Abstract
Molecular subclassification of endometrial carcinoma (EC) with Proactive Molecular Risk Classifier for Endometrial Cancer (ProMisE) identifies four subtypes [DNA polymerase epsilon (POLE) mutant, mismatch repair‐deficient, p53 wild‐type (wt), and p53 abnormal]. The aim of this study was to evaluate additional EC biomarkers in the context of these subtypes. Tissue microarrays encompassing 460 previously characterized ECs were assessed for L1‐cell adhesion molecule (L1CAM), progesterone receptor (PR), estrogen receptor (ER) alpha, stathmin, and phosphatase and tensin homolog (PTEN), by immunohistochemistry (IHC). Associations with clinicopathological parameters, molecular subtype, and outcomes were determined. About 413 ECs (75% endometrioid, >15% serous) had complete data. L1CAM overexpression was found in 16%, associated with older age, lower body mass index (BMI), advanced stage, grade 3 (97%), non‐endometrioid histology (84%), deep myometrial invasion, lymphovascular space invasion (LVSI), and ER‐negative, PR‐negative status. Tumours overexpressing L1CAM were associated with poor outcomes {hazard ratio (HR) [95% confidence interval (CI)] 3.35 [2.10–5.23] for disease‐specific survival [DSS], p < 0.0001}. PR positivity was associated with younger women, higher BMI, early stage (77% stage I), low grade (61%), endometrioid histology (90%) without LVSI or nodal disease, ER positivity (90%), p53wt tumours (55%), and favourable outcomes [HR (CI) 0.39 (0.25–0.62) for DSS, p < 0.0001]. ER positive tumours were early stage (73%), low grade, endometrioid histology, with improved DSS. Stathmin and PTEN IHC were not associated with outcomes. There was minimal agreement between IHC and mutation status for PTEN. L1CAM overexpression was significantly associated with the p53 abnormal molecular subtype, which accounted for more than 70% of the tumours overexpressing L1CAM. PR expression also correlated with molecular subtype, with most PR negative tumours being p53 abnormal. Multivariable analysis demonstrated that only ProMisE subtype [overall survival (OS), DSS, and progression‐free survival] and age (OS only) maintained an association with outcomes. The prognostic significance of the single biomarkers tested could be explained based on their being covariable with the ProMisE molecular subtype.
Keywords: endometrial cancer, prognosis, biomarker, TCGA, ProMisE, L1CAM, progesterone receptor, estrogen receptor
Introduction
Endometrial cancer (EC) is the most common gynaecological cancer and fourth most common cancer in women overall in the developed world. There were 320 000 new cases of EC diagnosed worldwide in 2012 1. Advances in clinical management and research have been hampered by an inability to reproducibly categorize endometrial tumours based on histomorphological features 2, 3. Risk stratification systems used to guide treatment rely on these parameters 4, 5, 6, in the absence of more robust markers. A push to more biologically informative molecular tools to classify tumours and stratify according to risk of metastases and recurrence has led to a multitude of studies interrogating the prognostic and predictive implications of one or more biomarkers or molecular features.
The most comprehensive of these investigations was the collaborative Cancer Genome Atlas project (TCGA) that characterized 373 endometrioid and serous endometrial carcinomas (ECs) through genomic, proteomic, and gene expression assays, identifying four prognostic subtypes with distinct prognostic outcomes 7. Subsequently, two research teams have pared down and simplified the molecular methods in TCGA to identify similar but not identical prognostic subtypes from formalin‐fixed paraffin embedded material 8, 9, 10, 11, 12. Proactive Molecular Risk Classifier for Endometrial Cancer (ProMisE) identifies these four TCGA‐based molecular subtypes using immunohistochemistry and sequencing of the POLE exonuclease domains (Figure 1), and has been developed according to the Institute of Medicine (IOM) guidelines for the development of ‘omics’ based tests, following a strict pathway of ‘discovery’ 9, ‘confirmation’ 10, and ‘validation’ (manuscript herein) steps. ProMisE is ready to cross the ‘bright line’ for application in the clinic 13, at which point all historical biomarkers (clinical, pathological, molecular) need to be re‐evaluated in the context of these molecular subtypes. ProMisE has also proven to work on diagnostic endometrial biopsy/curettage specimens with high concordance to hysterectomy specimens, enabling earlier biologically relevant information for patients and clinicians that may guide management from the earliest time point 8.
Figure 1.
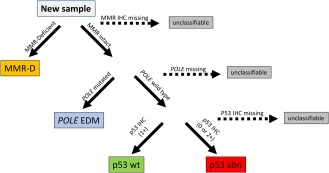
Algorithm for ProMisE molecular classifier. A sample is tested for MMR deficiency by IHC for the presence or absence of MSH6 and PMS2 proteins, sequencing is performed for exons 9–14 of the POLE exonuclease domain (EDM), and IHC for p53 performed to identify complete loss/null (IHC score 0), accumulation/missense (IHC score 2), or normal expression/wt (IHC score 1) yielding four molecular subtypes with distinct clinical outcomes. Reproduced with permission from 10.
Our objective in this study was to re‐evaluate select molecular markers of purported prognostic significance: L1‐cell adhesion molecule (L1CAM) 14, 15, 16, progesterone receptor (PR) 17, 18, 19, estrogen receptor alpha (ER) 18, 19, 20, stathmin (STMN) 21, 22, and phosphatase and tensin homolog (PTEN) 23, 24, assessing their value in the framework of modern molecular classification (by ProMisE).
Methods
Cohort
Following ethical review board approval, 460 cases of EC, encompassing the ‘discovery’ 9 and ‘confirmation’ 10 cohorts for the development of the ProMisE molecular classifier were represented in duplicate cores over six tissue microarrays (TMAs) (supplementary material, Figure S1). This cohort has been previously characterized, with demographic, clinical, pathological, and outcomes reported 9, 10. As performed previously 9, 10, to ensure random censoring and minimize ascertainment bias, observations were censored on December 31 on the fifth year of follow‐up.
Methods for ProMisE, including details on TMA construction, DNA extraction, sequencing, and interpretation have been described previously 9, 10, 25; these studies yielded four prognostic subtypes; ECs with mismatch repair deficiency based on immunohistochemical (IHC) testing for the presence/absence of MSH6 and PMS2 [mismatch repair‐deficient (MMR‐D)], ECs with DNA polymerase epsilon (POLE) exonuclease domain mutations detected through focused sequencing, or ECs without the previously described molecular changes who have normal versus aberrant expression of p53 on IHC; p53 wild‐type (p53wt) and p53 abnormal (p53abn), respectively. These four molecular EC subtypes (MMR‐D, POLE, p53wt, p53abn) correspond to the hypermutated/microsatellite instability, ultramutated, copy number low, and copy number high subtypes of TCGA, respectively.
Immunohistochemistry: Additional biomarker evaluation
Additional IHC was performed on previously constructed TMAs for each marker as outlined in Table 1. Scoring was performed by gynaecological pathologists (BG, AK, CL, FK, MK) who were all blinded to clinical outcome. For tumours with only one core present for evaluation, the final score for the tumour was the score given to that individual core. For tumours with two cores, the final score was determined as if both cores were a single larger core.
Table 1.
Methodological details for the five immunohistochemical marker
| Biomarker | Company | Catalogue | Titration | Staining protocol | Scoring criteria |
|---|---|---|---|---|---|
| L1CAM | Covance | SIG‐3911 | 1:25 |
Ventana Discovery Ultra ‐ 64 min CC1 ‐ 32 min primary at 37°C ‐ UltraMap DAB anti‐Ms kit |
Low = up to 50% High = >50% |
| PR | Ventana |
790–2223 (clone 1E2) |
Undiluted |
Ventana Discovery Ultra ‐ 64 min CC1 ‐ 16 min primary at 36°C ‐ DABMap kit |
Negative = up to 1% Positive = >1% |
| ER | Thermo |
RM‐9101 (clone SP1) |
1:25 |
Ventana Discovery Ultra ‐ 64 min CC1 ‐ 1 h primary at 37°C ‐ DABMap kit |
Negative = up to 5% Positive = >5% |
| STMN | Cell Signaling | 3352 | 1:50 |
Ventana Discovery XT ‐ Standard CC1 ‐ 1 h primary with heat ‐ DABMap kit |
Low = negative, weak, moderate (scores 0–4) High = strong (scores 6, 9) |
| PTEN | Cell Signaling | 9559 | 1:25 |
Ventana Discovery Ultra ‐ 64 min CC1 ‐ 1 h primary at room temp ‐ UltraMap DAB anti‐Rb kit |
Negative = no staining Positive = any staining |
L1 Cell Adhesion Molecule (L1CAM) scoring was based on Zeimet et al 16, with 0 = no epithelial staining, 1 = up to 10% staining, 2 = 11–50% staining, 3 = >50% of epithelial cells staining. A predefined cut point was then used to divide tumours into two groups: low (0,1+) versus high (2+, 3+, also referred to as ‘overexpressors’) (see supplementary material, Figure S2).
Progesterone Receptor (PR) positivity was per the cutoff used in breast cancer, with a predefined cut point of 1% of tumour cell nuclei showing staining (<1% versus ≥1%) 17, 26.
Estrogen Receptor (ER) scoring was determined with a predefined cut point of 5% of tumour cell nuclei staining (<5% ‘negative’, versus >5% encompassing weak intermediate or strongly positive), disregarding squamous metaplasia, based on a previous study on EC 27.
Stathmin (STMN) staining index (SI) was based on Trovik et al 21, which was the product of staining intensity (0, 1+, 2+, 3+) × percent of cells positive (0, 1 = <10%, 2 = 10–50%, 3 = >50%), resulting in a range of 0–9. This SI score was reduced to four categories: SI 0 = negative, SI 1,2 = weak, SI 3,4 = moderate, SI 6,9 = strong. A cut point was then applied [based on optimal separation of Kaplan Meier curves with disease‐specific survival (DSS) on the whole cohort], designating STMN low (negative, weak, and moderate) versus high (strong).
Phosphatase and tensin homolog (PTEN) was scored as positive (any staining) versus negative (complete absence of staining suggestive of PTEN mutation or silencing) 24.
Statistical analysis
We tested for associations of previously identified prognostic parameters in this cohort [e.g. age, stage, lymphovascular space invasion (LVSI)] with IHC biomarker. Chi‐squared test was used for binary and categorical variables, and Welch's t‐test (one way analysis of variance) was used for continuous variables [body mass index (BMI) and age]. Univariable cox regression models and Kaplan‐Meier survival curves for each biomarker were assessed both across the full cohort, and within MMR‐D, POLE, p53wt, and p53abn ProMisE molecular subtypes. Multivariable cox regression analysis was performed to look for parameters associated with outcomes after correction for adjuvant treatment and (1) prognostic parameters available at time of diagnosis (i.e. pre‐surgical variables: age, BMI, histotype, grade but not stage, each IHC biomarker) or (2) pre‐surgical plus post‐surgical parameters (stage, myometrial invasion, LVSI, nodal status).
Concordance (Cohen's kappa coefficient) between IHC and mutation status of PTEN/PTEN was compared in ∼400 of cases where both parameters were known.
In addition, we tested for any association between PTEN IHC expression or PTEN mutation and outcomes according to BMI category of obese (BMI ≥ 30) or non‐obese (BMI < 30) 23. Welch's t‐test (one‐way analysis of variance with no assumption on homogeneity of variances) was used to assess significance of BMI distribution between PTEN status. Statistical significance was set at p = 0.05 and no attempts were made to adjust for multiple comparisons. The proportional hazard assumption for Cox models was assessed by visual examination of Kaplan‐Meier plots and Schoenfeld residual plots. Proportional hazard assumption failed for L1CAM and ER. Therefore, Cox models were built separately for follow‐up time between 0 and 2 years and after 2 years for these two markers based on visual examination of Kaplan‐Meier plots (Figure 2A,B,F). All statistical analyses were performed using R project for statistical computing. The statistical analysis was carried out according to REMARK criteria (supplementary material, Table S1).
Figure 2.
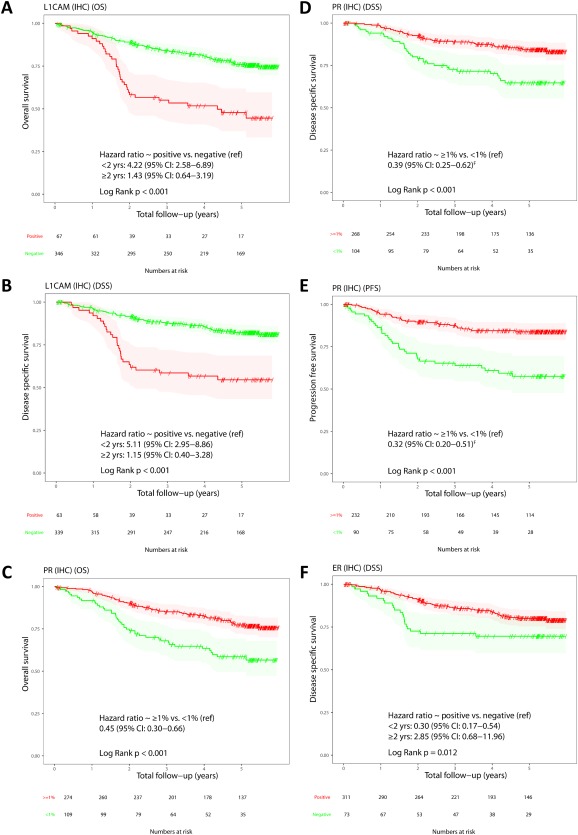
Kaplan‐Meier plots of IHC markers on whole cohort. (A) L1CAM OS. (B) L1CAM disease specific survival. (C) PR OS. (D) PR disease specific survival. (E) PR PFS. (F) ER disease specific survival. For L1CAM and ER, HRs are shown for follow‐up period 0–2 years and >2 years.
Results
Of the 460 cases evaluated, the proportion of uninterpretable cases (due to, e.g., no tumour or insufficient tumour in the tissue cores) varied by IHC marker; 10.2% for L1CAM, 16.7% for PR, 14.1% for ER, 10.8% for STMN, and 14.3% for PTEN. About 100% of cases had ProMisE molecular subtype previously assigned 9, 10, but a proportion of these cases had required whole sections to determine IHC for MMR status and/or p53, consistent with the known challenges of using TMAs. For the current investigation of additional IHC biomarkers, additional testing of whole sections was not performed, and we proceeded to evaluate only the subset interpretable on TMA.
A summary of the clinical and pathological features of the cohort in which these additional IHC parameters were tested is provided in Table 2. High L1CAM expression was seen in 16.2% of the cohort, PR positivity in 71.5%, ER positivity in 80.8%, high STMN in 42.2%, and PTEN loss in 46.2%. Univariable associations demonstrated L1CAM overexpression was associated with: higher age, lower BMI, more advanced stage, non‐endometrioid histotype, presence of LVSI, deep (>50%) myometrial invasion, positive nodal status, and requiring additional treatment. Of tumours that exhibited overexpression of L1CAM, 70% were categorized by ProMisE as p53abn, with 11.9% MMR‐D, 14.9% p53wt, and very rarely (2 cases, 3%) POLE subtype. However, approximately half the p53abn subtype (45/92, 49%) did not exhibit L1CAM overexpression indicating that this biomarker is not a consistent feature of this aggressive subgroup. Similarly, PR negativity was significantly associated with poor prognostic features listed for L1CAM. PR was negative in 29.1% of MMR‐D, 25% of POLE, 16.1% of p53wt, and 55.3% of p53abn subtypes. Of the PR negative ECs, the highest proportion resided within the p53abn subtype.
Table 2.
Descriptive statistics of cohort (n = 460); demographic, clinicopathological, and molecular parameters within the four ProMisE molecular subtypes
| Total | MMR‐D | POLE EDM | p53wt | p53 abn | |
|---|---|---|---|---|---|
| Age | |||||
| Mean (±SD) | 65.6 (± 12) | 65.6 ± 11.3 | 59.8 ± 11.8 | 63.6 ± 12.8 | 71.5 ± 8.8 |
| Median (IQR) | 66.1(57.4–74.4) | 66.1(56.6–73.0) | 58.4(51.8–65.9) | 63.3(54.9–72.9) | 71.5(65.8–78.0) |
| Missing | 3 | 1 | 0 | 2 | 0 |
| BMI | |||||
| Mean (±SD) | 31.8 (± 17.7) | 33 ± 31.1 | 26.6 ± 4.9 | 33.7 ± 12.5 | 29.4 ± 7.8 |
| Median (IQR) | 28.5(23.6–36.7) | 28.5(23.2–34.1) | 27.2(22.3–29.2) | 30.5(24.1–41.2) | 28.0(23.4–33.5) |
| Missing | 42 | 7 | 6 | 25 | 4 |
| Stage | |||||
| IA | 237 (52.0%) | 47 (45.2%) | 24 (58.5%) | 129 (64.5%) | 37 (33.3%) |
| IB | 85 (18.6%) | 24 (23.1%) | 15 (36.6%) | 31 (15.5%) | 15 (13.5%) |
| II | 31 (6.8%) | 11 (10.6%) | 0 (0.0%) | 15 (7.5%) | 5 (4.5%) |
| III | 71 (15.6%) | 16 (15.4%) | 2 (4.9%) | 20 (10.0%) | 33 (29.7%) |
| IV | 32 (7.0%) | 6 (5.8%) | 0 (0.0%) | 5 (2.5%) | 21 (18.9%) |
| Missing | 4 | 1 | 1 | 2 | 0 |
| Grade | |||||
| Grade 1 | 136 (29.6%) | 19 (18.1%) | 7 (16.7%) | 107 (53.0%) | 3 (2.7%) |
| Grade 2 | 74 (16.1%) | 17 (16.2%) | 7 (16.7%) | 45 (22.3%) | 5 (4.5%) |
| Grade 3 | 250 (54.3%) | 69 (65.7%) | 28 (66.7%) | 50 (24.8%) | 103 (92.8%) |
| Missing | 0 | 0 | 0 | 0 | 0 |
| Histological Subtype | |||||
| Endometrioid | 332 (72.2%) | 83 (79.0%) | 35 (83.3%) | 187 (92.6%) | 27 (24.3%) |
| Serous | 104 (22.6%) | 11 (10.5%) | 3 (7.1%) | 11 (5.4%) | 79 (71.2%) |
| Undifferentiated | 6 (1.3%) | 4 (3.8%) | 1 (2.4%) | 1 (0.5%) | 0 (0.0%) |
| Clear Cell | 5 (1.1%) | 1 (1.0%) | 1 (2.4%) | 1 (0.5%) | 2 (1.8%) |
| Mixed + small cell | 13 (2.9%) | 6 (5.9%) | 2 (4.8%) | 2 (1.0%) | 3 (2.7%) |
| Missing | 0 | 0 | 0 | 0 | 0 |
| LVSI | |||||
| No | 266 (60.9%) | 48 (47.5%) | 22 (55.0%) | 155 (80.7%) | 41 (39.4%) |
| Yes | 171 (39.1%) | 53 (52.5%) | 18 (45.0%) | 37 (19.3%) | 63 (60.6%) |
| Missing | 23 | 4 | 2 | 10 | 7 |
| Myometrial Invasion | |||||
| None | 75 (16.6%) | 4 (3.8%) | 4 (9.8%) | 49 (24.5%) | 18 (16.8%) |
| <50% | 205 (45.3%) | 56 (53.3%) | 20 (48.8%) | 93 (46.5%) | 36 (33.6%) |
| >50% | 173 (38.2%) | 45 (42.9%) | 17 (41.5%) | 58 (29.0%) | 53 (49.5%) |
| Missing | 7 | 0 | 1 | 2 | 4 |
| Nodal Status | |||||
| Not Tested | 150 (32.9%) | 22 (21.0%) | 11 (26.2%) | 101 (50.2%) | 16 (14.8%) |
| Tested Negative | 269 (59.0%) | 72 (68.6%) | 31 (73.8%) | 92 (45.8%) | 74 (68.5%) |
| Tested Positive | 37 (8.1%) | 11 (10.5%) | 0 (0.0%) | 8 (4.0%) | 18 (16.7%) |
| Missing | 4 | 0 | 0 | 1 | 3 |
| Treatment | |||||
| No Treatment | 241 (53.6%) | 52 (50.0%) | 18 (43.9%) | 140 (70.0%) | 31 (29.5%) |
| Chemotherapy only | 45 (10.0%) | 10 (9.6%) | 3 (7.3%) | 6 (3.0%) | 26 (24.8%) |
| Radiation therapy only | 75 (16.7%) | 18 (17.3%) | 8 (19.5%) | 31 (15.5%) | 18 (17.1%) |
| Both chemo and radiation | 81 (18.0%) | 23 (22.1%) | 10 (24.4%) | 19 (9.5%) | 29 (27.6%) |
| Vag.brachy.only | 8 (1.8%) | 1 (1.0%) | 2 (4.9%) | 4 (2.0%) | 1 (1.0%) |
| Missing | 10 | 1 | 1 | 2 | 6 |
| ESMO | |||||
| Low | 151 (33.0%) | 23 (21.9%) | 12 (29.3%) | 112 (55.7%) | 4 (3.6%) |
| Intermediate | 71 (15.5%) | 18 (17.1%) | 12 (29.3%) | 29 (14.4%) | 12 (10.8%) |
| High | 236 (51.5%) | 64 (61.0%) | 17 (41.5%) | 60 (29.9%) | 95 (85.6%) |
| Missing | 2 | 0 | 1 | 1 | 0 |
| L1CAM overexpr | 67 (16.2%) | 8 (11.9%) | 2 (3.0%) | 10 (14.9%) | 47 (70.1%) |
| PR positive | 274 (71.5%) | 61 (22.3%) | 24 (8.8%) | 151 (55.1%) | 38 (13.9%) |
| ER positive | 319 (80.8%) | 68 (21.3%) | 28 (8.8%) | 163 (51.1%) | 60 (18.8%) |
| STMN overexpr | 173 (42.2%) | 53 (30.6%) | 19 (11.0%) | 50 (28.9%) | 51 (29.5%) |
| PTEN loss | 182 (46.2%) | 63 (34.6%) | 12 (6.6%) | 91 (50.0%) | 16 (8.8%) |
| Total | 460 (100%) | 105 (22.8%) | 42 (9.1%) | 202 (43.9%) | 111 (24.1%) |
ESMO, European Society for Medical Oncology; IQR, interquartile range.
ER positive ECs were more likely to be stage I, low grade (95% of ER negative tumours were grade 3), endometrioid histology, without LVSI or nodal disease (thus not requiring adjuvant therapy). Although over 92% of p53wt tumours were ER positive, ER expression was common across all ECs subtypes [73.9% MMR‐Ds, 75.7% of POLE and even within the majority (67.4%) of p53abn tumours]. Stathmin overexpression was statistically significantly associated with lower BMI, higher grade, non‐endometrioid histotype, presence of LVSI (trend towards association with nodal disease), and requiring adjuvant treatment. Most p53wt tumours showed minimal stathmin expression (72.7% STMN negative), with overexpression in just over half of the MMR‐D, POLE, and p53abn tumours (53.5%, 51.4%, 56%, respectively) PTEN loss was associated with younger age, higher BMI, low grade, endometrioid histotype but the differences between ECs with PTEN loss versus expression within each of these clinicopathological parameters was small (e.g. median age difference 2.4 years). Although PTEN expression was associated with ProMisE subtypes this mainly reflects the very low proportion of ECs with PTEN loss that were p53abn. Even within the subgroup demonstrated to have POLE exonuclease domain mutations, where we would expect from the TCGA data that PTEN loss would be nearly ubiquitous, we saw PTEN loss of expression in only 12 of 35 tumours. Summary statistics for each marker and univariable associations with other clinicopathological parameters are found in supplementary material, Tables S2–S6.
Univariable survival analysis revealed statistically significant associations with outcome for L1CAM, PR, and ER. Women with tumours with L1CAM overexpression had a higher number of events (recurrence, death from disease). Hazard ratios (HRs) and 95% confidence intervals (CIs) for overall survival (OS), DSS, and progression free survival (PFS), for overexpression of L1CAM (0–2 years) were 4.22 (2.58–6.89), 5.11 (2.95–8.86), and 4.51 (2.64–7.73), respectively, for L1CAM (>2 years) were 1.43 (0.64–3.19), 1.15 (0.40–3.28), and 4.59 (1.75–12.09), respectively. In contrast, women with PR or ER positive tumours were much less likely to recur or die from their tumours. For PR positive tumours, OS, DSS, and PFS were 0.45 (0.3–0.66), 0.39 (0.25–0.62), and 0.32 (0.20–0.51), respectively. For ER positive tumours, OS, DSS, and PFS from 0 to 2 years were 0.40 (0.24–0.68), 0.30 (0.17–0.54), and 0.52 (0.29–0.93), respectively. However, for follow‐up period >2 years, the opposite association was observed; OS, DSS, and PFS for this follow‐up period were 2.90 (0.90–9.37), 2.85 (0.68–11.96), and 1.14 (0.33–3.93), respectively. PTEN loss was associated with PFS only (HR 0.60, 95% CI: 0.37–0.96). There was a trend towards worse DSS in tumours with STMN expression, but it did not reach statistical significance. As previously demonstrated, ProMisE molecular subtype was strongly associated with clinical outcomes, as were traditional prognostic parameters of age, stage, grade, histotype, LVSI, nodal disease, and treatment and degree of myometrial invasion. Full details, including HRs and log rank test (LRT) values are given in Table 3.
Table 3.
Univariable survival analysis with LRT values for each biomarker tested (IHC) and ProMisE molecular subgroups
| # of events/n | Hazard ratio (95% CI) | LRT P value | ||
|---|---|---|---|---|
| L1CAM (0–2 years) (Ref: negative) | ||||
| OS | 65/413 | 4.22 (2.58–6.89) | <0.001 | |
| DSS | 51/402 | 5.11 (2.95–8.86) | <0.001 | |
| PFS | 56/344 | 4.51 (2.64–7.73) | <0.001 | |
| L1CAM (>2 years) (Ref: negative) | ||||
| OS | 46/334 | 1.43 (0.64–3.19) | 0.41 | |
| DSS | 31/330 | 1.15 (0.40–3.28) | 0.80 | |
| PFS | 19/268 | 4.59 (1.75–12.09) | 0.006 | |
| STMN (Ref: negative) | ||||
| OS | 110/410 | 1.23 (0.85–1.79) | 0.28 | |
| DSS | 82/399 | 1.54 (1.00–2.37)a | 0.051 | |
| PFS | 73/339 | 1.44 (0.91–2.27)a | 0.12 | |
| ER (0–2 years) (Ref: negative) | ||||
| OS | 61/395 | 0.40 (0.24–0.68) | 0.001 | |
| DSS | 47/384 | 0.30 (0.17–0.54) | <0.001 | |
| PFS | 54/329 | 0.52 (0.29–0.93) | 0.035 | |
| ER (>2 years) (Ref: negative) | ||||
| OS | 45/321 | 2.90 (0.90–9.37) | 0.037 | |
| DSS | 30/317 | 2.85 (0.68–11.96) | 0.095 | |
| PFS | 19/256 | 1.14 (0.33–3.93) | 0.83 | |
| PR (Ref: <1%) | ||||
| OS | 100/383 | 0.45 (0.30–0.66) | <0.001 | |
| DSS | 71/372 | 0.39 (0.25–0.62)a | <0.001 | |
| PFS | 69/322 | 0.32 (0.20–0.51)a | <0.001 | |
| PTEN (Ref: positive) | ||||
| OS | 108/394 | 0.77 (0.52–1.13) | 0.17 | |
| DSS | 82/383 | 0.68 (0.43–1.06)a | 0.087 | |
| PFS | 73/325 | 0.60 (0.37–0.96)a | 0.03 | |
| ProMisE molecular subgroup | ||||
| OS | 120/460 | MMR‐D | 2.04 (1.25–3.33)a | <0.001 |
| POLE EDM | 0.59 (0.19–1.41)a | |||
| p53 abn | 3.73 (2.44–5.77)a | |||
| DSS | 89/447 | MMR‐D | 2.23 (1.23–4.01)a | <0.001 |
| POLE EDM | 0.49 (0.10–1.51)a | |||
| p53 abn | 4.68 (2.85–7.87)a | |||
| PFS | 89/387 | MMR‐D | 1.90 (1.04–3.44)a | <0.001 |
| POLE EDM | 0.26 (0.03–0.99)a | |||
| p53 abn | 5.09 (3.14–8.47)a | |||
| ESMO | ||||
| OS | 120/458 | Intermediate | 1.23 (0.55–2.57)a | <0.001 |
| High | 3.96 (2.46–6.73)a | |||
| DSS | 89/445 | Intermediate | 2.51 (0.87–7.45)a | <0.001 |
| High | 9.53 (4.63–23.59)a | |||
| PFS | 89/386 | Intermediate | 3.91 (1.04–16.85)a | <0.001 |
| High | 21.82 (8.66–78.69)a | |||
| Age | ||||
| OS | 118/457 | 1.04 (1.03–1.06) | <0.001 | |
| DSS | 87/444 | 1.03 (1.01–1.05) | 0.002 | |
| PFS | 89/386 | 1.02 (1.00–1.04) | 0.018 | |
| BMI | ||||
| OS | 104/418 | 0.98 (0.96–1.00) | 0.065 | |
| DSS | 76/405 | 0.98 (0.96–1.00) | 0.08 | |
| PFS | 84/357 | 0.98 (0.96–1.01) | 0.11 | |
| Stage (Ref stage I) | ||||
| OS | 118/456 | 3.90 (2.72–5.63)a | <0.001 | |
| DSS | 87/443 | 5.21 (3.40–8.09)a | <0.001 | |
| PFS | 89/386 | 6.71 (4.38–10.49)a | <0.001 | |
| Grade (Ref grade 1/2) | ||||
| OS | 120/460 | 2.85 (1.92–4.32)a | <0.001 | |
| DSS | 89/447 | 5.41 (3.18–9.89)a | <0.001 | |
| PFS | 89/387 | 7.09 (4.05–13.53)a | <0.001 | |
| Histology (Ref: Endometrioid) | ||||
| OS | 120/460 | 2.62 (1.82–3.75)a | <0.001 | |
| DSS | 89/447 | 3.43 (2.26–5.21)a | <0.001 | |
| PFS | 89/387 | 4.20 (2.77–6.39)a | <0.001 | |
| LVSI | ||||
| OS | 111/437 | 3.36 (2.30–4.97)a | <0.001 | |
| DSS | 82/425 | 4.44 (2.82–7.21)a | <0.001 | |
| PFS | 83/370 | 4.73 (3.00–7.66)a | <0.001 | |
| Any Nodes (Ref: Negative) | ||||
| OS | 118/456 | Not Tested | 0.81 (0.54–1.22) | 0.030 |
| Positive | 1.90 (1.10–3.29) | |||
| DSS | 87/443 | Not Tested | 0.82 (0.50–1.32)a | 0.003 |
| Positive | 2.68 (1.48–4.61)a | |||
| PFS | 89/385 | Not Tested | 0.23 (0.10–0.44)a | <0.001 |
| Positive | 2.72 (1.63–4.39)a | |||
| Any Treatment (No treatment) | ||||
| OS | 117/450 | 1.96 (1.36–2.86)a | <0.001 | |
| DSS | 86/437 | 2.98 (1.90–4.81)a | <0.001 | |
| PFS | 87/380 | 3.29 (2.10–5.30)a | <0.001 | |
| Myometrial invasion | ||||
| OS | 117/453 | <50 | 1.28 (0.67–2.68)a | <0.001 |
| >50 | 3.89 (2.13–7.90)a | |||
| DSS | 86/440 | <50 | 1.13 (0.49–2.97)a | <0.001 |
| >50 | 5.34 (2.56–13.28)a | |||
| PFS | 89/385 | <50 | 0.94 (0.46–2.11)a | <0.001 |
| >50 | 4.02 (2.13–8.54)a |
Reference (Ref.) parameters are shown. Of note, statistically significant associations of all other clinicopathological parameters tested (age, stage, histotype, myometrial invasion, nodal status, and any treatment given) with outcomes (OS/DSS/PFS) were noted and have been previously reported [10].
ESMO, European Society for Medical Oncology.
Indicates that the Firth's penalized maximum likelihood bias reduction method was used to estimate the HR.
Kaplan‐Meier survival analysis demonstrated statistically significant associations with outcomes (OS, DSS, and PFS) for both L1CAM and PR expression across the whole cohort (p < 0.001) with DSS curves shown in Figure 2.
Univariable and Kaplan‐Meier survival analyses were also performed within ProMisE subtypes for each IHC marker. In ECs classified as MMR‐D with L1CAM overexpression (∼7%), worse PFS was observed (Log Rank p = 0.005; Figure 3D). Within the p53wt subtype of ECs with L1CAM overexpression (∼6%), worse DSS and PFS were observed (Log Rank p = 0.006 and 0.004, respectively; Figure 3B,C). Within p53abn, of which half the tumours exhibited L1CAM positivity, no association with outcomes was demonstrated. Low number of events are apparent across all subgroups, particularly within POLE, precluding meaningful assessment. PR overexpression was associated with good outcomes only within the p53wt group (Log Rank p ≤ 0.01 for OS, DSS, and PFS; Figure 3E–G). ER positivity was also shown to be associated with OS, DSS, and PFS within the p53wt subtype (Log Rank p ≤ 0.04 for OS, DSS, and PFS; Figure 3H–J), although the majority of p53wt tumours were ER positive (91%). No association of outcomes within ProMisE subtype was seen for STMN or PTEN.
Figure 3.
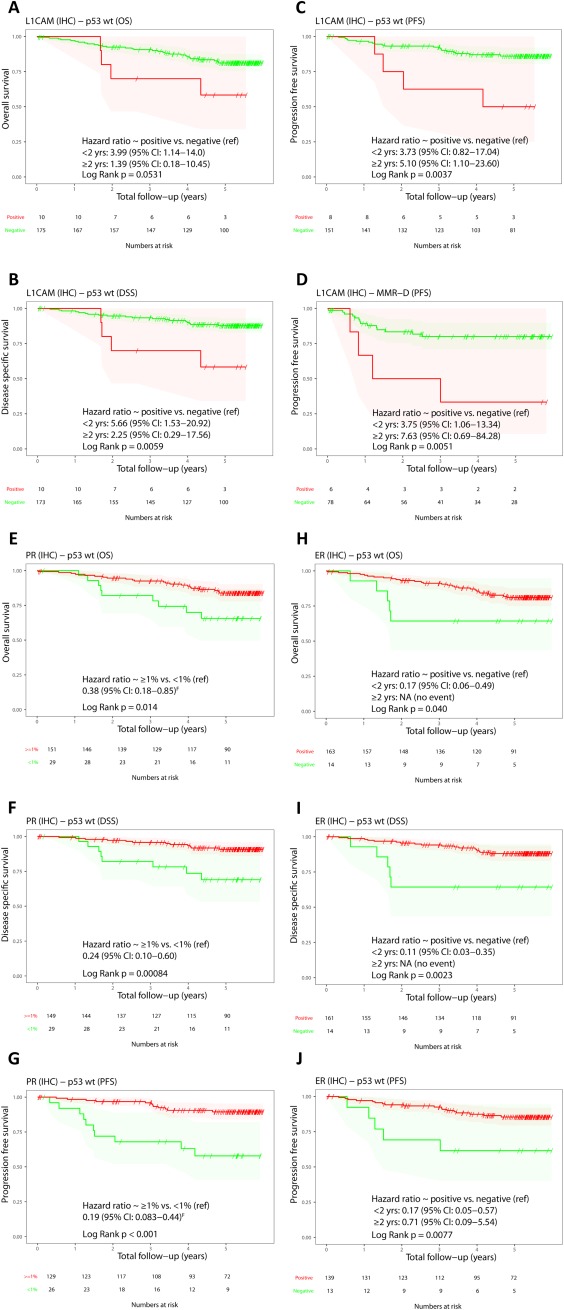
Kaplan‐Meier plots of IHC markers within selected ProMisE subtypes. (A) L1CAM OS within p53wt. (B) L1CAM DSS within p53wt. (C) L1CAM PFS within p53wt. (D) L1CAM PFS within MMR‐D. (E) PR OS within p53wt. (F) PR DSS within p53wt. (G) PR PFS within p53wt. (H) ER OS within p53wt. (I) ER DSS within p53wt. (J) ER PFS within p53wt. For L1CAM and ER, HRs are shown for follow‐up period 0–2 years and >2 years.
On multivariable survival analysis, ProMisE molecular subtype maintained prognostic significance for DSS and PFS when correcting for each IHC biomarker (except for L1CAM where ProMisE remained significant for PFS only) and for clinicopathological parameters available at time of diagnosis (age, grade, histotype, and BMI). Age maintained an association with OS only. Among all the biomarkers tested, only ER was significantly associated with outcome when corrected for other prognostic parameters. In particular, between 0 and 2 years after surgery, ER positivity was associated with better DSS (trend towards significance: p = 0.057), while after 2 years, high ER expression correlated with worse overall and disease specific survival (p ≤ 0.002). Age remained associated with OS when corrected for each additional IHC biomarker (L1CAM, PR, ER, STMN, or PTEN) and other clinicopathological parameters available at time of diagnosis (age, grade, histotype, ProMisE subtype, and BMI).
In testing for associations between IHC biomarkers tested, L1CAM expression was associated with STMN expression and ER/PR negative carcinomas. PR positive tumours were most commonly ER positive (>90%) with rare (7%) L1CAM overexpression and low STMN staining indices. PTEN expression revealed few correlations with other markers; PTEN was associated with PR expression (79% of ECs with PTEN loss were PR positive). One hundred forty cases had both IHC and mutational data for PTEN for concordance testing. PTEN IHC expression was demonstrated to be only in ‘slight agreement’ with PTEN mutation status, kappa statistic 0.19.
Testing for the prognostic impact of PTEN loss, by IHC or mutation status, in the context of BMI revealed no improved discernment of outcomes. As mentioned above, PTEN loss is associated with improved PFS across all ECs and there was a similarly reduced HR 0.47 (95% CI 0.21–0.97) (LRT‐P value 0.03) within women with BMI < 30 but not associated with outcome in women with BMI ≥ 30. When restricted to endometrioid histology only, all associations of PTEN and outcomes were lost in both BMI categories (univariable and Kaplan‐Meier survival analysis, p = ns).
Discussion
Molecular classification of EC enables reproducible categorization of tumours, identification of women who may have hereditary cancer syndromes, provides prognostic information, and has the potential to guide therapy from first diagnostic specimen 7, 8, 9, 10, 12. Optimal utilization of molecular classification, as through ProMisE, needs to be tested in prospective clinical trials. However, it is clear that the diagnostic inconsistency, and lack of biologically relevant information to inform EC risk stratification 6, 28, make the current histopathological classification system suboptimal for this disease site.
We anticipate that ProMisE, although now validated as a standalone prognostic test 9, 10, 29, may be further strengthened by the addition of select demographic, pathological, or molecular parameters. Focusing on molecular parameters of purported prognostic significance in EC that can be performed at relatively low cost (IHC), we investigated five protein biomarkers for their prognostic value in the context of post‐TCGA era of molecularly defined subtypes.
L1‐cell adhesion molecule (L1CAM) is a transmembrane protein implicated in driving tumour invasion, cell motility, and angiogenesis 30, 31. Overexpression of L1CAM has been associated with aggressive tumour features in multiple cancers, including head and neck, vulvar, ovarian and ECs. L1CAM overexpression in ECs has been demonstrated to be associated with worse outcomes, even in low stage tumours 14, 15, 16, 32. Whether these worse outcomes are secondary to L1CAM itself or if L1CAM is simply a marker for ECs with poor prognostic features (non‐endometrioid histology, LVSI, high grade) has not yet been definitively answered. Interestingly, RNA‐seq data from the TCGA cohort has been used to examine L1CAM overexpression but L1CAM status tested against traditional clinicopathological parameters and outcomes, not TCGA subtypes. The authors concluded that L1CAM was associated with multiple poor prognostic factors. Worse outcomes were seen across all L1CAM tumours; however, again, these cases were more likely advanced stage, grade, non‐endometrioid histotype so whether or not the worse outcomes were secondary to L1CAM or these other features was unclear. On multivariable analysis, looking within stage I tumours L1CAM overexpression trended towards worse survival but did not reach statistical significance 33. This is not the first study to look at this marker in the context of molecular subtypes; in the TransPORTEC series, L1CAM was the only molecular marker, of many tested, that was of prognostic significance independent of molecular subtypes 12. Together with our finding that L1CAM overexpression is associated with a worse outcome in the important MMR‐D and p53wt groups suggests it may have value within those subtypes, but further validation is required. It should be noted that there are very few tumours with L1CAM overexpression in the MMR‐D and p53wt groups (7 and 6%, respectively), which limits our power to validate previous studies demonstrating the prognostic significance of L1CAM among low‐risk tumours 12, 15, 16, 32.
Hormone receptor status has long been appreciated as prognostic in ECs; PR and estrogen receptor (ER) positivity are associated with low grade low‐risk ECs and more favourable outcomes 18, 19, 20, 34 in addition to providing therapeutic opportunities. A recent comprehensive review in over 200 ‘high‐risk’ ECs showed that although the proportion of cases with PR expression was lower in their high‐grade cohort, PR positive grade 3 endometrioid and serous carcinomas had improved OS 17. The utility of PR and ER IHC in the context of molecular subtypes was recently evaluated in the postoperative radiation therapy for endometrial carcinoma (PORTEC) cohorts 12; ER/PR status correlated with molecular subtype, and low ER and PR correlated with higher incidence of loco‐regional and recurrence and lower OS.
We confirmed the association of both hormone receptors with outcomes; however, on multivariable analysis this association was not maintained except for a trend towards significance (p = 0.057) for improved DSS for ER‐positive tumours (supplementary material, Tables S7 and S8). Furthermore, for patients surviving more than 2 years, we observed that ER‐positive tumours were associated with worse prognosis (Figure 2F). However, due to the limitation of our relatively small and heterogeneous study cohort, we cannot explore this finding further. Our data show that, within the p53wt subtype, the presence of either hormone receptor may add value in discerning improved outcomes (Figure 3E–J). A recent meta‐analysis showed that hormonal status is associated with a greater response to hormonal therapies in advanced ECs 35, so there may both a prognostic and predictive role of ER and PR in the appropriate clinical setting.
Stathmin (STMN) is an oncoprotein that has been linked to phosphoinositide‐4,5‐bisphosphate 3‐kinase (PI3K) pathway activation, aggressive clinicopathological features, and poor prognosis in several solid tumours including ECs 21, 36, 37, 38. Stathmin has also been suggested as a predictive biomarker, associated with resistance to antimicrotubule agents 39, and possibly identifying tumours that may benefit from PI3Kinase/mTOR (mammalian target of rapamycin) inhibitor therapy 40. In our series, although we demonstrated STMN was associated with several adverse prognostic features, no clear associations with outcomes were demonstrated. This may be a result of our simplified scoring system (9‐category, ultimately binarized to overexpression versus not) but even upon testing at several different thresholds, the lack of profound differences in clinicopathological parameters between ECs with stathmin overexpression versus low expression cases, the lack association with outcomes, and the somewhat tedious and complex initial scoring system dampens our enthusiasm for widespread use of this marker.
Mutations in the tumour suppressor gene PTEN are common in ECs. Publications on the prognostic implications of PTEN loss in ECs, as measured by IHC or mutation status, are discordant 23, 25, 41. The impact of functional PTEN loss on tumour behavior and clinical outcomes may need to be interpreted within the context of metabolic phenotype (BMI). Westin et al demonstrated that PTEN loss (by mutation status or IHC) was not associated with outcomes across a cohort of ∼190 ECs but, after restricting evaluation to obese individuals (BMI ≥ 30), PTEN loss was associated with improved PFS 23. Functional PTEN loss can arise by somatic mutations, epigenetic/post‐transcriptional events, or instability and degradation of the PTEN protein. Concordance between PTEN mutation status and IHC is poor and arguably PTEN IHC may better represent functional loss 24. However, difficulties with staining, heterogeneity of staining patterns, and interpretation have all hindered its use in clinical practice 42, 43. In this study, correlation between PTEN IHC and mutation status was poor, differences between clinicopathological variables for ECs with PTEN loss versus expression were minimal, and prognostic value of PTEN status limited; PTEN loss was associated with PFS only (p = 0.03). Although PTEN loss by IHC was associated with obesity, no differences in our ability to predict outcomes were seen within obese (BMI ≥ 30) individuals. That said, evaluation of PTEN loss might still be of use as a predictive biomarker for response to poly (ADP‐ribose) polymerase (PARP) inhibitors 44, 45.
In this series of over 400 ECs, survival analysis demonstrated that although these additional biomarkers evaluated may be associated with selected clinicopathological variables or outcome parameters, no single IHC biomarker outperformed ProMisE. The additional IHC parameters evaluated frequently segregated with specific molecular subtype (e.g. L1CAM overexpression with the p53abn subtype), suggesting these proteins may simply be a feature of an overriding genotype/phenotype that classification tools such as ProMisE provide. Although both L1CAM and PR were significantly associated with OS, DSS, and PFS (ER with DSS only, PTEN with PFS only), on multivariable survival analysis, only the ProMisE molecular classifier maintained a statistically significant association with OS, DSS, and PFS. In terms of clinicopathological parameters, only age maintained an association with outcomes and only for OS.
The polarity of outcomes in the p53abn subtype (poor) and POLE subtype (highly favourable) suggests that these subtypes will likely not benefit from further refinement through the application of additional prognostic parameters. Of greater clinical import would be discerning outcomes within the two ‘intermediate’ survival outcome molecular subtypes; both the p53wt and MMR‐D subtypes exhibit diversity of outcomes within them, with little indication as yet as to how to discriminate between these tumours. The greatest potential to add prognostic value within these intermediate outcome ProMisE subtypes was shown for L1CAM and PR. We demonstrated an improved ability to discern outcomes through the addition of L1CAM testing within the MMR‐D and p53wt subtypes but only a small proportion of these cases exhibited L1CAM overexpression. Nonetheless, for those MMR‐D or p53wt ECs that did have L1CAM overexpression outcomes were clearly worse. Some of the prior publications on L1CAM and prognosis, which suggested it helps discern a worse outcome group within low or intermediate risk tumours may actually reflect that these tumours were incorrectly assigned by histopathological features and may represent ‘missed’ serous or high grade aggressive endometrioid adenocarcinomas incorrectly classified. With molecular classification, as through ProMisE, serous carcinomas would be identified as p53abn and thus clearly identified as high risk for recurrence and managed appropriately. An additional benefit of L1CAM status within the p53abn subtype was not seen in our series. PR status improved the ability to discern outcomes within the p53wt subtype only, and if this association is validated, could be considered as an additional prognostic test within this select cohort.
Strengths of this study include utilization of a previously described large, well‐characterized EC cohort with mature outcome data. TMAs had proven successful for IHC in the ProMisE classifier and were again successfully utilized, with ∼10–15% of uninterpretable cases. However, TMAs do not reflect what would be used in clinical practice (e.g. whole sections) and heterogeneity of these tumours means TMA may not always provide the best representation of the tumour. For example, L1CAM expression can be heterogeneous and is often only expressed at the invasive tumour front, which is not typically represented on TMAs, so our data may underestimate L1CAM overexpression in our cohort. The usual challenges of IHC for PTEN were demonstrated again in this series, lessening its applicability in clinical practice. While the ProMisE classifier has been validated in a retrospective cohort 10, it awaits validation in a prospective setting. The need to sequence POLE could be considered a limitation to the widespread implementation of this molecular classifier in clinical practice.
In summary, testing L1CAM, PR, ER, STMN, and PTEN IHC biomarkers across all ECs does not appear to add prognostic value over ProMisE classification alone. Potential value added within molecular subtypes associated with intermediate outcomes may justify further studies on L1CAM and hormone receptor status specifically in the MMR‐D and p53wt subtypes.
Author contributions statement
JMc, BG: conceived of and designed study with DH and AK contributing; JMc: wrote the manuscript with BG and AK; AK, FK, MK, CHL, JM, BG: scored/interpreted the IHC on TMA, and contributed to writing and editing of paper; MMc, WY: did the molecular work including POLE testing and interpretation; SL, AT: designed and performed the statistical analyses; CC: created TMA and aided in all steps IHC with assistance JM.
Supporting information
Figure S1. Case selection
Figure S2. Immunohistochemistry for L1CAM demonstrating L1CAM low (up to 10%) versus high (>10%) expression
Table S1. REMARK criteria
Tables S2‐6 show summary statistics of clinicopathological variables of interest split by score of each IHC marker; L1CAM, STMN, PR ER,and PTEN. For categorical and binary variables, counts and corresponding column percentages are shown. For continuous variables, the mean, median, IQR, and range are calculated. In addition, the number of missing cases is shown at the bottom of each subsection in the table. Note that the missing cases do not contribute to the percentages.
Table S2. Clinicopathological Variables by IHC Marker Status. Univariable association of predictors with L1CAM IHC
Table S3. Clinicopathological Variables by IHC Marker Status. Univariable association of predictors with STMN IHC
Table S4. Clinicopathological Variables by IHC Marker Status. Univariable association of predictors with PR IHC
Table S5. Clinicopathological Variables by IHC Marker Status. Univariable association of predictors with ER IHC
Table S6. Clinicopathological Variables by IHC Marker Status. Univariable association of predictors with PTEN IHC
Table S7. Multivariable model with parameters available at diagnosis (adjusting for treatment)
Table S8. Multivariable model with parameters available after surgery (adjusting for treatment)
Acknowledgement
This study was supported by Canadian Institute of Health Research [Proof of Principal (POP2)] and BC Cancer Foundation (Clinical Investigator Award).
References
- 1. Ferlay J, Soerjomataram I, Dikshit R, et al Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–E386. [DOI] [PubMed] [Google Scholar]
- 2. Han G, Sidhu D, Duggan MA, et al Reproducibility of histological cell type in high‐grade endometrial carcinoma. Mod Pathol 2013; 26: 1594–1604. [DOI] [PubMed] [Google Scholar]
- 3. Gilks CB, Oliva E, Soslow RA. Poor interobserver reproducibility in the diagnosis of high‐grade endometrial carcinoma. Am J Surg Pathol 2013; 37: 874–881. [DOI] [PubMed] [Google Scholar]
- 4. Colombo N, Preti E, Landoni F, et al Endometrial cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol 2013; 24: vi33–vi38. [DOI] [PubMed] [Google Scholar]
- 5. Colombo N, Creutzberg C, Amant F, et al ESMO‐ESGO‐ESTRO Consensus Conference on Endometrial Cancer: Diagnosis, treatment and follow‐up. Int J Gynecol Cancer 2016; 26: 2–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paulsson G, Genell A, Jakobsen AM, et al Usefulness of current risk groups in the treatment of surgically staged endometrial carcinomas; a population‐based study from Western Sweden. Anticancer Res 2009; 29: 1585–1590. [PubMed] [Google Scholar]
- 7. Kandoth C, McLellan MD, Vandin F, et al Mutational landscape and significance across 12 major cancer types. Nature 2013; 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Talhouk A, Hoang LN, McConechy MK, et al Molecular classification of endometrial carcinoma on diagnostic specimens is highly concordant with final hysterectomy: earlier prognostic information to guide treatment. Gynecol Oncol 2016; 143: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Talhouk A, McConechy MK, Leung S, et al A clinically applicable molecular‐based classification for endometrial cancers. Br J Cancer 2015; 113: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Talhouk A, McConechy MK, Leung S, et al Confirmation of ProMisE: a simple, genomics‐based clinical classifier for endometrial cancer. Cancer 2017; 123: 802–813. [DOI] [PubMed] [Google Scholar]
- 11. Stelloo E, Bosse T, Nout RA, et al Refining prognosis and identifying targetable pathways for high‐risk endometrial cancer; a TransPORTEC initiative. Mod Pathol 2015; 28: 836–844. [DOI] [PubMed] [Google Scholar]
- 12. Stelloo E, Nout RA, Osse EM, et al Improved risk assessment by integrating molecular and clinicopathological factors in early‐stage endometrial cancer‐combined analysis of the PORTEC cohorts. Clin Cancer Res 2016; 22: 4215–4224. [DOI] [PubMed] [Google Scholar]
- 13. Micheel CM, Nass SJ, Omenn GS. (Eds). Evolution of Translational Omics: Lessons Learned and the Path Forward. National Academies Press (US): Washington, DC, 2012. [PubMed] [Google Scholar]
- 14. Van Gool IC, Stelloo E, Nout RA, et al Prognostic significance of L1CAM expression and its association with mutant p53 expression in high‐risk endometrial cancer. Mod Pathol 2016; 29: 174–181. [DOI] [PubMed] [Google Scholar]
- 15. Kommoss F, Kommoss F, Grevenkamp F, et al L1CAM: amending the “low‐risk” category in endometrial carcinoma. J Cancer Res Clin Oncol 2017; 143: 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zeimet AG, Reimer D, Huszar M, et al L1CAM in early‐stage type I endometrial cancer: results of a large multicenter evaluation. J Natl Cancer Inst 2013; 105: 1142–1150. [DOI] [PubMed] [Google Scholar]
- 17. Kobel M, Atenafu EG, Rambau PF, et al Progesterone receptor expression is associated with longer overall survival within high‐grade histotypes of endometrial carcinoma: a Canadian high risk endometrial cancer consortium (CHREC) study. Gynecol Oncol 2016; 141: 559–563. [DOI] [PubMed] [Google Scholar]
- 18. Shen F, Gao Y, Ding J, et al Is the positivity of estrogen receptor or progesterone receptor different between type 1 and type 2 endometrial cancer? Oncotarget 2017; 8: 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y, Zhao D, Gong C, et al Prognostic role of hormone receptors in endometrial cancer: a systematic review and meta‐analysis. World J Surg Oncol 2015; 13: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trovik J, Wik E, Werner HM, et al Hormone receptor loss in endometrial carcinoma curettage predicts lymph node metastasis and poor outcome in prospective multicentre trial. Eur J Cancer 2013; 49: 3431–3441. [DOI] [PubMed] [Google Scholar]
- 21. Trovik J, Wik E, Stefansson IM, et al Stathmin overexpression identifies high‐risk patients and lymph node metastasis in endometrial cancer. Clin Cancer Res 2011; 17: 3368–3377. [DOI] [PubMed] [Google Scholar]
- 22. Trovik J, Wik E, Stefansson I, et al Stathmin is superior to AKT and phospho‐AKT staining for the detection of phosphoinositide 3‐kinase activation and aggressive endometrial cancer. Histopathology 2010; 57: 641–646. [DOI] [PubMed] [Google Scholar]
- 23. Westin SN, Ju Z, Broaddus RR, et al PTEN loss is a context‐dependent outcome determinant in obese and non‐obese endometrioid endometrial cancer patients. Mol Oncol 2015; 9: 1694–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Djordjevic B, Hennessy BT, Li J, et al Clinical assessment of PTEN loss in endometrial carcinoma: immunohistochemistry outperforms gene sequencing. Mod Pathol 2012; 25: 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McConechy MK, Talhouk A, Leung S, et al Endometrial carcinomas with POLE exonuclease domain mutations have a favorable prognosis. Clin Cancer Res 2016; 22: 2865–2873. [DOI] [PubMed] [Google Scholar]
- 26. Terry J, Torlakovic EE, Garratt J, et al Implementation of a Canadian external quality assurance program for breast cancer biomarkers: an initiative of Canadian Quality Control in immunohistochemistry (cIQc) and Canadian Association of Pathologists (CAP) National Standards Committee/Immunohistochemistry. Appl Immunohistochem Mol Morphol 2009; 17: 375–382. [DOI] [PubMed] [Google Scholar]
- 27. Alkushi A, Clarke BA, Akbari M, et al Identification of prognostically relevant and reproducible subsets of endometrial adenocarcinoma based on clustering analysis of immunostaining data. Mod Pathol 2007; 20: 1156–1165. [DOI] [PubMed] [Google Scholar]
- 28. Bendifallah S, Canlorbe G, Collinet P, et al Just how accurate are the major risk stratification systems for early‐stage endometrial cancer? Br J Cancer 2015; 112: 793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Talhouk A, Kommoss S, McConechy MK, et al. Confirmation of a simple clinically-applicable molecular classifier of endometrial cancer: moving to prime time. International Gynecologic Cancer Society (IGCS): Lisbon, Portugal, 2017. [Google Scholar]
- 30. Gast D, Riedle S, Riedle S, et al L1 augments cell migration and tumor growth but not beta3 integrin expression in ovarian carcinomas. Int J Cancer 2005; 115: 658–665. [DOI] [PubMed] [Google Scholar]
- 31. Mechtersheimer S, Gutwein P, Agmon‐Levin N, et al Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J Cell Biol 2001; 155: 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bosse T, Nout RA, Stelloo E, et al L1 cell adhesion molecule is a strong predictor for distant recurrence and overall survival in early stage endometrial cancer: pooled PORTEC trial results. Eur J Cancer 2014; 50: 2602–2610. [DOI] [PubMed] [Google Scholar]
- 33. Dellinger TH, Smith DD, Ouyang C, et al L1CAM is an independent predictor of poor survival in endometrial cancer ‐ an analysis of The Cancer Genome Atlas (TCGA). Gynecol Oncol 2016; 141: 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jongen V, Briet J, de Jong R, et al Expression of estrogen receptor‐alpha and ‐beta and progesterone receptor‐A and ‐B in a large cohort of patients with endometrioid endometrial cancer. Gynecol Oncol 2009; 112: 537–542. [DOI] [PubMed] [Google Scholar]
- 35. Ethier JL, Desautels DN, Amir E, et al Is hormonal therapy effective in advanced endometrial cancer? A systematic review and meta‐analysis. Ethier Gynecol Oncol 2017;147:158–166. [DOI] [PubMed] [Google Scholar]
- 36. Salvesen HB, Carter SL, Mannelqvist M, et al Integrated genomic profiling of endometrial carcinoma associates aggressive tumors with indicators of PI3 kinase activation. Proc Natl Acad Sci U S A 2009; 106: 4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wik E, Birkeland E, Trovik J, et al High phospho‐Stathmin(Serine38) expression identifies aggressive endometrial cancer and suggests an association with PI3K inhibition. Clin Cancer Res 2013; 19: 2331–2341. [DOI] [PubMed] [Google Scholar]
- 38. Reyes HD, Miecznikowski J, Gonzalez‐Bosquet J, et al High stathmin expression is a marker for poor clinical outcome in endometrial cancer: an NRG oncology group/gynecologic oncology group study. Gynecol Oncol 2017; 146: 247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Werner HM, Trovik J, Halle MK, et al Stathmin protein level, a potential predictive marker for taxane treatment response in endometrial cancer. PLoS One 2014; 9: e90141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Del Campo JM, Birrer M, Davis C, et al A randomized phase II non‐comparative study of PF‐04691502 and gedatolisib (PF‐05212384) in patients with recurrent endometrial cancer. Gynecol Oncol 2016; 142: 62–69. [DOI] [PubMed] [Google Scholar]
- 41. Estrella JS, Broaddus RR, Mathews A, et al Progesterone receptor and PTEN expression predict survival in patients with low‐ and intermediate‐grade pancreatic neuroendocrine tumors. Arch Pathol Lab Med 2014; 138: 1027–1036. [DOI] [PubMed] [Google Scholar]
- 42. Pallares J, Bussaglia E, Martinez‐Guitarte JL, et al Immunohistochemical analysis of PTEN in endometrial carcinoma: a tissue microarray study with a comparison of four commercial antibodies in correlation with molecular abnormalities. Mod Pathol 2005; 18: 719–727. [DOI] [PubMed] [Google Scholar]
- 43. Mutter GL, Ince TA, Baak JP, et al Molecular identification of latent precancers in histologically normal endometrium. Cancer Res 2001; 61: 4311–4314. [PubMed] [Google Scholar]
- 44. Dedes KJ, Wetterskog D, Mendes‐Pereira AM, et al PTEN deficiency in endometrioid endometrial adenocarcinomas predicts sensitivity to PARP inhibitors. Sci Transl Med 2010; 2: 53ra75. [DOI] [PubMed] [Google Scholar]
- 45. Forster MD, Dedes KJ, Sandhu S, et al Treatment with olaparib in a patient with PTEN‐deficient endometrioid endometrial cancer. Nat Rev Clin Oncol 2011; 8: 302–306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Case selection
Figure S2. Immunohistochemistry for L1CAM demonstrating L1CAM low (up to 10%) versus high (>10%) expression
Table S1. REMARK criteria
Tables S2‐6 show summary statistics of clinicopathological variables of interest split by score of each IHC marker; L1CAM, STMN, PR ER,and PTEN. For categorical and binary variables, counts and corresponding column percentages are shown. For continuous variables, the mean, median, IQR, and range are calculated. In addition, the number of missing cases is shown at the bottom of each subsection in the table. Note that the missing cases do not contribute to the percentages.
Table S2. Clinicopathological Variables by IHC Marker Status. Univariable association of predictors with L1CAM IHC
Table S3. Clinicopathological Variables by IHC Marker Status. Univariable association of predictors with STMN IHC
Table S4. Clinicopathological Variables by IHC Marker Status. Univariable association of predictors with PR IHC
Table S5. Clinicopathological Variables by IHC Marker Status. Univariable association of predictors with ER IHC
Table S6. Clinicopathological Variables by IHC Marker Status. Univariable association of predictors with PTEN IHC
Table S7. Multivariable model with parameters available at diagnosis (adjusting for treatment)
Table S8. Multivariable model with parameters available after surgery (adjusting for treatment)


