SUMMARY
Reproductive isolation defines species divergence and is linked to adaptive evolution of hybrid incompatibility genes. Hybrids between Drosophila melanogaster and Drosophila simulans are sterile and phenocopy mutations in the piRNA pathway, which silences transposons and shows pervasive adaptive evolution, and Drosophila rhino and deadlock encode rapidly evolving components of a complex that binds to piRNA clusters. We show that Rhino and Deadlock interact and co-localize in simulans and melanogaster, but simulans Rhino does not bind melanogaster Deadlock, due to substitutions in the rapidly evolving Shadow domain. Significantly, a chimera expressing the simulans Shadow domain in a melanogaster Rhino backbone fails to support piRNA production, disrupts binding to piRNA clusters, and leads to ectopic localization to bulk heterochromatin. Fusing melanogaster Deadlock to simulans Rhino, by contrast, restores localization to clusters. Deadlock binding thus directs Rhino to piRNA clusters, and Rhino-Deadlock co-evolution has produced cross-species incompatibilities, which may contribute to reproductive isolation.
INTRODUCTION
Transposable elements (TEs) are ubiquitous genome constituents with the potential to mobilize and induce insertional mutations and double strand breaks (Belancio et al., 2008; Biemont and Vieira, 2006; Hedges and Deininger, 2007; Khurana and Theurkauf, 2010). Protecting the genome from these selfish elements is especially critical in the germline, which is dedicated to transmitting genetic information to the next generation. Germline transposon silencing is mediated by small PIWI interacting RNAs (piRNAs), which are bound by PIWI proteins and direct post-transcriptional and transcriptional silencing of target transposons (Brennecke et al., 2007; Gunawardane et al., 2007; Iwasaki et al., 2015; Kuramochi-Miyagawa et al., 2008; Sienski et al., 2012). Mutations that disrupt the piRNA pathway lead to sterility and transposon mobilization in worms, flies, fish and mice, and have been linked to human infertility (Batista et al., 2008; Carmell et al., 2007; Das et al., 2008; Gou et al., 2017; Gu et al., 2010; Heyn et al., 2012; Houwing et al., 2008; Lin and Spradling, 1997; Weick and Miska, 2014). In striking contrast, genes with essential functions in piRNA production and transposon silencing in established model systems are often very poorly conserved, and piRNA biogenesis and sequence composition show remarkable phylogenetic diversity (Chirn et al., 2015; Obbard et al., 2009; Simkin et al., 2013; Yi et al., 2014; Zanni et al., 2013). For example, the overwhelming majority of piRNAs in the Drosophila female germline map to transposons and are derived from heterochromatic domains that span hundreds of kilobases. These “clusters” produce long precursors that are processed into primary piRNAs, which are amplified by a ping-pong cycle driven by PIWI mediated RNA cleavage (Brennecke et al., 2007; Gunawardane et al., 2007). In C. elegans, by contrast, most piRNAs appear to target protein coding genes, each piRNA is produced from a single gene, and amplification involves piRNA primed generation of secondary siRNAs by RNA-dependent RNA polymerases (RdRPs) (Batista et al., 2008; Das et al., 2008; Weick and Miska, 2014).
The divergence of piRNA biogenesis mechanisms is not understood, but many piRNA pathway genes are evolving rapidly under positive selection, which is a hallmark of a “Red Queen” host-pathogen arms race (Daugherty and Malik, 2012; Duggal and Emerman, 2012; Obbard et al., 2009). In a simple Red Queen arms race, mutations that allow a pathogen to evade the host defense system will propagate, compromise host fitness, and lead to selection of host alleles that restore pathogen control. This positive selection cycle continues, driving rapid evolution of host and pathogen genes. Rapid evolution of piRNA genes could therefore reflect a Red Queen arms race with the transposons the system controls. Transposons are a very significant source of genome variation between closely related species (Warren et al., 2015), suggesting that piRNA pathway adaptation to mobile elements could produce genes that are unable to function across recently diverged species. Supporting this hypothesis, hybrids between the sibling species Drosophila melanogaster (melanogaster) and Drosophila simulans (simulans) are viable but sterile (Sturtevant, 1920), and show defects in transposon silencing, piRNA production, and organization of the piRNA biogenesis machinery (Kelleher et al., 2012). In addition, melanogaster and simulans share over 100 transposon families, but also show significant differences in total transposon content, and several families are unique to each species (Bartolome et al., 2009; Lerat et al., 2011). Together, these observations raise the possibility that adaptive evolution of piRNA pathway genes contributes to hybrid sterility (Kelleher et al., 2012).
The Drosophila melanogaster rhino (rhi) gene encodes an HP1 homolog that localizes to piRNA clusters and is required for piRNA biogenesis. The rhi gene shows elevated rates of non-synonymous substitution between melanogaster and simulans, consistent with adaptive evolution (Klattenhoff et al., 2009; Le Thomas et al., 2014; Mohn et al., 2014; Vermaak et al., 2005; Zhang et al., 2014). Rhino (Rhi) interacts with the linker protein Deadlock (Del) to promote piRNA precursor formation, and del, like rhi, is rapidly evolving. We show that rhi and del are co-evolving, and that this process has generated species-specific interactions that prevent function across the melanogaster-simulans species barrier. For simulans Rhi, this is reflected in a failure to bind melanogaster Del, or rescue melanogaster rhi mutations. This leads to significantly reduced binding of sim-Rhi to melanogaster piRNA clusters. Strikingly, fusing melanogaster Del to the C-terminal shadow domain of simulans Rhi restores cluster localization. Adaptive evolution thus targets a Rhi-Del interaction that directs assembly of cluster chromatin, and generates biochemical incompatibilities in the piRNA machinery. We speculate that these incompatibilities contribute to hybrid sterility, reproductive isolation, and speciation.
RESULTS
simulans Rhino does not function in melanogaster
In Drosophila, piRNAs are derived from large heterochromatic domains composed of complex arrays of nested transposon insertions, supporting a model for transposon adaptation in which invading mobile elements remain active until a copy inserts into a cluster, leading to sequence incorporation into piRNA precursors and trans-silencing (Bergman et al., 2006; Brennecke et al., 2007). Drosophila clusters are marked by histone H3 tri-methylated at lysine 9 (H3K9me3) and the HP1 homolog Rhi, which anchors a complex that includes Del and the Rai/DXO homolog Cutoff (Cuff), and all three proteins are required to suppress piRNA precursor splicing and for piRNA biogenesis (Chen et al., 2016; Le Thomas et al., 2014; Mohn et al., 2014; Zhang et al., 2014). Precursor transcripts from clusters are bound by the DEAD box protein UAP56, and transported across the nuclear pore for processing into piRNAs in the perinuclear nuage (Zhang et al., 2012a). Rhi thus anchors the core of the adaptive transposon silencing system. The rhi gene is also rapidly evolving under positive selection (Vermaak et al., 2005), raising the intriguing possibility that it is engaged in host-pathogen arms race with transposons. To determine the functional consequences of rhi divergence, we genetically replaced the rhi gene in melanogaster (mel-rhi) with rhi from the sibling species simulans (sim-rhi), assayed gene function, and characterized interacting proteins.
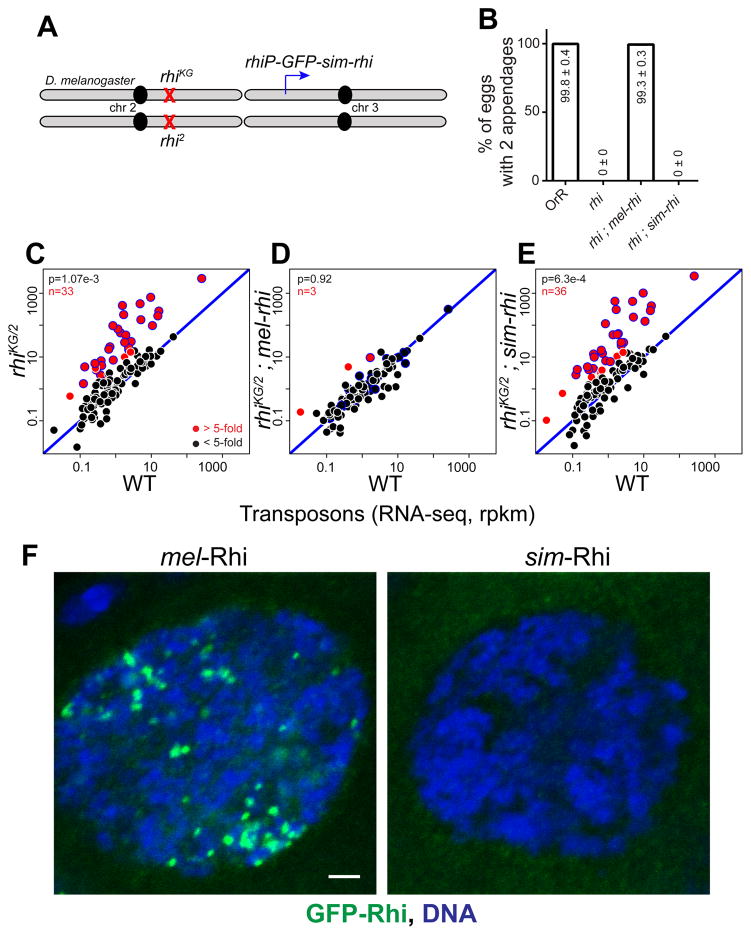
To functionally replace melanogaster rhi, we expressed GFP tagged sim-Rhi, under control of the melanogaster rhi promoter, in a melanogaster rhi null mutant background (Figure 1A). As a control, an analogous GFP:mel-Rhi fusion was expressed in the same rhi background. Mutations in rhi, and most other piRNA pathway genes, lead to female sterility and dorsal-ventral egg patterning defects (Klattenhoff et al., 2007; Klattenhoff et al., 2009). The patterning defects are secondary to DNA damage and Chk2 kinase activation, and serve as an easily quantifiable biological readout of germline genome instability (Klattenhoff et al., 2007). The mel-rhi control rescued both embryo viability as measured by hatch rate, and D-V patterning scored by dorsal appendage formation (Figures 1B and S1A). By contrast, sim-rhi failed to rescue D-V patterning or fertility, and by these assays was equivalent to a null rhi allelic combination.
Figure 1. simulans Rhi does not function in melanogaster.
(A) Genetic complementation strategy. The sim-rhi gene was expressed under endogenous rhi promoter in a melanogaster rhiKG/2 trans-heterozygous null background.
(B) Bar graphs showing percentages of eggs with normal dorsoventral patterning produce by OrR (wild type (WT) control), rhi mutants, and rhi mutants rescued by either mel-rhi or sim-rhi. The numbers in/above the bars show mean ± standard deviation of three biological replicates, with a minimum of 500 embryos scored per replicate, except for rhi mutants and rhi mutants rescued by sim-rhi where average of at least 30 eggs were scored.
(C–E) Transposon expression in rhi mutants (C), rhi mutant rescued by mel-rhi (D), and rhi mutants rescued by sim-rhi (E). RNA-seq was performed on ovaries, and each point on the scatterplots shows rpkm values for a transposons family in ovaries of the indicated mutant/transgene combination relative to WT control. Diagonal represents x=y. Points in red show y/x>5 (n is number of these transposons). Blue bordered points are over-expressed by 5 fold or more in rhi mutants and rhi mutants expressing sim-rhi. p value for differences obtained by Wilcoxon test.
(F) Localization of rhi promoter driven GFP tagged mel-Rhi and sim-Rhi in melanogaster germline. GFP-Rhi is in green and DNA is in blue. Scale bar: 2 μm
See also Figure S1.
Mutations in rhi lead to transposon over-expression in the ovary, which appears to be the primary cause of sterility and D-V patterning defects (Klattenhoff et al., 2009). We therefore measured transposon and gene expression by RNA sequencing (RNA-seq). The scatterplot in Figure 1C shows expression of TE families in the ovaries of rhi mutants vs. wild type (WT). Increased transposon expression is reflected in points that fall above the diagonal. Consistent with previous observations, rhi mutations led to over-expression of numerous transposon families (Figure 1C), but did not alter gene expression (Figure S1B). Transposon silencing is restored by expression of mel-rhi (Figure 1D), but not by expression of sim-rhi (Figure 1E). Strikingly, transposon expression in rhi mutants and rhi mutants carrying the sim-rhi transgene were essentially identical (Figure S1C). Consistent with these observations, small RNA sequencing demonstrates that the sim-rhi transgene does not restore piRNA production from germline clusters or target transposons. By contrast, the mel-rhi transgene restores essentially WT piRNA levels (see below). Therefore, by both biological and molecular measures, the sim-rhi gene, when placed within melanogaster, is equivalent to a null allele.
We next determined fusion protein localization by direct imaging of the GFP tags. Endogenous Rhino localizes to piRNA clusters and forms distinct foci in the germline nuclei, and this pattern is seen with GFP tagged mel-Rhi (Figure 1F). By contrast, GFP tagged sim-Rhi does not localize to distinct nuclear or cytoplasmic structures in germline cells. Western blotting shows that both the mel-Rhi and sim-Rhi proteins are expressed at similar levels (Figure S1D), indicating that the simulans protein is stable, but unable to localize to clusters or promote piRNA production in a melanogaster genetic background.
Evolution of the Shadow domain restricts cross-species function
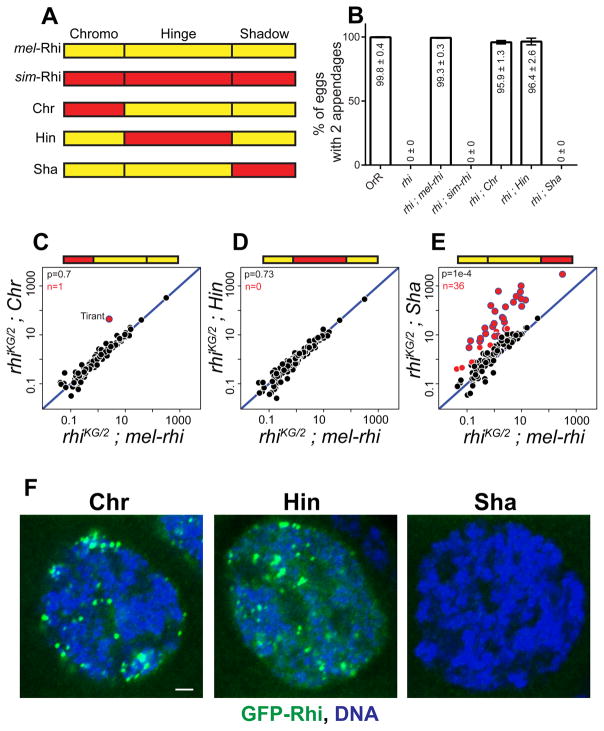
HP1 family proteins are composed of chromo, hinge and shadow domains (Vermaak and Malik, 2009). The Chromo domain of HP1a, the founding member of the family, binds to histone H3 tri-methylated at Lysine 9 (H3K9me3) (Bannister et al., 2001; Lachner et al., 2001). Structural and biochemical studies indicate that H3K9me3 binding is shared by the Chromo domain of Rhi (Le Thomas et al., 2014; Mohn et al., 2014; Yu et al., 2015). The Hinge domain is a variable linker that may also bind RNA or DNA, and the Shadow domain mediates protein-protein interactions (Meehan et al., 2003; Muchardt et al., 2002; Smothers and Henikoff, 2000). To localize changes in simulans Rhi that prevent function in melanogaster, we generated transgenes expressing chimeric proteins composed of individual functional domains from sim-Rhi in a mel-Rhi backbone (Figure 2A). All chimeric proteins were fused to GFP, and the native rhi promoter was used to drive expression. Both chromo and hinge domain chimeras were able to fully rescue D-V patterning of eggs (Figure 2B), but only partially rescued hatching (Figure S2A). However, the shadow domain chimera failed to rescue D-V patterning or hatching.
Figure 2. The Shadow domain of sim-Rhi does not function in sibling species melanogaster.
(A) Design of Rhino chimeras: Chromo (Chr), Hinge (Hin) and Shadow (Sha) domains are shown for mel-Rhi (yellow) and sim-Rhi (red). Each domain from sim-Rhi is placed in the mel-Rhi backbone and expressed as a GFP tagged transgene driven by the rhi promoter.
(B) Bar graphs showing percentages of eggs with normal dorsoventral patterning produced by OrR (WT control), rhi mutant, and rhi mutants expressing mel-rhi, sim-rhi or the chimeric Rhi variants. The numbers in/above the bars show mean ± standard deviation of three biological replicates, with a minimum of 500 embryos scored per replicate, except for rhi mutants and rhi mutants rescued by sim-rhi or Shadow chimera where average of at least 30 eggs were scored.
(C–E) Scatterplots showing transposon expression, measured by RNA-seq, in ovaries of rhi mutant expressing the chimeras vs. mel-rhi. Each point represents rpkm values for a different transposon family. Diagonal represents x=y. Points in red show y/x>5 (n is number of these transposons). Blue bordered points are over-expressed in rhi mutants, rhi mutants expressing sim-rhi, and rhi mutants expressing the Sha chimera. p value for differences obtained by Wilcoxon test.
(F) Localization of rhi promoter driven GFP tagged Rhino variants in melanogaster germline. GFP-Rhi is in green and DNA is in blue. Scale bar: 2 μm.
See also Figure S2.
To determine if the fertility and patterning defects are linked to transposon over-expression, used RNA-seq to analyze the transcriptome in rhi mutant flies expressing the chimeric Rhi proteins (Figures 2C, 2D and 2E). Consistent with our phenotypic data, the hinge domain chimera completely restored transposon silencing, and the chromo domain chimera silenced all transposon families with the exception of the Tirant retrotransposon. This does not reflect a defect in piRNA production (see below), and the mechanism leading to Tirant over-expression in this background remains to be explored. We speculated that over-expression of Tirant could contribute to the low hatch rate with the chromo-domain chimera, but forced expression of a full length element in WT, using the nanos-Gal4 driver and UASp promoter, did not reduce hatch rate or lead to D-V patterning defects (unpublished observation). In contrast to the chromo and hinge chimeras, the shadow chimera failed to restore silencing and was comparable to the rhi null allelic combination (Figure S2B). The chromo and hinge domain substitutions also localize to nuclear foci in germline cells (Figure 2F), while the shadow chimera is expressed but fails to localize. Changes in the Rhi shadow domain have therefore disrupted the ability of the protein to function across the melanogaster-simulans species barrier.
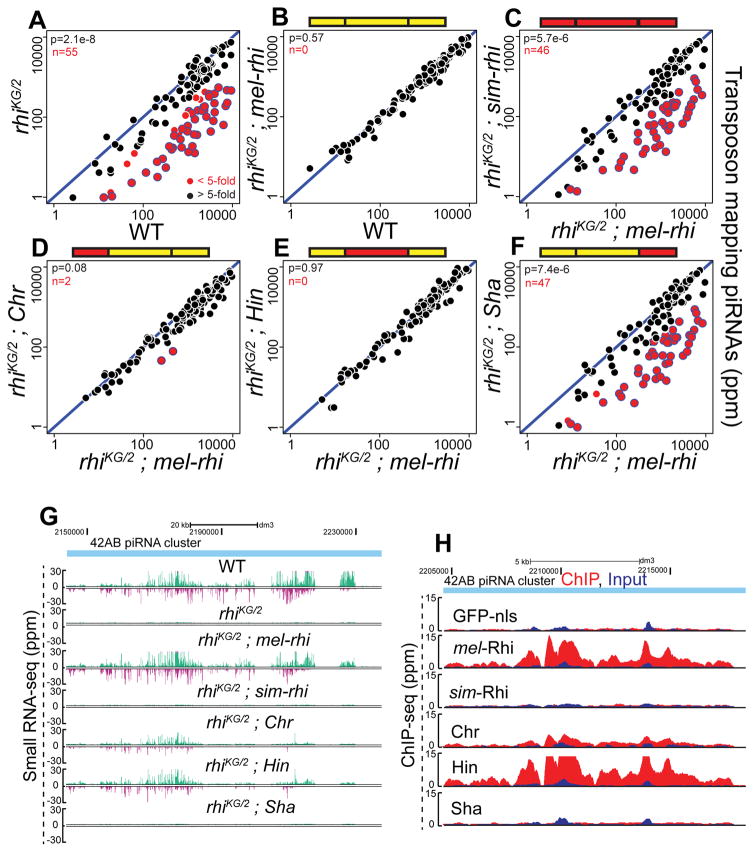
To determine if defects in transposon silencing are due to loss of piRNAs, we sequenced small RNAs from ovaries of rhi mutants expressing chimeric Rhino proteins. The mel-rhi, chromo and hinge domain chimeras were able to restore production of piRNAs mapping to transposon repeats (Figures 3A to 3F). By contrast, sim-rhi and the shadow chimera failed to rescue transposon mapping piRNA expression, and were comparable to the null allelic combination. The primary piRNAs that initiate piRNA biogenesis are derived from clusters, and 42AB is the major dual strand piRNA cluster in melanogaster germline cells. As shown in Figure 3G, 42AB piRNAs are lost in rhi mutants. mel-rhi and the chromo and hinge domain chimeras are able to rescue piRNA production, but sim-rhi and shadow domain chimera do not. Rhi functions with Del and Cuff to suppress splicing of piRNA cluster transcripts, and mel-Rhi and the chromo and hinge chimeras are able to suppress the splicing at clusters (Figure S3C). By contrast, sim-Rhi and the shadow domain chimera fail to suppress splicing of piRNA precursors.
Figure 3. sim-Rhi and Shadow chimera do not bind to piRNA clusters and fail to support piRNA production.
(A–F) Scatterplots showing abundance of transposon mapping piRNAs in ovaries of rhi mutant (A), rhi mutant expressing either mel-rhi (B) vs. WT control, sim-rhi (C) or the chimeras (D-F) vs. mel-rhi. Points in red show x/y>5 (n is number of these transposons). Blue bordered points have reduced expression in rhi mutants, rhi mutants expressing sim-Rhi, and rhi mutants expressing the Sha chimera. p value for differences is obtained by Wilcoxon test.
(G) Genome browser view showing abundance of piRNAs uniquely mapping to 42AB piRNA cluster in WT, rhi mutant and rhi mutants expressing mel-rhi, sim-rhi or chimeric proteins. The Watson strand is in green, and Crick strand in magenta.
(H) Genome browser view of ChIP-seq profiles at 42AB cluster for mel-Rhi, sim-Rhi, chimeras, and GFP-nls control. All ChIP done under identical conditions, using the same anti-GFP antibody. ChIP signal in red, input signal in blue.
See also Figure S3.
WT Rhi localizes specifically to piRNA clusters. We therefore performed ChIP-seq to determine if the chimeric proteins support cluster localization. To assay localization independent of the ability to promote cluster assembly, these studies were done in a WT genetic background. As shown in Figure 3H, full length mel-Rhi, the chromo and hinge chimeras bind to the 42AB piRNA cluster. By contrast, sim-Rhi and shadow chimera show signal comparable to GFP-nls control (Figure 3H). Intriguingly, the shadow domain shows the strongest signature of positive selection (Vermaak et al., 2005). Adaptive evolution of this domain thus prevents Rhi function across the simulans-melanogaster species barrier, and appears to alter sequences that direct localization to piRNA clusters.
Evolution of the Rhi-Del interaction
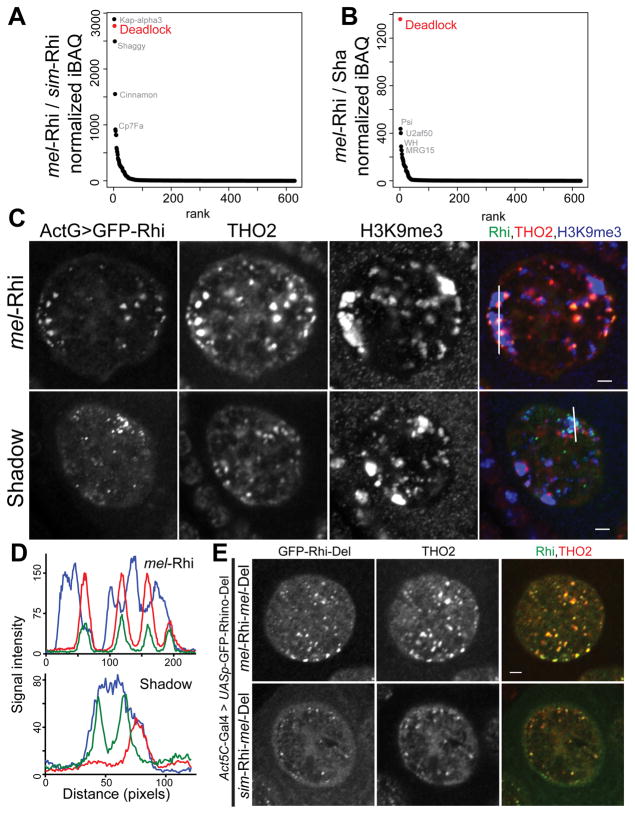
To determine if species-specific substitutions in Rhi alter interactions with partner proteins, we expressed GFP tagged full length simulans and melanogaster Rhi, and each of the chimeras, in WT melanogaster ovaries, immuno-precipitated the tagged proteins, and identified associated polypeptides by mass spectrometry. We expressed these transgenes using both the rhi promoter, and the inducible UASp promoter with the germline specific nanos-Gal4 driver. Rhi is expressed at low levels and peptide counts with the rhi promoter were low, making statistical analysis difficult. As the interacting proteins were very similar with two systems, we focused our analysis on the nanos-Gal4 driven transgenes (Figures 4A, 4B and S4A). Spectrum counts for GFP tag and the Rhi protein variants were similar, indicating that the fusion proteins were expressed at comparable levels, and precipitated with similar efficiencies. For each of the fusions, we quantified co-precipitating proteins (normalized iBAQ), and divided normalized protein levels in the mel-Rhi control by normalized protein levels with each experimental fusion (see Methods). Pseudocounts were used to avoid dividing by zero, and proteins with reduced binding relative to control produce a ratio greater than 1. Figures 4A and 4B show rank order of this ratio for full length sim-Rhi and the shadow chimera. The vast majority of proteins were present at similar levels in all samples, including the GFP control, and ratios clustered around 1. A single protein, Del, was not detected with sim-Rhi or the shadow chimera, and but showed essentially identical binding to full length mel-Rhi and the chromo and hinge chimeras (Figure S4A). We have been unable to obtain antibodies to Rhi or Del that work reliably on Western blots, and therefore confirmed these observations by expressing Rhi:GFP with Del:FLAG:mRFP fusions in melanogaster ovaries, and performing reciprocal co-immunoprecipitation and Western blotting. As shown in Figure S4B, melanogaster Del co-precipitated with mel-Rhi, and mel-Rhi precipitated with melanogaster Del. However, melanogaster Del did not co-IP with sim-Rhi, and sim-Rhi did not co-IP with melanogaster Del. Substitutions in sim-Rhi shadow thus prevent binding to Del from melanogaster. Previous studies indicate that Del is required for primary piRNA biogenesis and interacts with Rhi through the shadow domain (Mohn et al., 2014). In yeast two hybrid assays, Del also interacts with the Rai1/DXO homolog Cuff, which functions with Rhi and Del in piRNA precursor processing. Adaptive evolution has therefore targeted a Rhi-Del interaction that is essential to the assembly of the nuclear piRNA precursor processing machinery, and generated a barrier to function across closely related species.
Figure 4. Cross species incompatibility in Rhi-Del interaction.
(A, B) Mass spectrometric analysis of Rhi binding proteins. Graphs showing ratios of GFP normalized iBAQ values for mel-Rhi vs. sim-Rhi (A), and mel-Rhi vs. Sha chimera (B), ranked by ratio values. Transgenes were expressed in melanogaster using the germline specific nanos-Gal4 driver.
(C) Localization of THO2 (piRNA cluster marker), H3K9me3 marked chromatin in the germline nuclei expressing Act5C-Gal4 driven Rhi:GFP. Color assignments for merged image shown on top. Scale bar: 2μm. Fluorescence intensities are measured along the line shown in merged image for Rhi:GFP (green), THO2 (red) and H3K9me3 (blue) as depicted in (D).
(E) Localization of Act5C-Gal4 driven Rhi-Del fusion proteins with respect to THO2 (piRNA cluster marker) in the female germline nuclei. Scale bar: 2μm.
See also Figure S4.
Rhino localization to piRNA clusters
simulans Rhi fails to function in melanogaster and does not interact with melanogaster Del, which is required for Rhi localization to the nucleus (Mohn et al., 2014). To determine if the requirement for Del could be bypassed by driving Rhi into the nucleus by an independent mechanism, we generated a transgene expressing simulans Rhi with 3 copies of the nuclear localization signal (nls) from SV40 large T antigen (Kalderon et al., 1984). The sim-Rhi-nls protein localized to germline nuclei and formed foci (Figure S4C). However, it did not rescue rhi mutations (Figure S4D) and the TREX complex component THO2, which co-localizes with Rhino at clusters (Hur et al., 2016), did not co-localize with the foci formed by sim-Rhi-nls (Figure S4C). The Shadow domain chimera does not bind Del, but is not detected in nuclear foci when expressed by the rhi promoter and imaged using our standard confocal procedures. However, the GFP tagged shadow chimera, over-expressed using either the constitutive Act5C-Gal4 driver or the germline specific nanos-Gal4 driver, localizes to distinct nuclear foci. Significantly, these foci do not co-localize with THO2 (Figures 4C and 4D). By contrast, similarly expressed full-length melanogaster Rhi consistently co-localizes with downstream components of the piRNA machinery (Figure 4C). In addition, ChIP-seq confirms that over-expressed shadow chimera fails to localize to clusters, while over-expressed mel-Rhi shows normal cluster binding (Figure S4F). These data strongly suggest that binding to Del is required to direct Rhino to clusters.
The chromo domain of Rhi binds to H3K9me3, and this mark and Rhi show similar distributions over germline clusters. However, H3K9me3 also marks constitutive centromeric heterochromatin and a number of euchromatic loci, which do not bind Rhi or produce piRNAs. This restricted distribution is reflected in cytological localization of Rhi foci to the periphery of large H3K9me3 domains, but not within these domains (Figures 4C and 4D). By contrast, the foci formed by the shadow domain chimera are embedded within the prominent H3K9me3 domains, and do not accumulate to the periphery of these domains (Figures 4C and 4D). These findings strongly suggest that binding to Del directs Rhi to H3K9me3 marks on clusters, and away from bulk heterochromatin. To test this hypothesis, we generated a transgene expressing full length sim-Rhi fused through the C-terminal shadow domain to full length melanogaster Del. We included a flexible linker, to reduce conformational constraints. As a control, we generated an analogous transgene expressing mel-Rhi fused to mel-Del. In WT ovaries, both fusions localize to germline nuclei and formed distinct foci. Significantly, the foci formed by sim-Rhi fused to mel-Del also co-localized with THO2 (Figure 4E). Forced binding to Del is therefore sufficient to direct sim-Rhi to clusters.
To assay for fusion protein function, we crossed both transgenes into rhi, del, and rhi,del double mutants and assayed fertility. Surprisingly, neither fusion transgenes rescued rhi, del or the double mutant combination (Figure S4E), and the fusion proteins show very weak localization to nuclear foci. Directly linking Rhi to Del is therefore sufficient for cluster localization in WT ovaries, where these chromatin domains appear to be established by endogenous gene products. However, the fusions are not able to promote assembly of these domains. The fusion could disrupt the function of critical domains near the junction, but both proteins localize to clusters and a transgene expressing GFP fused to N-terminus of Del rescues del mutants. We therefore speculate that a dynamic interaction between Rhi and Del is critical for the function of both proteins, which may shuttle between biochemically distinct complexes during biogenesis. For example, Del could bind to Rhi before moving to a distinct complex with downstream piRNA components, including Cuff, UAP56 and THO. While the reason the fusion proteins fail to rescue full biological function remains to be determined, these data indicate that the interaction with Del is required for Rhi localization specifically to piRNA cluster chromatin.
Directional incompatibility in Rhi-Del interaction
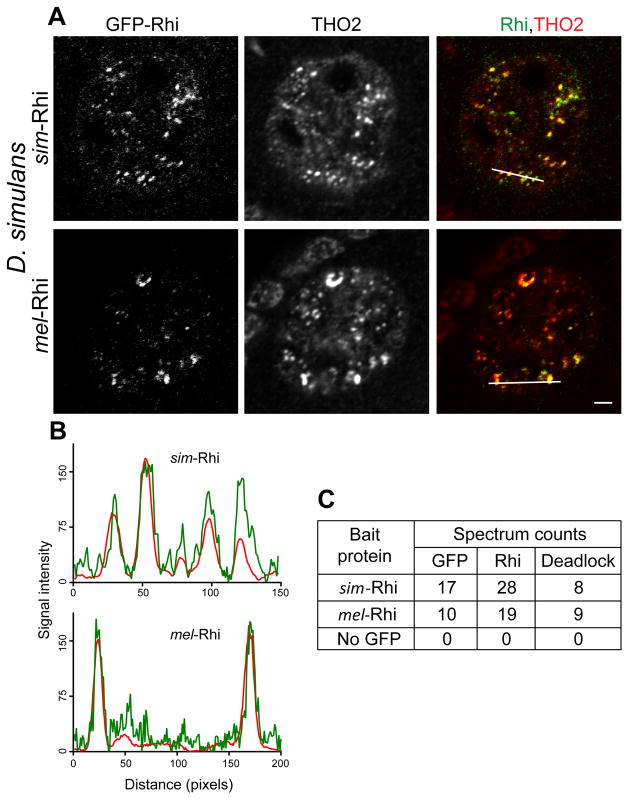
The inability of sim-Rhi to rescue rhi mutants or interact with melanogaster Del raised the possibility that Rhi-Del co-evolution had generated species-specific interaction interfaces that prevent cross-species heterodimer formation. Alternatively, the stable interaction of Rhi and Del could have evolved in the melanogaster lineage. To differentiate between these alternatives, we generated transgenic simulans lines expressing GFP tagged sim-Rhi and mel-Rhi, under control of the rhi promoter, and directly assayed subcellular localization and interactions with Del and other proteins. IP-mass spectrometry demonstrated that simulans Del co-precipitates with sim-Rhi, indicating that the interaction is conserved. Surprisingly, simulans Del also co-precipitated with mel-Rhi (Figure 5C). Consistent with these studies, sim-Rhi and mel-Rhi co-localized with THO2 in germline nuclear foci (Figures 5A and 5B). The interaction between Rhi and Del thus shows cross-species directionality: mel-Rhi is able to bind to Del from both simulans and melanogaster, and localizes in both species, while sim-Rhi binds to sim-Del and localizes to nuclear foci in simulans, but cannot interact with mel-Del or localize in the melanogaster germline.
Figure 5. mel-Rhi binds to Del in simulans and localizes to piRNA clusters.
(A) Localization of sim-Rhi:GFP and mel-Rhi:GFP in the simulans female germline. Egg chambers were double labeled for the piRNA cluster marker THO2. Both forms of Rhi co-localize to nuclear foci with THO2. Scale bar: 2μm. Fluorescence intensities are measured along the line shown in merged image for Rhi:GFP (green), THO2 (red) as depicted in (B).
(C) Total spectrum counts for GFP, Rhi and Del co-precipitating with sim-Rhi and mel-Rhi expressed in simulans. Expression was driven with the rhi promoter. Both sim-Rhi and mel-Rhi coprecipitate with the endogenous simulans Del ortholog.
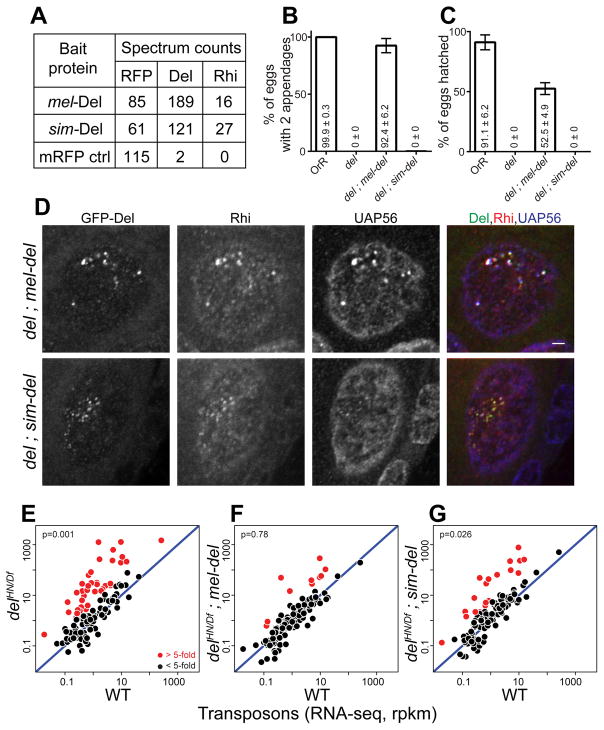
These observations suggested that the sim-del gene may function in melanogaster. We therefore expressed mRFP or GFP tagged sim-Del and mel-Del in melanogaster, and assayed interacting proteins, subcellular localization, and the ability to rescue del mutations. IP and mass spectrometry on the tagged proteins expressed in WT ovaries showed that mel-Del and sim-Del interact with mel-Rhi (Figure 6A). The mel-Del and sim-Del proteins also formed nuclear foci that co-localized with THO2 (Figure S5B). To directly assay biological function, we expressed both tagged proteins in a melanogaster del mutant background. Mutations in del disrupt oocyte/embryo D-V patterning, embryo viability, and transposon silencing (Mohn et al., 2014; Wehr et al., 2006). The mel-del transgene rescued all three of these defects (Figures 6B, 6C and 6F). By contrast, the sim-del was similar to a null allele by all three measures (Figure 6B, 6C and 6G). These findings were surprising, given the robust cluster localization of sim-Del in WT ovaries (Figure S5B). In the del mutant background, however, the sim-Del showed very weak localization to nuclear foci, which were present in only a subset of germline nuclei. These foci also showed very weak localization of Rhi, and the downstream proteins THO2 and UAP56 (Figures 6D, S5A and S5C). In WT ovaries, sim-Del thus appears to localize to clusters through endogenous Rhi, which functions with endogenous Del to promote cluster assembly. In the del mutants, by contrast, the mel-Rhi interacts with sim-Del, but the complex is unable to promote cluster assembly. This is likely due to defects in recruitment of downstream components of the pathway. We therefore speculate that Del has a minimum of two rapidly evolving functional domains, which mediate binding to Rhi and interactions with downstream proteins. Together, evolution of these domains creates a barrier to cross-species function.
Figure 6. sim-Del binds to mel-Rhi, but fails to function in melanogaster.
(A) Mass spectrometric analysis of Del binding proteins. Table shows total spectrum counts for the mRFP tag, Rhi and Del in immunoprecipitates of mel-Del, sim-Del and mRFP control, expressed in the melanogaster germline under nanos-Gal4 driver. sim-Del co-precipitates with mel-Rhi.
(B, C) Bar graphs showing percentages of eggs with normal dorsoventral patterning (B) and percentages of hatched eggs (C) produced by females of the following genotypes: OrR (WT control); del mutant; del mutants expressing either mel-del or sim-del. sim-del fails to rescue embryo patterning and hatching. The numbers in/above the bars show mean ± standard deviation of three biological replicates, with a minimum of 100 embryos scored per replicate, except for del mutants where average of 7.33 eggs were scored.
(D) Localization of Rhi and UAP56 in del mutants expressing either mel-Del or sim-Del. Scale bar: 2μm (all images at same scale).
(E-G) Scatterplots showing transposon expression levels measured by RNA-seq in ovaries of del mutant (E), del mutant rescued by either mel-del (F) or sim-del (G) vs. WT control. Each point represents rpkm values for a different transposon. Diagonal represents x=y. Points in red show y/x>5. p value for differences is obtained by Wilcoxon test. sim-del fails to rescue transposon silencing.
See also Figure S5.
DISCUSSION
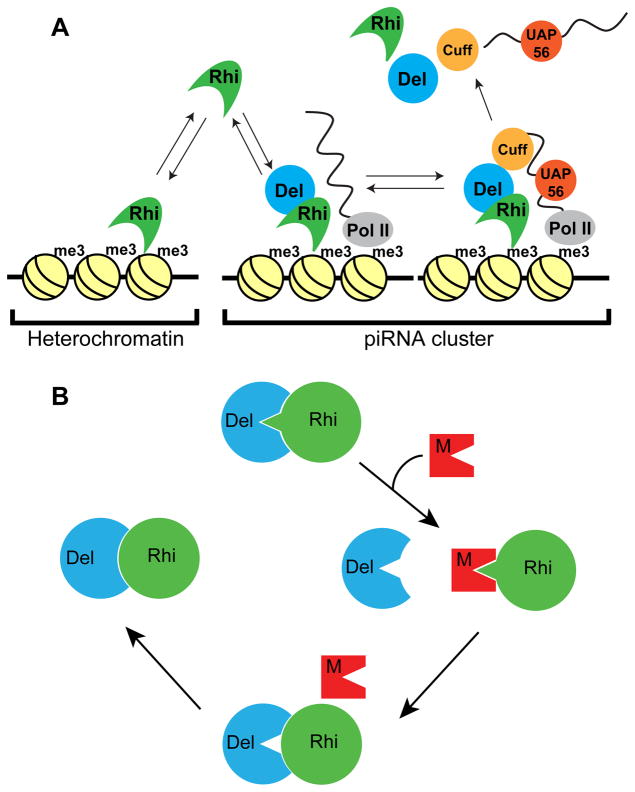
piRNA clusters determine sequence specificity for transposon silencing by the piRNA pathway, and thus function at the heart of the adaptive genome immune system (Brennecke et al., 2007). Rhi localizes specifically to piRNA clusters, where it promotes cluster transcription, suppresses cluster transcript splicing, and promotes piRNA production (Le Thomas et al., 2014; Mohn et al., 2014; Zhang et al., 2014). Our data indicate that co-evolution of this key component of the piRNA machinery, with its partner Del, prevents assembly of functional complexes across the sibling species barrier. The simulans rhi and del genes do not rescue melanogaster mutants, sim-Rhi does not interact with mel-Del, and sim-Del forms a non-functional complex with mel-Rhi. These studies also provide intriguing insights into the mechanisms that localize Rhi to clusters. Substituting the mel-Rhi rapidly evolving Shadow domain with the simulans Rhi shadow domain is sufficient to block piRNA production, transposon silencing, and binding to Del. Significantly this Shadow domain chimera does not localize to germline piRNA clusters and forms ectopic nuclear foci that do not co-localize with the downstream processing machinery. By contrast, a fusion between sim-Rhi and mel-Del localizes to clusters. Binding to Del, and likely subsequent recruitment of downstream piRNA biogenesis proteins, thus allows Rhi to discriminate between H3K9me3 marks at clusters and other repeats (Figure 7A).
Figure 7. Model for co-evolution of the Rhino-Deadlock interface.
(A) Model for Del function in Rhi localization to piRNA clusters. The Rhi Chromo domain interact with H3K9me3 marks throughout the genome, but most of these marks are in transcriptionally silent regions. At clusters, which are transcribed, Del interactions with Cuff, a putative RNA end binding protein, leads to formation of a chromatin bound complex, which recruits additional RNA binding components (i.e. UAP56). Assembly of these complexes leads to Rhi accumulation at clusters. We further propose that release of piRNA precursor complexes is accompanied by release and recycling of Rhi, Del and Cuff, which then re-initiate the cycle.
(B) A transposon mutation generates a protein that mimics the Del surface that binds to Rhi. Competition for productive Rhi-Del complex formation disrupts piRNA biogenesis and causes increased transposition. Reduced fertility leads to selection of Rhi mutations that reduce mimic binding, at the expense of Rhi-Del affinity. “Leaky” transposition leads to selection of Del mutations that completely restore Rhi binding. Pathogen mimicry thus leads to rapid evolution of Rhi-Del interface.
See also Figure S6.
How Del directs Rhi to clusters remains to be determined, but piRNA clusters are actively transcribed, while most heterochromatin marked by H3K9me3 is transcriptionally silent. In addition, many of the piRNA biogenesis factors that co-localize with Rhino at clusters, including Cuff, UAP56 and the THO complex, are RNA binding proteins that appear to be loaded on cluster transcripts co-transcriptionally. We therefore propose that Rhino, though it’s Chromo domain, samples available H3K9me3 marks, moving between centromeric heterochromatin and clusters. At clusters, however, interactions between Del and Cuff, a putative RNA end binding protein, lead to recruitment of UAP56 and THO, which bind single stranded RNA. We propose that Rhino in the resulting chromatin bound RNA complex does not exchange with the soluble pool, driving specific accumulation at clusters (Figure 7A). However, covalent binding of Rhi to Del prevents function. We therefore propose that as piRNA precursors are released from these complexes, Rhi and Del dissociate and reinitiate assembly of the chromatin bound RNPs, and that fusion of Del to Rhi prevents this recycling step. While speculative, this model provides a useful framework for future studies.
piRNA pathway evolution
The piRNA pathway has a conserved function in germline development and transposon silencing, but piRNA sequence composition and biogenesis mechanisms show remarkable phylogenetic diversity, and many genes in the piRNA pathway are evolving rapidly under positive selection, and are poorly conserved (Chirn et al., 2015; Khurana and Theurkauf, 2010; Obbard et al., 2009; Simkin et al., 2013; Yi et al., 2014; Zanni et al., 2013). We directly determined the functional consequences of piRNA gene divergence over a short evolutionary time scale, by expressing the simulans rhi and del genes in melanogaster and assaying protein interactions, subcellular localization, and the ability to rescue chromosomal mutations. Our data indicate that rapid co-evolution of rhi and del has generated orthologs that do not form functional complexes across the sibling species barrier.
How did this incompatibility arise? rhi and del are evolving rapidly under positive selection, characteristic of genes engaged in a “Red Queen” host-pathogen arms race, presumably with transposons, which function as mobile genome pathogens. A simple Red Queen system, however, leads to rapid co-evolution of host and pathogen genes that encode interacting proteins (Daugherty and Malik, 2012; Duggal and Emerman, 2012; Elde and Malik, 2009), and rhi and del encode interacting components of the host defense system. This could arise through pathogen mimicry. In this variant of an arms race, mutations in the pathogen generate a protein with a surface that structurally mirrors a host partner engaged in an interaction required for defense. Competition with the functional interaction leads to pathogen propagation, reduced host fitness, and selection of host mutations that reduce host binding to the mimic (Daugherty and Malik, 2012; Elde and Malik, 2009). Figure 7B outlines a speculative model for an evolutionary cycle driven by a mimic targeting the Rhi-Del interface. In this model, mutations in a transposon gene (retrotransposon gag, pol or env, for example) generate a mimic of the Del surface that interacts with Rhi. Mimic competition with Del for Rhi reduces productive dimer formation and piRNA production, leading to increased transposition. Reduced fertility then leads to selection of mutations in Rhi that reduce mimic binding and increase the Del interaction, but at a cost of biding affinity. “Leaky” transposon silencing could then lead to selection of Del mutations that restore high affinity binding to Rhi (Figure 7B). This “mimicry cycle” thus remodels the Rhi-Del interface. As diagrammed in Figure S6, this process also has the potential to produce “directional incompatibility” at the dimerization interface.
Our data, with the finding that melanogaster- simulans inter-species hybrids phenocopy piRNA mutations (Kelleher et al., 2012), raise the intriguing possibility that adaptive evolution of piRNA pathway genes directly contributes to the reproductive barrier between these sibling species. For example, we show that sim-Del binds to mel-Rhi, but is unable to rescue melanogaster del mutations or direct Rhino to cluster chromatin. In hybrids, non-functional complexes between sim-Del and mel-Rhi may therefore complete with productive complexes between proteins from the same species. It is unclear if the resulting reduction in functional Rhi-Del complexes would be sufficient to trigger the observed transposon silencing defects. However, our preliminary data indicate that adaptive evolution of additional piRNA genes prevents cross-species function (Parhad and Theurkauf, unpublished), and mimics could target any non-redundant interaction in the piRNA pathway. We therefore speculate that adaptive evolution of several piRNA pathway genes leads to multiple biochemical incompatibilities, which together disrupt piRNA function in hybrids.
It is difficult to directly test the hypothesis that transposon-encoded mimics drive piRNA pathway evolution, as mimics would arise over an evolutionary time scale (millions of years), and once bypassed by a compensating host mutation, provide no selective benefit and would be lost. However, mimics targeting piRNA biogenesis are predicted to mobilize all transposon families with functional copies in the genome, not just the transposon family that produced the mimic, leading to bursts of global transposon activity. Consistent with this prediction, melanogaster has retained active copies of almost all transposon families, while simulans has retained very few functional transposons (Lerat et al., 2011). Furthermore, global bursts of transposition are associated with species divergence in plants and animals (Belyayev, 2014; Fontdevila, 2005), which could reflect periodic relaxation of germline transposon silencing. We therefore speculate that an ongoing arms race between transposons and the piRNA pathway facilitates speciation by producing biochemical incompatibilities that help build reproductive barriers, and by triggering bursts of insertional mutations that serve as substrates for natural selection.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact William Theurkauf (william.theurkauf@umassmed.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experiments were performed on females of two Drosophila species: Drosophila melanogaster and Drosophila simulans. All flies were kept at 25°C on cornmeal medium. All D. melanogaster transgenic lines were generated by φC31 integration at 3L-68A4. All D. simulans transgenic lines in w501 strain were generated by random P-element mediated transformation. rhiKG and rhi2 alleles were described in (Klattenhoff et al., 2009; Volpe et al., 2001). delHN and delDf alleles were obtained from Trudi Schüpbach (Princeton University). F1 females from OregonR crossed to w1 were used as wild type (WT) control, unless mentioned otherwise.
METHOD DETAILS
Generation of transgenic flies
Gateway Technology from Invitrogen was used to generate the plasmids. φC31 attB was added to Drosophila gateway transformation vector pPGW to get attB-pPGW. φC31 attB was PCR amplified from pUASTattB plasmid (Bischof et al., 2007) (by using primers 5′-CGA TTA TGC ATG TCG ACG ATG TAG GTC ACG GTC TC -3′ and 5′-AAT CGA TGC ATG TCG ACA TGC CCG CCG TGA CCG TC -3′). Both the PCR product and pPGW vector were digested by NsiI, gel purified and ligated to get the final vector attB-pPGW. This serves as entry vector for expressing N′ GFP tagged proteins under UASp promoter. The procedure for generation of rhiP-attB-pPGW (for expressing N′ GFP tagged proteins under rhi promoter) is as follows: rhi promoter was PCR amplified by using primers (PCR1: 5′-CTC TCT TTC TCG AGG TCA TCA AGC TTA GGC ATG TAC CAA GTT GTT AAC TCT ATC GAA TTA-3′, 5′-GAA GAT TTC TCC TTG ACG TTT CGG ACA CCC AAG GTT AGC CCA AAT CGA TGG ATT TCT GGG ACA TGA TC -3′; PCR2: 5′-GAT CAT GTC CCA GAA ATC CAT CGA TTT GGG CTA ACC TTG GGT GTC CGA AAC GTC AAG GAG AAA TCT TC -3′, 5′-CTC ACC ATG GTG GCG GGC TTC TCT AGA CAG GAA CTT ATC CGC TCA CAG GAC GCC GAG CAA AAG -3′) to introduce STOP codons in the upstream Oxp gene, from w1 genomic DNA. PCR1, PCR2 and StuI digested attB-pPGW were ligated by Clontech In-Fusion® HD Cloning Kit. Same cloning strategy was used to express Rhino in D. simulans, except for rhi promoter cloned from D. simulans w501(by using primers: PCR1:5′-CTC TCT TTC TCG AGG TCA TCA AGC TTA GGC ATG TAC CGA GTT GTT AAC TCT ATC GAA TTA -3′, 5′-GAG GAT TTT TCC TTG ACG TTG CGG ACA CCA AGG GTT ATC CCA GAT CAA CTG ATT TCT TGG GCA TGA TC -3′; PCR2:5′-GAT CAT GCC CAA GAA ATC AGT TGA TCT GGG ATA ACC CTT GGT GTC CGC AAC GTC AAG GAA AAA TCC TC -3′, 5′-CTC ACC ATG GTG GCG GGC TTC TCT AGA CAG GAA CTT AAA CGC TGA AAG GAC GCC GAG CAA ATG -3′). For expression of N′ mRFP-FLAG tagged proteins, the vector attB-pPFVR was generated as follows: The FLAG tag was PCR amplified from pPFW (primers: CGG ACG AAT TTT TTT TTG AAA ACC GGT GAT AGA GCC TGA ACC AGA AAA G and GGA CTG GAA GTA CAG GTT CTC CTT GTC ATC GTC ATC CTT GTA ATC) and mRFP tag from pPRW (primers: GAG AAC CTG TAC TTC CAG TCC ATG GCC TCC TCC GAG GAC GTC ATC AAG and CAG CTT TTT TGT ACA AAC TTG TAT ACC GGT GGG CG). The FLAG and mRFP tag PCRs and AgeI digested attB-pPFW were ligated by Clontech In-Fusion® HD Cloning Kit. The resulting plasmid is gateway destination vector attB-pPFVR, having φC31 attB site and expressing N′ FLAG and mRFP tagged protein under UASp promoter. TEV protease site is cloned between FLAG and mRFP tag. mel-rhi (primers: CAC CAT GTC TCG CAA CCA TCA GCG ACC AAA TC and TTA CTT GGG CAC AAT GAT CCT CAA GCT C) from cDNA clone obtained from DGRC clone RE36324 (from Riken y; cn, bw, sp strain), sim-rhi (primers: CAC CAT GTC TCG CAA AAA TCA ACG ACC AAA TCT TG and TTA CTT GAG CAC AGT GGT CCT CAA GCT C) from cDNA from simulans C167.4 strain ovaries, mel-del (primers: CAC CAT GGA AAA GTT GGA CAA AAT AAG GAT G and TTA ATC AAA ATT ATG TAT ATT GAT CGC ATA TTC ATT GG) from OregonR genomic DNA, sim-del (primers: CAC CAT GGA AAA CTT GGC TAA AAT AAG GAT G and TTA ATC AAA ATG ATG TAT ATT GGT CGT A) from simulans C167.4 genomic DNA were cloned in the gateway entry vector with pENTR directional TOPO cloning kit (Invitrogen). (The sequences are provided in Mendeley Data). sim-Rhi-nls was made by PCR of sim-Rhi with reverse primer encoding for nls sequence (TTA CAC CTT GCG CTT CTT CTT TGG ATC CAC CTT GCG CTT CTT CTT TGG ATC CAC CTT GCG CTT CTT CTT TGG ATC AGC TCG GGA TCT GAG TCC GGA CTT GAG CAC AGT GGT CCT CAA GCT C) and forward primer mentioned above. Rhi and Del fusions were made by gene synthesis and cloned into pENTR vector. Del was at C′ end of Rhi, with a flexible linker in the middle. The plasmids obtained after LR gateway cloning reaction were sequenced and injected into respective fly strains.
Fertility assays
2–4 day old flies were kept on grape juice agar plates for 1 or 2 days. After removal of flies, the eggs were scored for fused appendages and the hatching was measured after 2 days. The bar graphs show mean and standard deviation for 3 biological replicates, with the indicated number of scored embryos.
Immuno-staining
Immuno-staining and image analysis was done as described in (McKim et al., 2009; Zhang et al., 2012a). In brief, 2–4 day old female ovaries were fixed with 4% formaldehyde, washed, incubated overnight with primary antibody, washed, incubated with secondary antibody with fluorophore overnight and mounted on slide. ChromoTek anti-GFP Booster (Atto-488) antibody added with secondary antibody to enhance GFP signal.
Immuno-precipitation
2–4 day old female ovaries were dissected in Robb’s buffer. The ovaries were washed once with lysis buffer with composition: HEPES (pH 7.5) 50mM, NaCl 150mM, MgCl2 3.2mM, NP40 0.5%, PMSF 1mM, Proteinase Inhibitor (Roche) 1X. The ovaries suspended in lysis buffer were homogenized, sonicated in bioruptor (5 min, 30sec on and 30 sec off), centrifuged at 13200 rpm for 30min at 40C. The supernatant was used as input and added to chromotek GFP-Trap®_A or RFP-Trap®_A beads suspended in lysis buffer. The lysate and beads were kept rotating at 40C for 3 hours and then washed 4 times with lysis buffer. The beads were resuspended SDS-PAGE lysis buffer. The procedure for mass spectrometry of IPed samples is descried in (Vanderweyde et al., 2016). In brief, the IPed samples were resolved on a 10% SDS-PAGE gel. The gel pieces were processed for trypsin digestion to get the peptides, which were further analyzed by LC-MS/MS. For Rhi-Del co-IP westerns, the samples were separated on a SDS-PAGE gel, transferred onto nitrocellulose membrane, incubated with anti-GFP and anti-FLAG antibodies and imaged with LI-COR Odyssey system.
Small RNA-seq
Small RNA libraries were prepared as described in (Zhang et al., 2014). In brief, total RNA prepared from 2–4 day old female ovaries by mirVANA kit (Ambion) were size selected for 18–30nt small RNAs by gel purification. They were further 3′ and 5′ ligated by adapters, reverse transcribed, PCR amplified and sequenced by Illumina platform.
RNA-seq
RNA-seq libraries were prepared as described in (Zhang et al., 2012b). In brief, ribosomal rRNA depleted (Ribo-Zero kit (Illumina)) RNA samples were fragmented, reverse transcribed, ligated by adapters and PCR amplified to make libraries. dUTP incorporation done for strand specificity. Sequenced by Illumina platform.
ChIP-seq
ChIP-seq libraries were prepared as described in (Zhang et al., 2014). In brief, ovaries were fixed with 2% formaldehyde, sonicated for 2 hours in Bioruptor. The lysate after centrifugation was added to Dynabeads Protein G (Invitrogen) bound by anti-GFP antibody (Invitrogen #A11122). After overnight incubation, the beads were washed, reverse crosslinked and DNA was purified for library preparation. The libraries for input and ChIP samples were prepared by adapter ligation and PCR amplification for sequencing by Illumina platform.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image analysis for immuno-staining
Image processing was done by Adobe Photoshop and ImageJ. For fluorescence intensity quantification in ImageJ, the GFP-Del foci were defined after background subtraction, thresholding. Using these foci as reference, the fluorescence intensity was quantified in other channels.
Bioinformatics analysis
Reads from small RNA-seq libraries were aligned to the genome (dm3) by bowtie (Langmead et al., 2009), after removing the 3′end linkers. The transcriptome annotations were collected from Flybase r5.50. The piRNA cluster coordinates were taken from (Brennecke et al., 2007). Reads that were mapped to known non-coding RNAs (ncRNAs, such as rRNAs, tRNAs, etc.) and miRNAs were excluded for the quantification of piRNA abundance of clusters. The counts of reads were obtained using BEDTools (Quinlan and Hall, 2010) and normalized by the total number of reads aligned to the genome excluding known ncRNAs. A read is counted proportionally if it has multiple mapping locations. RNA-seq reads were aligned to the genome by TopHat (Trapnell et al., 2009), and rRNA reads were removed before the quantification of expression levels of genes, piRNA clusters, and transposons. ChIP-seq reads were aligned by BWA (Li and Durbin, 2009), and duplicate reads were marked and removed by Picard tools.
Analysis of Proteome obtained by Mass spectrometry
The raw data was processed through Proteome Discoverer and Mascot Server before display on Scaffold Viewer (Proteome Software, Inc.). iBAQ values (Schwanhausser et al., 2011) of each IPed protein were normalized to corresponding GFP iBAQ values after adding pseudocount. The ratios of these normalized values were ranked and plotted with R.
Statistical analysis
The error bars in the bar graphs represent standard deviation for 3 biological replicates.
DATA AND SOFTWARE AVAILABILITY
High throughput sequencing files are available from NCBI short read archive (SRA) SRP111075. The scaffold files for mass spectrometry analysis and cloned gene sequences are available at Mendeley Data (http://dx.doi.org/10.17632/w2ym383bp8.1).
Supplementary Material
Acknowledgments
We would like to thank the members of Theurkauf and Weng labs and UMassmed RNA Biology community for their insightful discussions and critical comments throughout the project; Trudi Schüpbach for del stocks; Bloomington and UCSD Drosophila stock centers for various fly stocks, John Leszyk of UMassmed Proteomics facility for mass spectrometry. This work was supported by National Institute of Child Health and Human Development (R01HD049116 and P01HD078253).
Footnotes
AUTHOR CONTRIBUTIONS
S.S.P., W.E.T. and Z.W. conceived the project. S.S.P. performed all the experiments. S.S.P. and S.T. performed bioinformatics analysis. S.S.P. and W.E.T. wrote the paper with help of Z.W. and S.T.
References
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bartolome C, Bello X, Maside X. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 2009;10:R22. doi: 10.1186/gb-2009-10-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- Belyayev A. Bursts of transposable elements as an evolutionary driving force. J Evol Biol. 2014;27:2573–2584. doi: 10.1111/jeb.12513. [DOI] [PubMed] [Google Scholar]
- Bergman CM, Quesneville H, Anxolabehere D, Ashburner M. Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol. 2006;7:R112. doi: 10.1186/gb-2006-7-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemont C, Vieira C. Genetics: junk DNA as an evolutionary force. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Chen YC, Stuwe E, Luo Y, Ninova M, Le Thomas A, Rozhavskaya E, Li S, Vempati S, Laver JD, Patel DJ, et al. Cutoff Suppresses RNA Polymerase II Termination to Ensure Expression of piRNA Precursors. Mol Cell. 2016;63:97–109. doi: 10.1016/j.molcel.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirn GW, Rahman R, Sytnikova YA, Matts JA, Zeng M, Gerlach D, Yu M, Berger B, Naramura M, Kile BT, et al. Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals. PLoS Genet. 2015;11:e1005652. doi: 10.1371/journal.pgen.1005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- Duggal NK, Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol. 2012;12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl DF, Lorenz LJ, Melnick MB, Sood V, Lasko P, Perrimon N. A new enhancer of position-effect variegation in Drosophila melanogaster encodes a putative RNA helicase that binds chromosomes and is regulated by the cell cycle. Genetics. 1997;146:951–963. doi: 10.1093/genetics/146.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde NC, Malik HS. The evolutionary conundrum of pathogen mimicry. Nat Rev Microbiol. 2009;7:787–797. doi: 10.1038/nrmicro2222. [DOI] [PubMed] [Google Scholar]
- Fontdevila A. Hybrid genome evolution by transposition. Cytogenet Genome Res. 2005;110:49–55. doi: 10.1159/000084937. [DOI] [PubMed] [Google Scholar]
- Gou LT, Kang JY, Dai P, Wang X, Li F, Zhao S, Zhang M, Hua MM, Lu Y, Zhu Y, et al. Ubiquitination-Deficient Mutations in Human Piwi Cause Male Infertility by Impairing Histone-to-Protamine Exchange during Spermiogenesis. Cell. 2017;169:1090–1104. e1013. doi: 10.1016/j.cell.2017.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu A, Ji G, Shi X, Long Y, Xia Y, Song L, Wang S, Wang X. Genetic variants in Piwi-interacting RNA pathway genes confer susceptibility to spermatogenic failure in a Chinese population. Hum Reprod. 2010;25:2955–2961. doi: 10.1093/humrep/deq274. [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Hedges DJ, Deininger PL. Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res. 2007;616:46–59. doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Ferreira HJ, Bassas L, Bonache S, Sayols S, Sandoval J, Esteller M, Larriba S. Epigenetic disruption of the PIWI pathway in human spermatogenic disorders. PLoS One. 2012;7:e47892. doi: 10.1371/journal.pone.0047892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27:2702–2711. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Luo Y, Moon S, Ninova M, Marinov GK, Chung YD, Aravin AA. Splicing-independent loading of TREX on nascent RNA is required for efficient expression of dual-strand piRNA clusters in Drosophila. Genes Dev. 2016;30:840–855. doi: 10.1101/gad.276030.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu Rev Biochem. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kelleher ES, Edelman NB, Barbash DA. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol. 2012;10:e1001428. doi: 10.1371/journal.pbio.1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, Theurkauf W. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol. 2010;191:905–913. doi: 10.1083/jcb.201006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Stuwe E, Li S, Du J, Marinov G, Rozhkov N, Chen YC, Luo Y, Sachidanandam R, Toth KF, et al. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 2014;28:1667–1680. doi: 10.1101/gad.245514.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Burlet N, Biemont C, Vieira C. Comparative analysis of transposable elements in the melanogaster subgroup sequenced genomes. Gene. 2011;473:100–109. doi: 10.1016/j.gene.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- McKim KS, Joyce EF, Jang JK. Cytological analysis of meiosis in fixed Drosophila ovaries. Methods Mol Biol. 2009;558:197–216. doi: 10.1007/978-1-60761-103-5_12. [DOI] [PubMed] [Google Scholar]
- Meehan RR, Kao CF, Pennings S. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 2003;22:3164–3174. doi: 10.1093/emboj/cdg306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, Brennecke J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157:1364–1379. doi: 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Gordon KH, Buck AH, Jiggins FM. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci. 2009;364:99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Herold A, Gari K, Kocher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol. 2004;11:558–566. doi: 10.1038/nsmb759. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin A, Wong A, Poh YP, Theurkauf WE, Jensen JD. Recurrent and recent selective sweeps in the piRNA pathway. Evolution. 2013;67:1081–1090. doi: 10.1111/evo.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers JF, Henikoff S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH. Genetic Studies on DROSOPHILA SIMULANS. I. Introduction. Hybrids with DROSOPHILA MELANOGASTER. Genetics. 1920;5:488–500. doi: 10.1093/genetics/5.5.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderweyde T, Apicco DJ, Youmans-Kidder K, Ash PE, Cook C, Lummertz da Rocha E, Jansen-West K, Frame AA, Citro A, Leszyk JD, et al. Interaction of tau with the RNA-Binding Protein TIA1 Regulates tau Pathophysiology and Toxicity. Cell Rep. 2016;15:1455–1466. doi: 10.1016/j.celrep.2016.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Henikoff S, Malik HS. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 2005;1:96–108. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Malik HS. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu Rev Genet. 2009;43:467–492. doi: 10.1146/annurev-genet-102108-134802. [DOI] [PubMed] [Google Scholar]
- Volpe AM, Horowitz H, Grafer CM, Jackson SM, Berg CA. Drosophila rhino encodes a female-specific chromo-domain protein that affects chromosome structure and egg polarity. Genetics. 2001;159:1117–1134. doi: 10.1093/genetics/159.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren IA, Naville M, Chalopin D, Levin P, Berger CS, Galiana D, Volff JN. Evolutionary impact of transposable elements on genomic diversity and lineage-specific innovation in vertebrates. Chromosome Res. 2015;23:505–531. doi: 10.1007/s10577-015-9493-5. [DOI] [PubMed] [Google Scholar]
- Wehr K, Swan A, Schupbach T. Deadlock, a novel protein of Drosophila, is required for germline maintenance, fusome morphogenesis and axial patterning in oogenesis and associates with centrosomes in the early embryo. Dev Biol. 2006;294:406–417. doi: 10.1016/j.ydbio.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Weick EM, Miska EA. piRNAs: from biogenesis to function. Development. 2014;141:3458–3471. doi: 10.1242/dev.094037. [DOI] [PubMed] [Google Scholar]
- Yi M, Chen F, Luo M, Cheng Y, Zhao H, Cheng H, Zhou R. Rapid evolution of piRNA pathway in the teleost fish: implication for an adaptation to transposon diversity. Genome Biol Evol. 2014;6:1393–1407. doi: 10.1093/gbe/evu105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Cassani M, Wang M, Liu M, Ma J, Li G, Zhang Z, Huang Y. Structural insights into Rhino-mediated germline piRNA cluster formation. Cell Res. 2015;25:525–528. doi: 10.1038/cr.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanni V, Eymery A, Coiffet M, Zytnicki M, Luyten I, Quesneville H, Vaury C, Jensen S. Distribution, evolution, and diversity of retrotransposons at the flamenco locus reflect the regulatory properties of piRNA clusters. Proc Natl Acad Sci U S A. 2013;110:19842–19847. doi: 10.1073/pnas.1313677110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012a;151:871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Theurkauf WE, Weng Z, Zamore PD. Strand-specific libraries for high throughput RNA sequencing (RNA-Seq) prepared without poly(A) selection. Silence. 2012b;3:9. doi: 10.1186/1758-907X-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, Tu S, Vreven T, Zamore PD, Weng Z, Theurkauf WE. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell. 2014;157:1353–1363. doi: 10.1016/j.cell.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.