Abstract

Knowledge of the ionic strength in cells is required to understand the in vivo biochemistry of the charged biomacromolecules. Here, we present the first sensors to determine the ionic strength in living cells, by designing protein probes based on Förster resonance energy transfer (FRET). These probes allow observation of spatiotemporal changes in the ionic strength on the single-cell level.
The ionic strength influences a wide array of electrostatic interactions in the cell.1,2 To understand the role of the ionic strength in cell physiology, a sensor to quantify the ionic strength in vivo is needed. The ionic strength is the effective ion concentration and screens electrostatic interactions of (macro)molecules, which is obviously of crucial importance to the cell. For example, the ionic strength determines the structure of intrinsic disordered proteins,3 activity of enzymes,4 protein aggregation,5 quinary assemblies/phase separations,6 protein binding to (poly)nucleic acids,7 the catalytic function of riboswitches,8 and many other processes. The ionic strength governs the cell volume by activating the channels and transporters involved.9,10 The ionic strength can vary, however, depending on the extracellular environment, due to, for example, abrupt changes in medium osmolarity and fluctuations with intracellular events such as metabolite or polynucleotide synthesis.11
To quantify the ionic strength, one cannot simply use the concentration of ions, because the effective ion concentration (i.e., thermodynamic activity) is altered due to interaction of the ions with each other and the biomolecules. Indeed, a subset of ions has a higher affinity for macromolecules, such as magnesium, while others are more loosely associated. Their activity coefficient may approach that in the extracellular medium.12 Such effects result in a strong dependence of electrostatic screening on the identity of the ions, roughly following the Hofmeister series.13,14 Thus, although it is possible to determine the concentration of a particular ion with, for example, flame photometry of dry cell mass,15 this will not immediately reveal the ionic strength; that is, the effective concentration of an ion depends on its environment. In addition, these techniques will not yield information on population heterogeneity or rapid temporal changes in the ionic strength. Probes to infer the ionic strength from the activity of a membrane protein9 or the fluorescence of a fluorescein–BSA conjugate16 are complicated to use and interpret, and they have only been applied in vesicles and isolated mitochondria but not in living cells.
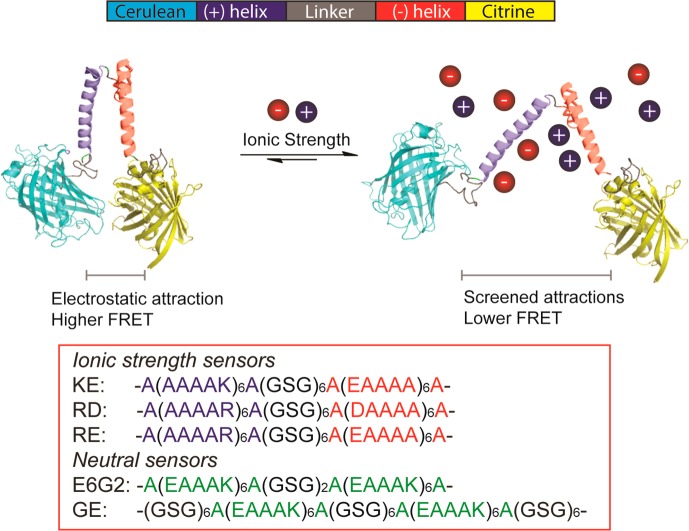
Here, we fill this void by designing the first probes that sense the ionic strength in living cells. The sensors are based on FRET and are genetically encoded, because these properties allow monitoring analytes with high spatiotemporal precision inside living cells. Fluorescent protein probes can be affected by ions nonspecifically,17 and FRET sensors selective for a specific ion have been developed, for example, zinc, calcium, or choride.18−20 We construct our sensors to contain a positively and a negatively charged α-helix, whose electrostatic attraction will depend on the ionic strength (Figure 1). At the N- and C-termini of the helices are two fluorescent proteins, mCerulean3 and mCitrine, that form a FRET pair.21 Increasing the ionic strength will decrease the attraction between the helices, which will result in a decrease in FRET efficiency.
Figure 1.
Design of ionic strength sensors and sensing concept. Ions screen the attraction between the positively and negatively charged helices, reducing the FRET efficiency.
To induce charge in the helices, we inserted six glutamates in an alanine background in one helix and six lysines in the other helix (Figure 1). The charged amino acids are in an i + 5 spacing, a staircase-like configuration, ensuring that all sides of the helix are covered with charges. This prevents the helix from having a charged patch as well as preventing metal ion chelation. We further reasoned that the high intramolecular concentration of the oppositely charged α-helix will outcompete electrostatic interactions with most of the charged cellular biomacromolecules. Because the presence of specific ion effects could be dependent on the identity of the amino acids in the helix, we designed sensors with glutamate or aspartate residues, and lysine or arginine residues, and thereby alter, e.g., salt-bridge stability.22 With these design elements, we avoid selectivity for specific ions in the sensors, allowing for ionic strength determination in living cells.
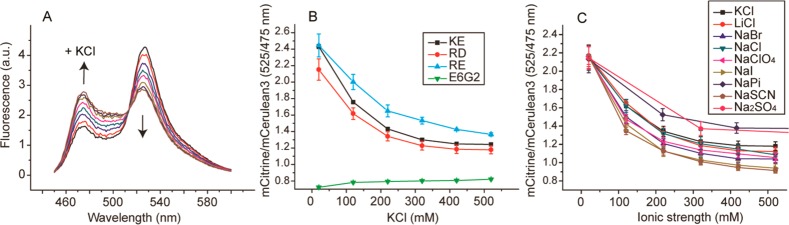
To determine the ionic strength sensitivity of the sensors, we performed extensive in vitro tests on isolated probes (Figure 2A, Supporting Information Figure S1). We tested the three ionic strength sensors, which are the KE (lysine-glutamate), the RE (arginine-glutamate), and the RD (arginine-aspartate) sensors, and compared the findings with a sensor that contains neutral helices, the E6G2 probe.23 We found that the FRET efficiencies of the ionic strength sensors in 10 mM NaPi buffer were much higher than that of the E6G2 probe, confirming that the helices attract each other. The addition of potassium chloride to the ionic strength sensors led to a decrease in FRET efficiency (Figure 2B), with the highest sensitivity between 0 and 300 mM KCl. The E6G2 probe was not sensitive, showing that we probe the ionic strength. Salt does not affect other neutral sensors with higher FRET efficiencies,23 and hence the FRET efficiency does not relate to the ionic strength dependence. We added a wide range of salts to test whether specific ion effects interfere with the readouts (Figure 2C). We find that the sensitivity to the cations of KCl, NaCl, MgCl2, and LiCl is the same when the ratios are plotted versus the ionic strength (Supporting Information Figure S3). In contrast, the readout had a dependence on the identity of the anion (Supporting Information Figure S2). The order of sensitivity followed the Hofmeister series, that is, the measure to what extent ions are hydrated, which is generally more dependent on the anion. This deviation from ideal behavior depended on the probe and qualitatively followed the intramolecular salt bridge strength between the two helices;22 the RD probe has the lowest salt-bridge strength and is least affected by the nonideal behavior, as judged from the spread in deviation between the different ions. Likely due to the lower salt-bridge strength, the RD probe also has a lower FRET efficiency. Together, the dependence on ion identity indicates that we probe the effective ion concentration (or ionic strength) rather than total ion concentration. In all cases, the E6G2 probe was insensitive to any ion.
Figure 2.
In vitro characterization of the ionic strength sensors. (a) Fluorescence emission spectra of the RD sensor titrated with potassium chloride. (b) Dependence of the mCitrine/mCerulean3 ratio of the EK, RD, RK, and E6G2 sensors on the potassium chloride concentration. (c) Dependence of the mCitrine/mCerulean3 ratio of the RD sensor on the ion identity. All experiments are in 10 mM NaPi, at pH 7.4. Error bars indicate the standard deviation of three biological replicates.
We investigated sensitivity to a wide variety of other parameters to make the transition to in vivo measurements. Similar to the E6G2 and GE crowding probes,21 mCitrine induced pH sensitivity in all the probes only below pH 7.0 (Supporting Information Figure S4). The zwitterionic glycine betaine and neutral small molecules sorbitol and sucrose hardly influenced the probes (Supporting Information Figure S5). The common intracellular organophosphates fructose bisphoshate, ATP, ADP, and AMP influenced the probes as can be expected from their charge, albeit the sensitivity to the nucleotides was somewhat higher (Supporting Information Figure S6). Glutathione influenced the probes according to its net negative charge (Supporting Information Figure S7). The sensitivity to potassium glutamate was less than that to potassium chloride, as expected from the Hofmeister series, with the RD probe again deviating the least (Supporting Information Figure S8). The sensors associated with the polyelectrolyte DNA in the absence of KCl, but not in the presence of 100 mM KCl (Supporting Information Figure S9). Macromolecular crowding induced by Ficoll 70 compressed all the probes (Supporting Information Figure S10) and hence should be taken into account when measuring in live cells. The ionic strength probes are temperature sensitive in the absence of salt, but the temperature sensitivity decreased with increasing salt concentration (Supporting Information Figure S11). Increasing the sensor concentration itself did not affect the ratio (Supporting Information Figure S12), ruling out interference from self-association. Hence, the sensors are sensitive to the ionic strength mainly, but other parameters such as macromolecular crowding, pH, and temperature need to be controlled for.
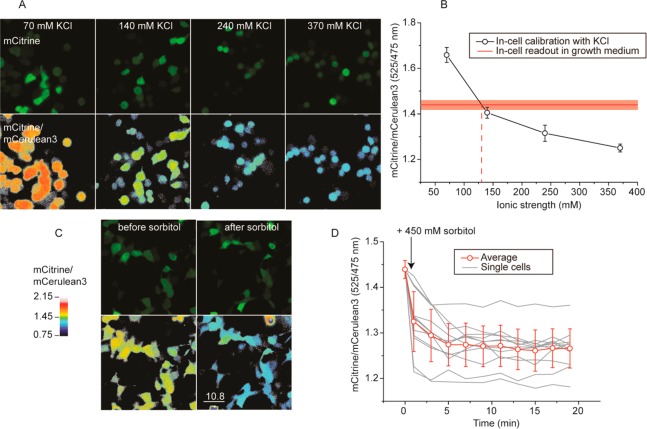
To demonstrate ionic strength sensing in vivo, we expressed the probes in the mammalian cell line HEK293 and imaged the cells by scanning confocal microscopy. The cells were subsequently imaged by excitation of the mCerulean3 at 405 nm, and the emission ratio of mCitrine (505–750 nm) over mCerulean3 (450–505 nm), after background subtraction, was monitored (Figure 3A,B). The ratios obtained were very homogeneous over all the cells. When comparing the different sensors, we found that RD gave lower ratios than the KE and RE probes, similar to what we observed in vitro (Supporting Information Figure S14). To quantify the readouts, we calibrated the probes in the cell by clamping the internal potassium concentration by titrating the external medium with potassium chloride in the presence of the ionophores valinomycin plus nigericin. These ionophores equilibrate the protons and potassium over the membrane, providing a calibration curve of the corresponding sensor with a known concentration of potassium. All the ion sensors sensed the potassium concentration in a similar manner to that observed in vitro. The ratios for the GE crowding sensor remained unchanged, indicating that with the calibration method we did not alter the crowding and prevent osmotic pressure differences over the membrane by ionophore-assisted ion equilibration. The cells did change shape during the calibration procedure (Figure 3A). We subsequently used the calibration curves to quantify the ionic strength from the readout in a regular growth medium and found that the ionic strength was comparable to ∼130 mM for the RD and RE sensors and ∼110 mM for the KE sensor (Figure 3B, Supporting Information Figure S14). These values are in the same range as the ion concentrations reported for the monovalent ions in various cells, and the concentration of “free” ions is somewhat lower.24 The total ion concentration would be 140 mM and 12 mM for potassium and sodium, respectively. Divalent cations were not monitored because they are mostly bound to the biopolymers. Organophosphates form complexes with magnesium and may bind nonspecifically to proteins,25−27 and the molecules thus contribute less to the ionic strength than is expected from their total concentrations. Single cell measurements as a function of time indicate that the ionic strength is determined with a precision better than 10 mM (that is, changes in mCitrine/mCerulean3 of less than 0.02). Thus, we conclude that the probes function in HEK293 cells, and we can determine the ionic strength in vivo.
Figure 3.
Ionic strength determination in vivo and observation of dynamic changes observed in scanning fluorescence confocal microscopy of HEK293 cells expressing the RD sensor. (a) Fluorescence and mCitrine/mCerulean3 ratio images of sensor calibration with KCl, nigericin, and valinomycin. (b) Calibration curve of the mCitrine/mCerulean3 ratio versus potassium concentration; the red line is the mCitrine/mCerulean3 ratio of cells in growth medium (red shading is the standard deviation within a population, n = 20). (c) Fluorescence and mCitrine/mCerulean3 ratio images of cells before and 10 min after addition of 450 mM sorbitol. See Supporting Figure S13 for more detail. (d) mCitrine/mCerulean3 ratio upon addition of sorbitol as a function of time. The data of 20 individual cells and the average are shown. Error bars are standard deviation within a single population of cells. The standard deviations of the averages of independent biological repeats are smaller (±0.03, n = 4).
To further demonstrate the potential of the sensors in vivo, we monitored changes in ionic strength over time after an osmotic upshift by adding 450 mM sorbitol to HEK293 cells (Figure 3C,D). We expected the short-term response to be an initial increase in crowding and ionic strength, followed by a regulatory volume increase by uptake of potassium and chloride from the medium, retaining the abnormally high ionic strength but decreasing the crowding.11 We assume that the response mechanism of the sensor is much faster than the biological events. We saw with the GE crowding probe that the crowding indeed increased with 450 mM sorbitol (Supporting Information Figure S15), as we observed previously, after which a slow decrease in crowding took place. In the time frame of ∼20 min, the crowding was not yet fully recovered. The RD sensor showed that the ionic strength increased with an osmotic upshift and remained at this level. It showed no apparent sensitivity to in vivo crowding, contrary to the KE and RE sensors that showed an initial increase in FRET directly after the osmotic upshift. Interestingly, an increase in ionic strength of ∼190 mM to 320 mM can be inferred with the RD calibration curve, which is similar to the expected increase in osmolarity simply based on equating the sum of K and Cl to the number of sorbitol molecules added. We could monitor single cells in time and found that the distribution of FRET values increased after the osmotic upshift and the adaptation process that followed. This shows that the cells are affected differently by the osmotic upshift, which could, for example, be related to the cell cycle15 or intrinsic variation in fitness in the population of cells that we analyzed. Hence, the probes can sense the ionic strength on the single-cell level during dynamic changes in the ionic strength.
In conclusion, we present here the first sensors to determine the ionic strength that function in living cells. Taking into account changes in macromolecular crowding and pH, the sensors allow facile determination of the intracellular ionic strength on the single-cell level in changing environments or intracellular conditions. Given the wide variety of processes that are influenced by the ionic strength, these sensors will aid future investigations on the importance of the ionic strength during a wide variety of conditions such as cell volume regulation, disease, and environmental stresses.
Methods
Expression and Purification of the Ionic Strength Sensors
Synthetic genes (Supporting Information) encoding the ionic strength sensors in pRSET-A were obtained from GeneArt. The plasmids were transformed into the E. coli strain BL21(DE3) pLysS (Promega). The cells were grown to an OD600 of 0.6 at 37 °C and shaking at 200 rpm in LB medium (1.0% bactotryptone, 0.5% yeast extract, 1% NaCl) with 1 mg mL–1 ampicillin, after which the cells were induced overnight with 0.1 mM isopropyl-β-d-thiogalactoside (IPTG) at 25 °C and shaking at 200 rpm. The cell lysate was cleared by centrifugation, supplemented with 10 mM imidazole, and purified by nickel-nitrilotriacetic acid Sepharose chromatography (wash/elution buffer: 20/250 mM imidazole, 50 mM NaPi, 300 mM NaCl, at pH 8.0). The sensor was further purified by Superdex 200 10/300 GL size-exclusion chromatography (Amersham Biosciences) in 10 mM NaPi, at pH 7.4. Fractions containing pure protein were aliquoted and stored at −80 °C.
In Vitro Characterization of the Sensors
A 300 μL solution containing the given salt and 10 mM NaPi (adjusted the pH to 7.4 after dissolution of the salts) was added to a 96-well plate (Greiner). The purified sensor was added, and the fluorescence intensities at 475 and 525 nm were recorded separately in a Spark 10 M microplate reader with excitation at 420 nm at RT. A 20 nm bandwidth for excitation and emission was applied, and the average of 10 measurements of a single well was taken. The background fluorescence, buffer without sensor, was subtracted.
Transfection and Imaging of HEK293 Cells
Transfection and imaging of HEK293 cells was performed as described,21 with some minor modifications. HEK293 cells (ATCC CRL-1573, tested for mycoplasma contamination) were cultured in DMEM (Gibco) supplemented with 10% (v/v) fetal calf serum, 2 mM l-glutamine (Gibco), 100 units mL–1 penicillin (Invitrogen), and 100 μg mL–1 streptomycin (Invitrogen). For transfection experiments, HEK293 cells were plated in eight-well Labtek glass chamber slides (Thermo Scientific) at 6 × 104 cells per well. One day after plating, the cells were transfected with plasmid DNA coding for the sensors as follows: lipoplexes composed of 1.5 μL of Lipofectamine 2000 (Invitrogen) and 0.5 μg of the pcDNA 3.1 vector carrying the corresponding sensor gene were prepared in 100 μL of serum-free DMEM, according to the manufacturer’s instructions. A total of 25 μL of lipoplex solution was added per well and incubated for 4 h at 37 °C and 5% CO2, after which the medium was refreshed. The next day, the medium was replaced by 200 μL of DMEM with HEPES without phenol red, and sensor expression in the HEK293 cells was subsequently analyzed by confocal fluorescence microscopy. The cells were imaged directly in the eight-well Labtek glass chamber slides. The slides were mounted on a laser-scanning confocal microscope (Zeiss LSM 710), at 37 °C. The sensor was excited using a 405 nm LED laser, and the emission was split into a 450–505 nm channel and 505–797 nm channel. The fluorescence intensity of the cells was determined in ImageJ for each channel. The backgrounds for each channel were subtracted and the mCitrine intensity divided by the mCerulean intensity for each cell. When the mCitrine intensity was plotted versus the mCerulean intensity, linear fits with R2 > 0.99 were generally obtained.
Acknowledgments
This work was supported by the China Scholarship Council grant to B.L., The Netherlands Organization for Scientific Research program grant TOP-PUNT (project number 13.006) and an ERC Advanced Grant (ABCVolume) to B.P., and The Netherlands Organization for Scientific Research Vidi grant (723.015.002) to A.J.B. P. Alvarez Sieiro is acknowledged for assistance with molecular cloning and B. Crielaard for the use of the mammalian cell culture facilities.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.7b00348.
Supporting Figures S1–S15, DNA sequences of the ionic strength sensors (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Warshel A.; Russell S. T. (1984) Calculations of electrostatic interactions in biological systems and in solutions. Q. Rev. Biophys. 17, 283–422. 10.1017/S0033583500005333. [DOI] [PubMed] [Google Scholar]
- Honig B.; Nicholls A. (1995) Classical electrostatics in biology and chemistry. Science 268, 1144–1149. 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- Konig I.; Zarrine-Afsar A.; Aznauryan M.; Soranno A.; Wunderlich B.; Dingfelder F.; Stuber J. C.; Pluckthun A.; Nettels D.; Schuler B. (2015) Single-molecule spectroscopy of protein conformational dynamics in live eukaryotic cells. Nat. Methods 12, 773–779. 10.1038/nmeth.3475. [DOI] [PubMed] [Google Scholar]
- Norby J. G.; Esmann M. (1997) The effect of ionic strength and specific anions on substrate binding and hydrolytic activities of na,K-ATPase. J. Gen. Physiol. 109, 555–570. 10.1085/jgp.109.5.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek P. J.; Patsalo V.; Green D. F.; Raleigh D. P. (2012) Ionic strength effects on amyloid formation by amylin are a complicated interplay among debye screening, ion selectivity, and hofmeister effects. Biochemistry 51, 8478–8490. 10.1021/bi300574r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S.; Kim Y.; Szczepaniak K.; Chen C. C.; Eckmann C. R.; Myong S.; Brangwynne C. P. (2015) The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U. S. A. 112, 7189–7194. 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M. T. Jr; Anderson C. F.; Lohman T. M. (1978) Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: The roles of ion association or release, screening, and ion effects on water activity. Q. Rev. Biophys. 11, 103–178. 10.1017/S003358350000202X. [DOI] [PubMed] [Google Scholar]
- Roth A.; Nahvi A.; Lee M.; Jona I.; Breaker R. R. (2006) Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA 12, 607–619. 10.1261/rna.2266506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemans-Oldehinkel E.; Mahmood N. A.; Poolman B. (2006) A sensor for intracellular ionic strength. Proc. Natl. Acad. Sci. U. S. A. 103, 10624–10629. 10.1073/pnas.0603871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syeda R.; Qiu Z.; Dubin A. E.; Murthy S. E.; Florendo M. N.; Mason D. E.; Mathur J.; Cahalan S. M.; Peters E. C.; Montal M.; Patapoutian A. (2016) LRRC8 proteins form volume-regulated anion channels that sense ionic strength. Cell 164, 499–511. 10.1016/j.cell.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E. K.; Lambert I. H.; Pedersen S. F. (2009) Physiology of cell volume regulation in vertebrates. Physiol. Rev. 89, 193–277. 10.1152/physrev.00037.2007. [DOI] [PubMed] [Google Scholar]
- Freedman J. C.; Hoffman J. F. (1979) Ionic and osmotic equilibria of human red blood cells treated with nystatin. J. Gen. Physiol. 74, 157–185. 10.1085/jgp.74.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Cremer P. S. (2006) Interactions between macromolecules and ions: The hofmeister series. Curr. Opin. Chem. Biol. 10, 658–663. 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Leirmo S.; Harrison C.; Cayley D. S.; Burgess R. R.; Record M. T. Jr. (1987) Replacement of potassium chloride by potassium glutamate dramatically enhances protein-DNA interactions in vitro. Biochemistry 26, 2095–2101. 10.1021/bi00382a006. [DOI] [PubMed] [Google Scholar]
- Jung C.; Rothstein A. (1967) Cation metabolism in relation to cell size in synchronously grown tissue culture cell. J. Gen. Physiol. 50, 917–932. 10.1085/jgp.50.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese J. D.; Voglino A. L.; Hackenbrock C. R. (1991) Ionic strength of the intermembrane space of intact mitochondria as estimated with fluorescein-BSA delivered by low pH fusion. J. Cell Biol. 113, 1331–1340. 10.1083/jcb.113.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussa R.; Baierl A.; Steffen V.; Kubitzki T.; Wiechert W.; Pohl M. (2014) An evaluation of genetically encoded FRET-based biosensors for quantitative metabolite analyses in vivo. J. Biotechnol. 191, 250–259. 10.1016/j.jbiotec.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Vinkenborg J. L.; Nicolson T. J.; Bellomo E. A.; Koay M. S.; Rutter G. A.; Merkx M. (2009) Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat. Methods 6, 737–740. 10.1038/nmeth.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L.; Hires S. A.; Mao T.; Huber D.; Chiappe M. E.; Chalasani S. H.; Petreanu L.; Akerboom J.; McKinney S. A.; Schreiter E. R.; Bargmann C. I.; Jayaraman V.; Svoboda K.; Looger L. L. (2009) Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods 6, 875–881. 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner T.; Augustine G. J. (2000) A genetically encoded ratiometric indicator for chloride: Capturing chloride transients in cultured hippocampal neurons. Neuron 27, 447–459. 10.1016/S0896-6273(00)00056-8. [DOI] [PubMed] [Google Scholar]
- Boersma A. J.; Zuhorn I. S.; Poolman B. (2015) A sensor for quantification of macromolecular crowding in living cells. Nat. Methods 12, 227–229. 10.1038/nmeth.3257. [DOI] [PubMed] [Google Scholar]
- White A. D.; Keefe A. J.; Ella-Menye J. R.; Nowinski A. K.; Shao Q.; Pfaendtner J.; Jiang S. (2013) Free energy of solvated salt bridges: A simulation and experimental study. J. Phys. Chem. B 117, 7254–7259. 10.1021/jp4024469. [DOI] [PubMed] [Google Scholar]
- Liu B.; Åberg C.; van Eerden F. J.; Marrink S. J.; Poolman B.; Boersma A. J. (2017) Design and properties of genetically encoded probes for sensing macromolecular crowding. Biophys. J. 112, 1929–1939. 10.1016/j.bpj.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire M. E.; Cowan J. A. (2002) Magnesium chemistry and biochemistry. BioMetals 15, 203–210. 10.1023/A:1016058229972. [DOI] [PubMed] [Google Scholar]
- DeMott C. M.; Majumder S.; Burz D. S.; Reverdatto S.; Shekhtman A. (2017) Ribosome mediated quinary interactions modulate in-cell protein activities. Biochemistry 56, 4117. 10.1021/acs.biochem.7b00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.; Mori T.; Ando T.; Harada R.; Jung J.; Sugita Y.; Feig M. (2016) Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm. eLife 5, e19274. 10.7554/eLife.19274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.; Malinovska L.; Saha S.; Wang J.; Alberti S.; Krishnan Y.; Hyman A. A. (2017) ATP as a biological hydrotrope. Science 356, 753–756. 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.