Abstract
Aberrant PD‐L1 (CD274) expression has been described in different types of tumour and linked to tumour aggressiveness and a poor prognosis. In primary colorectal carcinomas (CRCs), CD274 expression was reported to be associated with mismatch repair (MMR)‐deficiency, BRAF mutation, and “stem‐like” immunophenotype defined by down‐regulation of homeobox protein CDX2 and membranous expression of activated leukocyte cell adhesion molecule (ALCAM). However, the immunophenotype and genotype of CD274‐positive metastatic CRC have not been extensively analysed. In this study, 189 CRC metastases were evaluated immunohistochemically for CD274, MMR proteins, CDX2, and ALCAM expression. Immunostaining for CD4, CD8, and FOXP3 was also performed to characterize tumour‐associated immune cells. In addition, 34 arbitrarily selected lesions were genotyped using Sanger‐ and next‐generation sequencing. Univariate analyses showed no clear association between CD274 expression and clinicopathological parameters including MMR‐deficiency or “stem‐like” immunophenotype after adjustment for multiple testing. Comparison of the clinicopathological profiles of CD274‐positive primary and metastatic tumours revealed in the latter younger age of occurrence (60.9 ± 13.3 versus 72.6 ± 13.1 years, p = 0.001), cytoplasm‐dominant CD274 expression (p < 0.001), infrequent MMR‐deficiency (p < 0.001), and common KRAS mutations (54%, p < 0.001). In five cultured colon cancer cell lines, CD274 was expressed and modulated after exogenous exposure to IFNγ and TGF‐β1. Thus, CD274 regulation mechanisms might include tumour micro environmental factors. Based on significantly different characteristics in CD274‐positive metastatic and primary CRCs, evaluation of metastases should also be considered when planning immune checkpoint inhibitor therapy.
Keywords: metastatic colorectal carcinoma, immunohistochemistry, CD274 (PD‐L1), BRAF, KRAS, CDX2, ALCAM (CD166), IFNγ, TGF‐β1
Introduction
The PD‐Ls (programmed cell death ligands)/PDCD1 (programmed cell death 1, PD‐1) axis is crucial for the modulation of the immune system to reduce peripheral tissue damage from excess inflammatory responses. The PD‐Ls/PDCD1 axis has also been reported to play a pivotal role in the maintenance of self‐tolerance to avoid autoimmune diseases 1, 2.
CD274 (B7‐H1, PD‐L1) has been identified as a cell‐surface glycoprotein belonging to the B7 family. Physiologically, CD274 is primarily induced by interferon‐γ (IFNγ) from T helper 1 (TH1) cells under inflammatory conditions 3, 4, 5, 6.
Variable CD274 expression has been reported in different types of tumour, including oesophageal, gastric, colorectal, and lung cancers. In some cases, CD274 expression was linked to tumour aggressiveness and a poor prognosis 7, 8, 9, 10. In primary colorectal carcinomas (CRCs), CD274 expression was reported to be associated with mismatch repair (MMR)‐deficiency, BRAF mutation, and “stem‐like” immunophenotype defined by down‐regulation of CDX2 (caudal‐type homeobox transcription factor 2), an intestinal differentiation marker, and membranous expression of activated leukocyte cell adhesion molecule (ALCAM, CD166), a stem cell marker 11, 12, 13, 14. However, those characteristics have not been fully evaluated in metastatic colorectal cancer.
In this study, a cohort of metastatic CRCs was evaluated immunohistochemically for CD274, MMR‐proteins, CDX2, and ALCAM expression. Immune phenotypes of tumour‐infiltrating inflammatory cells were also assessed by CD4, CD8, and FOXP3 immunostaining. In addition, 34 arbitrarily selected metastases were screened for gain‐of‐function mutations in the BRAF, KRAS, NRAS, and PIK3CA oncogenes. Phenotypic and genotypic characteristics of the two CD274‐positive cohorts consisting of metastatic CRCs evaluated in this study and previously reported primary tumours 13 were compared. Furthermore, to evaluate the regulation mechanism(s) of CD274 in colon cancer cells, CD274 expression, and responsiveness to exogenous INFγ and TGF‐β1 were examined.
This study attempts to identify the clinicopathological profile of metastatic CRCs potentially targetable by immune checkpoint inhibitors against the CD274/PDCD1 axis.
Materials and methods
Tissue samples
A total of 189 anonymized metastatic CRCs were collected. This project was completed under Office of Human Subject Research Exemption with anonymized specimens. Tumours were assembled into multitumour blocks containing approximately 50 rectangular tissue samples as previously described 15. The size of tumour tissue samples was estimated to exceed the size of a single 0.6 mm2 core by a factor of 10–15.
All cases were extensively characterized. Tumour differentiation, mucus production, solid/sheet‐like proliferation, and existence of tumour‐associated immune cells (TAIs), at least 50 TAIs/high‐power field (HPF) within or adjacent tumour foci, were evaluated in haematoxylin and eosin (H&E) stained sections.
Immunohistochemistry
The antibodies used for immunohistochemistry are summarized in supplementary material, Table S1, available online. All immunostaining was performed with Leica Bond‐Max automation and the Leica Refine detection kit (Leica Biosystems, Bannockburn, IL, USA) as previously reported 10, 13, 16. Immunoreactivity of CD274 (membrane and/or cytoplasm) and ALCAM (membrane) was evaluated with a detection cut‐off of 5% according to our previous reports 10, 13. Representative photographs for CD274‐positive metastatic CRC cases are shown in Figure 1. CDX2 immune reactivity (nucleus) was classified into two categories; positive (same or stronger than the normal colonic mucosa) and down‐regulated (weaker than the normal colonic mucosa or loss of expression). “Stem‐like” immunophenotype was defined by CDX2‐down‐regulation and ALCAM‐positivity (representative photographs in supplementary material, Figure S1). Immunohistochemistry for MMR proteins (MLH1, MSH2, MSH6, and PMS2) was performed as previously reported 10, 13. A threshold of ≥ 50 positive TAIs/HPF was used to define CD4, CD8, and FOXP3‐positive cases.
Figure 1.
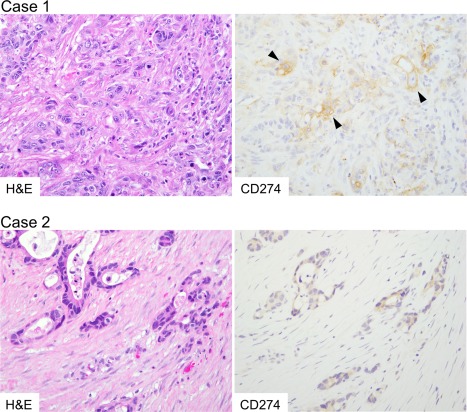
Histology and CD274 expression of metastatic CRCs. Case 1, a case of liver metastasis. Tumour tissue contained highly atypical tumour cells with CD274 expression on the cell membrane and in the cytoplasm (arrow heads). KRAS p.G12D (GGT to GAT) mutation was identified by gene mutation analysis. Case 2, a case of ovarian metastasis. Tumour cells are invading into desmoplastic stroma. Tumour cells showed cytoplasm‐dominant expression of CD274. In this case, BRAF p.V600E mutation was identified.
Genotyping
Sanger sequencing for BRAF, KRAS, and NRAS was performed on 34 arbitrarily selected metastatic lesions and cell lines as previously reported 13, 16. Primer sequences, PCR conditions and size of amplicons, are provided in supplementary material, Table S2. Nine metastases, carrying wild‐type BRAF, KRAS, and NRAS genes, were subsequently evaluated by next‐generation sequencing (NGS) using the Ion AmpliSeq™ Cancer Hotspot Panel v2 Kit (targeting 50 known oncogenes and tumour suppressor genes) and Ion Torrent™ platform following the manufacturer's instructions (Life Technologies/Thermo Fisher Scientific, Waltham, MA, USA) and previously published procedure 17. All experiments were carried out in a blinded manner.
Statistical analysis
Histopathological, immunohistochemical, and genetic data for primary CRCs were derived from our previous study 13. Chi‐square or Fisher's exact test was performed with EZR version 1.32 software 18 to analyse the statistical correlation between categorical data. Simple Bonferroni correction for multiple hypothesis testing was applied to generate an adjusted two‐sided alpha level. Cases with missing information were eliminated from the statistical analysis of that parameter.
Cell culture, reverse transcription quantitative polymerase chain reaction (RT‐qPCR), immunoblot assay, and Fluorescence‐ activated cell sorting (FACS) analysis
The human colon cancer cell lines COLO205, CW‐2, HCT116, and LoVo were obtained from the RIKEN BioResource Center (Tsukuba, Japan). SW480 was from American Type Culture Collection (ATCC, Manassas, VA, USA). BRAF and KRAS mutations were previously reported in these cell lines; BRAF p.V600E in COLO205, KRAS p.G13D in HCT116, KRAS p.G12V in SW480, and KRAS p.G13D and A14V in LoVo 19, 20. Identical BRAF and KRAS mutations were identified in the cell line cultures used in this study (data not shown).
Cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) with a supplement of 10% fetal bovine serum. IFNγ or TGF‐β1 (R&D systems/Thermo Fisher Scientific, Waltham, MA, USA) were added to the culture medium at the indicated concentration and duration. JAK inhibitor I (Calbiochem/Merck Millipore, Darmstadt, Germany) blockade was performed an hour prior to IFNγ addition. Total RNAs and whole‐cell lysates were prepared and subjected to RT‐qPCR and immunoblot analyses using a previously reported procedure 19, 20, 21. Details of the antibodies for immunoblot assays are summarized in supplementary material, Table S3. Signal intensity was measured using ImageJ software (NIH, Bethesda, MD, USA).
In FACS analyses, PE‐conjugated anti‐CD274 antibody (BioLegend, Inc., San Diego, CA, USA) was applied to harvested colon cancer cells at a dilution of 1:20 for an hour on ice. After washing and staining with 7‐AAD (Beckman Coulter, Inc., Brea, CA, USA), the cells were analysed using a FACSCanto II flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Collected data were analysed by FlowJo 7.6.5 software (Tomy Digital Biology Co., Ltd., Taito‐ku, Japan).
Results
Clinicopathological and immunohistochemical characteristics of metastatic CRCs
One hundred eighty‐nine metastatic CRCs including 110 (58%) liver metastases were evaluated in this study. Clinical data are summarized in Table 1, while Table 2 presents pathological and immunohistochemical characteristics according to CD274 expression. CD274‐positive metastatic CRCs had a tendency to occur in female patients, display poorly differentiated histology and mucus production with rare TAIs. Immunohistochemical analyses showed that CD274‐positive metastases contained fewer CD4‐positive lymphocytes. However, there was no significance after multiple testing adjustment.
Table 1.
Clinical characteristics of 189 metastatic CRCs
| Total no. | |
|---|---|
| 189 (100%) | |
| Sex (%) | |
| Male | 94 (53%) |
| Female | 86 (47%) |
| Age, years (median) | 30–91 (65) |
| Metastatic sites (%) | |
| Connective tissues | |
| Mesentery | 14 (7%) |
| Soft tissue | 11 (6%) |
| Digestive system | |
| Gastrointestinal tract | 3 (2%) |
| Liver | 110 (58%) |
| Pancreas | 2 (1%) |
| Haematopoietic and lymphatic systems | |
| Lymph nodes | 8 (4%) |
| Spleen | 2 (1%) |
| Neural and endocrine systems | |
| Adrenal gland | 1 (0.5%) |
| Brain | 10 (5%) |
| Respiratory system | |
| Lung | 13 (7%) |
| Urogenital system | |
| Kidney | 1 (0.5%) |
| Ovary | 14 (7%) |
Table 2.
Clinicopathological and immunohistochemical characteristics of 189 metastatic CRCs with or without CD274 expression
| CD274 | |||||||
|---|---|---|---|---|---|---|---|
| Total no. | Positive | Negative | |||||
| 189 | (100%) | 26 | (14%) | 163 | (86%) | P value | |
| Sex | 0.042 * | ||||||
| Male | 94 | [53%] | 8 | [32%] | 86 | [56%] | |
| Female | 84 | [47%] | 17 | [68%] | 67 | [44%] | |
| Age, years (mean ± SD) | 63.4 ± 11.6 | 60.9 ± 13.3 | 63.9 ± 11.3 | 0.24 † | |||
| Metastatic sites | 0.75 ‡ | ||||||
| Connective tissues | 25 | [13%] | 4 | [15%] | 21 | [13%] | |
| Digestive systems | 115 | [61%] | 15 | [58%] | 100 | [61%] | |
| Haematopoietic and lymphatic systems | 10 | [5%] | 1 | [4%] | 9 | [6%] | |
| Neural and endocrine systems | 11 | [6%] | 1 | [4%] | 10 | [6%] | |
| Respiratory system | 13 | [7%] | 1 | [4%] | 12 | [7%] | |
| Urogenital systems | 15 | [8%] | 4 | [15%] | 11 | [7%] | |
| Tumour differentiation | 0.032 ‡ | ||||||
| Well to moderate | 161 | [85%] | 18 | [69%] | 143 | [88%] | |
| Poor | 28 | [15%] | 8 | [31%] | 20 | [12%] | |
| Mucus production | 0.012 ‡ | ||||||
| 0–49% | 173 | [92%] | 20 | [77%] | 153 | [94%] | |
| 50%– | 16 | [8%] | 6 | [23%] | 10 | [6%] | |
| Solid/sheet‐like proliferation | 1 ‡ | ||||||
| 0–49% | 173 | [92%] | 24 | [92%] | 149 | [91%] | |
| 50%– | 16 | [8%] | 2 | [8%] | 14 | [9%] | |
| Tumour‐associated immune cells | 0.050 * | ||||||
| Present | 123 | [65%] | 12 | [46%] | 111 | [68%] | |
| Absent | 66 | [35%] | 14 | [54%] | 52 | [32%] | |
| CD4‐positive lymphocytes | 0.014 * | ||||||
| Present | 88 | [47%] | 6 | [23%] | 82 | [51%] | |
| Absent | 98 | [53%] | 20 | [77%] | 78 | [49%] | |
| CD8‐positive lymphocytes | 0.26 * | ||||||
| Present | 94 | [51%] | 10 | [38%] | 84 | [53%] | |
| Absent | 92 | [49%] | 16 | [62%] | 76 | [48%] | |
| FOXP3‐positive lymphocytes | 0.63 * | ||||||
| Present | 39 | [21%] | 4 | [15%] | 35 | [22%] | |
| Absent | 148 | [79%] | 22 | [85%] | 126 | [78%] | |
| MMR status | 0.42 ‡ | ||||||
| Deficient | 14 | [7%] | 3 | [12%] | 11 | [7%] | |
| Preserved | 175 | [93%] | 23 | [88%] | 152 | [93%] | |
| CDX2 | 0.075 ‡ | ||||||
| Down‐regulated | 28 | [15%] | 7 | [27%] | 21 | [13%] | |
| Positive | 161 | [85%] | 19 | [73%] | 142 | [87%] | |
| ALCAM | 0.23 * | ||||||
| Positive | 38 | [20%] | 8 | [31%] | 30 | [18%] | |
| Negative | 151 | [80%] | 18 | [69%] | 133 | [82%] | |
| “Stem‐like” immune phenotype | 0.63 ‡ | ||||||
| CDX2‐down‐regulated and ALCAM‐positive | 10 | [5%] | 2 | [8%] | 8 | [5%] | |
| CDX2‐positive and/or ALCAM‐negative | 179 | [95%] | 24 | [92%] | 155 | [95%] | |
The Bonferroni‐corrected P value for significance was p ≈ 0.0036 (0.05/14).
*P values were calculated using the Chi‐square test.
† t‐test was used to compare the means of age.
‡ P values were calculated using Fisher's exact test.
Comparison between CD274‐positive primary and metastatic CRCs
Table 3 outlines the clinicopathological characteristics of 26 CD274‐positive metastatic CRCs and previously reported 54 CD274‐positive primary colorectal tumours 13. The patient cohorts for metastatic and primary tumours were different. The mean age of the patients with metastatic lesions (60.9 ± 13.3 years) was significantly lower than those with primary lesions (72.6 ± 13.1 years, p = 0.001). Metastatic tumours showed dominant cytoplasmic CD274 expression (88% versus 44%, p < 0.001). Most CD274‐positive primary tumours (74%, [40/54]) carried a MMR‐deficient phenotype, while MMR‐deficiency was rarely detected (12%, [3/26], p < 0.001) in metastatic CD274‐positive lesions. Also, CRC metastases displayed solid/sheet‐like proliferation (8% versus 37%), CDX2‐down‐regulation (27% versus 54%), and “stem‐like” immune phenotype (8% versus 31%) less frequently than primary colorectal tumours. Although no significant difference was detected, ALCAM positivity was lower in metastatic CRCs (31% versus 48%).
Table 3.
Clinicopathological and immunohistochemical characteristics of CD274‐positive CRCs
| Total no. | Metastatic CRCs | Primary CRCs | |||||
|---|---|---|---|---|---|---|---|
| 80 | (100%) | 26 | (33%) | 54 | (68%) | P value | |
| Sex | 0.77 * | ||||||
| Male | 28 | [36%] | 8 | [32%] | 20 | [38%] | |
| Female | 49 | [64%] | 17 | [68%] | 32 | [62%] | |
| Age, years (mean ± SD) | 68.8 ± 14.2 | 60.9 ± 13.3 | 72.6 ± 13.1 | 0.001 † | |||
| Tumour differentiation | 0.13 * | ||||||
| Well to moderate | 44 | [56%] | 18 | [69%] | 26 | [48%] | |
| Poor | 35 | [44%] | 8 | [31%] | 28 | [52%] | |
| Mucus production | 0.37 ‡ | ||||||
| 0–49% | 66 | [83%] | 20 | [77%] | 46 | [85%] | |
| 50%– | 14 | [18%] | 6 | [23%] | 8 | [15%] | |
| Solid/sheet‐like proliferation | 0.013 * | ||||||
| 0–49% | 58 | [73%] | 24 | [92%] | 34 | [63%] | |
| 50%– | 22 | [28%] | 2 | [8%] | 20 | [37%] | |
| CD274 localization | <0.001 * | ||||||
| Membrane | 33 | [41%] | 3 | [12%] | 30 | [56%] | |
| Cytoplasm | 47 | [59%] | 23 | [88%] | 24 | [44%] | |
| MMR status | <0.001 * | ||||||
| Deficient | 43 | [54%] | 3 | [12%] | 40 | [74%] | |
| Preserved | 37 | [46%] | 23 | [88%] | 14 | [26%] | |
| CDX2 | 0.044 * | ||||||
| Down‐regulated | 36 | [15%] | 7 | [27%] | 29 | [54%] | |
| Positive | 44 | [85%] | 19 | [73%] | 25 | [46%] | |
| ALCAM | 0.22 * | ||||||
| Positive | 34 | [43%] | 8 | [31%] | 26 | [48%] | |
| Negative | 46 | [58%] | 18 | [69%] | 28 | [52%] | |
| “Stem‐like” immune phenotype | 0.039 * | ||||||
| CDX2‐down‐regulated and ALCAM‐positive | 19 | [24%] | 2 | [8%] | 17 | [31%] | |
| CDX2‐positive and/or ALCAM‐negative | 61 | [76%] | 24 | [92%] | 37 | [69%] | |
CRCs, colorectal carcinomas. The Bonferroni‐corrected P value for significance was p = 0.005 (0.05/10).
* P values were calculated using the Chi‐square test.
† t‐test was used to compare the means of age.
‡ P values were calculated using Fisher's exact test.
Mutation analyses of metastatic CRCs
Thirty‐four arbitrarily selected metastatic lesions including 13 CD274‐positive tumours were evaluated for BRAF, KRAS, and NRAS mutations using Sanger sequencing. The results are summarized in supplementary material, Table S4. In seven CD274‐positive metastases, KRAS mutations, p.G12V (n = 3), p.G12D (n = 2), p.G12C (n = 1), and p.G13D (n = 1) were identified. Three CD274‐expressing metastases were BRAF p.V600E mutants. Subsequently, nine BRAF‐ and RAS‐wild‐type metastases were evaluated by NGS. In two cases, PIK3CA p.H1047R mutation was detected whereas seven KRAS, NRAS, BRAF, and PIK3CA (quadruple negative) wild‐type lesions carried mutations affecting TP53 (n = 3), CDKN2A (n = 2), APC (n = 1), ATM (n = 1), FLT3 (n = 1), PTEN (n = 1), and STK11 (n = 1). The 13 CD274‐positive colorectal metastases included KRAS (n = 7), BRAF (n = 3), and PIK3CA (n = 1) mutants, and two quadruple negative tumours. One of the latter carried TP53 mutation.
Genotype comparison of primary and metastatic CD274‐positive CRCs
The gene mutation status of 13 CD274‐positive metastatic lesion evaluated in this study and 16 previously studied CD274‐positive primary CRCs 13 are summarized in Table 4. Most CD274‐expressing primary tumours (88%, [14/16]) were BRAF mutants. In contrast, CD274‐expressing metastatic CRCs frequently carried KRAS mutation (54%, [7/13]). The significance of this difference was confirmed statistically (p < 0.001). Detailed clinicopathological, immunohistochemical, and gene mutation data from the CRCs analysed are summarized in supplementary material, Tables S4 and S5.
Table 4.
BRAF and KRAS mutations in 16 primary and 13 metastatic CRCs
| Total no. | Metastatic CRCs | Primary CRCs | |||||
|---|---|---|---|---|---|---|---|
| 29 | (100%) | 13 | (45%) | 16 | (55%) | P value | |
| Gene mutation | <0.001* | ||||||
| BRAF mutant | 17 | [59%] | 3 | [23%] | 14 | [88%] | |
| KRAS mutant | 8 | [28%] | 7 | [54%] | 1 | [6%] | |
| Other mutations and wild type | 4 | [14%] | 3 | [23%] | 1 | [6%] | |
CRCs, colorectal carcinomas; *P value was calculated using Fisher's exact test.
Characterization of five colon cancer cell lines
Immunoblot assay revealed variable CD274 expression in all analysed colon cancer cell lines (Figure 2A). A significant fraction (3.2–32.0%) of colon cancer cells expressed CD274 on the cell membrane under culture conditions (Figure 2B). High‐level phospho‐STAT1 (Tyr701) expression indicating aberrant JAK‐STAT pathway activation was detected in CW‐2. CDX2 was silenced in COLO205 and HCT116. VIM expression suggesting a mesenchymal‐like phenotype was shown in SW480 and LoVo with lower CD274 expression levels (Figure 2C and supplementary material, Figure S2). Among the five analysed cell lines, COLO205 and SW480 appeared to be derived from microsatellite‐stable tumours based on the MMR protein expression status (Figure 2C). COLO205 was considered a consensus molecular subtype 1 (CMS1) tumour because of its BRAF mutation, CDX2‐negativity, and epithelial phenotype (Figure 2C and supplementary material, Figure S2) 22.
Figure 2.
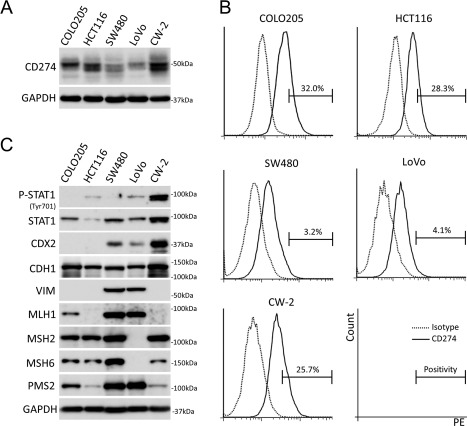
Basal CD274 expression and other characteristics of colon cancer cells under cultured conditions. (A) Immunoblot analysis for CD274 in colon cancer cells. (B) FACS analyses for cell surface CD274 in colon cancer cells. (C) Immunoblot analyses for the characterization of colon cancer cells. Note that VIM expression uniquely showed inverse correlation to CD274.
IFNγ up‐regulates CD274 through the JAK‐STAT pathway
Twenty‐four hours of IFNγ stimulation up‐regulated membranous CD274 expression on all of the colon cancer cells in a dose‐dependent manner (Figure 3A,B). Pre‐treatment with JAK inhibitor I blocked CD274‐ and phospho‐STAT1/3 up‐regulation induced by IFNγ (Figure 3C). Thirty‐six hours of JAK inhibitor I blockade slightly suppressed CD274 expression with down‐regulated phospho‐STAT1 in CW‐2 (Figure 3D).
Figure 3.
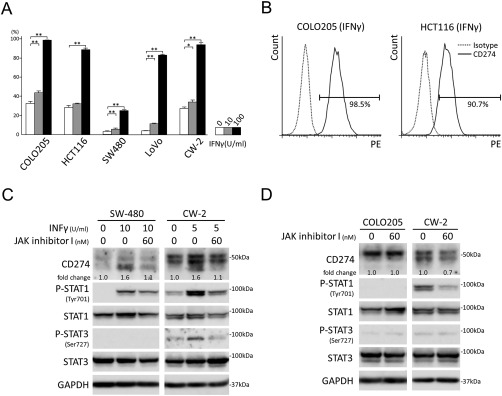
IFNγ up‐regulates CD274 through the JAK‐STAT pathway. (A) FACS analyses for cell surface CD274 in IFNγ‐stimulated colon cancer cells. The experiments were performed in triplicate. Columns, mean values; bars, SD. **p < 0.01; *p < 0.05. (B) Representative results of FACS analyses for cell surface CD274 in COLO205 and HCT116 stimulated with IFNγ (100 U/ml) for 24 h. (C) Immunoblot analyses of SW480 and CW‐2 cells with or without IFNγ and JAK inhibitor I treatment. Note that phospho‐STAT2 (Tyr690), phospho‐STAT3 (Tyr705), phospho‐STAT5 (Tyr694), and phospho‐STAT6 (Tyr641) were expressed at under‐detectable levels (data not shown). (D) Immunoblot analyses of COLO205 and CW‐2 cells with or without JAK inhibitor I blockade.
TGF‐β1 modulates CD274 regardless of EMT (epithelial‐mesenchymal transition) status
Forty‐eight hours of TGF‐β1 stimulation mildly up‐regulated cell surface CD274 in HCT116. On the other hand, CD274 was down‐regulated in Lovo and CW‐2 by TGF‐β1 (Figure 4A,B). SW480 uniquely showed a mild EMT phenotype (CDH1 down‐regulation with VIM up‐regulation) upon TGF‐β1 stimulation (Figure 4B). However, no clear correlation between EMT phenotype including VIM up‐regulation and CD274 down‐regulation was seen under our experimental conditions (Figure 4B).
Figure 4.
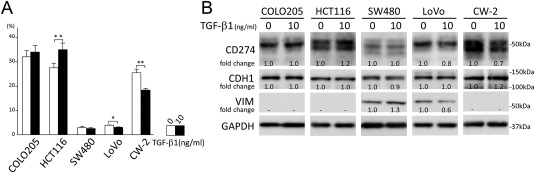
TGF‐β1 modulates CD274 regardless of EMT status. (A) FACS analyses for cell surface CD274 on colon cancer cells with 48 h of TGF‐β1 stimulation. The experiments were performed in triplicate. Columns, mean values; bars, SD. **p < 0.01; *p < 0.05. (B) Immunoblot analyses of colon cancer cells stimulated with TGF‐β1 for 48 h. Note that SW480 uniquely showed mild EMT. However, no correlation between EMT and CD274 expression was detected.
Discussion
In this study, 189 well characterized metastatic CRCs were examined immunohistochemically for CD274 (PD‐L1), MMR‐protein, CDX2, and ALCAM expression in order to assess the characteristics of CD274 positive metastatic tumours. Characterization of TAIs was also performed by CD4, CD8, and FOXP3 immunostaining. In addition, Sanger sequencing and NGS were performed in a representative subgroup of metastases. These characteristics of metastatic CRCs were compared to those of primary tumours from our previous study 13. However, a limitation in the comparison is that the patient cohorts are different. Finally, CD274 expression and responsiveness to exogenous INFγ or TGF‐β1 were examined in cultured colon cancer cells to assess the regulation mechanism(s) of CD274 expression.
Approximately 15% of primary colorectal cancers, recently classified as consensus molecular subtype 1 (CMS1), are believed to develop through the serrated neoplasia pathway, showing mucinous and/or poorly differentiated/medullary histology, CpG island methylator phenotype, MMR‐deficiency, mutational BRAF activation and, in some cases, CDX2‐negativity 14, 22, 23. In primary colorectal tumours, significant associations between CD274 expression and characteristics of serrated neoplasia pathway‐driven CRCs have been identified 11, 12, 13. In CMS1 colorectal cancers, similar to other tumour‐driving intrinsic factor/signalling pathway(s) such as PTEN‐loss, EGFR‐mutation, or ALK‐translocation 24, 25, 26, activated serrated neoplasia or JAK‐STAT pathways, have been suggested to be associated with high CD274 expression 22.
In metastatic CRCs, 14% of the cases analysed showed variable CD274 expression. In contrast with primary CRCs, univariate analyses failed to identify significant association between CD274 expression and tumour intrinsic factors including MMR‐deficiency (p = 0.42), ALCAM expression (p = 0.23), and “stem‐like” immunophenotype (p = 0.63) after multiple testing adjustments 13. The comparison of CD274‐positive metastatic and primary CRCs revealed earlier occurrence (p = 0.001), and decreased membranous CD274 expression (p < 0.001) and MMR‐deficiency (p < 0.001) in metastatic tumours. Also, different mutation status (common KRAS mutation in CD274‐positive metastatic tumours) was identified (p < 0.001). Because concordant gene mutation status and phenotype have been previously reported between primary and metastatic colorectal lesions 27, the higher incidence of KRAS mutants and lower frequency of MMR‐deficiency might point to different mechanisms triggering CD274 expression in metastatic and primary CRCs. Also, intra‐tumour heterogeneity or differential expression of CD274 in isogenic primary and metastatic tumours has been reported in several types of cancer such as lung cancer, breast carcinoma, and renal cell carcinoma 28, 29, 30.
The lines of evidence presented here strongly support previous observations that tumour micro environmental factor(s) rather than tumour‐intrinsic factors dominantly regulate CD274 expression 31. In the current study, CD274 up‐regulation mechanisms were studied “in vitro” using five colon cancer cell lines. All cultured colon cancer cells (5/5) expressed CD274 at variable levels with no significant correlation with BRAF/KRAS mutations, MMR‐deficiency, CDX2, or phospho‐STAT1 expression levels. In CMS1 colorectal tumours, aberrantly activated JAK‐STAT pathway was suggested 22. However, phospho‐STATs were not expressed at a detectable level in COLO205 cells under cultured conditions. Therefore, the activated JAK‐STAT pathway phenotype in CMS1 tumours might originate from the association with TAIs because prominent infiltration of T and NK cells was identified in this type of tumour 22. Conversely, CW‐2 showed aberrant expression of phospho‐STAT1 with an unclear mechanism. JAK inhibitor I uniquely suppressed CD274 expression in CW‐2 suggesting that aberrant JAK‐STAT pathway activation plays some role in CD274 up‐regulation. In the present study, all of the cell lines, regardless of their subtype, were responsive to exogenous INFγ for CD274 up‐regulation through the JAK‐STAT pathway. Although histological and immunohistochemical analyses showed no significant association between CD274 expression and TAIs, these experimental results suggest that INFγ from inflammatory cells within the tumour micro environment could modulate CD274 expression.
Inverse correlation between CD274‐ and VIM‐expression (one of the EMT phenotypes) was suggested in immunoblot analyses of colon cancer cells. Based on these observations, colon cancer cells were tested using TGF‐β1 stimulation, one of the notable EMT‐inducers, for CD274 down‐regulation. Contrary to our expectations, TGF‐β1 variably regulated CD274 expression in colon cancer cells regardless of the EMT phenotype. TGF‐β signalling in colon cancer cells is complicated. At first, it was reported that EMT can only be induced by TGF‐β1 in microsatellite‐stable colon cancer cells with wild‐type TGFBR2 through its normal canonical TGF‐β1‐SMAD2 signalling 32. However, a recent study showed that signals can be transduced through the canonical TGF‐β1‐SMAD2 pathway even in microsatellite‐unstable colon cancer cells with TGFBR2 frameshift mutation through the production of functional TGFBR2 protein by transcriptional slippage 33. Actually, HCT116 showed SMAD2 phosphorylation upon TGF‐β1 stimulation in the present study (supplementary material, Figure S3). Conversely, microsatellite‐stable COLO205 did not show EMT even with activation of the canonical TGF‐β1‐SMAD2 signalling (supplementary material, Figure S3). In the present study, bidirectional CD274 regulation by exogenous TGF‐β1 stimulation was identified. These results might indicate the involvement of not only canonical TGF‐β1‐SMAD2 signalling but also the SMAD2‐independent non‐canonical TGF‐β1 pathway and/or epigenetic mechanism due to the CpG island methylator phenotype for the regulation of EMT and CD274 expression in colon cancer cells 34. These complicated mechanism(s) should be examined further.
PD‐Ls/PDCD1 immune checkpoint inhibitors have been introduced as cancer treatment and have shown significant anti‐cancer effects in some cases 35, 36. At the time of submission of this manuscript, several clinical trials (such as NCT02060188 and NCT02437071) were in progress to assess CD274/PDCD1 immune checkpoint inhibitors in metastatic colorectal cancer patients. CD274 immunohistochemistry has been used as a potential biomarker to predict clinical response to PD‐Ls/PDCD1 immune checkpoint inhibitors as well as resistance to aspirin therapy 35, 36, 37, 38, 39. In the present study, a significant number of metastatic CRCs (88%, [23/26]) showed cytoplasm‐dominant CD274 expression. Future retrospective studies should address whether decreased membranous CD274 on metastatic CRC cells dampens the efficacy of PD‐Ls/PDCD1 immune checkpoint inhibitors. The discordance of CD274 expression between primary and metastatic lesions can result in non‐optimal use of CD274/PDCD1 axis inhibitors. Thus, CD274 immunohistochemistry of both primary and metastatic CRCs should be examined when planning anti‐PD‐Ls/PDCD1 immune checkpoint inhibitor treatment for colorectal cancer patients.
In summary, this study has identified immunohistological and genotypic characteristics of CD274‐positive metastatic CRCs, showing significant differences between CD274 positive primary and metastatic lesions and indicating epigenetic mechanisms, such as tumour micro environmental factors, in regulation of CD274 expression.
Author contributions statement
SI: conceived and supervised the overall study; SI, JL: designed the experiments; AFG, JL, AK, SZ, JK: performed the sequencing analyses; ZW, MM: performed tissue processing and immunohistochemical staining; SI: performed the molecular experiments, analysed the data, made the figures and tables, and wrote the manuscript; JL, HI, MM: critically reviewed the manuscript. All authors read and gave final approval to the submitted version.
Supporting information
Figure S1. ALCAM and CDX2 expression in metastatic colon cancer and normal colonic mucosa. A‐C: Histology (A), ALCAM (B) and CDX2 (C) immunostaining of metastatic colon cancer. D: CDX2 immunostaining of normal colonic mucosa
Figure S2. Characterization of colon cancer cells. RT‐qPCR analyses for CDH1 and VIM in cultured colon cancer cells. The data were normalized to those of HCT116 and are shown on a log2 scale
Figure S3. Immunoblot analyses for colon cancer cells with TGF‐β1 treatment. TGF‐β1 stimulation for 24 hours up‐regulated phospho‐SMAD2 expression even in microsatellite‐unstable HCT116 cells
Table S1. Antibodies and conditions for immunohistochemistry
Table S2. Primer sequences and PCR conditions used to amplify targets for Sanger sequencing
Table S3. Antibodies and dilutions for immunoblot assay
Table S4. Characterization of 34 metastatic colorectal carcinomas analysed for gene mutation
Table S5. Characterization of 66 primary colorectal carcinomas analysed for gene mutation
Acknowledgements
This research was supported as a part of National Cancer Institute's intramural research program. This research was also supported in part by a JSPS KAKENHI Grant‐in‐Aid for Scientific Research (C) (to SI, 17K08706). We thank Dr. Kenji Kasai (Aichi Medical University) for providing SW 480 color cancer cell line. We thank Mr. Makoto Naruse and Ms. Natsumi Kodama (Aichi Medical University) for the assistance in FACS analyses.
No conflicts of interest were declared.
References
- 1. Nishimura H, Honjo T. PD‐1: an inhibitory immunoreceptor involved in peripheral tolerance. Trends Immunol 2001; 22 : 265–268. [DOI] [PubMed] [Google Scholar]
- 2. Nishimura H, Nose M, Hiai H, et al Development of lupus‐like autoimmune diseases by disruption of the PD‐1 gene encoding an ITIM motif‐carrying immunoreceptor. Immunity 1999; 11 : 141–151. [DOI] [PubMed] [Google Scholar]
- 3. Dong H, Zhu G, Tamada K, et al B7‐H1, a third member of the B7 family, co‐stimulates T‐cell proliferation and interleukin‐10 secretion. Nat Med 1999; 5 : 1365–1369. [DOI] [PubMed] [Google Scholar]
- 4. Freeman GJ, Long AJ, Iwai Y, et al Engagement of the PD‐1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med 2000; 192 : 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Latchman Y, Wood CR, Chernova T, et al PD‐L2 is a second ligand for PD‐1 and inhibits T cell activation. Nat Immunol 2001; 2 : 261–268. [DOI] [PubMed] [Google Scholar]
- 6. Tseng SY, Otsuji M, Gorski K, et al B7‐DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J Exp Med 2001; 193 : 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao Q, Wang XY, Qiu SJ, et al Overexpression of PD‐L1 significantly associates with tumor aggressiveness and postoperative recurrence in human hepatocellular carcinoma. Clin Cancer Res 2009; 15 : 971–979. [DOI] [PubMed] [Google Scholar]
- 8. Ohigashi Y, Sho M, Yamada Y, et al Clinical significance of programmed death‐1 ligand‐1 and programmed death‐1 ligand‐2 expression in human esophageal cancer. Clin Cancer Res 2005; 11 : 2947–2953. [DOI] [PubMed] [Google Scholar]
- 9. Wu C, Zhu Y, Jiang J, et al Immunohistochemical localization of programmed death‐1 ligand‐1 (PD‐L1) in gastric carcinoma and its clinical significance. Acta Histochem 2006; 108 : 19–24. [DOI] [PubMed] [Google Scholar]
- 10. Inaguma S, Wang Z, Lasota J, et al Comprehensive immunohistochemical study of programmed cell death ligand 1 (PD‐L1): analysis in 5536 cases revealed consistent expression in trophoblastic tumors. Am J Surg Pathol 2016; 40 : 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Masugi Y, Nishihara R, Yang J, et al Tumour CD274 (PD‐L1) expression and T cells in colorectal cancer. Gut 2017; 66 : 1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rosenbaum MW, Bledsoe JR, Morales‐Oyarvide V, et al PD‐L1 expression in colorectal cancer is associated with microsatellite instability, BRAF mutation, medullary morphology and cytotoxic tumor‐infiltrating lymphocytes. Mod Pathol 2016; 29 : 1104–1112. [DOI] [PubMed] [Google Scholar]
- 13. Inaguma S, Lasota J, Wang Z, et al Clinicopathologic profile, immunophenotype, and genotype of CD274 (PD‐L1)‐positive colorectal carcinomas. Mod Pathol 2017; 30 : 278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dienstmann R, Vermeulen L, Guinney J, et al Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017; 17 : 79–92. [DOI] [PubMed] [Google Scholar]
- 15. Miettinen M. A simple method for generating multitissue blocks without special equipment. Appl Immunohistochem Mol Morphol 2012; 20 : 410–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lasota J, Kowalik A, Wasag B, et al Detection of the BRAF V600E mutation in colon carcinoma: critical evaluation of the immunohistochemical approach. Am J Surg Pathol 2014; 38 : 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kopczynski J, Kowalik A, Chlopek M, et al Oncogenic activation of the Wnt/beta‐catenin signaling pathway in signet ring stromal cell tumor of the ovary. Appl Immunohistochem Mol Morphol 2016; 24 : e28–e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanda Y. Investigation of the freely available easy‐to‐use software ‘EZR’ for medical statistics. Bone Marrow Transplant 2013; 48 : 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inaguma S, Kasai K, Ikeda H. GLI1 facilitates the migration and invasion of pancreatic cancer cells through MUC5AC‐mediated attenuation of E‐cadherin. Oncogene 2011; 30 : 714–723. [DOI] [PubMed] [Google Scholar]
- 20. Inaguma S, Riku M, Hashimoto M, et al GLI1 interferes with the DNA mismatch repair system in pancreatic cancer through BHLHE41‐mediated suppression of MLH1. Cancer Res 2013; 73 : 7313–7323. [DOI] [PubMed] [Google Scholar]
- 21. Inaguma S, Ito H, Riku M, et al Addiction of pancreatic cancer cells to zinc‐finger transcription factor ZIC2. Oncotarget 2015; 6 : 28257–28268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guinney J, Dienstmann R, Wang X, et al The consensus molecular subtypes of colorectal cancer. Nat Med 2015; 21 : 1350–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Je IJ, Vermeulen L, Meijer GA, et al Serrated neoplasia‐role in colorectal carcinogenesis and clinical implications. Nat Rev Gastroenterol Hepatol 2015; 12 : 401–409. [DOI] [PubMed] [Google Scholar]
- 24. Azuma K, Ota K, Kawahara A, et al Association of PD‐L1 overexpression with activating EGFR mutations in surgically resected nonsmall‐cell lung cancer. Ann Oncol 2014; 25 : 1935–1940. [DOI] [PubMed] [Google Scholar]
- 25. Marzec M, Zhang Q, Goradia A, et al Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD‐L1, B7‐H1). Proc Natl Acad Sci U S A 2008; 105 : 20852–20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parsa AT, Waldron JS, Panner A, et al Loss of tumor suppressor PTEN function increases B7‐H1 expression and immunoresistance in glioma. Nat Med 2007; 13 : 84–88. [DOI] [PubMed] [Google Scholar]
- 27. Mao C, Wu XY, Yang ZY, et al Concordant analysis of KRAS, BRAF, PIK3CA mutations, and PTEN expression between primary colorectal cancer and matched metastases. Sci Rep 2015; 5 : 8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Callea M, Albiges L, Gupta M, et al Differential expression of PD‐L1 between primary and metastatic sites in clear‐cell renal cell carcinoma. Cancer Immunol Res 2015; 3 : 1158–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cimino‐Mathews A, Thompson E, Taube JM, et al PD‐L1 (B7‐H1) expression and the immune tumor microenvironment in primary and metastatic breast carcinomas. Hum Pathol 2016; 47 : 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pinato DJ, Shiner RJ, White SD, et al Intra‐tumoral heterogeneity in the expression of programmed‐death (PD) ligands in isogeneic primary and metastatic lung cancer: implications for immunotherapy. Oncoimmunology 2016; 5 : e1213934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taube JM, Anders RA, Young GD, et al Colocalization of inflammatory response with B7‐h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med 2012; 4 : 127ra137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pino MS, Kikuchi H, Zeng M, et al Epithelial to mesenchymal transition is impaired in colon cancer cells with microsatellite instability. Gastroenterology 2010; 138 : 1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Miranda NF, van Dinther M, van den Akker BE, et al Transforming growth factor beta signaling in colorectal cancer cells with microsatellite instability despite biallelic mutations in TGFBR2. Gastroenterology 2015; 148 : 1427–1437.e8. [DOI] [PubMed] [Google Scholar]
- 34. Iwata J, Hacia JG, Suzuki A, et al Modulation of noncanonical TGF‐beta signaling prevents cleft palate in Tgfbr2 mutant mice. J Clin Invest 2012; 122 : 873–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Borghaei H, Paz‐Ares L, Horn L, et al Nivolumab versus Docetaxel in advanced nonsquamous non‐small‐cell lung cancer. N Engl J Med 2015; 373 : 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brahmer J, Reckamp KL, Baas P, et al Nivolumab versus Docetaxel in advanced squamous‐cell non‐small‐cell lung cancer. N Engl J Med 2015; 373 : 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ansell SM, Lesokhin AM, Borrello I, et al PD‐1 blockade with Nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med 2015; 372 : 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Motzer RJ, Escudier B, McDermott DF, et al Nivolumab versus Everolimus in advanced renal‐cell carcinoma. N Engl J Med 2015; 373 : 1803–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hamada T, Cao Y, Qian ZR, et al Aspirin use and colorectal cancer survival according to tumor CD274 (programmed cell death 1 ligand 1) expression status. J Clin Oncol 2017; 35 : 1836–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. ALCAM and CDX2 expression in metastatic colon cancer and normal colonic mucosa. A‐C: Histology (A), ALCAM (B) and CDX2 (C) immunostaining of metastatic colon cancer. D: CDX2 immunostaining of normal colonic mucosa
Figure S2. Characterization of colon cancer cells. RT‐qPCR analyses for CDH1 and VIM in cultured colon cancer cells. The data were normalized to those of HCT116 and are shown on a log2 scale
Figure S3. Immunoblot analyses for colon cancer cells with TGF‐β1 treatment. TGF‐β1 stimulation for 24 hours up‐regulated phospho‐SMAD2 expression even in microsatellite‐unstable HCT116 cells
Table S1. Antibodies and conditions for immunohistochemistry
Table S2. Primer sequences and PCR conditions used to amplify targets for Sanger sequencing
Table S3. Antibodies and dilutions for immunoblot assay
Table S4. Characterization of 34 metastatic colorectal carcinomas analysed for gene mutation
Table S5. Characterization of 66 primary colorectal carcinomas analysed for gene mutation


