Summary
Hexanucleotide repeat expansion in the C9ORF72 gene results in production of dipeptide repeat (DPR) proteins that may disrupt pre-mRNA splicing in ALS/FTD patients. At present, the mechanisms underlying this mis-splicing are not understood. Here, we show that addition of proline-arginine (PR) and glycine-arginine (GR) toxic DPR peptides to nuclear extracts blocks spliceosome assembly and splicing but not other types of RNA processing. Proteomic and biochemical analyses identified U2 snRNP as a major interactor of PR and GR peptides. In addition, U2 snRNP, but not other splicing factors, mislocalizes from the nucleus to the cytoplasm in C9ORF72 patient-iPSC derived motor neurons and also in HeLa cells treated with the toxic peptides. Bioinformatic studies support a specific role for U2 snRNP-dependent mis-splicing in C9ORF72 patient brains. Together, our data indicate that DPR-mediated dysfunction of U2 snRNP could account for as much as ~44% of the mis-spliced cassette exons in C9ORF72 patient brains.
Keywords: Pre-mRNA splicing, U2 snRNP, ALS, FTD, C9ORF72, toxic polydipeptide repeats, DPRs, polyGR, polyPR, iPSC-derived motor neurons
Introduction
Expansion of a hexanucleotide repeat (G4C2) in the first intron of the C9ORF72 gene is the most frequent cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (DeJesus-Hernandez et al., 2011; Renton et al., 2011; Rutherford et al., 2012). The expansion generates both sense and anti-sense repeat RNAs, which form aggregates in patient brains (Cooper-Knock et al., 2014; DeJesus-Hernandez et al., 2011; Renton et al., 2011). The repeat RNAs also generate five dipeptide repeat (DPR) proteins via translation of both the sense and anti-sense transcripts (Taylor et al., 2016). These DPR (GR, PR, GA, GP and PA) proteins are found in both the nucleus and cytoplasm of C9ORF72 patient tissues, including brain and spinal cord, as well as in patient-iPSC derived motor neurons (iPSC-MNs) (Ash et al., 2013; Mori et al., 2013; Wen et al., 2014). Of the DPR proteins, GR and PR are toxic in human, yeast, and Drosophila model systems (Freibaum et al., 2015; Jovicic et al., 2015; Mizielinska et al., 2014; Tran et al., 2015; Wen et al., 2014; Yang et al., 2015; Zhang et al., 2015). Understanding the mechanisms by which the repeat RNAs and/or the DPRs cause neurodegeneration is a focus of intense investigation. Among the possibilities being examined are that the repeat RNAs or DPR proteins interfere with essential cellular processes (Taylor et al., 2016), such as nucleocytoplasmic trafficking (Freibaum et al., 2015; Jovicic et al., 2015; Zhang et al., 2015), protein synthesis (Kanekura et al., 2016), mitochondria function (Lopez-Gonzalez et al., 2016) and pre-mRNA splicing (Conlon et al., 2016; Cooper-Knock et al., 2014; Kwon et al., 2014; Prudencio et al., 2015). In addition, pathogenesis may involve DPR protein-mediated disruption of the dynamics and assembly of membrane-less organelles such as nucleoli, stress granules, and nuclear speckle domains (Lee et al., 2016).
The focus of our study is to understand the mechanisms by which the C9ORF72 repeat expansion causes mis-splicing. Several mechanisms have been proposed. For example, the repeat RNAs may sequester essential splicing factors and thereby block splicing (Conlon et al., 2016; Cooper-Knock et al., 2015; Cooper-Knock et al., 2014; Haeusler et al., 2014). In addition, ~5000 mis-splicing events were observed when cultured astrocytes were treated with PR toxic peptides, indicating that DPR proteins may play a role in mis-splicing (Kwon et al., 2014). It was also reported that synthetic DPR proteins expressed in tissue culture cells interact with RNA binding proteins and, in particular, with those containing low complexity domains (LCDs) (Lee et al., 2016; Lin et al., 2016). The LCD proteins are thought to mediate the assembly of membrane-less cellular compartments, including nuclear speckle domains, which are where splicing components normally localize (Li et al., 2013; Ramaswami et al., 2013; Taylor et al., 2016). Thus, mis-splicing in C9ORF72 patient cells could be caused by disruption of nuclear speckles and/or to loss of RNA binding proteins essential for splicing due to their LCD-mediated aggregation.
Here, we report that both GR and PR toxic peptides block spliceosome assembly and splicing in nuclear extracts, and both peptides specifically associate with U2 snRNP. In C9ORF72 patient iPSC-MNs, U2 snRNP is depleted from nuclear speckle domains and is mislocalized to the cytoplasm. This mislocalization may be directly caused by the DPR proteins, as we observe U2 snRNP mislocalization to the cytoplasm in HeLa cells treated with PR peptide. In addition, bioinformatic analyses indicate that the toxic peptides interfere with U2 snRNP to cause mis-splicing in C9ORF72 patient brains. Together, these results provide evidence that DPR protein-mediated dysfunction of a major spliceosomal snRNP is a central mechanism underlying mis-splicing in C9ORF72 patients. The genes disrupted by this mechanism include those with mitochondrial, neuronal, and gene expression functions, and are candidate genes for contributing to ALS/FTD pathogenesis.
Results
GR and PR toxic dipeptides inhibit splicing in vitro
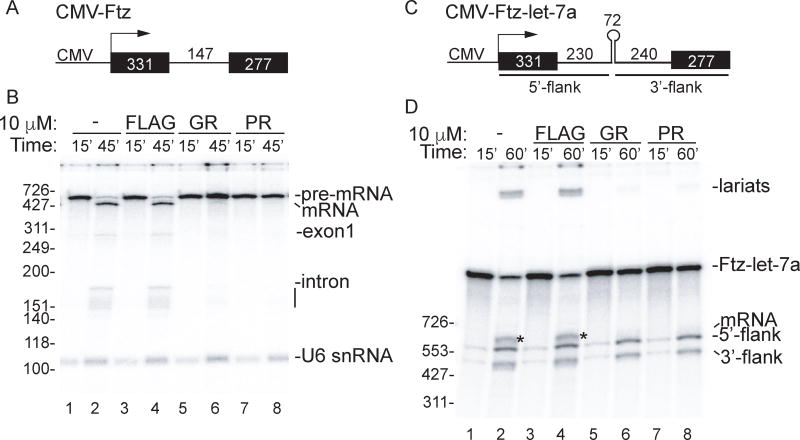
To investigate the mechanisms by which the DPR proteins and/or repeat RNAs encoded by the C9ORF72 expansion inhibit splicing, we used an RNAP II transcription-coupled splicing system (Das et al., 2006; Das et al., 2007; Folco et al., 2011). For this system, we used a DNA template driven by the CMV promoter and encoding the well characterized Ftz splicing substrate (Das et al., 2006; Das et al., 2007; Folco et al., 2011) (Figure 1A). This CMV-Ftz DNA template was incubated in HeLa cell nuclear extracts for 15 minutes to generate an RNAP II transcript, which is then spliced by the 45-minute time point (Figure 1B, lanes 1 and 2). As shown in Figure S1A, increasing amounts of an RNA containing ~60 copies of the C9ORF72 repeats does not affect transcription-coupled splicing. In contrast, a positive control RNA containing exonic splicing enhancers (Das et al., 2007) efficiently blocked splicing in this assay (Figure S1A).
Figure 1. GR and PR toxic peptides inhibit splicing in vitro.
(A) Schematic of CMV-Ftz DNA template used for transcription-coupled splicing showing the CMV promoter and sizes of the exons and intron. (B) CMV-Ftz DNA was incubated in transcription-coupled splicing reaction mixtures for 15 minutes with no peptide or in the presence of 10 µM GR, PR or FLAG peptides. α-Amanitin was then added to stop further transcription, and incubation was continued for 30 minutes to allow splicing. RNA species are indicated. The line below the intron marks intron breakdown products. (C) Schematic of CMV-Ftz-let-7a DNA template which contains a pri-let-7a sequence in the Ftz intron. (D) CMV-Ftz DNA was incubated in transcription/splicing/pri-miRNA processing reaction mixtures for 15 minutes with 10 µM of the indicated peptides. After adding α-Amanitin, incubation was continued for 45 minutes. The asterisks on the gel indicates the spliced mRNA. Markers in nts are shown. See also Figure S1.
We next tested the effects of the toxic repeat dipeptides on splicing in our system using synthetic FLAG-tagged peptides containing 20 repeats of GR or PR. FLAG peptide alone was used as a negative control. When these peptides were added to nuclear extracts, transcription occurred as efficiently as adding no peptide at all (Figure 1B, lanes 1, 3, 5, and 7). Strikingly, however, splicing was potently blocked by GR and PR toxic peptides, but not by FLAG peptide (Figure 1B, lanes 2, 4, 6, and 8). Moreover, splicing intermediates (exon 1 and lariat-exon 2) were not detected, indicating that the block occurs prior to or at the first catalytic step of splicing. We obtained the same results with another splicing substrate (CMV-AdML, Figure S1B and S1C), suggesting that the inhibitory effect of the toxic peptides on splicing is general.
The observation that the GR and PR peptides block splicing is consistent with previous data showing that these peptides cause extensive mis-splicing in cultured cells (Kwon et al., 2014). The inhibitory effect of GR and PR on splicing in vitro is dose-dependent, as we observed no effect with 0.1 or 1 µM of either GR or PR, ~50% inhibition with 5 µM of either peptide and complete inhibition with 10 µM of the peptides (Figure S1D). Although high levels of the peptides are required to block splicing in the in vitro system, whereas only low levels of DPR proteins are present in C9ORF72 patient cells, there are reasonable explanations for how these low levels might interfere with splicing. Specifically, mis-splicing in C9ORF72 patient cells occurs in alternatively spliced exons (Prudencio et al., 2015). Such exons typically contain weak splicing signals, which are known to be sensitive to minor dysfunction in splicing factors (Fu and Ares, 2014). Thus, low levels of DPR proteins in patient cells might be sufficient to cause disruptions of alternative splicing. In contrast to patient cells, in vitro systems require the use of highly efficient splicing substrates in order for splicing to occur, and splicing of these efficient substrates is not affected by small alterations in the functions/levels of splicing factors. Thus, it is not unexpected that low levels of GR/PR peptides have no effect on splicing the efficient substrates used for in vitro studies. Another notable difference between the patient cells and our in vitro model system is that patient cells typically harbor DPR proteins containing hundreds to thousands of repeats whereas the peptides that we use contain only 20 repeats. Longer peptides could not be tested because of the technical difficulties in synthesizing them (Kwon et al., 2014; Lin et al., 2016; Shi et al., 2017).
To determine whether other types of RNA processing are affected by GR or PR peptides, we added them to our transcription-coupled primary-microRNA (pri-miRNA) processing system (Yin et al., 2015). As shown in Figure S1E and S1F, no effect on pri-miRNA processing was observed when the peptides were present at the same levels that block splicing. We also developed an in vitro transcription/splicing/pri-miRNA processing system in which the pri-let-7a is located in the Ftz intron (Figure 1C). In the absence of peptides or in the presence of the FLAG peptide, both splicing and pri-miRNA processing products were observed (Figure 1D, lanes 2 and 4). However, when the toxic peptides were added to the reaction, splicing was blocked whereas no apparent effect on pri-miRNA processing was observed (Figure 1D, lanes 6 and 8). The peptides also did not affect other gene expression steps, including transcription, U6 snRNA processing, or tRNA processing (We note that processing of U6 and tRNAs results in their labeling by the 32P-UTP added to the nuclear extract (Das et al., 2007), and we do not detect any effect on the levels of either labeled RNA species when the toxic peptides were added to the nuclear extract (Figure 1B and Figure S1)). We conclude that GR and PR specifically block splicing in vitro.
GR and PR associate with U2 snRNP
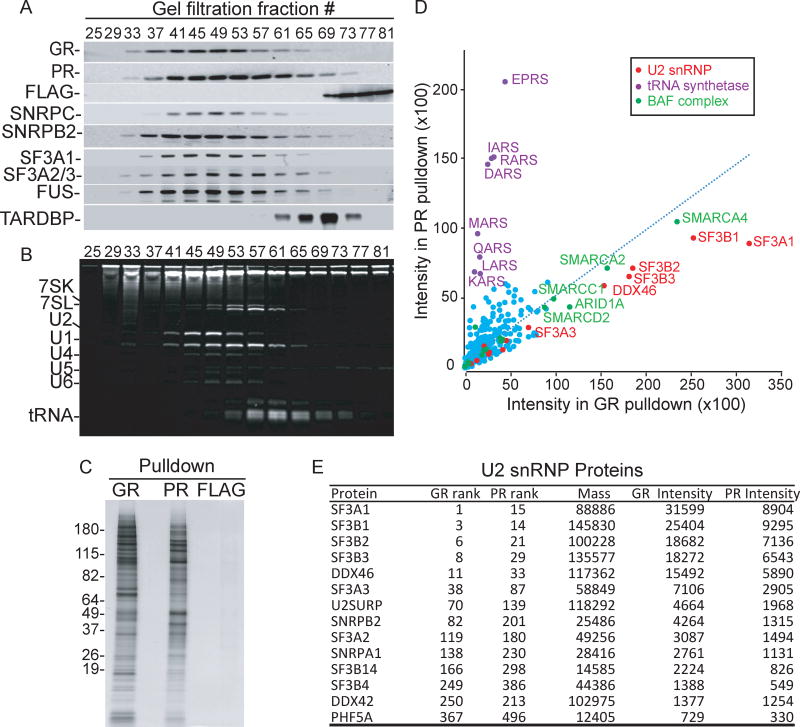
To investigate the mechanisms by which the toxic peptides block splicing we identified their interacting proteins in HeLa nuclear extract. As a first purification step, we added GR, PR, or FLAG peptides to the extract, and then size-separated these samples by gel filtration. This was followed by western blotting (Figure 2A) or ethidium bromide staining (Figure 2B) to detect the proteins and RNAs, respectively. Both GR and PR eluted in fractions containing high molecular weight RNP complexes, such as U1 and U2 snRNPs (Figures 2A and 2B). The toxic peptides also co-eluted with the ALS-causative protein FUS, but not with TDP-43 (Figure 2A). In contrast, FLAG peptide eluted in fractions containing free proteins. To identify proteins that interact with GR or PR, we individually pooled the fractions containing these peptides and used them for immunoprecipitations (IPs) with an antibody against FLAG. We found that these FLAG IPs were inefficient and thus tested pulldowns using avidin affinity selection of biotinylated peptides. As shown in Figure S1G, the biotinylated peptides inhibited splicing with similar efficiency as the non-biotinylated peptides. We then used the biotinylated peptides for pulldowns from the gel filtration fractions, and the set of proteins bound to them is shown on a silver-stained gel (Figure 2C). To identify the GR or PR-interacting proteins, we carried out quantitative mass spectrometry of the total proteins in each pulldown (Table S1). Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA) analyses of the proteins in both pulldowns revealed RNA splicing as the most enriched category (Figure S2 and Tables S2 and S3). Strikingly, further examination of the proteomic data revealed that six U2 snRNP components were among the highest hits shared between the GR and PR pulldowns (Figure 2D, red dots). The U2 snRNP component SF3A1 was the highest specific hit in the GR pulldown, and five out of the top seven hits in this pulldown were U2 snRNP proteins (Table S1). Moreover, all of the other known U2 snRNP proteins, including the full sets of SF3a and SF3b sub-complexes, were present in both pulldowns (Figure 2E–Table S1). In contrast to U2 snRNP, components of other snRNPs were not enriched in the pulldowns, and their complete sets of cognate proteins were not detected (Table S1). For example, U5 snRNP is known to contain eight specific components. We only detected three of them, two of which are ~200 kD and are found as common contaminants in many non-related pulldowns. These results suggest that U2 snRNP specifically associates with GR and PR. In addition to U2 snRNP, an enrichment of the BAF chromatin remodeling complex was seen in both pulldowns (Figure 2D, green dots and Table S1). We also detected a group of aminoacyl-tRNA synthetases that was specifically enriched in the PR, but not in the GR, pulldown (Figure 2D, purple dots and Table S1). We note that our silver-stained gel of the pulldowns revealed different patterns of proteins associated with the two peptides (Figure 2C). These differences can be ascribed to different levels of each protein pulled down by GR and PR (e.g. see GR and PR ranks in Table S1).
Figure 2. GR and PR associate with U2 snRNP.
(A) Nuclear extract reaction mixtures were incubated with 10 µM of either GR, PR, or FLAG peptide followed by separation on Sephacryl-S500 columns. The indicated fractions from each column were used for Western blots with an antibody against FLAG (rows GR, PR, FLAG). The GR gel filtration fractions were then used for Western blotting analysis with antibodies against the U1 snRNP component SNRPC, the U2 snRNP components SNRPB2, SF3A1, SF3A2 and SF3A3 (the latter two proteins co-migrate on the SDS gel), FUS, and TARDBP. We obtained similar results when the PR or FLAG gel filtration fractions were used for the Western blotting (data not shown). Fraction 25 is the void volume, and low molecular weight factors, such as free proteins, elute in fractions ~70–80. (B) Same as (A) except that total RNA from the GR fractions was run on an 8% denaturing gel and stained with ethidium bromide. RNA species are indicated. (C) Silver stained gel showing total proteins in GR, PR or FLAG pulldowns from gel filtration fractions 40–60. (D) Scatter plot depicting the protein intensity identified in GR or PR pulldowns. Red, purple, and green dots indicate U2 snRNP components, tRNA synthetases, and BAF complex components, respectively. (E) Table showing quantitative mass spectrometry data for the U2 snRNP components in GR or PR pulldowns. The rank of each protein in the total pulldowns sorted by mass spectrometry intensity, the calculated mass, and the mass spectrometry intensity are shown. See also Figure S2 and Tables S1–S3.
Importantly, only a few components of other multi-component complexes, such as the TREX or SMN complexes, were detected, indicating that GR and PR are not generally associated with all cellular complexes. Previous studies found that the translation machinery was the most abundant interactor of GR and PR expressed in HEK293 cells as GFP-fusions from C9ORF72 repeat-containing RNA (Kanekura et al., 2016; Lee et al., 2016; Lopez-Gonzalez et al., 2016). We also detected most of the components of the large and small ribosomal subunits in our GR and PR pulldowns, but they were not top hits. One of the possible reasons for this is that we used nuclear extract whereas the other studies used whole cell lysates, which are rich in ribosomes (Kanekura et al., 2016; Lee et al., 2016; Lopez-Gonzalez et al., 2016).
GR and PR bind to U2 snRNP and disrupt spliceosome assembly
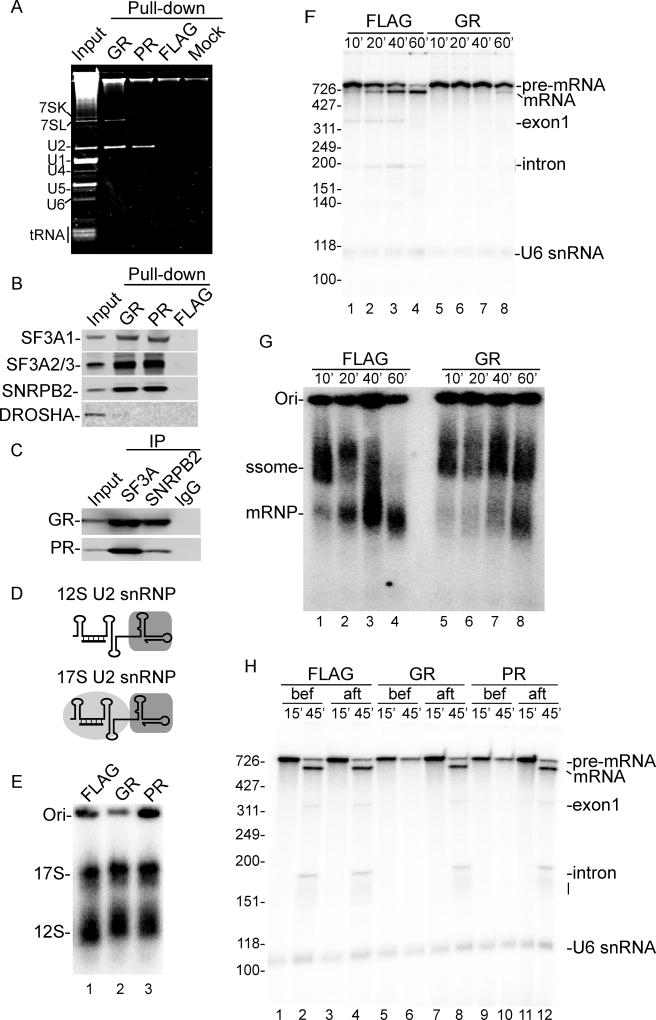
We next analyzed the RNAs present in the pulldowns on an ethidium bromide-stained gel (Figure 3A). Consistent with the proteomics results, U2 snRNA, but not the other spliceosomal snRNAs, was enriched in both GR and PR pulldowns, supporting a specific interaction between GR/PR and U2 snRNP. This was further confirmed by western blotting, which showed that the SF3a subunits (SF3A1-3) and SNRPB2 were enriched in the GR and PR pulldowns (Figure 3B). In contrast, the pri-miRNA processing factor DROSHA was not detected by western in the pulldowns, consistent with the observation that GR/PR does not affect pri-miRNA processing (Figure 1C and D, Figure S1E and S1F). We also performed reverse IPs using antibodies to SF3a or SNRPB2, which confirmed that GR/PR associate with U2 snRNP (Figure 3C).
Figure 3. GR and PR bind to U2 snRNP and inhibit spliceosome assembly.
(A) Ethidium bromide stained gel showing total RNA isolated from GR, PR, FLAG or beads alone (mock) pulldown from gel filtration fractions. A low level of the RNA component of signal recognition particle, 7SL RNA, was seen in the GR pulldown. However, the protein components of this complex were either low hits or not detected (Table S1). Thus, SRP is unlikely to be a genuine GR interactor. (B) Pulldowns were analyzed by western blotting using indicated antibodies (C) Antibodies against SF3A1-3, SNRPB2 or IgG were used for IPs from nuclear extract followed by western blotting with antibodies against GR or PR. (D) Schematic of 12S and 17S U2 snRNP showing the U2 2’O-Methyl oligo that base pairs to the 5’ end of U2 snRNA. The gray boxes indicate the Sm core, SNRPB2 and SNRPA1, and the gray circle indicates the SF3a and SF3b complexes. (E) Nuclear extracts were incubated with 10 µM of the indicated peptides under splicing conditions for 5 minutes followed by a 5 minute incubation with 32P-labeled U2 2’O-Methyl oligo. Samples were fractionated on a native agarose gel and detected by PhosphorImager. The 17S and 12S U2 snRNPs are indicated. Ori marks the gel origin. (F, G) CMV-Ftz DNA template was incubated in transcription-coupled splicing mixtures in the presence of FLAG or GR for the indicated times. An aliquot from each time point was analyzed on an 8% denaturing polyacrylamide gel (panel F) or on a native agarose gel (panel G) to detect total RNA or RNP complexes, respectively. The spliceosome (ssome) and spliced mRNP complex are indicated, and Ori marks the gel origin. (H) CMV-Ftz DNA template was incubated for 15 minutes in transcription-coupled splicing mixtures followed by addition of α-Amanitin and continued incubation for 30 minutes. The indicated peptides were added either prior to (bef) or after (aft) transcription.
Previous studies showed that the functional form of U2 snRNP in nuclear extracts is a 17S particle consisting of the SF3a and SF3b sub-complexes and a 12S U2 snRNP core particle, which contains U2 snRNA, the Sm proteins, SNRPB2, and SNRPA1 (Wahl et al., 2009). Both the 12S and 17S U2 snRNP can be detected on a native agarose gel using a 32P-labeled 2’ OMethyl oligonucleotide (U2 oligo) that base-pairs to the 5’ portion of U2 snRNA (Figure 3D and Figure 3E, lane 1) (Folco et al., 2011; Ruby et al., 1993). We next used this assay to examine the effect of GR and PR on U2 snRNP. This analysis revealed that addition of either toxic peptide resulted in a quantitative mobility shift of the 12S and 17S U2 snRNP particles (Figure 3E, lanes 2 and 3), consistent with the observation that both peptides associated with this snRNP in the pulldown assay. These results raise the possibility that GR and PR disrupt splicing by affecting normal U2 snRNP function.
As 17S U2 snRNP is essential for spliceosome assembly, we next examined the effects of the toxic peptides on this process. To do this, either FLAG or GR were added to nuclear extract followed by a transcription-coupled splicing reaction using the CMV-Ftz DNA substrate. The RNAs within the spliceosomal complexes were analyzed on a denaturing gel (Figure 3F), and the complexes were analyzed on a native agarose gel (Figure 3G). In the presence of FLAG peptide, the kinetics of splicing and spliceosome assembly were the same as with no peptide at all. Specifically, after 10 minutes of transcription, nascent transcripts were detected (Figure 3F, lane 1) and efficiently assembled into the spliceosome (Figure 3G, lane 1). Over time, the spliced mRNA and the spliced mRNP complex continued to accumulate (Figure 3F and G, lanes 2–4). In contrast, when GR was included in the reaction, splicing was inhibited (Figure 3F, lanes 5–8), and a complex with slower mobility than the spliceosome was detected by 10 minutes of incubation, and was not significantly changed over time (Figure 3G, lanes 5–8; note that a low level of splicing (Figure 3F, lane 8) and spliced mRNP (Figure 3G, lane 8) are detected by 60 minutes because the reaction is leaky by this late time point). We conclude that GR inhibits spliceosome assembly, thereby resulting in defective splicing. Previously, we showed that the spliceosome assembles on nascent transcripts as rapidly as they are synthesized by RNAP II (Das et al., 2006). Consistent with these data, we observed that addition of GR or PR only blocked splicing when added before, but not after, transcription (Figure 3H). This order of addition experiment further demonstrates that the effect of the toxic peptides is specific as they only cause a block to splicing at a specific step in the splicing pathway, and do not interfere with splicing after transcription and spliceosome assembly occur.
U2 snRNP mislocalizes to the cytoplasm in C9ORF72 patient iPSC-MNs
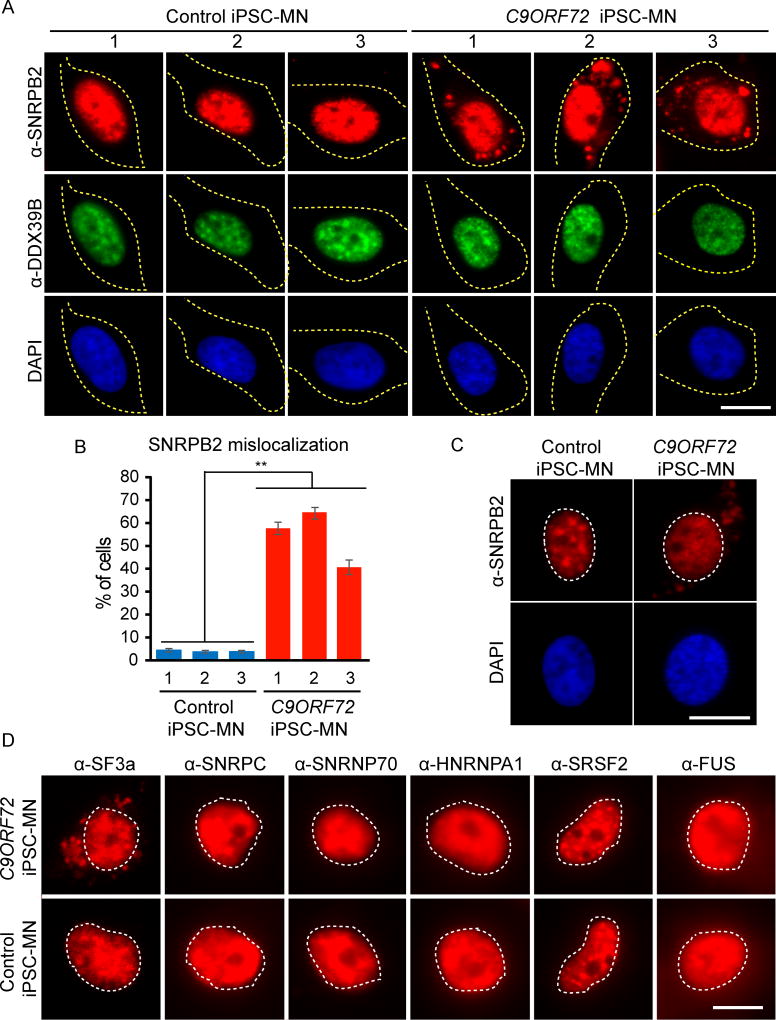
A critical unanswered question is whether U2 snRNP is affected in C9ORF72 patient cells. To investigate this possibility, we examined high-yield motor neuron cultures differentiated from 3 C9ORF72 patient-derived iPSC lines (Lopez-Gonzalez et al., 2016). These iPSC-MNs are known to contain the C9ORF72 repeat expansion as well as DPRs generated by translation of the repeat RNAs (Almeida et al., 2013; Ash et al., 2013; Donnelly et al., 2013; Lopez-Gonzalez et al., 2016; Mori et al., 2013; Su et al., 2014). iPSC-MNs derived from 3 non-C9ORF72 subjects were used as controls. Immunofluorescent (IF) staining showed that, in the controls, SNRPB2 and DDX39B were properly localized to the nucleus (Figure 4A). In contrast, we found a striking mislocalization of SNRPB2 to the cytoplasm in the C9ORF72 iPSC-MNs. This was not the case for DDX39B, which was properly localized to the nucleus (Figure 4A). Notably, mislocalization of SNRPB2 in patient iPSC-MNs occurs in the same cells in which DDX39B is properly localized, indicating that the effect is on SNRPB2 rather than a general disruption of the nucleus. Moreover, quantitation revealed mislocalization of SNRPB2 in a large fraction of the patient iPSC-MNs (~40–60%) (Figure 4B), and this was not seen in controls (p<0.001, two-way ANOVA, see Figure S3 for fields). Notably, lighter exposures of our iPSC-MN IF staining data indicate that in C9ORF72 patient cells in which SNRPB2 was mislocalized to the cytoplasm, the level of SNRPB2 in nuclear speckle domains was lower than in controls, and the nucleoplasmic level of SNRPB2 was likewise reduced (Figure 4C, and see Figure S4 for fields). Together, these data raise the possibility that U2 snRNP is mislocalized from its normal cellular location in C9ORF72 patient cells.
Figure 4. U2 snRNP is mislocalized to the cytoplasm in C9ORF72 iPSC-MNs.
(A) IF staining of SNRPB2 (red), DDX39B (green) and DAPI (blue) in iPSC-MNs from control subjects (N=3) or patients with C9ORF72 repeat expansion (N=3). Dotted line outlines the cell body. See Figure S3 for fields. (B) The percentage of cells with cytoplasmic SNRPB2 signals in each cell line. Mean ± SEM of 3 biological replicates is shown. Approximately 300 cells were counted for each line. **, p<0.001, two-way ANOVA. (C) Light exposure of IF staining using SNRPB2 (red) and DAPI (blue) in C9ORF72 iPSC-MNs. Dotted line outlines the cell nucleus. See Figure S4 for fields. (D) IF staining in patient or control iPSC-MNs using antibodies against SF3a, SNRPC, SNRNP70, HNRNPA1, SRSF2, and FUS. Dotted line outlines the cell nucleus. Scale bars: 20 µm.
To further investigate the significance of SNRPB2 mislocalization, we carried out additional IF staining in the iPSC-MNs. As shown in Figure 4D, SF3a is mislocalized to the cytoplasm only in patient iPSC-MNs and not in control iPSC-MNs. In contrast, the U1 snRNP components, SNRPC and SNRNP70, are properly localized to the nucleus in both patient and control iPSC-MNs. In addition, HNRNPA1, SRSF2, and FUS remain properly localized to the nucleus in the patient and control cells. Together, these data support the conclusion that U2 snRNP, a major component of the splicing machinery, is mislocalized to the cytoplasm in C9ORF72 iPSC-MNs, and this mislocalization may play a role in the mis-splicing observed in these patient cells.
U2 snRNP mislocalizes to the cytoplasm in HeLa cells treated with PR
To determine whether the toxic peptides could cause the mislocalization of U2 snRNP, we used IF staining to examine HeLa cells treated with PR or FLAG peptides (GR peptide is unstable in cell culture media (Kwon et al., 2014) and was not tested in our study). As shown in Figure S5, both PR and FLAG peptides were distributed in both the nucleus and cytoplasm. Significantly, U2 snRNP components SNRPB2 and SF3a were mislocalized to the cytoplasm in the PR-treated, but not control-treated, cells (Figure S5A, B and see S5E for fields). In contrast, DDX39B and SRSF2 were properly localized to the nucleus in both the PR-treated and control-treated cells (Figure S5C, D and see S5E for fields). These data are consistent with our results showing that U2 snRNP is mislocalized in C9ORF72 patient iPSC-MNs and suggest a direct role of C9ORF72 toxic peptides in causing U2 snRNP mislocalization.
U2-dependent exons are preferentially mis-spliced in C9ORF72 patient brains
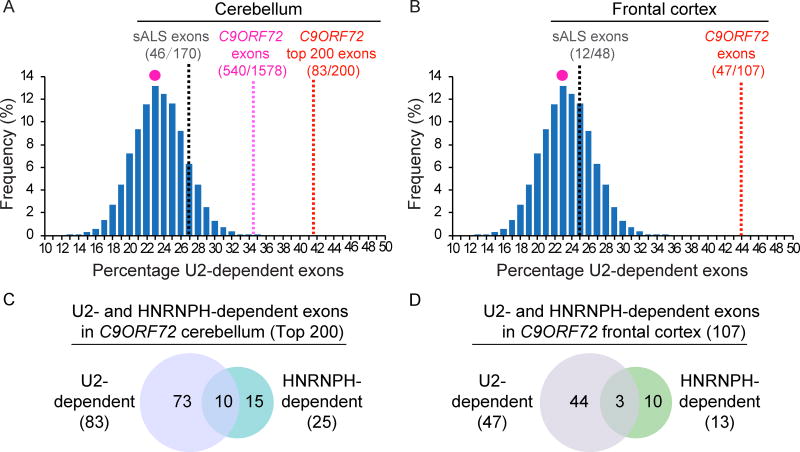
Previous RNA-seq analyses found that 23% of cassette exons out of the whole genome (9,878 out of 42,216) were skipped when the U2 snRNP component SF3B1 was knocked down in HeLa cells (Kfir et al., 2015). If U2 snRNP dysfunction plays an important role in mis-splicing in C9ORF72 patients, these “U2-dependent” exons might be preferentially mis-spliced in patients. To investigate this question, we reanalyzed RNA-seq datasets for cassette exons in C9ORF72, sporadic ALS (sALS), and control brains (Prudencio et al., 2015). In their study, they examined eight cerebellums and eight frontal cortexes, and we used these data for our analysis. In conjunction, we performed a neutral simulation to estimate the extent of U2-dependency expected by chance in the whole genome. This analysis revealed that in the random sampling the highest frequency of the percentage of U2 dependent exons in the genome was 23% (marked by red dot in Figures 5A and B). In contrast, in C9ORF72 cerebellums, U2-dependent exons account for 35% (540/1,578) of the total skipped cassette exons, significantly higher than expected by chance (X2=98.3, P<10−10, chi-square test, Figure 5A). Notably, as shown in Figure 5A, an even greater enrichment of U2-dependent exons (42%, 83/200) was observed in the top 200 most significantly mis-spliced exons in C9ORF72 cerebellum (X2=36.2, P<10−10, chi-square test). This was not the case for mis-spliced exons in sALS cerebellums, of which only 27% were U2-dependent (46 out of 170; X2=1. 3, P=0.26, chi-square test, Figure 5A). We obtained similar results when we reanalyzed the RNA seq data from the frontal cortex in which a total of 107 cassette exons were skipped (Prudencio et al., 2015). In this case, our analysis revealed that U2-dependent exons were significantly enriched in C9ORF72 (44%, 47/107, X2=25.1, P=1.0E-6, chi-square test, Figure 5B), but not in sALS (25%, 12/48, X2=0.07, P=0.8, chi-square test, Figure 5B). Together, our results lend further support to our proteomic, biochemical, and U2 snRNP cellular localization studies, indicating that U2 snRNP plays an important role in the mis-splicing in C9ORF72 patients.
Figure 5. U2-dependent exons are preferentially mis-spliced in C9ORF72 patient brain.
(A, B) Histogram showing U2-dependent exons present in 10,000 random samplings. The red dot indicates the percentage (23%) of the U2-dependent exons in the genome that occur with the highest frequency. The percentage of U2-dependent exons that are mis-spliced in C9ORF72 or sALS patient cerebellums (A) or frontal cortex (B) are indicated by the lines. For C9ORF72, U2-dependent exons for total mis-spliced (1,578) and top 200 mis-spliced cassette exons (FDR ranging from 7E-124 to 2E-12) are shown. C, D. Venn diagrams showing U2- and HNRNPH-dependent mis-spliced exons in C9ORF72 cerebellums (top 200 exons) (C) and frontal cortex (107 exons) (D). See also Tables S4–S6.
To further investigate whether mis-splicing in C9ORF72 was specific to U2 snRNP, we repeated the same analysis for “U1-dependent exons” using data from knockdown of the U1 snRNP component SNRNPC in HeLa cells (Rosel-Hillgartner et al., 2013). Our results showed that 15% (232/1,578) of all cassette exons or 17% of the top 200 cassette exons altered in C9ORF72 patient cerebellums were U1-dependent, which was not significantly different from that of the global average of total exons (13%, p=0.11 and p=0.12, chi-square test). Similarly, no enrichment of U1-dependent exons was observed for C9ORF72 patient frontal cortex (14%, 15/107, X2=0.05, p=0.8, chi-square test).
In a recent study, it was reported that HNRNPH plays a role in mis-splicing in C9ORF72 patient brains (Conlon et al., 2016). Accordingly, we next examined the contribution of U2 snRNP versus HNRNPH to mis-splicing using the top 200 mis-spliced exons from C9ORF72 patient cerebellum and the 107 exons from frontal cortex (Figure 5C and D). This analysis revealed that mis-splicing of 83 exons was U2-dependent, 25 exons were HNRNPH-dependent, and 10 exons were both U2- and HNRNPH-dependent in cerebellum (Figure 5C). Similar results were obtained in the frontal cortex (Figure 5D). These data indicate that U2 snRNP dysfunction has a greater contribution than HNRNPH to mis-splicing in C9ORF72 patient brain.
U2-dependent mis-splicing occurs in PR treated cells
In light of our multiple lines of evidence showing that GR/PR peptides associate with and affect U2 snRNP, we next investigated whether the U2-dependent mis-splicing observed in the bioinformatics analyses could be caused by the toxic peptides. A previous RNA-seq analysis showed that treatment of cultured astrocytes with PR resulted in splicing dysfunction, involving mis-splicing events in 4,298 genes (Kwon et al., 2014). We used these data to compare PR-dependent mis-spliced genes with the U2-dependent genes that we identified in C9ORF72 patient cerebellum and frontal cortex. This analysis revealed that 39% and 40% of the U2-dependent genes altered in the C9ORF72 cerebellum and frontal cortex, respectively, were also PR-dependent, significantly more than expected by chance (20%, P<10−10 and P=1.0E-6, chi-square test). These data support the model that the toxic peptides directly lead to dysfunction of U2 snRNP and mis-splicing in C9ORF72 patient cells. Further evidence for the importance of the toxic peptides in causing U2-dependent mis-splicing came from a genome-wide comparison of U2-dependent genes with PR-dependent genes. We found that 42% of the genes affected by PR are U2-dependent (1,797 out of 4,298), significantly higher than expected by chance (24%, X2=292.8, P<10−10, chi-square test). Thus, there is a high level of correlation between U2-dependent mis-splicing and PR-dependent mis-splicing events in the whole genome. Together, these analyses provide evidence that DPR proteins disrupt normal U2 snRNP function, and this could be an important mechanism underlying mis-splicing in C9ORF72 patient cells.
Discussion
Previous studies reported that mis-splicing occurs in C9ORF72 patient brains but the underlying mechanisms are not understood. Here, we obtained multiple lines of evidence that dysfunction of the general splicing factor U2 snRNP plays an important role in this mis-splicing. Our in vitro studies showed that C9ORF72 toxic dipeptides (GR and PR) block spliceosome assembly and splicing, and specifically associate and interfere with U2 snRNP. To investigate whether the effects on U2 snRNP are relevant to the mis-splicing observed in C9ORF72 patient cells, we examined iPSC-MNs generated from these patients. We observed a dramatic mislocalization of U2 snRNP components, but not other snRNPs or splicing factors, to the cytoplasm in C9ORF72 patient iPSC-MNs. Our data indicate that this mislocalization is due to the toxic peptides because we observed specific mislocalization of U2 snRNP components to the cytoplasm in HeLa cells treated with PR peptide. We also carried out bioinformatic analyses, which revealed that U2-dependent exons are preferentially mis-spliced in C9ORF72 patient brain tissues, accounting for as much as ~44% of the most significantly mis-spliced exons. Of these, PR-dependent mis-splicing accounts for ~40% of them. Together, these data are consistent with the possibility that the mis-splicing observed in patients with C9ORF72 repeat expansion is due, in part, to DPRs interfering with U2 snRNP. An important question for future work is to determine whether and how these U2-dependent mis-splicing events are involved in the pathogenesis of C9ORF72 repeat expansion in ALS/FTD patients.
One obvious mechanism by which mis-splicing of U2-dependent exons could contribute to disease is by affecting genes that are essential for normal motor function. We listed the 81 U2-dependent genes that are affected by mis-splicing in C9ORF72 cerebellum in Table S4 and the 46 U2-dependent genes affected by mis-splicing in the frontal cortex in Table S5. (Note that there are fewer genes than exons with mis-splicing because some of the genes have more than one mis-splicing event). We extracted potentially relevant information about the genes from the literature, and this information is shown in the tables. One of the largest categories of mis-spliced U2-dependent genes has mitochondrial functions (labeled red in Tables S4 and S5), an especially interesting finding in light of the known links between mitochondrial dysfunction and ALS resulting from multiple different causes (Cozzolino and Carri, 2012; Lopez-Gonzalez et al., 2016). It was also recently reported that C9ORF72 patient DPRs compromise mitochondrial function and cause oxidative stress (Lopez-Gonzalez et al., 2016). The U2-dependent genes related to mitochondria include HIF1A, UQCRH, DNM1L, COX16, TMEM126, NDUFAF5, PDHA1, TIMM9, and two mitochondrial ribosomal proteins MRPL52 and MRPS31 (Tables S4 and S5). HIF1A is interesting as it is a transcription factor that has a role in the distribution of mitochondria in axons. Genes with some relationship to neurons or neuronal disease were another large category in the U2-dependent mis-spliced genes (designated in blue, Tables S4 and S5). Notable among these are PARD3 and MARK2, which modulate neuronal polarity and neurite outgrowth, respectively. Finally, we found that several U2-dependent genes have functions in different steps of gene expression. These include transcription and splicing factors (marked in gray and yellow, respectively, Tables S4 and S5). Three of the splicing factors are members of the U2AF family (RBM39, U2AF1 and RBM23). These proteins are essential for recruiting U2 snRNP to the pre-mRNA, and it is possible that their mis-splicing further exacerbates the mis-splicing observed in C9ORF72 patient brains. We also found that GEMIN7 was mis-spliced (Table S5). This protein is a component of the SMN complex, which functions in snRNP biogenesis (Battle et al., 2006). Thus, defective snRNP biogenesis could also contribute to the mis-splicing observed in C9ORF72 patients. Notably, mutation of the SMN protein causes the childhood motor neuron disease Spinal Muscular Atrophy, and previous work revealed that this disease shares a biochemical pathway with ALS (Gama-Carvalho et al., 2017; Shan et al., 2010; Yamazaki et al., 2012). Our bioinformatic study also revealed that 40% of the U2-dependent genes in C9ORF72 patient brains are also PR-dependent. These genes are listed in Table S6, which reveals that they are enriched in genes with mitochondrial and pre-mRNA splicing functions. Together, our results raise the possibility that DPR-mediated dysfunction of U2 snRNP, a major component of the splicing machinery, causes mis-splicing of genes involved in cellular functions that have previously been associated with ALS/FTD pathogenesis.
Several mechanisms may underlie DPR-mediated U2-dependent mis-splicing in C9ORF72 patients. For example, our in vitro data showing that U2 snRNP binds to GR/PR toxic peptides and disrupts splicing and spliceosome assembly raises the possibility that interactions between the DPRs and U2 snRNP directly disrupt the function of this essential splicing factor. Another possibility is that the loss of U2 snRNP to the cytoplasm that we observed in C9ORF72 patient-iPSC derived motor neurons may decrease the levels of nuclear U2 snRNP required for proper splicing. In addition, we observed loss of U2 snRNP from nuclear speckle domains and this may disrupt normal U2 snRNP function. In previous studies, overexpression of tagged-GR/PR showed that it is primarily in the cytoplasm (Kwon et al., 2014; Lee et al., 2016) and we found that PR peptide localizes to the cytoplasm in HeLa cells. In addition, immunohistochemistry of C9ORF72 patient brains showed that DPRs are present in both the nucleus and cytoplasm (Mori et al., 2013; Wen et al., 2014). These observations may explain how U2 snRNP is mislocalized to the cytoplasm in C9ORF72 patient cells. Specifically, it is known that snRNP biogenesis takes place in the cytoplasm (Battle et al., 2006). Thus, it is possible that interactions between the DPRs and U2 snRNP may sequester U2 snRNP in the cytoplasm.
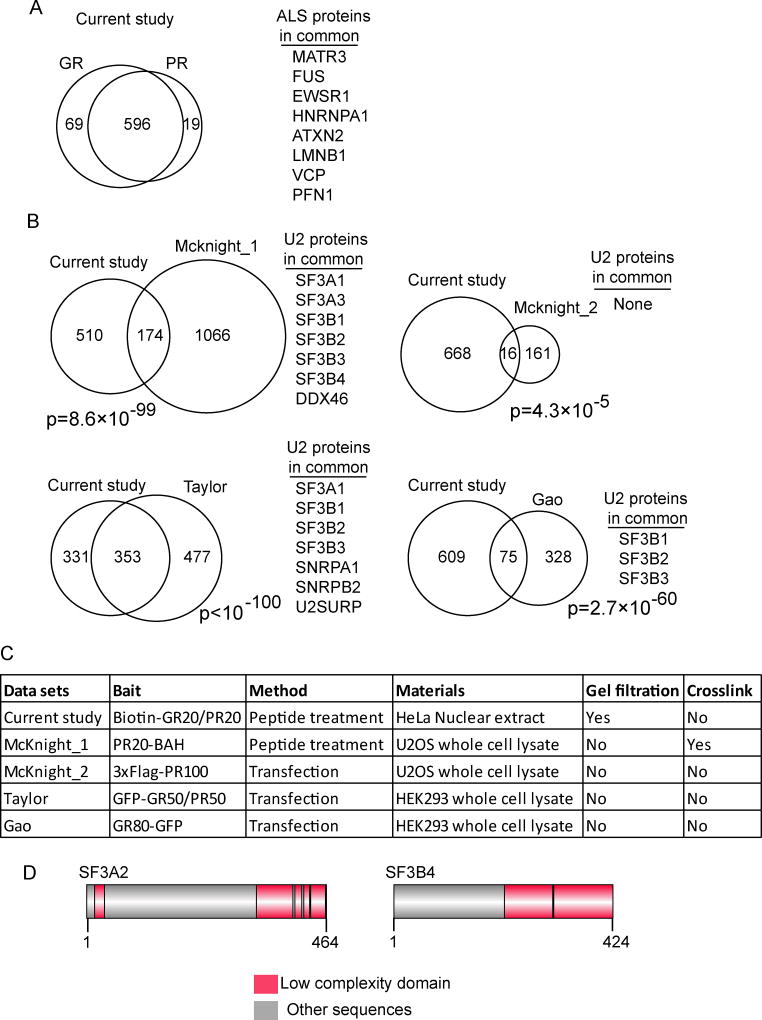
In recent studies, GR or PR peptides over-expressed in cultured cells were found to associate with numerous cellular proteins (Lee et al., 2016; Lin et al., 2016; Lopez-Gonzalez et al., 2016). Among these were RNA binding proteins that contain low complexity domains (LCDs), and of these, several are known ALS-causative proteins (e.g. FUS, TARDBP, HNRNPA1, and MATR3). Interactions between GR/PR and the LCD proteins disrupt the assembly and dynamics of membrane-less organelles, which may contribute to C9ORF72 pathogenesis (Lee et al., 2016). Consistent with these studies, our proteomic analysis of GR/PR-interacting proteins revealed a large overlap with numerous ALS-causative proteins, including those known to contain LCDs (Figure 6A). In addition, the GR/PR interactors identified in our study overlap significantly with those identified in previous studies (Lee et al., 2016; Lopez-Gonzalez et al., 2016) (Figure 6B). Several observations suggest that at least some of the GR/PR interactors are physiologically relevant. Specifically, multiple U2 snRNP proteins were not only identified as GR/PR interactors in our work, but also in the recent studies (Lee et al., 2016; Lopez-Gonzalez et al., 2016) (Figure 6B). In addition, these common interactors were identified using a variety of cell types and pulldown strategies, and are independent of repeat protein lengths (Figure 6C). Many of these U2 snRNP proteins contain LCDs (Figure 6D and Figure S6), and the interaction between GR/PR and these LCD-containing U2 snRNP proteins may explain why U2 snRNP is lost from its normal location in the membrane-less organelle (nuclear speckle domains) and is mis-localized to the cytoplasm.
Figure 6. Comparison of toxic peptide interactomes in different datasets.
(A) Venn diagram showing overlap between GR and PR-interacting proteins identified in this study. Known ALS proteins identified in both datasets are shown. (B) Venn diagrams showing overlap between interacting proteins identified in this study and in 4 published datasets (Mcknight_1 and Mcknight_2 datasets are described in (Lin et al., 2016), Taylor dataset is described in (Lee et al., 2016) and Gao dataset is described in (Lopez-Gonzalez et al., 2016)). (C) Table showing methods used to generate the DPR interactome datasets. (D) Examples of U2 snRNP proteins showing their low complexity domains. See Figure S6 for this analysis with all of the U2 snRNP components.
Our data indicate that U2-dependent mis-splicing accounts for a large fraction of the mis-splicing observed in patients with C9ORF72 repeat expansion, which is the most frequent mutation in familial ALS/FTD. In addition, mis-splicing is present in other forms of familial ALS (e.g. those due to FUS or TDP-43 mutation). Sporadic ALS, which is the most common form of all types of ALS, also exhibits mis-splicing. Thus, correction of mis-splicing is a potential therapeutic approach for multiple forms of ALS. In this regard, splicing modulator compounds that can correct mis-splicing are emerging as potential therapies for other diseases, and these or related compounds may have efficacy for treatment of ALS/FTD.
Experimental Procedures
Plasmids
CMV-Ftz plasmid was described (Das et al., 2006). CMV-Ftz-let-7a plasmid was constructed by inserting a 395-nt pri-let-7a fragment into the BamHI site in the Ftz intron. CMV-DNA templates were amplified by PCR using forward (5’-TGGAGGTCGCTGAGTAGTGC-3’) and reverse (5’- TAGAAGGCACAGTCGAG GCT-3’) primers.
Transcription-coupled RNA processing
For in vitro studies, we used HeLa cell nuclear extracts prepared according to (Krainer et al., 1984). Transcription-coupled splicing reactions were performed as described (Das et al., 2007; Folco et al., 2011) except that pre-initiation complexes (PICs) were assembled on the CMV-DNA templates prior to transcription (Yu et al., 2010). Briefly, CMV-DNA templates were incubated in 15 µl nuclear extract containing 3.2 mM MgCl2, and 5 µl polyvinyl alcohol for 20 minutes at 30°. The indicated peptides (10 µM) were then added in a final reaction mixture volume of 25 µl containing 0.5 mM ATP, 20 mM creatine phosphate (di-Tris salt), and 1 µl 32P-UTP (250 Ci/mmol; Perkin Elmer Life Sciences). Reaction mixtures were incubated for 15 minutes at 30°C to allow transcription followed by addition of α-Amanitin (200 ng). Incubation was continued at 30°C for times indicated. For transcription/splicing/pri-miRNA processing, PICs were assembled using the same conditions as for transcription-coupled splicing. No peptide or 10 µM of the indicated peptides were then added and incubation was continued for 15 minutes to allow transcription in the presence of 0.5 mM ATP, 20 mM creatine phosphate (di-Tris salt), 1 µl 32P-UTP, and an additional 3.2 mM MgCl2 was added to bring the final concentration to 6.4 mM in a final reaction mixture volume of 25 µl. α-Amanitin (200 ng) was added and incubation was continued for 45 minutes to allow processing. Total RNA was fractionated on 8% denaturing polyacrylamide gels and detected by PhosphorImager.
Proteomic analysis of GR and PR interacting proteins
GR, PR, or FLAG peptides were biotinylated using the EZ-Link TFPA-PEG3-Biotin kit (Thermo Fisher). Splicing reaction mixtures containing these peptides (10 µM) were separated on Sephacryl-S500 gel filtration columns in a buffer containing 20 mM Tris, pH 7.8, 60 mM KCl, 0.1% Triton X100, and 0.2 mM PMSF. Proteins in the gel filtration fractions were analyzed on NuPAGE Novex 4–12% Bis-Tris Gels (Thermo Fisher) followed by western blotting. Total RNAs from the fractions were analyzed on an 8% denaturing polyacrylamide gel stained with ethidium bromide. The data for rows FLAG, SNRPC, SNRPB2, SF3A1, SF3A2/3, FUS and TARDBP shown in Figure 2A and the RNA gel shown in Figure 2B were from the gel filtration column containing the GR peptide. Similar results were obtained with gel filtration fractions containing PR and FLAG peptides. Pulldowns were carried out from fractions 33–69 in the gel filtration buffer using streptavidin magnetic particles (Roche) at 4°C for 2 hours and washed 5 times with 1X PBS/0.1% Triton X100. Proteins associated with biotinylated peptides were labeled using tandem mass tag (TMT) (McAlister et al., 2012) and analyzed with an Orbitrap Fusion mass spectrometer coupled to a Proxeon EASY-nLC 1000 liquid chromatography (LC) pump (Thermo Fisher Scientific).
Immunoprecipitations (IPs)
A rabbit polyclonal antibody that recognizes the three SF3a complex subunits or a mouse monoclonal antibody against SNRPB2 were described (Das et al., 2000). Rabbit polyclonal antibodies against DROSHA and a control IgG were from Abcam. For protein IPs, antibodies were crosslinked to protein Sepharose beads with dimethylpymelimidate (Sigma). Prior to IPs, 250 µl reaction mixtures containing 75 µl nuclear extract, 3.2 mM MgCl2, 0.5 mM ATP, and 20 mM creatine phosphate (di-Tris salt) were incubated for 20 minutes at 30° and then spun at 14,000 rpm in a microfuge for 5 minutes at 4°C. The supernatant was mixed with 150 µl PBS/0.1% Triton X-100/0.2 mM PMSF), protease inhibitor EDTA free (Roche), and spun in a microfuge at 14,000 rpm for 5 minutes at 4°C. IPs were carried out at 4°C for 2 hours and washed 5 times with 1X PBS/0.1% Triton X. IPs were mixed with protein gel loading buffer without DTT at room temperature to elute the proteins. DTT (2 µM final) was then added, samples were boiled for 2 minutes, and run on 4–12% SDS-PAGE gels.
U2 snRNP and spliceosome assembly assays
A U2 2’OMethyl oligo that base pairs to U2 snRNA (5’-mCmAmGmAmUmAmCmUmAmCmAmCmUm UmGmA-3’) was 32P-labeled with γ-ATP (250 Ci/mmol; Perkin Elmer Life Sciences) and T4 polynucleotide kinase. Reaction mixtures (25 µl) containing 15 µl nuclear extract were incubated in the presence of 0.5 mM ATP, 3.2 mM MgCl2, 20 mM creatine phosphate (di-Tris salt), 10 µM peptides and 0.4 µM U2 oligo for 5 minutes. For spliceosome assembly, transcription-coupled splicing reactions with or without the peptides were incubated for the times indicated and then run on G-50 micro columns (Amersham Biosciences) to remove unincorporated 32P-UTP. Heparin (0.65 mg/ml final concentration) was added to the G-50 column-purified reactions before loading on a 1.2% low melting point agarose gel.
Generation of iPSC-Derived Motor Neurons
Generation of iPSC-derived Neurons was described (Lopez-Gonzalez et al., 2016). Briefly, iPSCs were expanded in Matrigel-coated wells and at 60% confluency were split with Accutase into Matrigel-coated wells. 24 hours after plating, culture medium was replaced with neuroepithelial progenitor (NEP) medium, DMEM/F12, Neurobasal medium at 1:1, 0.5X N2, 0.5X B27, 0.1 mM ascorbic acid (Santa Cruz Biotechnology), 1X Glutamax (Invitrogen), 3 µM CHIR99021 (Tocris Bioscience), 2 µM DMH1 (Tocris Bioscience), and 2 µM SB431542 (Stemgent) for 6 days. NEPs were dissociated with dispase and split 1:6 into Matrigel-coated wells. NEPs were cultured in motor neuron progenitor (MNP) induction medium (the same medium as described above with 0.1 µM RA (Stemgent) and 0.5 µM Purmorphamine (Stemgent)) for 6 days. MNPs were dissociated with dispase to generate suspension cultures. After 6 days in culture, cells were dissociated into single cells, plated on laminin-coated plates/coverslips in motor neuron differentiation medium containing 0.5 µM RA, 0.1 µM Purmorphamine, and 0.1 µM Compound E (Calbiochem), and cultured for 1 month.
Immunofluorescent (IF) staining
For IF, iPSC-MNs were fixed with 4% paraformaldehyde in PBS for 15 minutes and permeabilized with 0.1% Triton X-100 in PBS for 15 minutes. After incubation in 5% FBS for 1 hour at room temperature, cells were incubated overnight at 4°C in custom primary antibodies SNRPB2 (1:100), DDX39B (1:1000), SF3a (1:1000), FUS (1:1000), or in commercial antibodies SNRPC (Santa Cruz Biotechnology, 1:200), SNRNP70 (Millipore,1:200), HNRNPA1 (Santa Cruz Biotechnology, 1:200), SRSF2 (1:1000, Invitrogen). After 3 washes in PBS, cells were incubated with mouse Alexa-647 (for SNRPB2, SNRNP70 and SRSF2) or rabbit Alexa-488 (for HNRNPA1, DDX39B, SNRPC, SF3a, FUS) secondary antibodies (Invitrogen, 1:1000) for 1 hour at room temperature, followed by 3 washes in PBS. Images were captured with a Nikon TE2000U inverted microscope.
Bioinformatic analysis and statistics
Brain transcriptome profiles of C9ORF72 ALS and sALS were described in GSE67196 (Prudencio et al., 2015). Processed RNA-seq data was provided by the authors, and a cut-off of Bonferroni corrected FDR<0.05 was used to specify significantly altered exons. RNA-seq data of SF3B1 knockdown was from GSE65644 (Kfir et al., 2015). RNA-seq data of HNRNPH or control knockdown in 293T cells was from GSE16642 (Xiao et al., 2009). RNA-seq data of SNRNPC or control knockdown in HeLa cells was from GSE42485 (Rosel-Hillgartner et al., 2013). Mis-spliced genes in cultured astrocytes treated with PR peptide were from (Kwon et al., 2014). Low complexity domain prediction was done using SMART online tool (Letunic et al., 2015). Statistical analyses were done with GraphPad Prism (GraphPad Software, La Jolla California). Differences between groups were analyzed with Anova, t-test or chi-square test. P<0.05 was considered significant.
Supplementary Material
The proteins were sorted by GR intensity. Among the ~800 proteins detected in the pulldowns, 10 of the high hits ranging in size from 185-630 kD (indicated by * at the bottom of the table), were not considered in our analyses. These proteins were either high hits in the FLAG control pulldown, and/or are known to exist in distinct multi-component complexes but other members of their respective complexes were either not detected in the pulldowns or were low hits.
Genes involved in splicing (yellow), mitochondria (red), transcription (gray) and neuronal (blue) functions are indicated. Genes with multiple color codes have multiple functions.
Genes involved in splicing (yellow), mitochondria (red), transcription (gray) and neuronal (blue) functions are indicated. Genes with multiple color codes have numtiple functions.
Acknowledgments
We are grateful to the Nikon Imaging Center and the Image and Data Analysis Core at Harvard Medical School for help with microscopy and image analysis. GR and PR peptides were provided by A.D. Gitler. We thank R. Batra for the RNA-seq data from C9ORF72 and sALS patients. This work was supported by National Institutes of Health Grants GM043375 (to R.R.), NS057553 and NS079725 (to F.-B.G.), ALS Therapy Alliance Grant 2013-S-006 (to R.R.) and ALS Association grant 2179 (to F.-B.G.).
Footnotes
Author contributions
S.Y. and R.R. conceived the project and S.Y. carried out most of the experiments. F.-B.G. directed iPSC-MN generation, which was carried out by R.L.-G. Imaging analyses were carried out by S.Y. and J.G. Splicing and U2 snRNP native gel analysis were done C.B. Mass spectrometry was performed by R.K. The data were analyzed by R.K., S.P.G., S.Y. and R.R. The manuscript was written by S.Y. and R.R with input from F.-B.G. and R.L.-G and assistance from all authors.
References
- Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta neuropathologica. 2013;126:385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, Rademakers R, 3rd, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. The SMN complex: an assembly machine for RNPs. Cold Spring Harbor symposia on quantitative biology. 2006;71:313–320. doi: 10.1101/sqb.2006.71.001. [DOI] [PubMed] [Google Scholar]
- Conlon EG, Lu L, Sharma A, Yamazaki T, Tang T, Shneider NA, Manley JL. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. eLife. 2016;5 doi: 10.7554/eLife.17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Bury JJ, Heath PR, Wyles M, Higginbottom A, Gelsthorpe C, Highley JR, Hautbergue G, Rattray M, Kirby J, et al. C9ORF72 GGGGCC Expanded Repeats Produce Splicing Dysregulation which Correlates with Disease Severity in Amyotrophic Lateral Sclerosis. PloS one. 2015;10:e0127376. doi: 10.1371/journal.pone.0127376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Walsh MJ, Higginbottom A, Robin Highley J, Dickman MJ, Edbauer D, Ince PG, Wharton SB, Wilson SA, Kirby J, et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain : a journal of neurology. 2014;137:2040–2051. doi: 10.1093/brain/awu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzolino M, Carri MT. Mitochondrial dysfunction in ALS. Progress in neurobiology. 2012;97:54–66. doi: 10.1016/j.pneurobio.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Das R, Dufu K, Romney B, Feldt M, Elenko M, Reed R. Functional coupling of RNAP II transcription to spliceosome assembly. Genes & development. 2006;20:1100–1109. doi: 10.1101/gad.1397406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Das R, Zhou Z, Reed R. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Molecular Cell. 2000;5:779–787. doi: 10.1016/s1097-2765(00)80318-4. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p–linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco EG, Coil KE, Reed R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes & development. 2011;25:440–444. doi: 10.1101/gad.2009411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nature reviews. Genetics. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Carvalho M, M LG-V, F RP, Besse F, Weis J, Voigt A, Schulz JB, De Las Rivas J. Linking amyotrophic lateral sclerosis and spinal muscular atrophy through RNA-transcriptome homeostasis: a genomics perspective. Journal of neurochemistry. 2017;141:12–30. doi: 10.1111/jnc.13945. [DOI] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW, Sun S, 3rd, Herdy JR, Bieri G, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nature neuroscience. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekura K, Yagi T, Cammack AJ, Mahadevan J, Kuroda M, Harms MB, Miller TM, Urano F. Poly-dipeptides encoded by the C9ORF72 repeats block global protein translation. Human molecular genetics. 2016;25:1803–1813. doi: 10.1093/hmg/ddw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kfir N, Lev-Maor G, Glaich O, Alajem A, Datta A, Sze SK, Meshorer E, Ast G. SF3B1 association with chromatin determines splicing outcomes. Cell reports. 2015;11:618–629. doi: 10.1016/j.celrep.2015.03.048. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Maniatis T, Ruskin B, Green MR. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, et al. C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell. 2016;167:774–788. e717. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic acids research. 2015;43:D257–260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. The Journal of cell biology. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Mori E, Kato M, Xiang S, Wu L, Kwon I, McKnight SL. Toxic PR Poly-Dipeptides Encoded by the C9orf72 Repeat Expansion Target LC Domain Polymers. Cell. 2016;167:789–802. e712. doi: 10.1016/j.cell.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gonzalez R, Lu Y, Gendron TF, Karydas A, Tran H, Yang D, Petrucelli L, Miller BL, Almeida S, Gao FB. Poly(GR) in C9ORF72-Related ALS/FTD Compromises Mitochondrial Function and Increases Oxidative Stress and DNA Damage in iPSC-Derived Motor Neurons. Neuron. 2016;92:383–391. doi: 10.1016/j.neuron.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD, et al. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Analytical chemistry. 2012;84:7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ, Murray ME, Overstreet KK, Piazza-Johnston AE, Desaro P, et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nature neuroscience. 2015;18:1175–1182. doi: 10.1038/nn.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosel-Hillgartner TD, Hung LH, Khrameeva E, Le Querrec P, Gelfand MS, Bindereif A. A novel intra-U1 snRNP cross-regulation mechanism: alternative splicing switch links U1C and U1-70K expression. PLoS genetics. 2013;9:e1003856. doi: 10.1371/journal.pgen.1003856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby SW, Chang TH, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes & development. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- Rutherford NJ, Heckman MG, Dejesus-Hernandez M, Baker MC, Soto-Ortolaza AI, Rayaprolu S, Stewart H, Finger E, Volkening K, Seeley WW, et al. Length of normal alleles of C9ORF72 GGGGCC repeat do not influence disease phenotype. Neurobiology of aging. 2012;33:2950, e2955–2957. doi: 10.1016/j.neurobiolaging.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Chiang PM, Price DL, Wong PC. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16325–16330. doi: 10.1073/pnas.1003459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang S, Wu LC, Ding M, Yu Y, Gall JG, et al. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E1111–E1117. doi: 10.1073/pnas.1620293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83:1043–1050. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Brown RH, Jr, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Almeida S, Moore J, Gendron TF, Chalasani U, Lu Y, Du X, Nickerson JA, Petrucelli L, Weng Z, et al. Differential Toxicity of Nuclear RNA Foci versus Dipeptide Repeat Proteins in a Drosophila Model of C9ORF72 FTD/ALS. Neuron. 2015;87:1207–1214. doi: 10.1016/j.neuron.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Wang Z, Jang M, Nutiu R, Wang ET, Burge CB. Splice site strength-dependent activity and genetic buffering by poly-G runs. Nature structural & molecular biology. 2009;16:1094–1100. doi: 10.1038/nsmb.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Chen S, Yu Y, Yan B, Haertlein TC, Carrasco MA, Tapia JC, Zhai B, Das R, Lalancette-Hebert M, et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell reports. 2012;2:799–806. doi: 10.1016/j.celrep.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Abdallah A, Li Z, Lu Y, Almeida S, Gao FB. FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions. Acta neuropathologica. 2015;130:525–535. doi: 10.1007/s00401-015-1448-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Yu Y, Reed R. Primary microRNA processing is functionally coupled to RNAP II transcription in vitro. Scientific reports. 2015;5:11992. doi: 10.1038/srep11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Das R, Folco EG, Reed R. A model in vitro system for co-transcriptional splicing. Nucleic acids research. 2010;38:7570–7578. doi: 10.1093/nar/gkq620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The proteins were sorted by GR intensity. Among the ~800 proteins detected in the pulldowns, 10 of the high hits ranging in size from 185-630 kD (indicated by * at the bottom of the table), were not considered in our analyses. These proteins were either high hits in the FLAG control pulldown, and/or are known to exist in distinct multi-component complexes but other members of their respective complexes were either not detected in the pulldowns or were low hits.
Genes involved in splicing (yellow), mitochondria (red), transcription (gray) and neuronal (blue) functions are indicated. Genes with multiple color codes have multiple functions.
Genes involved in splicing (yellow), mitochondria (red), transcription (gray) and neuronal (blue) functions are indicated. Genes with multiple color codes have numtiple functions.