Abstract
In insect brains, the mushroom bodies (MBs) are a higher-order center for sensory integration and memory. Honeybee (Apis mellifera L.) MBs comprise four Kenyon cell (KC) subtypes: class I large-, middle-, and small-type, and class II KCs, which are distinguished by the size and location of somata, and gene expression profiles. Although these subtypes have only been reported in the honeybee, the time of their acquisition during evolution remains unknown. Here we performed in situ hybridization of tachykinin-related peptide, which is differentially expressed among KC subtypes in the honeybee MBs, in four hymenopteran species to analyze whether the complexity of KC subtypes is associated with their behavioral traits. Three class I KC subtypes were detected in the MBs of the eusocial hornet Vespa mandarinia and the nidificating scoliid wasp Campsomeris prismatica, like in A. mellifera, whereas only two class I KC subtypes were detected in the parasitic wasp Ascogaster reticulata. In contrast, we were unable to detect class I KC subtype in the primitive and phytophagous sawfly Arge similis. Our findings suggest that the number of class I KC subtypes increased at least twice – first with the evolution of the parasitic lifestyle and then with the evolution of nidification.
Introduction
Hymenoptera comprise many species that exhibit various behavioral traits1. Members of the most basal lineages are solitary and phytophagous2. Subsequently, a novel parasitoid lifestyle mode evolved, giving rise to prosperous groups of parasitic wasps2. In general, adult female parasitic wasps lay their eggs in the host, and their larvae grow inside the host by feeding on the host body2,3. From one of their lineages arose Aculeata2,4,5, the clade to which all of the eusocial Hymenoptera, such as hornets, ants, and honeybees, belong6. The neural mechanisms that underlie the variety of behavioral traits of the hymenopteran insects remain largely unknown.
Insect brains comprise several regions, including the mushroom bodies (MBs, a higher-order center), optic lobes (OLs, a center for visual information processing), antennal lobes (ALs, a center for olfactory and mechanosensory information processing), and subesophageal ganglion (SOG, a center for tasting and feeding behaviors). Centuries ago, Dujardin reported that social hymenopteran species possess large MBs7, drawing attention to the role of MBs in the development of sociality ever since. The MBs are a higher brain center conserved across almost all arthropods, including insecta8,9, that participate in learning and memory in a wide range of insect species10–17. In Drosophila, genetic studies revealed that some genes involved in cAMP signaling are expressed preferentially in the MBs, and mutants with defects in the function of these genes exhibit impaired learning ability18–20. In addition, in some insect species, including the honeybee, the MBs are also involved in sensory integration21–23.
Honeybee MBs are paired structures, each of which has two cup-shaped calyces. Somata of the MB intrinsic neurons, termed Kenyon Cells (KCs), are distributed to the outer surface and inside of the MB calyces8,24. MB calyces are composed of KC dendrites. The KCs of adult honeybees are categorized into two classes, class I and II, based on the timing of their development, location of their somata, and projection patterns of their dendrites24–26. Class I KC development starts in the prepupal stage27 and their somata fill the inside of the calyces, while class II KC development begins during the larval stage, and these KCs have characteristic “clawed” dendrites with their somata surrounding the outer surface of the calyces24–26. Categorization into these two classes based on similar criteria is also possible in insects more primitive than Hymenoptera, such as cockroaches and crickets, suggesting that KCs are highly conserved components of the MBs25,26. In the honeybee, class I KCs are further divided into three subtypes based on the location and size of their somata, and their gene expression28, for review, see refs29,30. Class I small-type KCs (sKCs) have small somata (5–7 µm) that occupy the innercore of the calyces, while class I large-type KCs (lKCs) have large somata (7–9 µm) that are located at the inner peripheral region of the calyces, and class I middle-type KCs (mKCs) have middle-sized somata that are located between the lKC and sKC somata28.
Class I lKCs, mKCs and sKCs have distinct gene expression profiles for review, see refs29,30; many genes involved in Ca2+-signaling, which participates in learning and memory in various organisms, are preferentially expressed in the lKCs in the honeybee brain31–35, implying their roles in learning and memory. Class I sKCs express hormone receptor-like 38 (HR38), whose expression varies with the division of labor of the workers, implying their involvement in the regulation of worker behaviors36. Both class I sKCs and some mKCs are active in the brains of workers engaged in foraging, suggesting their roles in visual information-processing during foraging flights37,38. Among KCs, mKCs selectively express middle-type Kenyon cell-preferential arrestin-related protein (mKast)28. As the honeybee is the only organism known to have such KC subtypes, these subtypes are assumed to contribute to the regulation of the highly advanced behaviors of the honeybee.
Farris and Schulmeister (2011) reported, based on anatomic comparison, that the elaborate hymenopteran MB structure arose in parallel with acquisition of the parasitoid lifestyle, and proposed that MB elaboration was driven by the cognitive demands of host-finding behavior39, rather than sociality as suggested centuries ago7. This led us to the question of what behavioral traits are associated with the elaboration of KC subtypes.
We addressed this question by cloning the orthologs of Tachykinin-related peptide (Trp) identified in the honeybee40,41 as a subtype marker gene, and performing expression analyses in the brains of various hymenopteran insects. Tachykinins are neuropeptides approximately 11 amino acids long that are conserved in metazoa and have a wide range of functions42–47. Tachykinin mRNA encodes a prepro peptide that contains several mature tachykinin peptides. In the honeybee brain, Trp is preferentially expressed in the MBs and in some cells scattered in various brain regions, such as the OLs, ALs, and SOG41. In the MB calyces, Trp expression is strong in the sKCs, moderate in the lKCs, and scarce in the mKCs28,41. Therefore, all three class I KC subtypes can be distinguished by in situ hybridization analysis of this one gene.
In the present study, we analyzed four hymenopteran insect species: the solitary and phytophagous sawfly Arge similis (Argidae, Tenthredinidae, Phytophagous groups), which belongs to one of the most basal hymenopteran lineages; the parasitic wasp Ascogaster reticulata (Braconidae, Ichneumonoidea, Apocrita); the eusocial hornet Vespa mandarinia (Vespidae, Aculeata, Apocrita); and the solitary hairy flower wasp Campsomeris prismatica (Scolioidea, Aculeata, Apocrita). The species above are listed in order of phylogeny from basal lineages to lineages close to the honeybee. Phylogenetic relationships are based on previous studies6,39.
Materials and Methods
Animals
Male and female adults of C. prismatica were captured at the Hongo campus of the University of Tokyo (Bunkyo-ku, Tokyo, Japan). Worker, queen and male adults of V. mandarinia were obtained from a matured nest collected at Bando-shi, Ibaraki Prefecture on 6th November 2016. Male and female adults of A. reticulata were from a stock culture at the Laboratory of Applied Entomology and Zoology, University of Tsukuba (Tsukuba, Ibaraki, Japan). The rearing of A. reticulata was based on the method described by Kainoh48, and that of host Adoxohyes honmai on Tamaki49. Emerged adults were fed with honey and water in a plastic container (1.8 L) under the laboratory conditions (25 ± 1 °C, 60 ± 10% RH, andL16:D8 photoperiod), and 2- to 3-day-old wasps were used for experiments. Male and female adults of A. similis were captured at the Hongo campus of the University of Tokyo. Eggs of A. similis were also captured at the Hongo campus of the University of Tokyo and fed azalea leaves at 25 °C with a 16-h light/8-h dark cycle in the laboratory. Adults were collected within 3 and more than10 days after emergence for further analysis.
Cloning of partial Trp orthologs and RNA probes synthesis
Adults of C. prismatica, V. mandarinia, A. reticulata, and A. similis were anesthetized on ice, and their heads were dissected with fine scissors. Total RNA extracted from the brains of each species was treated with DNase I Amp Grade (Invitrogen) and reverse-transcribed using SuperScript III (Invitrogen) with random primers. The cDNA was partially amplified by polymerase chain reaction (PCR) using Ex Taq (Takara) with primers designed for relatively conserved regions of hymenopteran preproTrp alignment. Nested PCR was performed for C. prismatica, A. reticulata, and A. similis. For primer sequences and PCR conditions, see Supplementary Table 1. The PCR products were cloned using a pGEM-T Easy (Promega) vector. The lengths of the cloned sequences were as follows: C. prismatica Trp, 462 bp; V. mandarinia Trp, 530 bp; A. reticulata Trp, 519 bp; A. similis Trp, 448 bp; and A. similis Elf-1 alfa, 529 bp. PCR was performed to obtain templates for in vitro transcription using Takara Ex Taq with M13 forward and reverse primers. The digoxigenin (DIG)-labeled sense and antisense RNA probes were synthesized by in vitro transcription using a DIG-RNA-labeling Kit (Roche).
Phylogenetic tree analysis
Trp cDNA sequences of hymenopteran insects and of Drosophila melanogaster, Periplaneta americana, and Tribolium castaneum, were obtained from the NCBI genome database. The sequences were aligned with clustalW using MEGA7 software. Based on this alignment, a maximum likelihood tree was constructed as implemented in MEGA7, using the Tamura-Nei model. The initial tree was obtained using the Neighbor-Join and BioNJ algorithms. The bootstrap test was performed with 200 replicates. The tree was rooted using Periplaneta americana as the most external out-group.
In situ hybridization
Whole brains were embedded in Tissue-Tek OCT compound (Sakura Finetek Japan) without fixation and immediately frozen. In situ hybridization was performed essentially as described previously50. For A. reticulata samples with Trp probes, signal amplification using the TSA biotin system (PerkinElmer) was performed between washing and color development. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Images were captured with a light microscope BX-50 (Olympus), merged using GIMP2.8 software (http://www.gimp.org/), and then the brightness and contrast was adjusted using ImageJ 64.
Cell body size measurement
The merged and adjusted images of V. mandarinia and C. prismatica were further analyzed with ImageJ 64 software to test if the three regions in the Class I KCs with different Trp signal intensity also differ in the respect of cell diameter. The three regions were manually marked. To randomize, a grid was overlaid using a plugin tool. Outlines of the cell somata under the intersections were hand-traced, and the sizes of the enclosed area were measured. Cell diameters were estimated based on the area size by approximating the cell as a true circle. To verify the cell diameter of the three regions differ each other, F-tests were performed first to test the equality of the variances. For two populations with the same variance, Student’s t-tests were performed. For the others, Welch’s t-tests were performed.
Data Availability
The sequence data generated in this study can be found in the DDBJ databases under the following accession numbers: LC271568 (C. prismatica), LC271567 (V. mandarinia), LC271570 (A. reticulata), LC271569 (A. similis).
Results
Identification of partial Trp nucleotide sequences from four hymenopteran insects
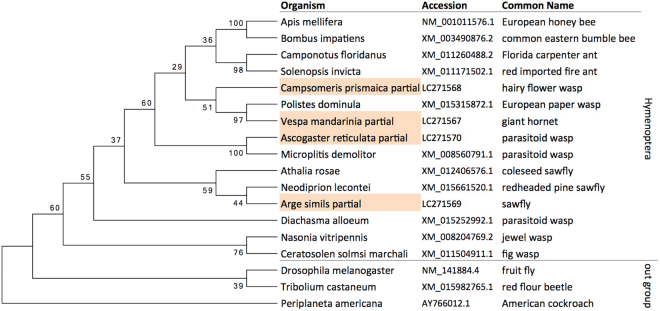
None of the four hymenopteran species we used have a genome database. We first aligned Trp nucleotide sequences of various hymenopteran insects with the genome database and designed primers to amplify a region relatively conserved among Hymenoptera. We then obtained partial Trp nucleotide sequences by PCR on cDNAs synthesized from total RNAs extracted from the brains of each of the four species. BLAST searches of the cloned sequences resulted in hymenopteran tachykinin genes as the top hits. Next, we compared the similarity of the cloned partial sequences and Trp mRNAs of other insects by constructing a phylogenetic tree. All of the cloned sequences belonged to the same group as the other hymenopteran Trp (Fig. 1), indicating they are indeed Trp orthologs. Based on our findings from genome databases, Trps are single-copy in all hymenopteran insects (data not shown).
Figure 1.
Phylogenetic tree analysis of Trp genes of the hymenopteran insect species. Maximum Likelihood tree of cDNA sequences of tachykinin gene of various hymenopteran insects and others: Drosophila melanogaster, Periplaneta americana, Tribolium castaneum, and the cloned partial sequences. Bootstrap values of 200 replicates are provided at the nodes. The cloned sequences are indicated with background color orange.
Expression analysis of Trp in the brain of the solitary wasp Campsomeris prismatica
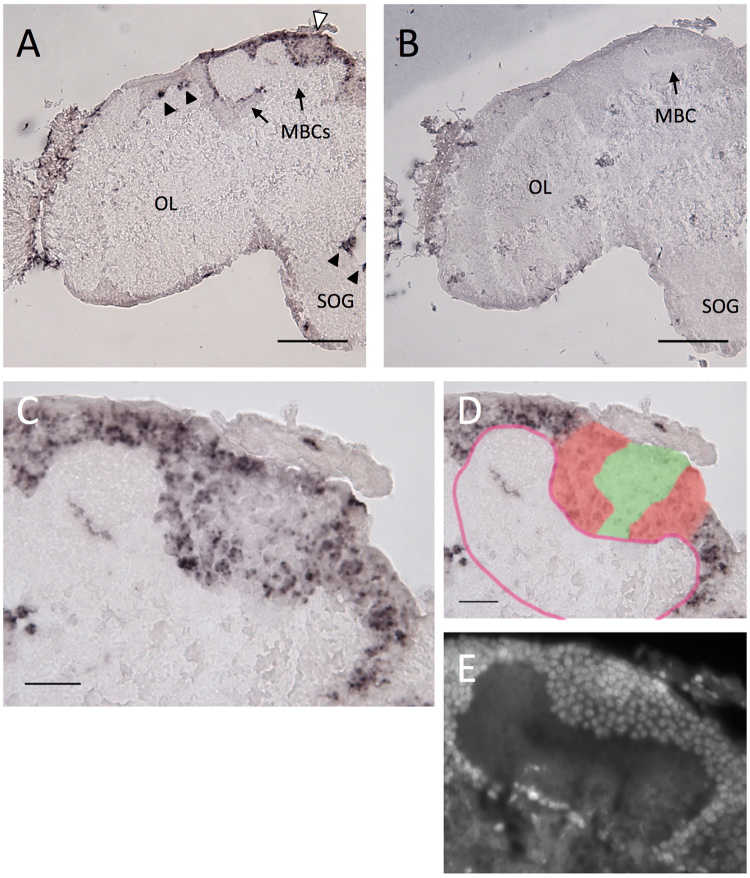
Next, we performed in situ hybridization analysis using a DIG-labeled RNA probe and 10-μm brain sections of the solitary hairy flower wasp C. prismatica (Scolioidea, Aculeata, Apocrita), which is most closely related to the honeybee of the four insect species we analyzed in this study. Strong signals were detected with antisense probe in the MBs (Fig. 2A). In addition, there were scattered signals in some cells in the other brain regions: ALs, OLs (Fig. 2A), and SOG (Fig. S1). In contrast, no signal was detected in sections hybridized with sense probe as a control (Fig. 2B), indicating that these signals represent Trp expression. Essentially the same results were obtained for another two female and three male individuals, and no clear difference in Trp expression was observed between male and female brains (Fig. S1). The MB-preferential as well as the scattered expression pattern of Trp in the other brain regions resembles that of A. mellifera 41.
Figure 2.
In situ hybridization of Trp in hairy flower wasp C. prismatica adult female brain. (A,B) Frontal section of the brain hemisphere hybridized with antisense (A) and sense (B) Trp probes. Trp signals scattered outside MBs are marked with arrowheads in (A). (C) Magnified view of the MB shown in (A). (D) Illustration indicating the Trp signal strength and the calyx is overlaid to (C). Kenyon cells were divided into three regions based on Trp signal intensity and the location of the somata: the Inner core (with strong Trp signal, blue), middle (with weak Trp signal, yellow), and inner peripheral (with moderate Trp signal, red) regions. The calyx is outlined with red line. (E) Distributions of the diameter of the somata of the each region are visualized with beanplot package60. Weak, Moderate and Strong correspond to regions colored in yellow, red, and blue in (D). MBC, mushroom body calyx; OL, optic lobe; AL, antennal lobe; “n = ”, number of the cells measured; asterisk, p-value < 0.01 (Welch’s t-test for Strong-Moderate, Student’s t-test for Weak-Moderate). Bars indicate 500 μm in (A) and (B), 100 μm in (C) and (D).
The MBs of C. prismatica had deep cup-shaped calyces similar to those of A. mellifera. Cells with strong Trp signal occupied the outer bottom region of the calyces as well as the inner core and inner periphery of the calyces, whereas no significant expression was detected in the regions between the inner periphery and the inner core (Fig. 2C and D). As the location of the somata and the relative Trp expression levels of these KCs closely resemble those of the honeybee, it is plausible that they correspond to class II KCs, class I lKCs, sKCs and mKCs, respectively. The somata of honeybee class I subtypes differ in diameter8,28. Therefore, we divided the KCs inside of the calyx into three regions based on the intensity of the Trp signal (Fig. 2D), and measured their sizes on the image shown. The somata of KCs that probably correspond to class I lKCs, mKCs and sKCs had relatively large, medial, and small diameters, respectively, further suggesting that the four KC subtypes in C. prismatica are homologous to those in the honeybee.
Expression analysis of Trp in the brains of eusocial hornet Vespa mandarinia
In situ hybridization analysis of the hornet V. mandarinia (Vespidae, Aculeata, Apocrita) revealed strong Trp expression in the MBs, and scattered expression in the other brain regions (Fig. 3A and B), similar to C. prismatica (Fig. 2A and B) and A. mellifera. No significant differences in Trp expression were observed in the brains of male, worker, and queen hornets (Fig. S2).
Figure 3.
In situ hybridization of Trp in eusocial hornet V. mandarinia adult worker brain. (A,B) Frontal section of the brain hemisphere hybridized with antisense (A) and sense (B) Trp probes. Trp signals scattered outside MBs are marked with arrowheads in (A). (C) Magnified view of the MB in a serial section of (A) and (B). (D) Illustration indicating the Trp signal strength and the calyx is overlaid to (C). Kenyon cells were divided into three regions based on the Trp signal intensity and the location of the somata: the Inner core (with moderate Trp signal, blue), middle (with weak Trp signal, yellow), and inner peripheral (with strong Trp signal, red) regions. The calyx is outlined with red line. (E) Distributions of the diameter of the somata of the each region are visualized with beanplot package60. Weak, Moderate and Strong correspond to regions colored in yellow, blue and red in (D). MBC, mushroom body calyx; OL, optic lobe; AL, antennal lobe; “n = ”, number of the cells measured; asterisk, p-value < 0.01(Welch’s t-test for Moderate-Weak, Student’s t-test for Moderate-Strong). Bars indicate 500 μm in (A) and (B), 100 μm in (C) and (D).
The MB calyces resembled those of C. prismatica and A. mellifera in sense of deep-cup shape, but the well developed lip (subcompartment of the calyx around the rim region) overhanging inward makes their appearance notably different. This structure is consistent with previous observations in another hornet species51. Cells with Trp signals occupied the outer bottom, inner core, and inner periphery of the calyces, and no signal was detected in the regions between the inner core and inner periphery as in C. prismatica and A. mellifera, although the signal level of the inner core and inner periphery were reversed (Fig. 3C and D). Essentially the same results were obtained for another worker, queen, and male individuals (Fig. S2). The cells in the inner core (with moderate Trp signal), middle (weak), and inner peripheral (strong) regions had relatively small, medial, and large diameters, respectively (Fig. 3E). We assumed that these three cell groups are probably homologous to class I sKCs, mKCs, and lKCs, and the outer bottom cells to class II KCs of A. mellifera, based on the similarities in the location and size of the somata despite the reversed relative Trp signal level between putative lKCs and sKCs. We also assume that gene expression levels are much more susceptible to the environmental effects and/or individual differences compared to somata location and size36,40.
Expression analysis of Trp in the brains of the parasitic wasp Ascogaster reticulata
In the brains of the parasitic wasp A.reticulata, we detected strong Trp expression in the MBs and scattered Trp expression in the other brain regions: in a part of OLs and SOG regions(Fig. 4A and B) and ALs (data not shown), as in A. mellifera L41, C. prismatica (Fig. 2A and B) and V. mandarinia (Fig. 3A and B). No significant differences were observed between males and female brains (Fig. S3).
Figure 4.
In situ hybridization of Trp in parasitoid wasp A. reticulata adult female brain. (A,B) Frontal section of the brain hemisphere hybridized with antisense (A) and sense (B) Trp probes. Black arrowheads indicate Trp signals scattered outside MBs, and white arrowhead indicates the regions of Kenyon cells inside the calyx with a relatively weak Trp signal in (A). (C) Magnified view of the MB shown in (A). (D) Illustration indicating the Trp signal strength and the calyx is overlaid to (C). Regions with strong and weak Trp expression are colored with red and green, respectively. The calyx is outlined with red line. (E) DAPI staining of a serial section of (C,D). MBC, mushroom body calyx; OL, optic lobe; SOG, suboesophageal ganglion. Bars indicate 100 μm in (A) and (B), 20 μm in (C) and (D).
The MBs of A. reticulata also had deep cup-shaped calyces similar to those of A. mellifera L., C. prismatica and V. mandarinia, as reported before39. This anatomic similarity suggests that the somata that fill the calyx are class I KCs and those that surround the outer surface are class II KCs, as in the other three species. The Trp expression in the MBs differed from that in the three species above: cells with Trp signals continuously occupied the outside and inner periphery of the calyces, whereas the signal was weak at the inner core (Fig. 4C and D). Measurement of the diameters of these somata proved quite difficult due to vague boundaries of the stained KCs. However, DAPI staining of the serial section demonstrated that while there was no clear difference between the nuclear size of KCs that reside outside and inner periphery of calyces, those of KCs that occupy the inner core of the calyces were more compact and smaller than those of the others (Fig. 4E). Essentially the same results were obtained for another two female and two male individuals (Fig. S3). These observations suggest that although the boundary between class I and II KCs was indistinguishable in this study, there are two class I KC subtypes distinguished on the same basis as the subtypes of the honeybee and the two described species above: the diameter, location of the somata, and Trp expression.
Expression analysis of Trp in the brains of the sawfly Arge similis
In the brains of the solitary and phytophagous sawfly Arge similis (Argidae, Tenthredinoidae, Phytophagous groups), the Trp expression patterns differed: some showed stronger Trp expression in the MBs than in the other brain regions(Fig. 5A,B), whereas the others showed ubiquitous Trp expression throughout the brain cortex (Fig. 5C and D). Stronger Trp expression in the MBs was observed in three of five younger (within 3 days after emergence) individuals, whereas ubiquitous Trp expression was observed in all four of the older (older than 10 days) individuals (Fig. S4), although whether stronger Trp expression in the MBs is unique to the younger individuals is not certain at present. To confirm that the stronger Trp signals in some individuals were not due to compacted KCs in their brains, we further performed in situ hybridization of the housekeeping gene Elongation factor-1 alfa (EF-1alfa) on the serial sections of the same brain used for the in situ hybridization of Trp. Ubiquitous Ef1-alfa expression was detected in all seven individuals, both young and old (Fig. S5), confirming that the stronger Trp signals in some individuals indeed reflect stronger Trp expression.
Figure 5.
In situ hybridization of Trp in sawfly Arge similis adult female brain. (A,B,C,D,) Frontal section of the brain hemisphere hybridized with antisense (A,C) and sense (B,D) Trp probes. (A,B) An individual younger than three days after emergence. (C,D) An individual older than ten days after emergence. (E) Magnified view of the MB of a serial section of (A,B). Schematic illustration of the measurements are overlaid. One soma is marked with black circle as an example. Diameter of the somata and the distances (black straight line) from the center of the calyx (black dot) were measured. Somata within the region enclosed with grey line are used for measurements. (F) A scattered plot graph of the measurements explained in (E). Each dot represents one soma. “n = ”, number of the cells measured; MBC, mushroom body calyx; OL, optic lobe; SOG, suboesophageal ganglion. Bars indicate 200 μm in (A,B,C,D), 50 μm in (E).
The MBs had simple ovoid-shape calyces with a small depression at the top, consistent with the previous observation in other sawfly species (Fig. 5A and C)39. Trp expression in the KCs was ubiquitous in both old and young individuals, and therefore subtypes were indistinguishable in this experiment. We verified the uniformity of the size of the somata by analyzing the correlation between the size and distance of the somata from the center of the depression. There was no significant relationship between them, suggesting that there are no KC subtypes detectable on the basis of the diameter, location of the somata, or Trp expression. Albeit undetectable, class I and II KCs are likely to be present in the sawfly because these two classes are assumed to be highly conserved across insecta25,26.
Discussion
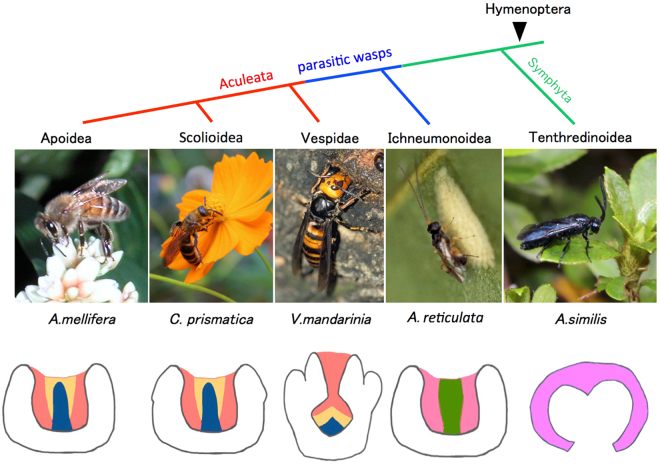
Our results demonstrated that KC subtypes became more diverse with the evolution of Hymenoptera: while a member of the basalmost Hymenoptera, A. similis, has only one class I subtype; parasitic wasp A. reticulata has two; and Aculeata wasps, V. mandarinia, C. prismatica, and A. mellifera have three (Fig. 6). Then what factors had been the background of the acquisition of novel KC subtypes in hymenopteran evolution?
Figure 6.
Phylogenic tree and schematic illustration of KC subtypes of hymenopteran species. (Upper panel) Phylogenic tree of honeybee and the four hymenopteran species examined in this study. (Lower panel) Schematic illustration of the KC subtypes. Three Class I KC subtypes that correspond to the lKCs, mKCs and sKCs in the MB are colored with red, yellow and blue for A. mellifera L., C. prismatica and V. mandarinia, respectively, Two putative Class I KC subtypes in the MB of A. reticulata are colored with pink and green, respectively. Single putative class I KC subtype in the MB of A. similis is colored with pale pink.
MB calyces commonly receive direct olfactory and mechanosensory inputs from ALs in most insect species9. In addition, they also receive direct inputs from OLs in Apocrita and possibly Orussoidea, but not in the phytophagous lineage39. The rise of the new subtype in the parasitic wasp might have been triggered by the gain of this novel sensory input into the MB calyx.
Which of the three subtypes of aculeate MBs correspond with class I KCs of A. similis and A. reticulata? We speculate that class I lKCs of the MBs of aculeate Hymenoptera and class I KCs of A. similis are homologous for two reasons. First, class I lKCs of the honeybee receive inputs from ALs24 as do the KCs of A. similis 39. The honeybee MB calyces comprise three subcompartments (neuropils): lip, collar, and basal ring. Olfactory information from the ALs projects to the lip, whereas visual information from the OLs projects to the collar, and both olfactory and visual information project to the basal rings. On the other hand, class I lKCs are likely to have their dendrites in both the lips and collars, whereas class I sKCs have their dendrites in the basal rings24. Therefore, the KCs that exclusively receive calycate AL input are lKCs. As for mKCs, their projections have yet to be clarified.
Second, lKCs are assumed to participate in learning and memory because they preferentially express many genes involved in Ca2+-signaling31–35, for review, see refs29 and30, which underlies learning and memory in many metazoan species. In insect species more primitive than Hymenoptera, such as cockroaches and locusts, the MBs also function as a center for learning and memory10,16. Therefore, KCs present in sawflies presumably resemble the lKCs in honeybee with regard to function. As for the two subtypes present in A. reticulata, we think that the cells expressing Trp whose somata occupy the inner peripheral region correspond to lKCs, because lKCs likewise occupy the inner peripheral region, express Trp, and have relatively large somata. The KCs that do not express Trp, which reside in the inner core of the calyces, may correspond to either mKCs or sKCs, or common ancestors of both mKCs and sKCs. This assumption is consistent with the notion that new KCs arose with the direct input of visual information, which mKCs and sKCs are thought to process during foraging flights. Further expression analysis with many other subtype marker genes for review, see refs29,30 will confirm the homologous relationships between the class I subtypes in honeybees and A. similis or A. reticulata.
What is the significance of a certain hymenopteran insect acquiring a novel KC subtype? The acquisition of a novel KC subtype may correlate with the development of novel behavioral traits. The sawfly A. similis, which has only one class I KC subtype, has a rather simple lifestyle: it lays eggs on the host plant, azalea, and the larvae sustain themselves by eating azalea leaves52. The sawfly pupates in the soil under the azalea tree, and the emerged adults live on nectars. This lifestyle does not likely require highly advanced learning and memory ability. In contrast, parasitic wasps, which acquired two KC subtypes and direct input of visual information into the calyces, have extensive learning abilities. They remember the odor, shape, color, and location of the host. A. reticulata is also reported to learn plant odor related to the host53,54. Consistent with a previous proposal that cognitive demands for host-finding behavior drove the anatomic elaboration of MBs with direct input of visual information to the calyces39, the subdivision of KCs may likewise underlie the evolution of parasitic behavior.
The aculeate hymenopteran insects that have three class I KC subtypes, share a behavioral trait to build nests that house their offspring. More precise learning and memory ability is likely required to remember the location and correctly return to their nests. Indeed, aculeate species are known to have sophisticated visual memories55–58. We speculate that acquisition of the three KC subtypes with distinct molecular characteristics conferred the aculeate hymenopteran insects the abilities needed for nidification, possibly by functional specialization of each subtype. Considering that all the eusocial Hymenoptera belong to Aculeata as well as a previous report on the possible involvement of the sKCs in the division of labor of workers in the honeybees36, it is also plausible that the acquisition of the three KC subtypes is a preadaptation for eusociality.
Lastly, the roles that Trp plays in hymenopteran brains remain unknown. In Drosophila, a genetic study demonstrated that Trp promotes male-specific aggression47. As Trp is a secretory neuropeptide and thought to affect various neural circuits42–47, it is difficult to deduce its role in Hymenoptera. We expect that a combination of the analyses of projection patterns, more detailed molecular characteristics, and functional analysis using genome-editing technology, which was recently established in the honeybee59, of each KC subtype will provide important clues to enhance our understanding of the neural bases of spatial memory and social behaviors, and their evolution.
Electronic supplementary material
Acknowledgements
This work was supported by the Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
S.O. and T.K. designed the experiments. S.O. performed all investigation (experiments and curation of the data). All S.O., H.K. and T.K. collected the scoliid (hairy flower) wasp (Campsomeris prismatica) and the sawfly (Arge similis). Y.K. and M.O. prepared the parasitic wasp (Ascogaster reticulata) and the giant hornet (Vespa mandarinia), respectively. All S.O., H.K., Y.K., M.O. and T.K. discussed on the results. S.O. drafted the manuscript and all S.O., H.K., Y.K., M.O. and T.K. wrote the paper. T.K. acquired the funds and supervised the study.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14174-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Huber, J. T. Biodiversity of Hymenoptera, in Insect Biodiversity: Science and Society (eds R. G. Foottit and P. H. Adler) 303–323 (Blackwell Publishing, Oxford, UK, 2009).
- 2.Whitfield JB. Phylogenetic insights into the evolution of parasitism in Hymenoptera. Adv. Parasitol. 2003;54:69–100. doi: 10.1016/S0065-308X(03)54002-7. [DOI] [PubMed] [Google Scholar]
- 3.Pennacchio F, Strand MR. Evolution of developmental strategies in parasitic Hymenoptera. Annu. Rev. Entomol. 2006;51:233–258. doi: 10.1146/annurev.ento.51.110104.151029. [DOI] [PubMed] [Google Scholar]
- 4.Klopfstein S, Vilhelmsen L, Heraty JM, Sharkey M, Ronquist F. The hymenopteran tree of life: evidence from protein-coding genes and objectively aligned ribosomal data. PLoS ONE. 2013;8:e69344. doi: 10.1371/journal.pone.0069344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao M, Gibson T, Dowton M. Higher-level phylogeny of the Hymenoptera inferred from mitochondrial genomes. Mol. Phyl. Evol. 2015;84:34–43. doi: 10.1016/j.ympev.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Johnson BR, et al. Phylogenomics resolves evolutionary relationships among ants, bees, and wasps. Curr. Biol. 2013;23:2058–2062. doi: 10.1016/j.cub.2013.08.050. [DOI] [PubMed] [Google Scholar]
- 7.Dujardin FM. ‘moire sur le syste ‘me nerveux des insects. Ann. Sci. Nat.l Zool. 1850;14:195–206. [Google Scholar]
- 8.Mobbs PG. The brain of the honeybee Apis mellifera. I. The connections and spatial organization of the mushroom bodies. Phil. Trans. R. Soc. London B. 1982;298:309–354. doi: 10.1098/rstb.1982.0086. [DOI] [Google Scholar]
- 9.Strausfeld NJ, Hansen L, Li Y, Gomez RS, Ito K. Evolution, discovery, and interpretations of Arthropod mushroom bodies. Learning & Memory. 1998;5:11–37. [PMC free article] [PubMed] [Google Scholar]
- 10.Mizunami M, Weibrecht JM, Strausfeld NJ. Mushroom bodies of the cockroach: Their participation in place memory. J. Comp. Neurol. 1998;402:520–537. doi: 10.1002/(SICI)1096-9861(19981228)402:4<520::AID-CNE6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 11.Erber J, Masuhr T, Menzel R. Localization of short-term memory in the brain of the bee, Apis mellifera L. Physiol. Entomol. 1980;5:343–358. doi: 10.1111/j.1365-3032.1980.tb00244.x. [DOI] [Google Scholar]
- 12.Hammer M, Menzel R. Multiple sites of associative odor learning as revealed by local brain microinjections of octopamine in honeybees. Learn. Mem. 1998;5:146–156. [PMC free article] [PubMed] [Google Scholar]
- 13.Locatelli F, Bundrock G, Müller U. Focal and temporal release of glutamate in the mushroom bodies improves olfactory memory in Apis mellifera. J. Neurosci. 2005;25:11614–11618. doi: 10.1523/JNEUROSCI.3180-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zars T, Fischer M, Schulz. R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- 15.Pascual A, Preat T. Localization of long-term memory within the Drosophila mushroom body. Science. 2001;294:1115–1117. doi: 10.1126/science.1064200. [DOI] [PubMed] [Google Scholar]
- 16.Perez-Orive J, et al. Oscillations and sparsening of odor representations in the mushroom body. Science. 2002;297:359–365. doi: 10.1126/science.1070502. [DOI] [PubMed] [Google Scholar]
- 17.Ito I, Ong RC, Raman B, Stopfer M. Sparse odor representation and olfactory learning. Nat. Neurosci. 2008;11:1177–1184. doi: 10.1038/nn.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han PL, Levin LR, Reed RR, Davis RL. Preferential expression of the Drosophila rutabaga gene in mushroom bodies, neural centers for learning in insects. Neuron. 1992;9:619–627. doi: 10.1016/0896-6273(92)90026-A. [DOI] [PubMed] [Google Scholar]
- 19.Nighorn A, Healy MJ, Davis RL. The cyclic AMP phosphodiesterase encoded by the Drosophila dunce gene is concentrated in the mushroom body neuropil. Neuron. 1991;6:455–467. doi: 10.1016/0896-6273(91)90253-V. [DOI] [PubMed] [Google Scholar]
- 20.Skoulakis EMC, Kalderon D, Davis RL. Preferential expression in mushroom bodies of the catalytic subunit of protein kinase A and its role in learning and memory. Neuron. 1993;11:1–14. doi: 10.1016/0896-6273(93)90178-T. [DOI] [PubMed] [Google Scholar]
- 21.Rybak J, Menzel R. Integrative properties of the Pe1 neuron, a unique mushroom body output neuron. Learn. Mem. 1998;5:133–145. [PMC free article] [PubMed] [Google Scholar]
- 22.Schildberger K. Multimodal interneurons in the cricket brain: properties of identified extrinsic mushroom body cells. J Comp Physiol A. 1984;154:71–79. doi: 10.1007/BF00605392. [DOI] [Google Scholar]
- 23.Li Y-S, Strausfeld NJ. Multimodal efferent and recurrent neurons in the medial lobes of cockroach mushroom bodies. J. Comp. Neurol. 1999;409:647–663. doi: 10.1002/(SICI)1096-9861(19990712)409:4<647::AID-CNE9>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Strausfeld NJ. Organization of the honey bee mushroom body: Representation of the calyx within the vertical and gamma lobes. J. Comp. Neurol. 2002;450:4–33. doi: 10.1002/cne.10285. [DOI] [PubMed] [Google Scholar]
- 25.Fahrbach SE. Structure of the mushroom bodies of the insect brain. Annu. Rev. Entomol. 2006;51:209–232. doi: 10.1146/annurev.ento.51.110104.150954. [DOI] [PubMed] [Google Scholar]
- 26.Farris SM. Evolution of insect mushroom bodies: Old clues, new insights. Arthropod Str. Dev. 2005;34:211–234. doi: 10.1016/j.asd.2005.01.008. [DOI] [Google Scholar]
- 27.Farris SM, Robinson GE, Davis RL, Fahrbach SE. Larval and pupal development of the mushroom bodies in the honey bee, Apis mellifera. J. Comp. Neurol. 1999;414:97–113. doi: 10.1002/(SICI)1096-9861(19991108)414:1<97::AID-CNE8>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko K, et al. Novel middle-type Kenyon cells in the honeybee brain revealed by area-preferential gene expression analysis. PLoS ONE. 2013;8:e71732. doi: 10.1371/journal.pone.0071732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko K, Suenami S, Kubo T. Gene expression profiles and neural activities of Kenyon cell subtypes in the honeybee brain: identification of novel “middle-type” Kenyon cells. Zool. Lett. 2016;2:14. doi: 10.1186/s40851-016-0051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubo, T. Neuroanatomical dissection of the honeybee brain based on temporal and regional gene expression patterns. In Honeybee neurobiology and Behavior. A Tribute to Randolf Menzel (Eds Galizia, C. G., Eisenhardt, D. & Giulfa, M.) 341–358 (Springer 2012).
- 31.Kamikouch A, et al. Preferential expression of the gene for a putative inositol 1,4,5-trisphosphate receptor homologue in the mushroom bodies of the brain of the worker honeybee Apis mellifera L. Biochem. Biophys. Res. Commun. 1998;242:181–186. doi: 10.1006/bbrc.1997.7870. [DOI] [PubMed] [Google Scholar]
- 32.Kamikouchi A, Takeuchi H, Sawata M, Natori S, Kubo T. Concentrated expression of Ca2+/calmodulin-dependent protein kinase II and protein kinase C in the mushroom bodies of the brain of the honeybee Apis mellifera L. J. Comp. Neurol. 2000;417:501–510. doi: 10.1002/(SICI)1096-9861(20000221)417:4<501::AID-CNE8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi H, et al. Identification of genes expressed preferentially in the honeybee mushroom bodies by combination of differential display and cDNA microarray. FEBS Lett. 2002;513:230–234. doi: 10.1016/S0014-5793(02)02319-0. [DOI] [PubMed] [Google Scholar]
- 34.Uno Y, Fujiyuki T, Morioka M, Kubo T. Mushroom body-preferential expression of proteins/genes involved in endoplasmic reticulum Ca2+-transport in the worker honeybee (Apis mellifera L.) brain. Insect Mol. Biol. 2012;22:52–61. doi: 10.1111/imb.12002. [DOI] [PubMed] [Google Scholar]
- 35.Sen Sarma. M, Rodriguez-Zas SL, Hong F, Zhong S, Robinson GE. Transcriptomic profiling of central nervous system regions in three species of honey bee during dance communication behavior. PLoS ONE. 2009;4:e6408. doi: 10.1371/journal.pone.0006408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamazaki Y, et al. Differential expression of HR38 in the mushroom bodies of the honeybee brain depends on the caste and division of labor. FEBS Lett. 2006;580:2667–2670. doi: 10.1016/j.febslet.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 37.Kiya T, Kunieda T, Kubo T. Increased neural activity of a mushroom body neuron subtype in the brains of forager honeybees. PLoS ONE. 2007;2:e371. doi: 10.1371/journal.pone.0000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiya T, Kunieda T, Kubo T. Inducible- and constitutive-type transcript variants of kakusei, a novel non-coding immediate early gene, in the honeybee brain. Insect Mol. Biol. 2008;17:531–536. doi: 10.1111/j.1365-2583.2008.00821.x. [DOI] [PubMed] [Google Scholar]
- 39.Farris SM, Schulmeister S. Parasitoidism, not sociality, is associated with the evolution of elaborate mushroom bodies in the brains of hymenopteran insects. Proc. R. Soc. B. 2011;278:940–951. doi: 10.1098/rspb.2010.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi H, Yasuda A, Yasuda-Kamatani Y, Kubo T, Nakajima T. Identification of a tachykinin-related neuropeptide from the honeybee brain using direct MALDI-TOF MS and its gene expression in worker, queen and drone heads. Insect Mol. Biol. 2003;12:291–298. doi: 10.1046/j.1365-2583.2003.00414.x. [DOI] [PubMed] [Google Scholar]
- 41.Takeuchi H, et al. Prepro-tachykinin gene expression in the brain of the honeybee Apis mellifera. Cell Tissue Res. 2004;316:281–293. doi: 10.1007/s00441-004-0865-y. [DOI] [PubMed] [Google Scholar]
- 42.Ikeda T, Minakata H, Nomoto K. The importance of C-terminal residues of vertebrate and invertebrate tachykinins for their contractile activities in gut tissues. FEBS Lett. 1999;461:201–204. doi: 10.1016/S0014-5793(99)01457-X. [DOI] [PubMed] [Google Scholar]
- 43.Johard HA, Muren JE, Nichols R, Larhammar DS, Nässel DR. A putative tachykinin receptor in the cockroach brain: molecular cloning and analysis of expression by means of antisera to portions of the receptor protein. Brain Res. 2001;919:94–105. doi: 10.1016/S0006-8993(01)03004-9. [DOI] [PubMed] [Google Scholar]
- 44.Kwok RN, Nässel DR, Lange AB, Orchard I. Locustatachykinin isoforms in the locust: distribution and quantification in the central nervous system and action on the oviduct muscle. Peptides. 1999;20:687–694. doi: 10.1016/S0196-9781(99)00051-0. [DOI] [PubMed] [Google Scholar]
- 45.Nässel DR. Tachykinin-related peptides in invertebrates. Peptides. 1999;20:141–158. doi: 10.1016/S0196-9781(98)00142-9. [DOI] [PubMed] [Google Scholar]
- 46.Siviter RJ, et al. Expression and functional characterization of a Drosophila neuropeptide precursor with homology to mammalian prepro-tachykinin A. J. Biol. Chem. 2000;275:23273–23280. doi: 10.1074/jbc.M002875200. [DOI] [PubMed] [Google Scholar]
- 47.Asahina K, et al. Tachykinin-expressing neurons control male-specific aggressive arousal in Drosophila. Cell. 2014;156:221–235. doi: 10.1016/j.cell.2013.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kainoh Y. Some factors influencing sex ratio in Ascogaster reticulatus Watanabe (Hymenoptera: Braconidae) Appl. Entomol. Zool. 1988;23:35–40. doi: 10.1303/aez.23.35. [DOI] [Google Scholar]
- 49.Tamaki Y. Mass rearing of the smaller tea tortrix, Adoxophyes orana Fischer von Röslerstamm, on a simplified artificial diet for successive generations (Lepidoptera: Tortricidae) Appl. Entomol. Zool. 1966;1:120–124. doi: 10.1303/aez.1.120. [DOI] [Google Scholar]
- 50.Suenami S, et al. Analysis of the differentiation of Kenyon cell subtypes using three mushroom body-preferential genes during metamorphosis in the honeybee (Apis mellifera L.) PLoS ONE. 2016;11:e0157841. doi: 10.1371/journal.pone.0157841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Couto A, Lapeyre B, Thiéry D, Sandoz JC. Olfactory pathway of the hornet Vespa velutina: New insights into the evolution of the hymenopteran antennal lobe. J. Comp. Neurol. 2016;524:2335–2359. doi: 10.1002/cne.23975. [DOI] [PubMed] [Google Scholar]
- 52.Li N. Life history and behavior of Arge similis, a pest of Rhododendron. Oyo-Dobutsugaku-Zasshi. 1934;6:273–289. [Google Scholar]
- 53.Seino H, Kainoh Y. Associative learning and discrimination of 10 plant species by the egg-larval parasitoid, Ascogaster reticulate Watanabe (Hymenoptera: Braconidae) Appl. Entomol. Zool. 2008;43:83–90. doi: 10.1303/aez.2008.83. [DOI] [Google Scholar]
- 54.Seino H, Shoji K, Kainoh Y. Utilization of learned plant chemicals in host searching behavior by the egg-larval parasitoid Ascogaster reticulata Watanabe (Hymenoptera: Braconidae) Appl. Entomol. Zool. 2010;45:339–345. doi: 10.1303/aez.2010.339. [DOI] [Google Scholar]
- 55.Toh Y, Okamura J. Foraging navigation of hornets studied in natural habitats and laboratory experiments. Zool. Sci. 2003;20:311–324. doi: 10.2108/zsj.20.311. [DOI] [PubMed] [Google Scholar]
- 56.Rosenheim JA. Host location and exploitation by the cleptoparasitic wasp Argochrysis armilla: the role of learning (Hymenoptera: Chrysididae) Behav. Ecol. Sociobiol. 1987;21:401–406. doi: 10.1007/BF00299935. [DOI] [Google Scholar]
- 57.Tibbetts EA. Visual signals of individual identity in the wasp Polistes fuscatus. Proc. R. Soc. B. 2002;269:1423–1428. doi: 10.1098/rspb.2002.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Menzel R, et al. Honey bees navigate according to a map-like spatial memory. Proc. Natl Acad. Sci. USA. 2005;102:3040–3045. doi: 10.1073/pnas.0408550102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kohno H, Suenami S, Takeuchi T, Sasaki T, Kubo T. Production of knockout mutants by CRISPR/Cas9 in the European honeybee, Apis mellifera L. Zool. Sci. 2016;33:505–512. doi: 10.2108/zs160043. [DOI] [PubMed] [Google Scholar]
- 60.Kampstra P. Beanplot: A boxplot alternative for visual comparison of distributions. J. Stat. Software, Code Snippets. 2008;28:1–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data generated in this study can be found in the DDBJ databases under the following accession numbers: LC271568 (C. prismatica), LC271567 (V. mandarinia), LC271570 (A. reticulata), LC271569 (A. similis).