a major event, involving 19% of SM proteins. Other events in this group included metastasis (16%, p-value: 7.3E-10), abnormality (11%, p-value: 7.0E-5), congenital malformation (9%, p-value: 1.8E-8), neoplastic transformation (8%, p-value: 5.0E-8), neurodegeneration (8%, p-value: 1.5E-5), malignant transformation (5%, p-value: 5.9E-5), and neurodegenerative disease (5%, p-value: 6.3E-5). Although carcinogenesis, metastasis, and abnormality owned similar shares in both groups, cancer state and neurodegenerative disease were major disorders which were specifically associated with the NSCLC and SM groups, respectively. The percentage indicates the ratio of involved proteins to the total number of proteins in each dataset.
Gene Ontology (GO) and pathway enrichment analysis of NSCLC and SM protein datasets
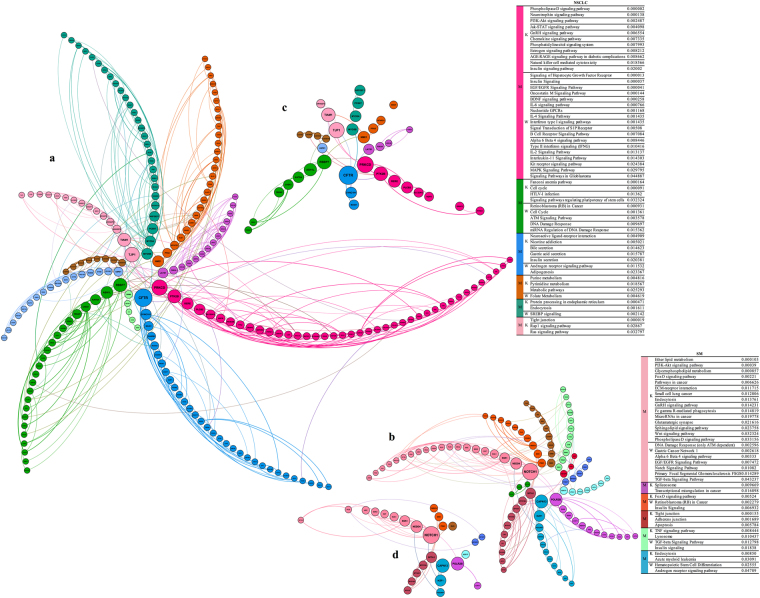
The NSCLC network constructed via Gephi contained 316 nodes and 513 edges, while the SM network consisted of 179 nodes and 255 edges (Fig. 3a). GO enrichment of the top six NSCLC- and SM-related modules by Metascape exhibited terms such as transmembrane receptor protein tyrosine kinase SP (GO:0007169), phospholipid metabolic process (GO:0006644), intrinsic apoptotic SP in response to oxidative stress (GO:0008631), collagen trimer (GO:0005581), DNA metabolic process (GO:0006259), signal transduction by p53 class mediator (GO:0072331), chromatin remodelling (GO:0006338), integrin-mediated SP (GO:0007229), cellular response to organic cyclic compound (GO:0071407), O-glycan processing (GO:0016266), regulation of GTPase activity (GO:0043087), phospholipid binding (GO:0005543), and apical junction complex (GO:0043296) in the NSCLC group, as well as regulation of cell migration (GO:0030334), inositol lipid-mediated signalling (GO:0048017), regulation of canonical Wnt SP (GO:0060828), DNA repair (GO:0006281), mitotic cell cycle process (GO:1903047), regulation of small GTPase-mediated signal transduction (GO:0051056), kinase activity (GO:0016301), calmodulin binding (GO:0005516), and toll-like receptor SP (GO:0002224) in the SM group. Furthermore, there were some terms related to cardiovascular diseases and diabetes which are highly prevalent disorders among Iranian population (as also confirmed among the participants of this study)25,26.
Figure 3.
Network visualization, modularity and pathway analysis of NSCLC and SM. The node size demonstrates the betweenness centrality and the edge thickness shows the combined score of STRING. Pathway enrichment analysis of the top six modules (M) of (a) NSCLC and (b) SM was performed using Enrichr (based on the KEGG and WikiPathways databases). (c) NSCLC- and (d) SM-specific hubs are also displayed, respectively.
Expectedly, pathways enrichment analysis based on the KEGG and WikiPathways databases in Enrichr presented a group of important pathways in the top six modules of NSCLC, including phospholipase D (PLD) SP, phosphoinositide 3-kinase-Akt (PI3K-Akt) SP, JAK-signal transducer and activator of transcription (JAK-STAT) SP, gonadotropin-releasing hormone (GnRH) SP, chemokine SP, natural killer cell (NKC) mediated cytotoxicity, pathways in cancer, signalling of hepatocyte growth factor receptor (HGFR), EGF/EGFR SP, IL-4 SP, IL-6 SP, type II interferon signalling (IFNγ), mitogen-activated protein kinase (MAPK) SP, signalling pathways regulating pluripotency of stem cells, ataxia telangiectasia mutated (ATM) SP, Fanconi anaemia pathway, nicotine addiction, gamma-aminobutyric acid (GABAergic) synapse, and Ras SP (Fig. 3a). Similarly, critical pathways were found in the SM group, such as PI3K-Akt SP, pathways in cancer, extracellular matrix (ECM)-receptor interaction, Fc gamma R-mediated phagocytosis, microRNAs in cancer, glutamatergic synapse, Wnt SP, PLD SP, DNA damage response (DDR) only ATM dependent, alpha 6 Beta 4 SP, EGF/EGFR SP, neurogenic locus notch homolog protein (NOTCH) SP, transforming growth factor beta (TGF-β) SP, transcriptional misregulation in cancer, Forkhead box O (FoxO) SP, tight junction, and tumour necrosis factor (TNF) SP (Fig. 3b). Table S3 summarizes the information on some significant pathways identified via data mining.
CFTR, PRKCD, PTK2B, CHEK1, and NOTCH1 as functional hubs
According to features such as betweenness centrality and degree implemented in Gephi, several NSCLC- and SM-specific hubs were identified (Table S4a). Proteins such as cystic fibrosis transmembrane conductance regulator (CFTR), PRKCD, tight junction protein ZO-1 (TJP1), PTK2B, TIAM1, nucleoside diphosphate kinase A (NME1), phospholipase C beta 2 (PLCB2), brefeldin A-inhibited guanine nucleotide-exchange protein 1 (ARFGEF1), and checkpoint kinase 1 (CHEK1) were NSCLC-specific hubs. Furthermore, NOTCH1, calpain-13 (CAPN13), DNA-directed RNA polymerase II subunit RPB2 (POLR2B), E3 ubiquitin-protein ligase NEDD4 (NEDD4), serine/threonine-protein kinase Sgk1 (SGK1), serine/threonine-protein kinase Sgk3 (SGK3), APC, PLD2, ARFGEF1, and regulatory-associated protein of mTOR (RPTOR) were highly ranked proteins in the SM group.
Assessment of NSCLC-specific hubs in the WebGestalt database demonstrated disorders such as lung neoplasms (p-value: 0.014), epithelial cancers (p-value: 0.014), lung diseases (p-value: 0.015), and carcinoma (p-value: 0.025), as well as several malignancies and disorders related to cancer. Evaluation of SM-specific hubs revealed confirmative results such as precancerous conditions (p-value: 0.003), substance-related disorders (p-value: 0.008), chromosome aberrations (p-value: 0.016), and stress (p-value: 0.021), in addition to adenocarcinoma and neoplasms.
Potential drugs, which could modulate NSCLC hubs, were determined by searching DrugBank and Reaxys® (Table S4b). Among NSCLC-specific hubs, a group of drugs was found to target PRKCD, PTK2B, and CHEK1, as indicated in several types of malignancies. In this regard, drugs such as bosutinib, dasatinib, axatinib, and lapatinib have been developed to concurrently inhibit these three kinases. Astonishingly, PTK2B inhibitors, crizotinib and mereletinib, are prescribed for patients with metastatic NSCLC showing specific molecular characteristics27,28.
Ability of the selected hubs for discrimination of NSCLC, SM, and control sera
Based on features such as betweenness centrality and degree, PTK2B and NOTCH1 were selected from the NSCLC and SM groups, respectively. The data exhibited specific binding of two hubs to the corresponding NSCLC and SM sera (p-value < 0.01). In contrast, they showed poor binding to the sera of age-matched healthy subjects (Fig. 4).
Figure 4.
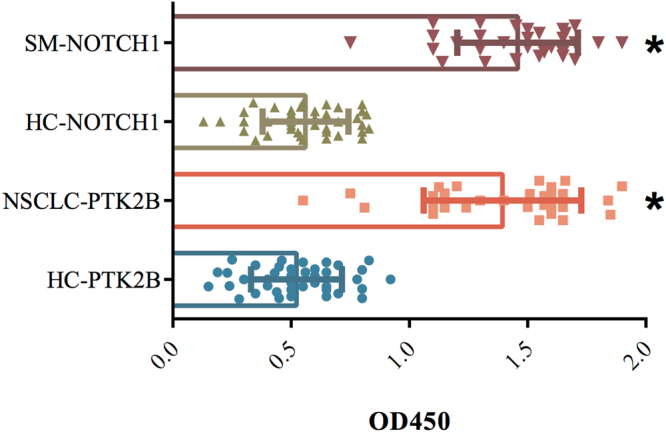
Binding assessment of the selected hubs to the NSCLC and SM sera. The results of PTK2B and NOTCH1 binding to the sera of 30 NSCLC patients (peach), 30 SM (red), and 40 age-matched healthy subjects (HC, as the controls) are presented (*p-value < 0.01).
The PLD SP and inflammatory cytokines network as overriding events in the NSCLC and SM groups
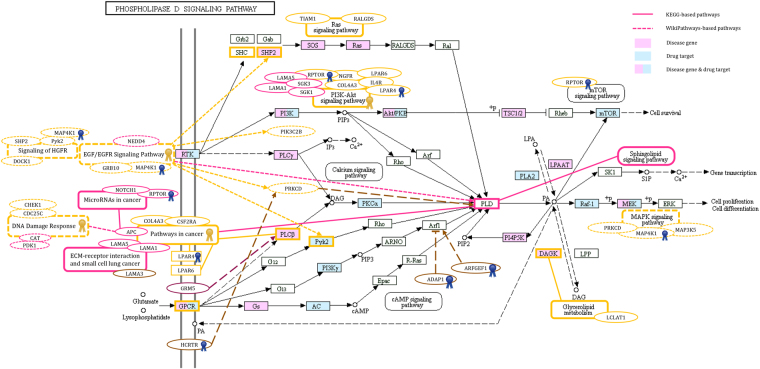
The PLD SP was undoubtedly the most significant NSCLC pathway. Remarkably, PLD was found among SM-specific proteins. Furthermore, phospholipid metabolic process and phospholipid binding were overrepresented GO terms identified through NSCLC-related module analysis. According to the literature review and evaluation of pathways involved in tumour formation and development, the PLD SP took centre stage in the pathogenesis of NSCLC in this study. Curation of the PLD SP derived from the KEGG database with the information attained through data-mining and Pathway Studio® led to the formation of a super network constructed with several NSCLC (e.g. PRKCD, PTK2B, PLCB2, and ARFGEF1) and SM (e.g. PLD2, ARFGEF1, and RPTOR) hubs. Among the mentioned hubs, RPTOR, a SM-specific hub, was also found in the NSCLC protein dataset. As exhibited in Fig. 5, a group of NSCLC proteins such as SHC-transforming protein 4 (SHC4), SHP2, mitogen-activated protein kinase kinase kinase kinase 1 (MAP4K1), phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit beta (PIK3C2B), diacylglycerol kinase theta (DGKQ), laminin subunit alpha-3 (LAMA3), arf-GAP with dual PH domain-containing protein 1 (ADAP1), ral guanine nucleotide dissociation stimulator (RALGDS), and G-protein-coupled receptors (GPCRs, including lysophosphatidic acid receptor 4 (LPAR4), lysophosphatidic acid receptor 6 (LPAR6), and orexin receptor type 1 (HCRTR1)) linked to the PLD SP, were observed in this network. Among the mentioned proteins, LPAR4, HCRTR1, and MAP4K1 were present in the SM protein dataset, as well. In addition, there were several key proteins such as NOTCH1, APC, pyruvate dehydrogenase (acetyl-transferring) kinase isozyme 1, mitochondrial (PDK1), laminin subunit alpha-1 (LAMA1), laminin subunit alpha-5 (LAMA5), and metabotropic glutamate receptor 5 (GRM5) which were only found in the SM protein dataset. Based on the KEGG and WikiPathways databases, significant SPs such as EGF/EGFR SP, HGFR SP, MAPK SP, PI3K-Akt SP, Ras SP, glycerophospholipid metabolism, glutamatergic synapse, ECM-receptor interaction, small cell lung cancer, microRNAs in cancer, DDR, and pathways in cancer identified via analysis of NSCLC and SM modules-related pathways, were also integrated in this super network. The members of this super network were linked to each other, resulting in a cascade of events. Binding of lysophosphatidic acid (LPA) to its receptors, LPAR4 and LPAR6, activates GPCR-related pathways which stimulate PLD29,30. Two other GPCRs, HCRTR1 activated by neuropeptide orexin and GRM5 activated by glutamate (released due to the indirect action of HIFs31 like HIF3A observed in the SM group), promote various pathways leading to PLD activation32,33. Another remarkable receptor in this network, EGFR, stimulates PLD via multiple routes containing proteins, some of which were found in this study, as well (Fig. 5)30. Among proteins involved in pathways leading to PLD activation, ARF1 acts as a key protein activated and inactivated by ARFGEF1 and ADAP1, respectively34. Several pathways such as glycerophospholipid metabolism, sphingolipid, mechanistic target of rapamycin (mTOR), and MAPK SPs, are induced through phosphatidic acid (PA) produced by the enzymatic activity of PLD30,35,36. Components of ECM (LAMA1, 3, and 5) induce the synthesis of PA by activation of PLD which stimulates matrix metalloproteinases (MMPs) and enhances the invasive capacity of tumor cells37. Among pathways connected to the PLD SP, pathways in cancer was one of the critical events observed in both NSCLC and SM groups and was linked to other significant pathways such as microRNAs in cancer, DDR, and small cell lung cancer. A set of proteins, including LPAR4, LPAR6, collagen alpha-3(IV) chain-like (COL4A3), PLCB2, and granulocyte-macrophage colony-stimulating factor receptor subunit alpha (CSF2RA) from the NSCLC protein dataset as well as APC, LAMA1, LAMA5, and PLD1 from the SM protein dataset, were involved in pathways in cancer. All NSCLC- and SM-specific proteins playing roles in the above-mentioned SPs are presented in Fig. 5.
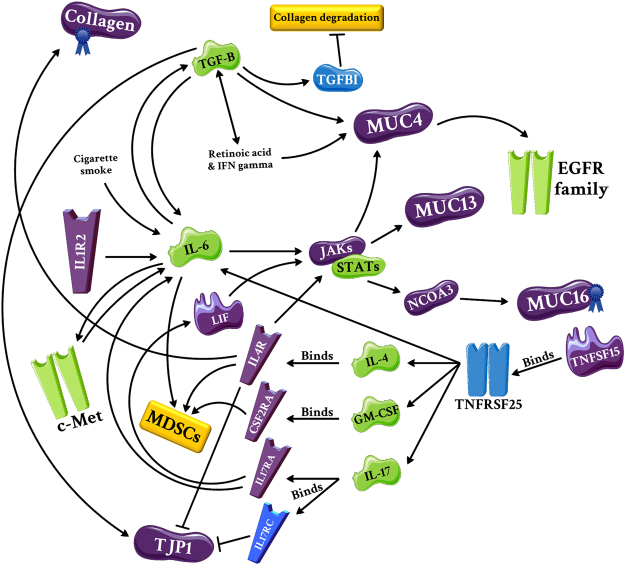
Figure 5.
The super network constructed based on the PLD SP. The network was established based on the PLD SP in the KEGG database30 and was expanded by the addition of some pathways and neighbours via WikiPathways and data mining in the literature. As the identified pathways are shown in this figure, the FcεRI SP has been deleted from the PLD SP. LPAR4, LPAR6, GRM5, HCRTR1, and EGFR are the known receptors in this network. Different genes involved in the PLD SP and other related pathways are presented as coloured boxes. Yellow and pink colours indicate NSCLC- and SM-specific proteins in significant pathways found in this study, respectively. Brown and plum colours correspond to proteins from the NSCLC and SM protein datasets, respectively. Additionally, blue and yellow ribbons show common proteins and pathways between the two groups, respectively.
The second super network was constructed to describe how cytokines alter the tumour microenvironment (e.g. ECM proteins) and how tumour recruits them to fabricate the immune system (Fig. 6). This network was originated from multiple significant pathways such as JAK-STAT SP, chemokine SP, NKC-mediated cytotoxicity, signalling of HGFR (c-Met), EGF/EGFR SP, IL-4 SP, IL-6 SP, and type II interferon signalling obtained from the NSCLC module analysis and ECM-receptor interaction, Fc gamma R-mediated phagocytosis, TGF- β SP, tight junction, and TNF SP obtained from the SM module analysis. It consisted of various proteins, including TJP1 (NSCLC hub), leukemia inhibitory factor (LIF), IL-4R, IL-1 receptor type 2 (IL-1R2), IL-17 receptor A (IL-17RA), CSF2RA, TNF ligand superfamily member 15 (TNFSF15), TYK2, PIK3C2B, nuclear receptor coactivator 3 (NCOA3), mucin-4 (MUC4), MUC13, MUC16, COL4A3, collagen alpha-1(XXVI) chain (COL26A1), and collagen alpha-1(XXVII) chain (COL27A1) which were present in the NSCLC protein dataset. SM-specific proteins such as TNF receptor superfamily member 25 (TNFRSF25), IL-17 receptor C (IL-17RC), TGF-beta-induced protein ig-h3 (TGFBI), MUC16, collagen alpha-1(III) chain (COL3A1), collagen alpha-1(XVI) chain (COL16A1), and COL27A1, were also integrated into the network. A remarkable relationship was found among the predicted proteins in this network. One of the most important pathways in the NSCLC group was IL-6 SP. IL-6 production is induced by IL-1R238, IL-1739,40 and cigarette smoking41. There is a two-way interaction between IL-6 and HGF, resulting in increased invasiveness of a lung cancer cell line42 and an autocrine loop between IL-6 and TGF-β1 which its dysregulation causes an array of inflammatory disorders43. As shown in Fig. 6, IL-6 can augment the expression of MUC4 and MUC13 through JAK/STAT SP44,45. Production of LIF, an IL-6 family cytokine, by synovial fibroblasts is also induced by IL-1739. Another interleukin, IL-4, can increase the transcription of MUC4 (via JAK/STAT) and MUC16 (through JAK/STAT and NCOA3)44,46,47. Furthermore, IL-4 triggers M2 macrophages to produce much more collagen a3 (VI)48. Expression of TJP1 is also decreased by IL-4, and its distribution at the interface of adjacent cells is disrupted by IL-17 receptor C (IL-17RC)49,50. The major cytokine in this network, TGF-β, contributes to the modification of tumour microenvironment via MUC4 upregulation in an interdependent manner with retinoic acid and interferon type II (IFNγ)51. TGF-β increases TJP1 expression52 and inhibits collagens degradation through activation of TGFBI53. Locally-secreted cytokines not only modulate the structure of tumour microenvironment, but also disrupt the protective function of the immune system. TGF-β signalling plays critical roles in the inhibition of tumour-specific CD8 T cell- and NK cell-mediated cytotoxicity through regulatory T cells54,55. In addition, IL-4, IL-6, and granulocyte-macrophage colony-stimulating factor (GM-CSF) increase the population of myeloid-derived suppressor cells (MDSCs) which interrupt the identification of tumour antigens by T cells through induction of nitric oxide synthase and consequent nitration of T cell receptor56. The final link is related to the TNF family which seems to be involved in tumour suppression57. However, the binding of TNFSF15 to its receptor, TNFRSF25, amplifies the expression of IL-4, IL-6, GM-CSF, and IL-1758,59, which may explain the overactivity of these tumour-promoting cytokines in our network.
Figure 6.
The super network constructed based on the relationship between inflammatory cytokines and tumour microenvironment. Similar to the first super network, the second network was created by proteins, particularly inflammatory cytokines, involved in significant NSCLC and SM pathways. IL-4, IL-6, IL-17, GM-CSF, and TGF-β are the most important cytokines in this network. Purple, blue, and green colours indicate presence of protein in the NSCLC protein dataset, presence of protein in the SM dataset, and unavailability of proteins in the datasets, respectively. Blue ribbons also show the presence of protein in the SM protein dataset.
Discussion
A combination of events such as cigarette smoking, nicotine addiction, delayed diagnosis, and resistance development during chemotherapy, has led to major concerns about lung cancer. Therefore, it can be useful to compare the AAb repertoires of NSCLC patients with SM to discover proteins involved in lung cancer pathogenesis60,61. According to the literature, AAbs exist in the plasma of patients several months to years before the emergence of any clinical signs, reflecting the functional immunoreactivity against tumour antigens62. AAbs against tumour-specific antigens generated due to antigen mutation, overexpression, or altered structure, have been recruited to discover cancer biomarkers63. Aside from the important role of AAbs in the discovery of tumour antigens for the development of T cell-based vaccines, they can act as therapeutic antibodies by either blocking tumour antigens or recruiting the immune system63. Various studies have assessed cancer-specific antibodies in different types of malignancies63. Similarly, several studies have examined NSCLC patients and have identified a panel of AAbs binding to rho-associated protein kinase 1, protein kinase C beta, complement factor H, c-myc, cyclin A, B1 and D1, cyclin-dependent kinase 2, MUC1, Dickkopf-related protein 1, p53, NY-ESO-1, and annexins (I and II)8–14. Furthermore, Yao, et al. introduced four combined AAb biomarkers identifying nucleolar and coiled-body phosphoprotein 1, metastasis-associated lung adenocarcinoma transcription 1, hyaluronan mediated motility receptor, and spermine oxidase for early detection of NSCLC7.
In the present study, classification of the predicted proteins into significant cellular processes revealed some overlapped areas as well as dissimilarities in the cellular behaviour of NSCLC and SM. Network enrichment analysis of NSCLC- and SM-related modules resulted in the identification of remarkable pathways which play key roles in cancer initiation, progression, or suppression. NSCLC-specific pathways included MAPK64,65, PI3K-Akt66, PLD36, Ras67, EGF/EGFR68–71, c-Met72–74, LPARs75, IL-476 and IL-677–79 SPs which play significant roles in tumorigenesis and cancer development. Remarkably, we found tumour suppressor pathways such as Fanconi anaemia pathway80 and ATM SP81 in the NSCLC group, which might be activated as feedbacks to the overactivated pathways in cancer cells.
Considerably, analysis of SM-related modules revealed multiple tumour-promoting pathways such as PI3K-Akt, PLD, and EGF/EGFR SPs which were commonly found in the NSCLC group. Conversely, activation of tumour suppressor pathways, including DDR and FoxO82 SPs as well as negative regulation of Wnt SP83 in the SM group, show activation of cellular defensive mechanisms against the carcinogenic compounds of cigarette smoke. Moreover, we found TGF-β and TNF SPs in this group, known as “double-edged swords”, which can promote or suppress cancers84,85. As the results demonstrate, carcinogenic events are strongly significant in SM and may accelerate the emergence of disorders such as cancer.
Among NSCLC-specific hubs, there are four proteins which can be considered as the key players involved in lung cancer development. The first hub is CFTR which its dysregulation has been reported in a group of cancers such as breast, gastric, prostate, lung, and colorectal cancers (CRC)86. Moreover, a significant correlation has been reported between CFTR downregulation and lung cancer progression and metastasis86,87. In contrast to studies reporting CFTR suppression in epithelial cells due to exposure to cigarette smoke, Li, et al. showed CFTR upregulation through nicotine-derived nitrosamine ketone (NNK), as one of the most toxic tobacco-specific carcinogens in cigarette smoke88. CFTR as an ABC transporter promotes protection against tobacco-specific carcinogens in cigarette smoke88. On the other hand, AAbs can lead to the development of lung cancer by blocking CFTR.
The second hub, PTK2B, is a part of protein complexes found in this study. This hub plays major roles in cancer cells due to its connections to critical pathways such as PLD, EGFR, and c-Met SPs30. It has been reported that PTK2B is upregulated in NSCLC. In fact, its higher expression and greater activity lead to the progression of NSCLC through modulating extracellular signal-regulated kinase 1/2 (ERK1/2)89. Furthermore, wound-induced activation of PTK2B stimulates EGFR activation in the epithelial cells of lung lesions90. PTK2B was also detected as a sunitinib-specific target in NSCLC patients with kirsten rat sarcoma viral oncogene (KRAS) mutations91. Likewise, prescription of crizotinib and merelotinib in NSCLC patients underscores the importance of PTK2B in the pathogenesis of lung cancer. KRAS-activated PRKCD promotes growth, invasion, and migration of cancer cells, through MAPK signalling in NSCLC cell lines (and mouse model), whereas PRKCD activation in cells without active KRAS leads to tumour suppression92. In this regard, the PRKCD inhibitor, rottlerin, can effectively potentiate chemotherapy-induced apoptosis, particularly if it is used concomitantly with trastuzumab93. Liu, et al. found that CHEK1 overexpression is associated with poor overall survival in NSCLC patients, and inhibition of CHEK1 by miR-195 suppresses tumour cell growth, migration, and invasion94. In this regard, Xiaojie, et al. revealed that miR-195 synergizes with microtubule-targeting agents (eribulin and paclitaxel) via CHEK1 regulation, leading to the inhibition of NSCLC cells growth95. In agreement with other studies, we confirmed the involvement of CFTR, PTK2B, PRKCD, and CHEK1 in NSCLC pathogenesis.
In terms of major cancer-related proteins among SM-specific hubs, NOTCH1, ARFGEF1, PLD2, and RPTOR were strongly aligned with hallmarks of NSCLC. NOTCH1 contributes to cell proliferation, invasion, and chemoresistance. Its involvement in tumor progression and prognosis of NSCLC has been demonstrated in different studies96,97. Mutations and upregulation of Notch1 have been reported in 10% and 30% of NSCLC patients, respectively97,98. Konishi, et al. showed that suppression of NOTCH signaling inhibits the growth of NSCLC99. Furthermore, NOTCH signaling can crosstalk with other SPs such as Wnt and TGF-β driving cancer development98. Therefore, targeting NOTCH signaling by therapeutic agents like antibodies which can inhibit delta-like protein 4 (Dll4) binding to NOTCH1, may be advantageous for NSCLC patients96,98. In this regard, a humanized anti-Dll4 antibody, demcizumab, is currently under evaluation in three Phase 1b studies on pancreatic cancer, CRC, and NSCLC98.
As illustrated in the results section, three interconnected proteins, ARFGEF1, PLD2, and RPTOR, were successively activated, stimulating the cancer driver mTOR SP. Intriguingly, mutations in ARFGEF1, an activator of ARF1, have been reported in lung cancer100. Among these three proteins, PLDs have been garnering attention as potential contributors to cellular signalling of tumours101. Surprisingly, PLD represented an important intersection for other pathways in this study, and the first super network was constructed based on this enzyme. The existence of PLD isozymes (PLD1 and PLD2) in the SM protein dataset indicates the role of these junction boxes in the initiation of tumorigenesis. PLD is activated through a variety of signals such as neurotransmitters, hormones, and growth factors, transducing signals into pivotal cellular events, including proliferation, secretion, respiratory burst, and actin cytoskeletal reorganization102. As mentioned in the results section, PLD network involves proteins such as LPAR4, LPAR6, HCRTR1, and GRM5, as well as pathways like EGF/EGFR SP, which their roles have been proven in initiation and progression of several cancers. Phosphatidic acid, the product of PLD enzymatic activity, can be also converted to LPA and form a positive loop of LPA-LPARs30. LPA is involved in various cancer-related processes such as proliferation, growth, and survival75. Even though LPAR4 and LPAR6 are newly-discovered members of LPAR family, their contributions to hepatocellular carcinoma103 and thyroid cancer104 have been demonstrated, respectively. Although HCRTR1 stimulation with high doses of orexin causes apoptosis in colon and prostate cancers105,106, elevated serum orexin level has not been detected in cancer patients107. It seems that the normal serum level of orexin is not sufficient to induce apoptosis in cancer cells. Physiologically activated HCRTR1 may activate some pathways other than apoptosis, such as the PLD SP which contributes to tumour progression33,105. Therefore, pharmacologically activated HCRTR1 may be promising in cancer prevention or treatment.
Hypoxia, as a common feature of tumour microenvironment, increases the transcription of glutamate receptors, promotes the activation of GRM1, 3, 4, and 5, and supports proliferation and survival of several types of tumours through MAPK and PI3K/Akt SPs31,107. Noteworthy, GRM4 and 8 have been reported as susceptibility genes in NSCLC31. One of the critical pathways in both NSCLC and SM groups is the EGFR SP. More than half of lung tumours express receptors from EGFR family and one-fifth of lung tumours carry EGFR mutations, causing proliferation, migration, and metastasis68,71. Despite the preliminary response of all mutated lung tumours to tyrosine kinase inhibitors, they essentially acquire resistance68. Activation of mTOR, a master regulator of cell growth and proliferation, is one of the most important consequences of PLD activation. Accordingly, combination therapy of mTOR antagonists with standard regimens for lung cancer has enhanced treatment efficacy in early-stage trials108. Similarly, the MAPK SP, triggered by both PA and PLD, is associated with P53 degradation and MMPs induction, leading to cell survival and motility, respectively36. Overall, the PLD super network clarifies why treatment fails in this type of lung cancer and presents new insights into NSCLC chemotherapy. Moreover, this network underscores the pivotal function of PLDs in the occurrence of lung cancer in smokers.
Another super network constructed in this study is a complicated linkage between inflammatory cytokines and tumour microenvironment. Inflammation is a mixed blessing in cancer and the T helper 2 response deteriorates the state of cancer through suppression of cellular immunity56. Incidentally, the significance of IL-4 and IL-6 SPs in the present study may suggest inappropriate activation of this unfavourable arm of immunity. IL-4 SP promotes metastasis by changing the microenvironment of epithelial tumours76. It has been reported that serum IL-6 levels in NSCLC patients (especially metastatic cases) are higher than healthy subjects77,78, and blockade of IL-6/STAT3 signalling results in the inhibition of lung tumour growth109. Besides, STAT3 is persistently activated in GPCR C5a-knockout (KO) mice, a spontaneous lung cancer developing model, and the use of an antibody against LIF leads to STAT3 inactivation in this model110. The IL-6 and c-Met loop causes cell invasion through MMP activation in A549 cells, as well42,72. Proteins such as IL-6 and c-Met, in cooperation with IL-17 and TGF-β, play various roles in the regulation of proteins such as mucins, collagens, and TJP1. It has been demonstrated that mucins such as MUC4 and MUC13 are associated with cancer development; the circulating level of MUC16 is even used to monitor patients with ovarian cancer111–113. Regarding collagens, their increased and decreased expressions are correlated with malignancy114; accordingly, collagen XXIII has been suggested as a biomarker for the diagnosis of NSCLC115. Another study showed that COL4A3 cleavage by MMP-9 produces tumstatin which is a circulating anti-angiogenic factor116. Hence, AAb against COL4A3 may worsen the state of cancer by preventing the generation of tumstatin. Additionally, COL27A1 is aberrantly expressed in idiopathic pulmonary fibrosis117, a condition associated with lung cancer118. Ni and colleagues found that the expression of TJP1 was decreased in NSCLC tissue in comparison to adjacent non-tumour tissue and its overexpression was correlated with better prognosis in NSCLC patients119. Considerably, increased MDSCs population as well as dysfunction of NKCs and CD8+ T cells highlight an inefficient immune system in NSCLC120–122. Notably, our network exhibits how IL-4, IL-6, GM-CSF, and TGF-β predispose these phenomena.
In conclusion, given the rapid emergence of lung cancer in smoker populations across the world, new aspects of NSCLC pathogenesis and effects of cigarette smoke should be addressed. Tailoring the AAb profiling with a systems biology approach provides an excellent image of the network of AAbs targeting proteins which may be key drivers of cancer-interacting SPs such as PLD, PI3K-Akt, MAPK, EGF/EGFR, c-Met, Ras, DDR, IL-4, IL-6, and TGF-β SPs in the NSCLC and SM groups. Based on the PLD SP and inflammatory cytokines network, two super networks were constructed with NSCLC- and SM-specific proteins such as PLD1/2, PRKCD, PTK2B, LPARs, RPTOR, NOTCH1, TJP1, laminins, mucins, and collagens as well as pathways found in this study. Focusing on these super networks will help to design novel therapeutic agents targeting proteins involved in the pathogenesis of lung cancer.
Methods
Sample collection
For obtaining a repertoire of NSCLC AAbs, 67 blood samples of untreated newly-diagnosed adenocarcinoma and squamous cell carcinoma patients (stages I, Ib, II, and IIIa) were collected from different centres, including Baqiyatallah and Masih Daneshvari hospitals123, Tehran, Iran, as well as Kurdistan Hospital in Sanandaj, Iran. The patients aged 22–77 years (mean, 50.7 years) were all confirmed for NSCLC via histopathological tests and were classified into different stages based on the tumour/node/metastasis scale (TNM) staging system. Furthermore, 57 subjects (age range, 20–72 years; mean, 40.0 years) were allocated to the SM group according to the complete blood cell count, blood level of C-reactive protein (≤6 mg/dL), erythrocyte sedimentation rate (≤32 mm/h), rheumatoid factor (negative), and chest X-ray imaging to evaluate their overall health. All the subjects in the SM group were current smokers with different smoking patterns, ranging from a few cigarettes per week to heavy smoking.
A control group of 92 healthy non-SM volunteers (range, 11–80 years; mean, 38.9 years) were also analysed, based on the same criteria considered for the subjects in the SM group and were enrolled in the study. All healthy participants avoided anti-inflammatory drugs for three days prior to blood collection. The demographic information was collected from the subjects, and they were interviewed for potential risk factors for cancers and autoimmune diseases, including personal and family medical history. This study was done in accordance with the Helsinki Declaration124,125 and was approved by the Ethics Committee of Baqiyatallah University of Medical Sciences, the Ethics Committee of Masih Daneshvari Hospital123, and the Ethics Committee of Kurdistan University of Medical Sciences. All participants provided written informed consent before enrollment. The characteristics of all three groups are presented in Table S5.
IgG purification from the sera of healthy control (HC), NSCLC patients, and SM
The sera of each group (HC, NSCLC, and SM) were mixed separately and IgG purification was performed using Melon™ Gel IgG Purification Kit (Pierce, Rockford, IL) according to the manufacturer’s instructions. The purification accuracy was confirmed by reducing SDS-PAGE analysis.
Biopanning
Using Ph.D.TM-C7C Phage Display Peptide Library Kit (New England Biolabs, Beverly, MA), SU and SL biopanning were carried out for three successive cycles on the purified IgG of NSCLC and SM groups. For SU biopanning, 96-well Nunc MaxiSorp flat-bottom plates were coated with IgG of HC, NSCLC and SM groups. In addition, for SL biopanning, Protein G Mag Sepharose Xtra (GE healthcare, US) was employed to expose IgG, according to the manufacturer’s instructions. In both procedures, the phages of three inputs were subtracted on HC IgG before being transferred to the wells coated with IgG (NSCLC or SM IgG) or microtubes containing IgG (NSCLC or SM IgG) according to the instructions of Ph.D.TM-C7C Phage Display Peptide Library Kit and Protein G Mag Sepharose Xtra).
Polyclonal and monoclonal phage ELISA
Polyclonal phage ELISA was conducted on phages obtained from three rounds of SU and SL biopanning on NSCLC and SM IgG. The specificity of peptides displayed on 60 selected phages (30 clones from SU biopanning and 30 clones from SL biopanning) to NSCLC and SM IgG was individually assayed by monoclonal phage ELISA according to the manufacturer’s instructions (Ph.D.TM-C7C kit) (see Supplementary information).
DNA sequencing
To extract the single-stranded phage DNAs, 20 and 10 clones obtained from SU and SL biopanning on NSCLC IgG, along with 15 and 10 clones obtained from SU and SL biopanning on SM IgG, were selected, respectively, according to the Ph.D.TM-C7C kit instructions. After sequencing and inference of amino acid sequences by Gene Runner version 5.0, the peptides were analysed in the Biopanning Data Bank (MimoDB) (http://immunet.cn/bdb/) to discard target-unrelated peptides126. Based on default parameters for the Blastp (score ≥18.5), the selected peptides were blasted against the Refseq_protein database for the Homo sapiens 127.
Determination of protein complexes in the NSCLC and SM protein datasets
NSCLC- and SM-specific proteins obtained through blasting (score ≥18.5) and preliminary analysis of the literature and UniProt database, were assessed in CPDB (http://cpdb.molgen.mpg.de) to find significant protein complexes with at least two members in the NSCLC or SM protein dataset (p-value < 0.05).
Primary evaluation of NSCLC and SM protein datasets in Pathway Studio®
Pathway Studio® 11.2.5.9 (https://mammalcedfx.pathwaystudio.com) was used to exhibit the top cellular processes and diseases in two datasets. For this purpose, the NSCLC and SM protein datasets were separately introduced to Pathway Studio® and the Summary Analysis function was used to present a quick analytical summary of the datasets.
GO and pathway-enrichment analysis of predicted proteins
For visualizing molecular interaction networks, the NSCLC and SM protein datasets were introduced to STRING 10.0 (http://string-db.org)128 and the nodes (proteins) and edges (protein-protein interactions) were extracted and transferred to Gephi 0.9.1129. The combined scores (from STRING) were defined as the edge weights. Modularity function implemented in Gephi was used for clustering, and the nodes were coloured differentially based on their modules. To identify GO terms and pathways related to the top six NSCLC and SM modules, Metascape (http://metascape.org)130 and Enrichr web tools (http://amp.pharm.mssm.edu/Enrichr/)131 were recruited. Disease Association Analysis in the WebGestalt database (http://webgestalt.org)132 was performed for 25 NSCLC- and 26 SM-specific nodes (hubs) which showed the highest scores of betweenness centrality and degree. Target-based search were also implemented with DrugBank 5.0 (https://drugbank.ca)133 and Reaxys® (https://reaxys.com) to identify marketed drugs affecting NSCLC-specific hubs.
To present a final image of NSCLC pathogenesis in this study, two super networks were constructed by combining pathways obtained from the KEGG PATHWAY and WikiPathways databases. Considering the PLD pathway as a central event in the NSCLC group, a super network was built on the PLD SP taken from the KEGG database30. This super network was developed by trimming the unrelated branches and adding connections based on the literature review.
Another super network was constructed, considering the effects of inflammatory cytokines on tumour microenvironment. For this purpose, proteins involved in GO terms and pathways related to inflammation or ECM, were introduced to Pathway Studio® for generating a primary network. Using “the properties function” defined in Pathway Studio®, each network component was assessed, leading to the generation of a secondary network expanded with NSCLC key proteins. Data mining was also carried out to verify this super network with the deliberated connections.
Assessment of the selected hubs by ELISA
PTK2B (ORIGENE) and NOTCH1 (R&D systems), as the representatives of NSCLC and SM groups, were selected for ELISA, respectively (see Supplementary information).
Statistical analysis
The statistical analyses were carried out using GraphPad prism®. The data were stated as the mean ± SD and the statistical significance (p-value < 0.05) was determined by two-tailed Student’s t-test.
Electronic supplementary material
Acknowledgements
The authors would like to thank the personnel of Baqiyatallah, Masih Daneshvari, and Kurdistan hospital for their assistance with sample collections. This work was supported by grants from the Iran national science foundation (grant number 92002095).
Author Contributions
F.R.J. and M.G. conceived and designed the experiments. F.M. helped to design the study. F.R.J. and M.P. performed the data analysis and interpretation. F.R.J., F.S., T.B., and F.A. performed the experiments. A.F.K., E.V., M.M., N.S., and E.T. contributed to the acquisition and interpretation of data. F.R.J. and M.P. wrote the paper. S.D.S., F.V., and A.F. were involved in the manuscript preparation. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14195-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mostafa Ghanei, Email: m.ghanei@bmsu.ac.ir.
Fatemeh Rahimi Jamnani, Email: Rahimi@pasteur.ac.ir.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Gridelli, C. et al. Non-small-cell lung cancer. Nat Rev Dis Primers 15009, 10.1038/nrdp.2015.9 (2015). [DOI] [PubMed]
- 3.Hecht SS, Szabo E. Fifty years of tobacco carcinogenesis research: from mechanisms to early detection and prevention of lung cancer. Cancer Prev Res (Phila) 2014;7:1–8. doi: 10.1158/1940-6207.CAPR-13-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu L, et al. Development of autoantibody signatures as novel diagnostic biomarkers of non-small cell lung cancer. Clin Cancer Res. 2010;16:3760–3768. doi: 10.1158/1078-0432.CCR-10-0020. [DOI] [PubMed] [Google Scholar]
- 5.Chang A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung Cancer. 2011;71:3–10. doi: 10.1016/j.lungcan.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 6.Zaenker P, Gray ES, Ziman MR. Autoantibody Production in Cancer–The Humoral Immune Response toward Autologous Antigens in Cancer Patients. Autoimmun Rev. 2016;15:477–483. doi: 10.1016/j.autrev.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 7.Yao Y, et al. Potential application of non-small cell lung cancer-associated autoantibodies to early cancer diagnosis. Biochem Biophys Res Commun. 2012;423:613–619. doi: 10.1016/j.bbrc.2012.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leidinger P, et al. Toward an early diagnosis of lung cancer: An autoantibody signature for squamous cell lung carcinoma. Int J Cancer. 2008;123:1631–1636. doi: 10.1002/ijc.23680. [DOI] [PubMed] [Google Scholar]
- 9.Amornsiripanitch N, et al. Complement Factor H Autoantibodies Are Associated with Early Stage NSCLC. Clin Cancer Res. 2010;16:3226–3231. doi: 10.1158/1078-0432.CCR-10-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rom WN, et al. Identification of an autoantibody panel to separate lung cancer from smokers and nonsmokers. BMC cancer. 2010;10:1. doi: 10.1186/1471-2407-10-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirasawa Y, et al. Natural Autoantibody to MUC1 Is a Prognostic Indicator for Non–Small Cell Lung Cancer. Am J Respir Crit Care Med. 2000;161:589–594. doi: 10.1164/ajrccm.161.2.9905028. [DOI] [PubMed] [Google Scholar]
- 12.Yao X, et al. Dickkopf-1 autoantibody is a novel serological biomarker for non-small cell lung cancer. Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals. 2010;15:128–134. doi: 10.3109/13547500903325662. [DOI] [PubMed] [Google Scholar]
- 13.Chapman CJ, et al. Autoantibodies in lung cancer: possibilities for early detection and subsequent cure. Thorax. 2008;63:228–233. doi: 10.1136/thx.2007.083592. [DOI] [PubMed] [Google Scholar]
- 14.Brichory FM, et al. An immune response manifested by the common occurrence of annexins I and II autoantibodies and high circulating levels of IL-6 in lung cancer. Proceedings of the National Academy of Sciences. 2001;98:9824–9829. doi: 10.1073/pnas.171320598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YC, Chen BS. A network-based biomarker approach for molecular investigation and diagnosis of lung cancer. BMC Med Genomics. 2011;4:2. doi: 10.1186/1755-8794-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li XJ, et al. A blood-based proteomic classifier for the molecular characterization of pulmonary nodules. Sci Transl Med. 2013;5:207ra142. doi: 10.1126/scitranslmed.3007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu B, et al. Screening of diagnostic biomarkers for lung cancer by bioinformatics analysis. International Journal Of Clinical And Experimental Medicine. 2017;10:2737–2745. [Google Scholar]
- 18.Wang LQ, Zhao LH, Qiao YZ. Identification of potential therapeutic targets for lung cancer by bioinformatics analysis. Mol Med Rep. 2016;13:1975–1982. doi: 10.3892/mmr.2015.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young JH, et al. Computational discovery of pathway-level genetic vulnerabilities in non-small-cell lung cancer. Bioinformatics. 2016;32:1373–1379. doi: 10.1093/bioinformatics/btw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huan T, et al. A whole-blood transcriptome meta-analysis identifies gene expression signatures of cigarette smoking. Hum Mol Genet. 2016;25:4611–4623. doi: 10.1093/hmg/ddw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao C, et al. Bioinformatics Analysis of the Effects of Tobacco Smoke on Gene Expression. PLoS One. 2015;10:e0143377. doi: 10.1371/journal.pone.0143377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elamin A, et al. Quantitative proteomics analysis using 2D-PAGE to investigate the effects of cigarette smoke and aerosol of a prototypic modified risk tobacco product on the lung proteome in C57BL/6 mice. J Proteomics. 2016;145:237–245. doi: 10.1016/j.jprot.2016.05.037. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, et al. Identification of feature genes for smoking-related lung adenocarcinoma based on gene expression profile data. Onco Targets Ther. 2016;9:7397–7407. doi: 10.2147/OTT.S114230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, Y., Qian, W. & Yuan, B. A graphical model of smoking-induced global instability in lung cancer. IEEE/ACM Trans Comput Biol Bioinform, 10.1109/TCBB.2016.2599867 (2016). [DOI] [PubMed]
- 25.Finegold JA, Asaria P, Francis DP. Mortality from ischaemic heart disease by country, region, and age: statistics from World Health Organisation and United Nations. Int J Cardiol. 2013;168:934–945. doi: 10.1016/j.ijcard.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majeed A, et al. Diabetes in the Middle-East and North Africa: an update. Diabetes Res Clin Pract. 2014;103:218–222. doi: 10.1016/j.diabres.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Poon, C. C. & Kelly, J. J. Development of crizotinib, a rationally designed tyrosine kinase inhibitor for non‐small cell lung cancer. Int J Cancer (2016). [DOI] [PubMed]
- 28.Wang S, Cang S, Liu D. Third-generation inhibitors targeting EGFR T790M mutation in advanced non-small cell lung cancer. J Hem Onc. 2016;9:1. doi: 10.1186/s13045-015-0229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi M, Zheng Y, Garcia A, Xu L, Foster DA. Phospholipase D provides a survival signal in human cancer cells with activated H-Ras or K-Ras. Cancer Lett. 2007;258:268–275. doi: 10.1016/j.canlet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu H, et al. Hypoxia-inducible factors enhance glutamate signaling in cancer cells. Oncotarget. 2014;5:8853–8868. doi: 10.18632/oncotarget.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res, gkv1070 (2015). [DOI] [PMC free article] [PubMed]
- 33.Jantti MH, Putula J, Somerharju P, Frohman MA, Kukkonen JP. OX1 orexin/hypocretin receptor activation of phospholipase D. Br J Pharmacol. 2012;165:1109–1123. doi: 10.1111/j.1476-5381.2011.01565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cherfils J, Zeghouf M. Regulation of Small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 35.Sun Y, Chen J. mTOR signaling: PLD takes center stage. Cell Cycle. 2008;7:3118–3123. doi: 10.4161/cc.7.20.6881. [DOI] [PubMed] [Google Scholar]
- 36.Bruntz RC, Lindsley CW, Brown HA. Phospholipase D signaling pathways and phosphatidic acid as therapeutic targets in cancer. Pharmacol Rev. 2014;66:1033–1079. doi: 10.1124/pr.114.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reich R, Blumenthal M, Liscovitch M. Role of phospholipase D in laminin-induced production of gelatinase A (MMP-2) in metastatic cells. Clin Exp Metastasis. 1995;13:134–140. doi: 10.1007/BF00133618. [DOI] [PubMed] [Google Scholar]
- 38.Mar A-C. Abstract 1361: The decoy receptor interleukin-1 receptor type 2 acts as an angiogenic factor in human colorectal cancer. Cancer Res. 2015;75:1361–1361. doi: 10.1158/1538-7445.AM2015-1361. [DOI] [Google Scholar]
- 39.Chabaud M, Fossiez F, Taupin J-L, Miossec P. Enhancing Effect of IL-17 on IL-1-Induced IL-6 and Leukemia Inhibitory Factor Production by Rheumatoid Arthritis Synoviocytes and Its Regulation by Th2 Cytokines. J Immunol. 1998;161:409–414. [PubMed] [Google Scholar]
- 40.Wang L, et al. IL-17 can promote tumor growth through an IL-6–Stat3 signaling pathway. J Exp Med. 2009;206:1457–1464. doi: 10.1084/jem.20090207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee H, et al. Cigarette smoke-mediated oxidative stress induces apoptosis via the MAPKs/STAT1 pathway in mouse lung fibroblasts. Toxicol Lett. 2016;240:140–148. doi: 10.1016/j.toxlet.2015.10.030. [DOI] [PubMed] [Google Scholar]
- 42.To Y, et al. A two-way interaction between hepatocyte growth factor and interleukin-6 in tissue invasion of lung cancer cell line. Am J Respir Cell Mol Biol. 2002;27:220–226. doi: 10.1165/ajrcmb.27.2.4804. [DOI] [PubMed] [Google Scholar]
- 43.Liton PB, Li G, Luna C, Gonzalez P, Epstein DL. Cross-talk between TGF-β1 and IL-6 in human trabecular meshwork cells. Mol Vis. 2009;15:326–334. [PMC free article] [PubMed] [Google Scholar]
- 44.Carraway, K. L. & Carothers Carraway, C. A. MUC4 and membrane receptors in cancer. Mucins and Cancer 68–81, 10.2217/fmeb2013.13.69 (2013).
- 45.Gupta BK, et al. Functions and regulation of MUC13 mucin in colon cancer cells. J Gastroenterol. 2014;49:1378–1391. doi: 10.1007/s00535-013-0885-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arimura A, van Peer M, Schröder AJ, Rothman PB. The transcriptional co-activator p/CIP (NCoA-3) is up-regulated by STAT6 and serves as a positive regulator of transcriptional activation by STAT6. Journal of Biological Chemistry. 2004;279:31105–31112. doi: 10.1074/jbc.M404428200. [DOI] [PubMed] [Google Scholar]
- 47.Kumar, S. et al. NCOA3-mediated upregulation of mucin expression via transcriptional and post-translational changes during the development of pancreatic cancer. Oncogene (2014). [DOI] [PMC free article] [PubMed]
- 48.Chen P, Cescon M, Bonaldo P. Collagen VI in cancer and its biological mechanisms. Trends Mol Med. 2013;19:410–417. doi: 10.1016/j.molmed.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y, Yang P, Li F, Kijlstra A. The Effects of Th17 Cytokines on the Inflammatory Mediator Production and Barrier Function of ARPE-19 Cells. PLoS ONE. 2011;6:e18139. doi: 10.1371/journal.pone.0018139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahdieh M, Vandenbos T, Youakim A. Lung epithelial barrier function and wound healing are decreased by IL-4 and IL-13 and enhanced by IFN-γ. Am J Physiol Cell Physiol. 2001;281:C2029–C2038. doi: 10.1152/ajpcell.2001.281.6.C2029. [DOI] [PubMed] [Google Scholar]
- 51.Chaturvedi P, Singh AP, Batra SK. Structure, evolution, and biology of the MUC4 mucin. FASEB J. 2008;22:966–981. doi: 10.1096/fj.07-9673rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee SH, et al. Tight junction protein 1 is regulated by transforming growth factor-β and contributes to cell motility in NSCLC cells. BMB Rep. 2015;48:115–120. doi: 10.5483/BMBRep.2015.48.2.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen G, et al. TGFBI expression reduces in vitro and in vivo metastatic potential of lung and breast tumor cells. Cancer Lett. 2011;308:23–32. doi: 10.1016/j.canlet.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen M-L, et al. Regulatory T cells suppress tumor-specific CD8 T cell cytotoxicity through TGF-β signals in vivo. Proc Natl Acad Sci USA. 2005;102:419–424. doi: 10.1073/pnas.0408197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wahl SM, Wen J, Moutsopoulos NM. The kiss of death: interrupted by NK-cell close encounters of another kind. Trends Immunol. 2006;27:161–164. doi: 10.1016/j.it.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 56.O’Callaghan DS, O’Donnell D, O’Connell F, O’Byrne KJ. The Role of Inflammation in the Pathogenesis of Non-small Cell Lung Cancer. J Thorac Oncol. 2010;5:2024–2036. doi: 10.1097/JTO.0b013e3181f387e4. [DOI] [PubMed] [Google Scholar]
- 57.Wang X, Lin Y. Tumor necrosis factor and cancer, buddies or foes? Acta Pharmacol Sin. 2008;29:1275–1288. doi: 10.1111/j.1745-7254.2008.00889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ślebioda, T. J. & Kmieć, Z. Tumour necrosis factor superfamily members in the pathogenesis of inflammatory bowel disease. Mediators Inflamm2014 (2014). [DOI] [PMC free article] [PubMed]
- 59.Jin S, et al. TL1A/TNFSF15 directly induces proinflammatory cytokines, including TNFα, from CD3+ CD161+ T cells to exacerbate gut inflammation. Mucosal Immunol. 2013;6:886–899. doi: 10.1038/mi.2012.124. [DOI] [PubMed] [Google Scholar]
- 60.Kim, E. S. In Lung Cancer and Personalized Medicine: Current Knowledge and Therapies (eds Aamir Ahmad & Shirish Gadgeel) 189–209 (Springer International Publishing, 2016).
- 61.Alberg AJ, Brock MV, Ford JG, Samet JM, Spivack SD. Epidemiology of lung cancer: Diagnosis and management of lung cancer, 3rd ed: american college of chest physicians evidence-based clinical practice guidelines. Chest. 2013;143:e1S–e29S. doi: 10.1378/chest.12-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zaenker P, Ziman MR. Serologic autoantibodies as diagnostic cancer biomarkers–a review. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2013;22:2161–2181. doi: 10.1158/1055-9965.EPI-13-0621. [DOI] [PubMed] [Google Scholar]
- 63.Järås K, Anderson K. Autoantibodies in cancer: prognostic biomarkers and immune activation. Expert Rev Proteomics. 2011;8:577–589. doi: 10.1586/epr.11.48. [DOI] [PubMed] [Google Scholar]
- 64.Vicent S, et al. ERK1/2 is activated in non-small-cell lung cancer and associated with advanced tumours. Br J Cancer. 2004;90:1047–1052. doi: 10.1038/sj.bjc.6601644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 66.Yip PY. Phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin (PI3K-Akt-mTOR) signaling pathway in non-small cell lung cancer. Transl Lung Cancer Res. 2015;4:165–176. doi: 10.3978/j.issn.2218-6751.2015.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mebratu Y, Tesfaigzi Y. How ERK1/2 Activation Controls Cell Proliferation and Cell Death Is Subcellular Localization the Answer? Cell cycle (Georgetown, Tex.) 2009;8:1168–1175. doi: 10.4161/cc.8.8.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siegelin MD, Borczuk AC. Epidermal growth factor receptor mutations in lung adenocarcinoma. Lab Invest. 2014;94:129–137. doi: 10.1038/labinvest.2013.147. [DOI] [PubMed] [Google Scholar]
- 69.Cortas T, et al. Activation state EGFR and STAT-3 as prognostic markers in resected non-small cell lung cancer. Lung cancer. 2007;55:349–355. doi: 10.1016/j.lungcan.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 70.Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. The FEBS journal. 2010;277:301–308. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 71.Normanno N, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 72.Landi L, Minuti G, D’Incecco A, Cappuzzo F. Targeting c-MET in the battle against advanced nonsmall-cell lung cancer. Curr Opin Oncol. 2013;25:130–136. doi: 10.1097/CCO.0b013e32835daf37. [DOI] [PubMed] [Google Scholar]
- 73.Liu X, Yao W, Newton RC, Scherle PA. Targeting the c-MET signaling pathway for cancer therapy. Expert Opin Investig Drugs. 2008;17:997–1011. doi: 10.1517/13543784.17.7.997. [DOI] [PubMed] [Google Scholar]
- 74.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 75.Leblanc R, Peyruchaud O. New insights into the autotaxin/LPA axis in cancer development and metastasis. Exp Cell Res. 2015;333:183–189. doi: 10.1016/j.yexcr.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 76.Bankaitis KV, Fingleton B. Targeting IL4/IL4R for the treatment of epithelial cancer metastasis. Clin Exp Metastasis. 2015;32:847–856. doi: 10.1007/s10585-015-9747-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kayacan O, et al. Impact of TNF-alpha and IL-6 levels on development of cachexia in newly diagnosed NSCLC patients. Am J Clin Oncol. 2006;29:328–335. doi: 10.1097/01.coc.0000221300.72657.e0. [DOI] [PubMed] [Google Scholar]
- 78.Yanagawa H, et al. Serum levels of interleukin 6 in patients with lung cancer. Br J Cancer. 1995;71:1095–1098. doi: 10.1038/bjc.1995.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo Y, Xu F, Lu T, Duan Z, Zhang Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat Rev. 2012;38:904–910. doi: 10.1016/j.ctrv.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 80.Moldovan G-L, D’Andrea AD. How the Fanconi Anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cremona CA, Behrens A. ATM signalling and cancer. Oncogene. 2014;33:3351–3360. doi: 10.1038/onc.2013.275. [DOI] [PubMed] [Google Scholar]
- 82.Coomans de Brachène A, Demoulin J-B. FOXO transcription factors in cancer development and therapy. Cell Mol Life Sci. 2016;73:1159–1172. doi: 10.1007/s00018-015-2112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stamos JL, Weis WI. The beta-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5:a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mocellin S, Nitti D. TNF and cancer: the two sides of the coin. Front Biosci. 2007;13:2774–2783. doi: 10.2741/2884. [DOI] [PubMed] [Google Scholar]
- 85.Neuzillet C, et al. Targeting the TGFbeta pathway for cancer therapy. Pharmacol Ther. 2015;147:22–31. doi: 10.1016/j.pharmthera.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 86.Tu Z, et al. CFTR is a potential marker for nasopharyngeal carcinoma prognosis and metastasis. Oncotarget. 2016;7:76955–76965. doi: 10.18632/oncotarget.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li J, et al. The cystic fibrosis transmembrane conductance regulator as a biomarker in non-small cell lung cancer. Int J Oncol. 2015;46:2107–2115. doi: 10.3892/ijo.2015.2921. [DOI] [PubMed] [Google Scholar]
- 88.Li C, Schuetz JD, Naren AP. Tobacco carcinogen NNK transporter MRP2 regulates CFTR function in lung epithelia: implications for lung cancer. Cancer Lett. 2010;292:246–253. doi: 10.1016/j.canlet.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang S, Qiu X, Gu Y, Wang E. Up-regulation of proline-rich tyrosine kinase 2 in non-small cell lung cancer. Lung Cancer. 2008;62:295–301. doi: 10.1016/j.lungcan.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 90.Block ER, Tolino MA, Klarlund JK. Pyk2 Activation Triggers Epidermal Growth Factor Receptor Signaling and Cell Motility after Wounding Sheets of Epithelial Cells. J Biol Chem. 2010;285:13372–13379. doi: 10.1074/jbc.M109.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gridling M, et al. Identification of kinase inhibitor targets in the lung cancer microenvironment by chemical and phosphoproteomics. Mol Cancer Ther. 2014;13:2751–2762. doi: 10.1158/1535-7163.MCT-14-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Symonds JM, et al. Protein kinase C delta is a downstream effector of oncogenic K-ras in lung tumors. Cancer Res. 2011;71:2087–2097. doi: 10.1158/0008-5472.CAN-10-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clark AS, West KA, Blumberg PM, Dennis PA. Altered Protein Kinase C (PKC) Isoforms in Non-Small Cell Lung Cancer Cells PKCδ Promotes Cellular Survival and Chemotherapeutic Resistance. Cancer Res. 2003;63:780–786. [PubMed] [Google Scholar]
- 94.Liu B, et al. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget. 2015;6:9445–9456. doi: 10.18632/oncotarget.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu X, Zhao Z, Ma X, Du L, Pertsemlidis A. miR-195 represses the tumorigenesis of non-small cell lung cancer and synergizes with microtubule targeting agents. Cancer Res. 2016;76:1063–1063. doi: 10.1158/1538-7445.AM2016-1063. [DOI] [Google Scholar]
- 96.Yuan X, et al. Notch signaling: an emerging therapeutic target for cancer treatment. Cancer Lett. 2015;369:20–27. doi: 10.1016/j.canlet.2015.07.048. [DOI] [PubMed] [Google Scholar]
- 97.Kikuchi H, et al. Expression of Notch1 and Numb in small cell lung cancer. Oncotarget. 2017;8:10348–10358. doi: 10.18632/oncotarget.14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang YL, Jablons D, You L. An alternative way to initiate Notch1 signaling in non-small cell lung cancer. Transl Lung Cancer Res. 2014;3:238–241. doi: 10.3978/j.issn.2218-6751.2013.12.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Konishi J, et al. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007;67:8051–8057. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 100.Lanzetti L, Di Fiore PP. Endocytosis and cancer: an ‘insider’ network with dangerous liaisons. Traffic. 2008;9:2011–2021. doi: 10.1111/j.1600-0854.2008.00816.x. [DOI] [PubMed] [Google Scholar]
- 101.Gomez-Cambronero J, Carman GM. Thematic minireview series on phospholipase D and cancer. J Biol Chem. 2014;289:22554–22556. doi: 10.1074/jbc.R114.593137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ha SH, et al. PLD2 forms a functional complex with mTOR/raptor to transduce mitogenic signals. Cell Signal. 2006;18:2283–2291. doi: 10.1016/j.cellsig.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 103.Mazzocca A, et al. Lysophosphatidic acid receptor LPAR6 supports the tumorigenicity of hepatocellular carcinoma. Cancer Res. 2015;75:532–543. doi: 10.1158/0008-5472.CAN-14-1607. [DOI] [PubMed] [Google Scholar]
- 104.Pan, W. et al. Whole exome sequencing identifies lncRNA GAS8-AS1 and LPAR4 as novel papillary thyroid carcinoma driver alternations. Hum Mol Genet, doi:10.1093/hmg/ddw056 (2016). [DOI] [PubMed]
- 105.Alexandre D, et al. The orexin type 1 receptor is overexpressed in advanced prostate cancer with a neuroendocrine differentiation, and mediates apoptosis. Eur J Cancer. 2014;50:2126–2133. doi: 10.1016/j.ejca.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 106.Laburthe M, Voisin T. The orexin receptor OX(1)R in colon cancer: a promising therapeutic target and a new paradigm in G protein-coupled receptor signalling through ITIMs. Br J Pharmacol. 2012;165:1678–1687. doi: 10.1111/j.1476-5381.2011.01510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Prickett TD, Samuels Y. Molecular pathways: dysregulated glutamatergic signaling pathways in cancer. Clin Cancer Res. 2012;18:4240–4246. doi: 10.1158/1078-0432.CCR-11-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pal SK, Figlin RA, Reckamp KL. The role of targeting mammalian target of rapamycin in lung cancer. Clin Lung Cancer. 2008;9:340–345. doi: 10.3816/CLC.2008.n.049. [DOI] [PubMed] [Google Scholar]
- 109.Lee JJ, et al. A high-affinity protein binder that blocks the IL-6/STAT3 signaling pathway effectively suppresses non-small cell lung cancer. Mol Ther. 2014;22:1254–1265. doi: 10.1038/mt.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen Y, et al. Gprc5a Deletion Enhances the Transformed Phenotype in Normal and Malignant Lung Epithelial Cells by Eliciting Persistent Stat3 Signaling Induced by Autocrine Leukemia Inhibitory Factor. Cancer Res. 2010;70:8917–8926. doi: 10.1158/0008-5472.CAN-10-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chauhan SC, et al. Expression and Functions of Transmembrane Mucin MUC13 in Ovarian Cancer. Cancer Res. 2009;69:765–774. doi: 10.1158/0008-5472.CAN-08-0587. [DOI] [PubMed] [Google Scholar]
- 113.Lakshmanan I, et al. Mucins in lung cancer: diagnostic, prognostic, and therapeutic implications. J Thorac Oncol. 2015;10:19–27. doi: 10.1097/JTO.0000000000000404. [DOI] [PubMed] [Google Scholar]
- 114.Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol. 2010;22:697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Spivey KA, et al. Collagen XXIII: a potential biomarker for the detection of primary and recurrent non-small cell lung cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19:1362–1372. doi: 10.1158/1055-9965.EPI-09-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hamano Y, et al. Physiological levels of tumstatin, a fragment of collagen IV α3 chain, are generated by MMP-9 proteolysis and suppress angiogenesis via αVβ3 integrin. Cancer Cell. 2003;3:589–601. doi: 10.1016/S1535-6108(03)00133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Selman M, et al. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. Am J Respir Crit Care Med. 2006;173:188–198. doi: 10.1164/rccm.200504-644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Samet JM. Does idiopathic pulmonary fibrosis increase lung cancer risk? Am J Respir Crit Care Med. 2000;161:1. doi: 10.1164/ajrccm.161.5.16154. [DOI] [PubMed] [Google Scholar]
- 119.Ni S, et al. Increased ZO-1 expression predicts valuable prognosis in non-small cell lung cancer. Int J Clin Exp Pathol. 2013;6:2887–2895. [PMC free article] [PubMed] [Google Scholar]
- 120.Domagala-Kulawik J. The role of the immune system in non-small cell lung carcinoma and potential for therapeutic intervention. Transl Lung Cancer Res. 2015;4:177–190. doi: 10.3978/j.issn.2218-6751.2015.01.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Carrega P, et al. Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56 bright CD16(−) cells and display an impaired capability to kill tumor cells. Cancer. 2008;112:863–875. doi: 10.1002/cncr.23239. [DOI] [PubMed] [Google Scholar]
- 122.Prado-Garcia H, Romero-Garcia S, Aguilar-Cazares D, Meneses-Flores M, Lopez-Gonzalez JS. Tumor-Induced CD8+ T-Cell Dysfunction in Lung Cancer Patients. Clin Dev Immunol. 2012;2012:11. doi: 10.1155/2012/741741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tafsiri E, Darbouy M, Shadmehr MB, Cho WC, Karimipoor M. Abberent expression of oncogenic and tumor-suppressive microRNAs and their target genes in human adenocarcinoma alveolar basal epithelial cells. J Cancer Res Ther. 2016;12:395–400. doi: 10.4103/0973-1482.148673. [DOI] [PubMed] [Google Scholar]
- 124.Morton LM, et al. Etiologic heterogeneity among non-Hodgkin lymphoma subtypes. Blood. 2008;112:5150–5160. doi: 10.1182/blood-2008-01-133587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zwick C, et al. Autoantigenic targets of B-cell receptors derived from chronic lymphocytic leukemias bind to and induce proliferation of leukemic cells. Blood. 2013;121:4708–4717. doi: 10.1182/blood-2012-08-447904. [DOI] [PubMed] [Google Scholar]
- 126.Vodnik M, Zager U, Strukelj B, Lunder M. Phage display: selecting straws instead of a needle from a haystack. Molecules. 2011;16:790–817. doi: 10.3390/molecules16010790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu X, et al. Serum Antibody Repertoire Profiling Using In Silico Antigen Screen. PloS one. 2013;8:e67181. doi: 10.1371/journal.pone.0067181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Szklarczyk D, et al. STRINGv10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. ICWSM. 2009;8:361–362. [Google Scholar]
- 130.Tripathi S, et al. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell host & microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research, gkw377 (2016). [DOI] [PMC free article] [PubMed]
- 132.Wang J, Duncan D, Shi Z, Zhang B. WEB-based gene set analysis toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wishart DS, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.