Abstract
A quantitative real-time PCR assay was developed that detects genomic RNA from reference strains representing the six major genotypes of hepatitis C virus (HCV) with equal sensitivity and accurately measured HCV RNA in JFH1 HCV-infected Huh7.5 cells. The method is indirectly calibrated to the first international (WHO 96/790) HCV standard preparation and has a linear dynamic range of 102.6–106.5 IU/ml. In addition, the inter- and intra-assay precision were ~3% CV and <2% CV, respectively. Comparison with results obtained by commercially available HCV RNA Nucleic Acid Technology kits (Versant HCV RNA 3.0 b-DNA and Amplicor HCV Monitor), that also employ the WHO standard, allowed validation of the TaqMan assay against all major HCV genotypes. Both commercial methods detected HCV RNA over a wide dynamic range, but showed a consistent difference of about 0.3 log10 when evaluating samples of different HCV genotypes. The genome titers obtained with the three methods correlated with the infectivity titers previously determined for the HCV reference strains. TaqMan assays have become an essential tool to follow viral load in clinical samples and cell culture-based experiments and this technology offers significant advantages in linear dynamic range, sensitivity and customization.
Keywords: HCV, viral load, infectious pools, TaqMan, genotypes
INTRODUCTION
HCV is an important human pathogen that produces, in a majority of infected individuals, chronic liver disease that can lead to end stage liver disease, including liver cancer [Alter et al., 1989; Bukh et al., 1998; Saldanha et al., 1999]. The virus has a positive-sense single-stranded RNA genome approximately 9.6 kb in length and is classified as a separate genus in the family Flaviviridae [Thiel et al., 2005]. The genome includes a ~340 nucleotide (nt) 5′ untranslated region (UTR), a single 9 kb open reading frame (ORF) that translates into at least 10 structural and non-structural proteins and a ~300 nt 3′ UTR [Major et al., 2001]. The 5′ UTR is a highly conserved region, suitable as a target for primers and probes in diagnostic assays [Bukh et al., 1992]. Currently, six major genotypes and more than 50 subtypes of HCV have been recognized, which presents challenges for the development of RNA Nucleic Acid Technology (NAT) assays for HCV that aim to detect all the genotypes equally [Lee et al., 2000]. Existing versions of commercially available assays appear to have met this challenge [Major and Feinstone, 1997; Puig et al., 2002; Ross et al., 2002] and in addition, some have adopted the first international HCV WHO 96/790 [Saldanha et al., 1999] standard for calibration. This allows for direct comparison of results from different methods and potentially from in-house assays, in particular the new generation of quantitative TaqMan real-time PCR assays. Commercial HCV TaqMan assays have already adopted the WHO standard [Halfon et al., 2006a].
Recent reports of robust HCV infections in vitro [Lindenbach et al., 2005; Wakita et al., 2005; Zhong et al., 2005] increase the need for accurate and dynamically comparable quantitative assays, such as those described herein. We evaluated the ability of an in-house TaqMan real-time PCR method to detect the six major genotypes and selected subtypes of HCV by comparing it to two commercial quantitative NAT assays. Initially, the ability of two commercial NATs to quantify HCV reference samples of genotypes 1a, 1b, 2a, 2b, 3a, 4a, 5a, and 6a were evaluated, and serial serum samples taken from chimpanzees infected experimentally with HCV genotypes 1a, 1b, and 6a were examined. These HCV reference samples were then used to validate our in-house TaqMan real-time PCR assay. We included an evaluation of viral titers of an HCV strain that was capable of growth in cell culture [Wakita et al., 2005].
MATERIALS AND METHODS
HCV IU Standards
Two standard NAT preparations were used. The first WHO 96/790 HCV standard [Saldanha et al., 1999] is a serum preparation containing HCV, genotype 1, distributed in vials containing 50,000 IU per 0.5 ml and can be obtained from the National Institute for Biological Standards and Control (NIBSC, Tel: 01707 641000, http://www.nibsc.ac.uk/). In order to conserve valuable reagent, we prepared a 1:10 dilution of the WHO standard with nuclease-free water to make up our working standard. We also used the PeliSpy S2176 (VQC, the Netherlands) HCV RNA run control, a serum preparation containing 38,000 GE/ml (approximately 10,000 IU/ml) of genotype 1 HCV.
Prototype H Strain of HCV
A pedigreed acute phase serum sample from patient H who had post transfusion hepatitis C was tested. This sample contains a well-characterized prototype genotype 1a strain [Feinstone et al., 1981]. It contains 106.5 50% chimpanzee infectious doses (CID50) per ml of HCV and has been widely used as a reference strain [Alter et al., 1989; Farci et al., 1994; Major et al., 2001] and as challenge virus in HCV vaccine studies in chimpanzees [Forns et al., 2002].
HCV Infectious Plasma Pools
HCV infectious plasma pools were generated by infecting experimentally chimpanzees with HCV strains representing different genotypes [Bukh et al., 1998; Forns et al., 2000; Sakai et al., 2007] and collecting acute phase samples prior to seroconversion. The range in infectivity titer was 103 to 105 CID per ml (Table I). The infectivity titers of the pools were determined by reverse titration in additional chimpanzees (Bukh, et al., unpublished work). The pools tested in this study contained polyclonal strains HC-TN (genotype 1a), HC-J4 (genotype 1b), HC-J6 (genotype 2a), HC-J8 (genotype 2b), S52 (genotype 3a), ED43 (genotype 4a), SA13 (genotype 5a), HK-6a (genotype 6a), and monoclonal strain H77C (genotype 1a).
TABLE I.
Comparison of Three NAT Assays for the Quantitation of HCV RNA in Chimpanzee-Derived Plasma Pools of the Six Major Genotypes and of Selected Subtypes
| Genotype | Virus strain | log10 chimpanzee infectious doses/mla | log10 IU/ml of HCV (N, SD)
|
||
|---|---|---|---|---|---|
| HCV Monitor 2.0 | HCV b-DNA 3.0 | HCV TaqMan | |||
| 1a | H77C | 3.5 | 4.8 (2, 0.04) | 4.6 (2, 0.00) | 4.4 (2, 0.07) |
| 1a | HC-TN | 5 | 5.3 (2, 0.05) | 5.0 (2, 0.01) | 4.7 (2, 0.06) |
| 1b | HC-J4 | 3 | 4.7 (3, 0.05) | 4.4 (2, 0.00) | 4.4 (4, 0.12) |
| 2a | HC-J6 | 4 | 5.0 (3, 0.07) | 4.5 (2, 0.02) | 4.7 (4, 0.10) |
| 2b | HC-J8 | 4 | 4.6 (2, 0.13) | 4.0 (3, 0.04) | 4.0 (4, 0.42) |
| 3a | S52 | 3 | 4.3 (3, 0.02) | 3.8 (2, 0.00) | 4.0 (4, 0.49) |
| 4a | ED43 | 5 | 5.6 (3, 0.05) | 5.6 (2, 0.04) | 5.4 (4, 0.30) |
| 5a | SA13 | 4 | 5.1 (2, 0.00) | 4.8 (2, 0.01) | 4.4 (4, 0.57) |
| 6a | HK-6a | 3 | 4.9 (2, 0.05) | 4.5 (2, 0.00) | 4.2 (3, 0.41) |
Plasma pools were evaluated in two or more independent tests by the HCV Monitor 2.0, the Versant HCV RNA 3.0 b-DNA and our in-house HCV TaqMan assays and yielded results that were not significantly different (P = 0.069) based on analysis of variance. The plasma pools, representing the six major genotypes of HCV, all yielded values that were within the linear testing ranges of all three assays.
Except for HCV strain H77C, titers were obtained by reverse titration in single chimpanzees and are therefore approximate.
Chimpanzee Sera
Weekly serial serum samples collected from three chimpanzees experimentally infected with genotypes 1a (HC-TN strain; polyclonal), 1b (Con1 strain; monoclonal) and 6a (HK strain; polyclonal), respectively, were evaluated with quantitative HCV assays. The National Institutes of Health guidelines for the humane use of animals were followed throughout these studies [National Research Council, 1996].
Cell Culture Infection
We inoculated 500 focus-forming units (ffu) of wild type JFH1 [Wakita et al., 2005] into 1 million Huh7.5 [Blight et al., 2002] cells (m.o.i. = 0.0005) and maintained the cultures for 21 days. Virus replication was monitored based on HCV RNA levels in the culture medium.
Commercial Quantitative HCV Assays
The quantitative Amplicor HCV Monitor™ Test version 2.0 (Roche Diagnostics, Branchburg, NJ) was used following all manufacturers’ recommendations. Input sample volume was 100 μl. This is a target amplification test that has a linear dynamic range from 102.8 to 105.9 IU/ml without dilution (Table II). The dynamic range can be expanded upward by dilution of samples. The Versant HCV RNA 3.0 b-DNA (Bayer, Tarrytown, NY) is a signal amplification assay that can accurately quantify from 103.0 to 106.9 IU/ml without sample dilution. Input volume for the b-DNA was 50 μl per replicate and samples were run in duplicate (Table II). Both methods use the WHO international standard preparation 96/790 as a reference and results are expressed in IU/ml, allowing direct comparison with other assays.
TABLE II.
Key Features of Three NAT Assays Used in This Study
| Method | HCV b-DNA 3.0
|
HCV Monitor 2.0
|
HCV TaqMan
|
|---|---|---|---|
| Signal amplification | Target amplification (RT-PCR) | Target amplification (RT-QPCR) | |
| Measurement | Fluorescence by luminometry | End-point colorimetry | Real time fluorescence |
| Sample input volume | 50 μl | 100 μl | 10–140 μl |
| Linear range | 103.0–106.9 IU/ml | 102.8–105.9 IU/ml | 102.6–106.5 IU/ml |
| Samples per run | 48 (8) duplicates | 12 | 48 (8) duplicates |
| Approximate cost per sample | $70 | $70 | $8 |
The key features of each NAT employed in this study are listed above. The sample input for TaqMan can be adjusted depending on the nature of the sample. The number of duplicate standards and controls per run for b-DNA and TaqMan are shown in parenthesis. TaqMan chemistry allows the detection of amplification in the early stages of PCR, adding precision, whereas end-point measurements are taken at the end of the PCR. Both Monitor and TaqMan require manual total RNA extraction.
In House Quantitative TaqMan
RNA was extracted from 140 μl of serum or plasma with a commercially available kit (QIAamp Viral RNA Mini Kit, Qiagen #52906; Qiagen, Valencia, CA), and collected RNA in 60 μl. Primers and probe were selected from a region of the 5′ UTR that is highly conserved among the six major genotypes (Fig. 1) [Bukh et al., 1992]. These consisted of a forward primer (HCV-Con259F-AGY GTT GGG TYG CGA AAG) and a reverse primer (HCV-Con312R-CAC TCG CAA GCR CCC T), and a nonfluorogenic quencher (NFQ) Minor Groove Binding (MGB) probe (HCV-278MGB-6FAM CCT TGT GGT ACT GCC TGA). Two bases in the forward primer and one base in the reverse primer were degenerate to ensure identity with the major genotypes and subtypes. The NFQ MGB probe permitted the use of shorter primer/probe fragments. We optimized the forward primer, reverse primer and probe to be used at 500, 1,000, and 250 nM, respectively. The RT and PCR reactions were performed using Applied Biosystems TaqMan RT and PCR Master Mix Reagents (ABI, Foster City, CA) and each reaction included 10 μl of sample. The test was performed using an ABI 7900HT Real-Time PCR System. The manufacturer’s standard recommendations were followed with regard to cycling conditions. In order to generate a standard line for our real-time TaqMan assay, we used the 5 × 106 IU/ml panel member of the OptiQuant HCV RNA (Acrometrix, Benicia, CA) NAT reference panel. The panel members are made up of naturally occurring HCV diluted with a normal human plasma matrix. This standard was subjected to the same extraction procedure as the samples to be tested and a 1:5 dilution of the resulting total RNA was made with nuclease free water. Serial 10-fold dilutions (105–101) of the 106 IU/ml working standard were made and duplicates of each were tested in every TaqMan run, along with negative, positive and no-template controls. We included positive samples in the testable range to control for extraction efficiency; the PeliSpy, an Acrometrix panel member (104.7 IU/ml) or both were included in each test run. Data analyses were carried out using ABI’s SDS version 2.2.
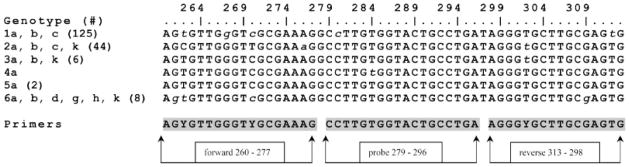
Fig. 1.
Sequence alignment of target region for TaqMan primers and probe. The highly conserved target area on the HCV 5′ UTR that was used for constructing the TaqMan primers and probe is shown. This alignment shows consensus sequences for each of the six major genotypes and subtypes. The number of sequences represented per genotype in the alignment is shown in parenthesis. The 5′ UTR of 186 full-length confirmed (as defined by the LANL HCV database) sequences were queried. Lower case letters represent mismatches in reference to the consensus sequence. Mismatches at positions 261, 267, 276, 280, 284, 309, and 312 are present in no more than two isolates per subtype and were not accounted for in the probe or primer sequences. In contrast, single nucleotide differences at positions 262 (T/C), 270 (C/T) and 302 (T/C) for genotypes 2a, 2b, 3a, and 6a were accounted for by the use of degenerate bases at those positions. Arrows and boxes highlight the selected primers and probe.
The HCV sequence data base at Los Alamos National Laboratory (http://hcv.lanl.gov/) [Kuiken et al., 2005] was consulted for initial sequence alignments during our search for suitable primers and probe. In addition, we used the Bioedit [Hall, 1999] and Accelrys DS Gene (Accelrys, Inc., San Diego, CA) suites of tools for sequence alignments. The resulting primers and probe were back-checked against 186 full-length genome sequences representing all major genotypes (Fig. 1) using the LANL HCV database tools. ABI’s Primer Express 2.3 was used to test and confirm primer and probe selections. Statistics were performed using Micro-soft Excel routines and GraphPad Prism.
RESULTS
Testing of IU Standards With b-DNA, Amplicor Monitor, and TaqMan
We designed primers and probes that are conserved among the different HCV genotypes (Fig. 1), and used them in a TaqMan assay to test samples that had been quantified with the b-DNA and Amplicor Monitor tests (Figs. 2 and 3; Table I). A 1:10 dilution of the WHO 96/790 standard, with an expected titer of 104.0 IU/ml, yielded a TaqMan titer of 103.8 IU/ml (0.33SD, 7% CV, N = 6). This was most similar to the titer of 104.0 IU/ml (N = 1) obtained with b-DNA; the titer with Amplicor Monitor was 104.3 IU/ml (0.23SD, 5% CV, N = 4). A similar pattern with a slightly higher Amplicor Monitor titer was found when we tested the PeliSpy S2176 NAT control with an expected titer of 104.0 IU/ml; we obtained titers of 104.0 (0.12SD, 3% CV, N = 6), 104.1 (0.07SD, 2% CV, N = 6) and 103.6 (0.13SD, 4% CV, N = 5) IU/ml with TaqMan, Amplicor Monitor and b-DNA, respectively.
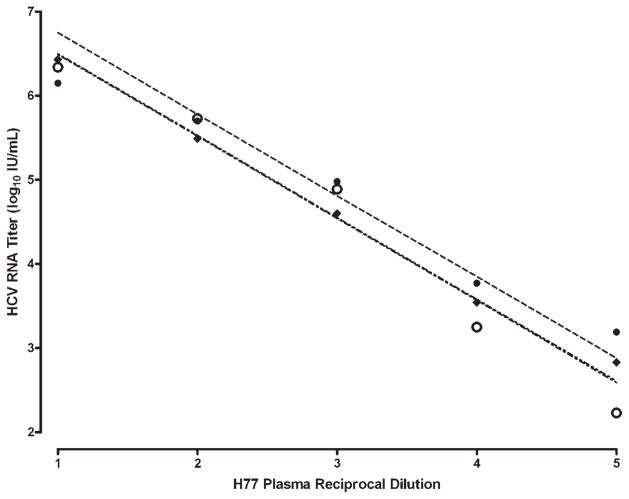
Fig. 2.
Titration of a prototype HCV H77 strain using three NAT assays for quantifying HCV RNA. The HCV H strain was tested by TaqMan (open circles), Versant HCV RNA 3.0 b-DNA (diamonds) and the HCV Monitor 2.0 (solid circles). Based on linear regression and titration values, the calculated quantity for the HCV H strain was 107.4, 107.5, and 107.7 IU/ml respectively by b-DNA, TaqMan and Amplicor Monitor, which correlated with the infectious titer (106.5 CID50/ml) of this strain as described by Feinstone et al. [1981] and Purcell (unpublished work). The slopes for b-DNA, Monitor and TaqMan (−0.975, −0.968, and −0.974) respectively, were not significantly different (P = 0.25).
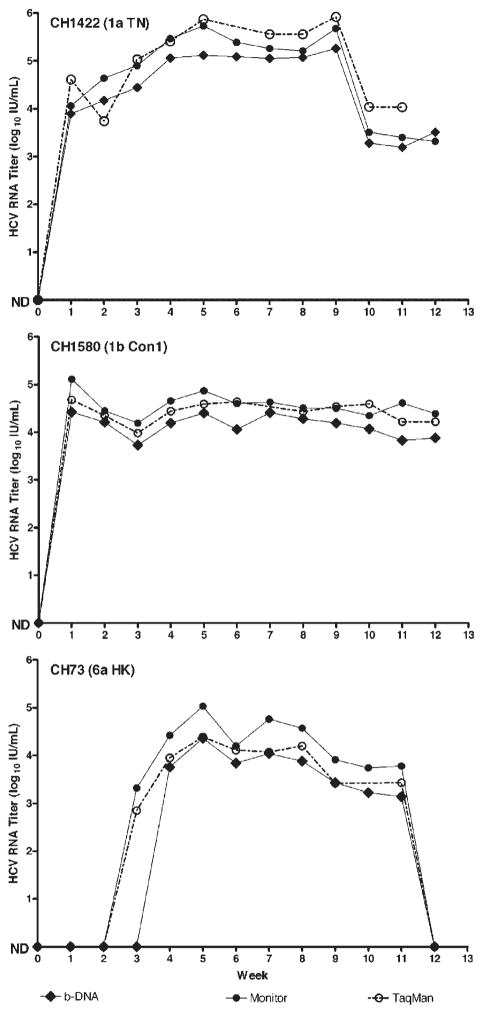
Fig. 3.
Comparison of three NAT assays for quantifying HCV RNA. Weekly samples from chimpanzees experimentally infected with HCV genotypes 1a (TN), 1b (Con1 strain), or 6a (HK strain) were tested with the Versant HCV RNA 3.0 b-DNA assay (solid diamonds), HCV Monitor 2.0 (solid circles) and TaqMan (open circles). Results are presented as HCV RNA log10 IU/ml. The Roche Monitor assay generated values approximately 0.3 log10 IU/ml higher than those of the Versant b-DNA assay throughout the dynamic range of each test.
Analysis of HCV Reference Genotype Pools and Serial Samples From Experimentally Infected Chimpanzees by b-DNA, Amplicor Monitor and TaqMan
Since both NAT standards are genotype 1 preparations, we also tested other HCV reference strains with known infectivity titers representing the six major genotypes and several subtypes of HCV to ascertain if the commercial tests and our TaqMan assay yielded comparable results. Serial 10-fold dilutions of the H77 acute-phase plasma, which is a genotype 1a prototype strain, were tested in duplicate and values within the dynamic range of the three tests generated straight-line formulas used to calculate the quantity of HCV (Fig. 2). There was a 0.3 log10 difference between the commercial assays at every quantifiable point. However the slopes were not significantly different (P = 0.25) and the lines’ goodness of fit (R2) were 0.99, 0.98, and 0.97 for b-DNA, Amplicor Monitor and TaqMan, respectively. The tight distribution of b-DNA points on its calculated line supported the consistency of this method, notwithstanding the single outlier below the b-DNA linear range (102.7 IU/ml at 10−5 dilution). Conversely, the Amplicor Monitor showed more variability that was partially corrected by removing a point above the upper linear range limit of this assay (106.7 IU/ml at 10−1 dilution). The TaqMan line superimposed the Amplicor Monitor line. Overall, the b-DNA, Amplicor Monitor, and Taq-Man yielded calculated titers of 107.4 IU/ml, 107.7 IU/ml and 107.5 IU/ml, respectively, for the H77 challenge virus. The H77 plasma HCV genome titer correlated with its infectivity titer of 106.5 CID50/ml.
The HCV titers determined by the two commercial methods and by our TaqMan assay correlated with infectivity titers in nine HCV plasma pools that represented all six major genotypes (Table I). The b-DNA and Amplicor Monitor quantities generated with genotype 1a and 4a pools showed the closest test-to-test agreement (104.6 vs. 104.8 and 105.6 vs. 105.6). Otherwise a 100.3 to 100.6 range of variation was observed, with the 2b genotype showing the greatest difference, but such differences can be attributed to a reported 0.5 log10 margin of error for these types of assays [Caliendo et al., 2006]. The TaqMan titers obtained on these HCV pools, based on two to four independent tests, were close to those obtained in the b-DNA assay with an average difference of only 100.2 IU/ml (Table I). The genotype 5a plasma pool showed the greatest difference at 100.4 IU/ml. TaqMan yielded lower values than Amplicor Monitor in every case, averaging a 100.5 IU/ml difference and with genotypes 5a and 6a showing the largest difference (100.7 IU/ml). However, one-way analysis of variance (ANOVA) of the three methods’ quantitations for the infectious HCV genotype pools showed no significant difference (P = 0.069).
Weekly samples from chimpanzees experimentally infected with HCV genotypes 1a (TN strain), 1b (Con1 strain) and 6a (HK strain) were also tested with both commercial methods and TaqMan (Fig. 3). The Amplicor Monitor generated IU values about 0.3 log10 higher than b-DNA for all three genotypes. Two-tailed t tests revealed that the means for genotypes 1b and 6a, respectively, were not different (P = 0.237 and 0.217) with the two tests, but they were significantly different for 1a (P = 0.002). The initial appearance of HCV at week three in the chimpanzee infected with genotype 6a was detected by Amplicor Monitor, suggesting a higher sensitivity. The TaqMan quantitation of these serial serum samples yielded the same pattern as seen with the two commercial tests (Fig. 3), and as with Monitor, included earlier detection at week 3 of the genotype 6a infection. Two-tailed t tests demonstrated that TaqMan versus Amplicor Monitor results were not significantly different for the 1a, 1b or 6a infected chimpanzees (P = 0.289, 0.719, and 0.445), respectively. TaqMan versus b-DNA values for the 1b and 6a infected chimpanzees were also not significantly different (P = 0.295 and 0.548, respectively). However the 1a results showed a significant difference (P = 0.014) between b-DNA and TaqMan.
Assessment of HCV RNA Titers In Virus-Infected Huh7.5 Cell Cultures
Samples taken every other day from the Huh7.5 cell cultures inoculated with 500 ffu of JFH1 HCV were assayed with TaqMan and Amplicor Monitor (Fig. 4). Residual input HCV RNA was detected up to day 4 by both methods. Rising titers from day 4 to 21 ranged from 103.7 to 106.0 IU/ml when measured by Amplicor Monitor and from 103.1 to 106.7 IU/ml when measured by Taq-Man. The peak HCV titer at day 21 fell above the linear range of Amplicor Monitor, but within the linear dynamic range of TaqMan. Nevertheless, pair-wise comparison of the parallel data points showed no significant difference (P = 0.5).
Fig. 4.
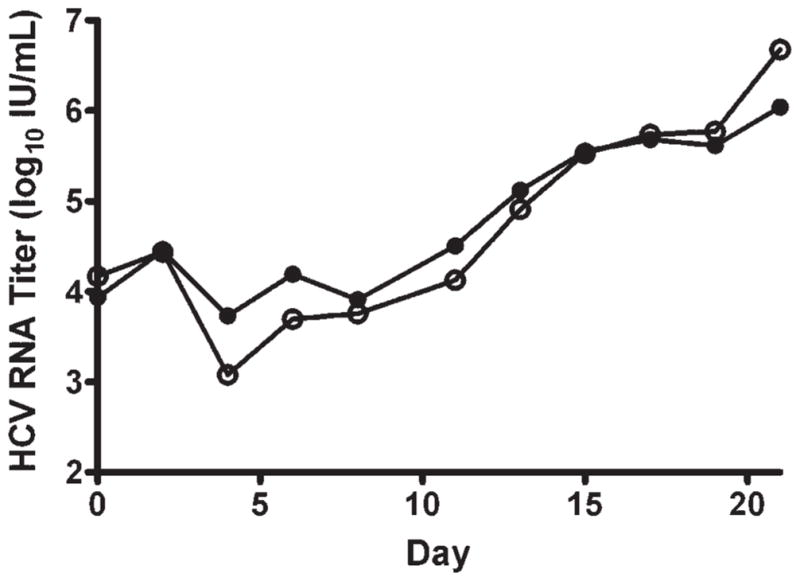
Production of HCV in Huh7.5 cells. Samples from cultures inoculated with 500 ffu of JFH1 into 1 million Huh7.5 cells (m.o.i. = 0.0005) and maintained for 21 days were assayed with TaqMan (open circles) and HCV Monitor (solid circles). Pairwise comparison of the parallel data points showed no significant difference (P = 0.5).
Using a secondary cell-culture derived HCV control calibrated to 106.5 IU/ml we tested six 10-fold dilutions in duplicate over four independent TaqMan runs to evaluate a linear dynamic range and assay precision. We determined the candidate linear range to be 106.5–102.6 IU/ml (slope = −0.9455, R2 = 0.994) and the inter-assay precision averaged 3% CV. Intra-assay precision was <2% CV.
DISCUSSION
In this study we confirmed performance of two commercial NAT methods (Versant HCV RNA 3.0 b-DNA and Amplicor HCV Monitor) for quantifying HCV RNA in two well-characterized genotype 1 HCV IU standards. In addition, we demonstrated that these methods produced HCV RNA titers that correlated with the known infectivity titers of plasma pools that represented the major HCV genotypes 1a, 1b, 2a, 2b, 3a, 4a, 5a, and 6a. This correlation was not reported previously for the different genotypes of HCV. Both assays performed well within the limits set by each manufacturer. For example, there was little intra-assay variation, irrespective of genotype. We observed some variation between methods, approximately 0.3 log10 to 0.6 log10 IU/ml, which agrees with findings by others that included testing HCV isolates of genotypes 1–5, and observing that the Amplicor Monitor yields higher values than b-DNA [Bresters et al., 1994; Ross et al., 2002; Caliendo et al., 2006]. In contrast, Halfon et al. [2006b] tested patients infected with genotypes 1–3 and reported that despite these methods’ being calibrated to the 96/790 WHO standard, they generated different results, regardless of genotype, with the b-DNA assay yielding higher titers than the Amplicor Monitor. In the same report, a Roche TaqMan assay produced higher titers than either b-DNA or Amplicor Monitor early during antiviral treatment of chronic infection and lower values 4 and 12 weeks after initiation of treatment. It is likely that some of the differences between methods are linked to secondary standards prepared by each manufacturer for assay quantitation, since in our hands the two commercial assays yielded a similar difference when tested against two different IU standards.
Overall both end-point methods are useful and accurate for the study of HCV [Saito et al., 1990; Gretch, 1997; Castro et al., 2001; Anderson et al., 2003; Caliendo et al., 2006; Halfon et al., 2006a]. Commercial TaqMan assays are also available; they perform at higher sensitivity than the analogous end-point assays [Halfon et al., 2006a,b] and they can detect the major genotypes. For example both Abbott laboratories and Roche HCV TaqMan assays claim to detect the major genotypes of HCV with high sensitivity. However, a report indicated that these assays underestimate the titer of genotypes 3 and 4 [Caliendo et al., 2006]. Konnick et al. reported on the Cobas HCV Analyte Specific Reagent (ASR) Taq-Man assay, which increases the sensitivity of TaqMan and uses an automated extraction process. The ASR performed well across genotypes 1–5 with regard to linearity and precision, but performed poorly on a sample-to-sample basis when compared with results from b-DNA and Amplicor Monitor [Konnick et al., 2002]. Anderson et al. described an inter-laboratory testing survey that included samples representing genotypes 1a, 1b, 2b, 3a, and 3b. The comparison of viral load as measured by a TaqMan method, b-DNA and Amplicor Monitor yielded only good to moderate correlation [Anderson et al., 2003]. Some authors [Anderson et al., 2003; Halfon et al., 2006a] discouraged the use of results from two different tests in the same analysis, in view of the differences seen in quantitative results. In spite of a common IU standard, quantitative results still vary between methods. Therefore, when following viral load during treatment or evaluating kinetics of in vitro infection it is advisable to use the same method.
The nature of b-DNA; namely specific and multiple hybridization steps, chemiluminescence and no PCR; results in more consistency than PCR-based assays. In contrast, Amplicor HCV Monitor can be more sensitive—in part due to PCR—but has a shallow upper dynamic range. Additionally, Monitor’s labor-intensive manual extraction step adds to the potential for variability. Nevertheless, it is recognized that the precision of b-DNA and Amplicor Monitor is superior to most other assays and in the absence of assay gold standards for the different HCV genotypes, we found it valid to use the combined power of these two commercial assays to help evaluate our new TaqMan method.
Although our TaqMan assay suffers from the same problems inherent in PCR assays, such as extraction efficiency and variability at the low end of sensitivity, it offers advantages, such as standardized extraction procedures, straightforward closed-tube RT and PCR reactions and ability to include internal or external standards. Our results when testing HCV plasma pools and serial samples from HCV-infected chimpanzees were not significantly different from either of the commercial end-point methods. Our TaqMan inter-assay precision, as reflected by higher standard deviations for some of the plasma pools encourage us to monitor more closely quality control measures, particularly during extraction. On the other hand, close intra-assay precision suggests the RT/PCR element of our TaqMan assay is highly reproducible.
TaqMan can be customized, for example, to perform multiplex assays with the same sensitivity of Amplicor Monitor and the same linear dynamic range of b-DNA. Of particular importance to us was the ability to quantify all the major HCV genotypes and subtypes. A clear advantage of the TaqMan assay is the lower cost associated with development and testing by this methodology compared with the two commercial assays used. However each of these assays is useful for evaluation of HCV infections and the decision of which method to employ rests on understanding the limitations of each assay.
In this study, we validated a new TaqMan assay that can quantify isolates of all six genotypes of HCV. The TaqMan assay performed well and results matched those obtained with the commercial tests and will provide a useful tool in HCV infectivity studies, and as documented in in vitro studies using the JFH1 based cell culture systems.
Acknowledgments
Grant sponsor: NIAID, NIH.
We thank Harvey Alter (NIH, USA) for providing the H strain of HCV, Takaji Wakita (National Institute of Infectious Diseases, Japan) for providing the JFH1 construct and Charles Rice (The Rockefeller University, New York) for providing the Huh7.5 cells. RSR is a recipient of a Fellowship from the Canadian Institutes of Health Research. This research was supported by the Intramural Research Program of the NIAID, NIH.
References
- Alter HJ, Purcell RH, Shih JW, Melpolder JC, Houghton M, Choo QL, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494–1500. doi: 10.1056/NEJM198911303212202. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Simonetti J, Fisher DG, Williams J, Yamamura Y, Rodriguez N, Sullivan DG, Gretch DR, McMahon B, Williams KJ. Comparison of different HCV viral load and genotyping assays. J Clin Virol. 2003;28:27–37. doi: 10.1016/s1386-6532(02)00235-4. [DOI] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresters D, Cuypers HT, Reesink HW, Mauser-Bunschoten EP, van den Berg HM, Schaasberg WP, Wilber JC, Urdea MS, Neuwald P, Lelie PN. Comparison of quantitative cDNA-PCR with the branched DNA hybridization assay for monitoring plasma hepatitis C virus RNA levels in haemophilia patients participating in a controlled interferon trial. J Med Virol. 1994;43:262–268. doi: 10.1002/jmv.1890430313. [DOI] [PubMed] [Google Scholar]
- Bukh J, Purcell RH, Miller RH. Importance of primer selection for the detection of hepatitis C virus RNA with the polymerase chain reaction assay. Proc Natl Acad Sci USA. 1992;89:187–191. doi: 10.1073/pnas.89.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh J, Apgar CL, Engle R, Govindarajan S, Hegerich PA, Tellier R, Wong DC, Elkins R, Kew MC. Experimental infection of chimpanzees with hepatitis C virus of genotype 5a: Genetic analysis of the virus and generation of a standardized challenge pool. J Infect Dis. 1998;178:1193–1197. doi: 10.1086/515683. [DOI] [PubMed] [Google Scholar]
- Caliendo AM, Valsamakis A, Zhou Y, Yen-Lieberman B, Andersen J, Young S, Ferreira-Gonzalez A, Tsongalis GJ, Pyles R, Bremer JW, Lurain NS. Multilaboratory comparison of hepatitis C virus viral load assays. J Clin Microbiol. 2006;44:1726–1732. doi: 10.1128/JCM.44.5.1726-1732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro FJ, Sauleda S, Esteban JI, Viladomiu L, Martell M, Dragon E, Esteban R, Guardia J. Evaluation of hepatitis C virus RNA RT/PCR qualitative and quantitative second generation assays. J Virol Methods. 2001;91:51–58. doi: 10.1016/s0166-0934(00)00243-3. [DOI] [PubMed] [Google Scholar]
- Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, Shapiro M, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstone SM, Alter HJ, Dienes HP, Shimizu Y, Popper H, Blackmore D, Sly D, London WT, Purcell RH. Non-A, non-B hepatitis in chimpanzees and marmosets. J Infect Dis. 1981;144:588–598. doi: 10.1093/infdis/144.6.588. [DOI] [PubMed] [Google Scholar]
- Forns X, Payette PJ, Ma X, Satterfield W, Eder G, Mushahwar IK, Govindarajan S, Davis HL, Emerson SU, Purcell RH, Bukh J. Vaccination of chimpanzees with plasmid DNA encoding the hepatitis C virus (HCV) envelope E2 protein modified the infection after challenge with homologous monoclonal HCV. Hepatology. 2000;32:618–625. doi: 10.1053/jhep.2000.9877. [DOI] [PubMed] [Google Scholar]
- Forns X, Bukh J, Purcell RH. The challenge of developing a vaccine against hepatitis C virus. J Hepatol. 2002;37:684–695. doi: 10.1016/s0168-8278(02)00308-2. [DOI] [PubMed] [Google Scholar]
- Gretch DR. Diagnostic tests for hepatitis C. Hepatology. 1997;26:43S–47S. doi: 10.1002/hep.510260708. [DOI] [PubMed] [Google Scholar]
- Halfon P, Bourliere M, Penaranda G, Khiri H, Ouzan D. Real-time PCR assays for hepatitis C virus (HCV) RNA quantitation are adequate for clinical management of patients with chronic HCV infection. J Clin Microbiol. 2006a;44:2507–2511. doi: 10.1128/JCM.00163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon P, Penaranda G, Bourliere M, Khiri H, Masseyeff MF, Ouzan D. Assessment of early virological response to antiviral therapy by comparing four assays for HCV RNA quantitation using the international unit standard: Implications for clinical management of patients with chronic hepatitis C virus infection. J Med Virol. 2006b;78:208–215. doi: 10.1002/jmv.20529. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Konnick EQ, Erali M, Ashwood ER, Hillyard DR. Performance characteristics of the COBAS Amplicor Hepatitis C Virus (HCV) Monitor, Version 2.0, International Unit assay and the National Genetics Institute HCV Superquant assay. J Clin Microbiol. 2002;40:768–773. doi: 10.1128/JCM.40.3.768-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken C, Yusim K, Boykin L, Richardson R. The Los Alamos hepatitis C sequence database. Bioinformatics. 2005;21:379–384. doi: 10.1093/bioinformatics/bth485. [DOI] [PubMed] [Google Scholar]
- Lee SC, Antony A, Lee N, Leibow J, Yang JQ, Soviero S, Gutekunst K, Rosenstraus M. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: Calibration to international units, enhanced genotype reactivity, and performance characteristics. J Clin Microbiol. 2000;38:4171–4179. doi: 10.1128/jcm.38.11.4171-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Major ME, Feinstone SM. The molecular virology of hepatitis C. Hepatology. 1997;25:1527–1538. doi: 10.1002/hep.510250637. [DOI] [PubMed] [Google Scholar]
- Major ME, Rehermann B, Feinstone SM. Hepatitis C viruses. In: Knipe DM, Howley PM, editors. Fields virology. 4. Ch 34 Philadelphia: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- National Research Council. Guide for the care and use of laboratory animals. National research council; Washington, D.C: National Academy Press; 1996. [Google Scholar]
- Puig M, Mihalik K, Yu MY, Feinstone SM, Major ME. Sensitivity and reproducibility of HCV quantitation in chimpanzee sera using TaqMan real-time PCR assay. J Virol Methods. 2002;105:253–263. doi: 10.1016/s0166-0934(02)00119-2. [DOI] [PubMed] [Google Scholar]
- Ross RS, Viazov S, Sarr S, Hoffmann S, Kramer A, Roggendorf M. Quantitation of hepatitis C virus RNA by third generation branched DNA-based signal amplification assay. J Virol Methods. 2002;101:159–168. doi: 10.1016/s0166-0934(01)00433-5. [DOI] [PubMed] [Google Scholar]
- Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y, et al. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Takikawa S, Thimme R, Meunier JC, Spangenberg HC, Govindarajan S, Farci P, Emerson SU, Chisari FV, Purcell RH, Bukh J. In vivo study of the HC-TN strain of hepatitis C virus recovered from a patient with fulminant hepatitis: RNA transcripts of a molecular clone (pHC-TN) are infectious in chimpanzees but not in Huh7.5 cells. J Virol. 2007;81:7208–7219. doi: 10.1128/JVI.01774-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha J, Lelie N, Heath A. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. WHO Collaborative Study Group. Vox Sang. 1999;76:149–158. doi: 10.1159/000031040. [DOI] [PubMed] [Google Scholar]
- Thiel HJ, Collett MS, Gould EA, Heinz FX, Meyers G, Purcell RH, Rice CM, Houghton M. Virus taxonomy. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus taxonomy. 8. San Diego: Elsevier; 2005. pp. 993–998. [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]