Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a deadly malignancy that affects nearly 50,000 patients each year. The overall 5-year survival rate for this malignancy remains the lowest of any cancer at around 7% due to limited diagnostic methods, disease aggressiveness and a lack of targeted therapeutic interventions. This review highlights the successes achieved over the past several decades as well as the significant cellular and molecular hurdles that remain in combatting this deadly disease at a translational level.
Keywords: Pancreatic cancer, GEMMs, Kinases, Cancer progression, Therapy resistance, Metabolism and desmoplasia
1. Pancreatic cancer statistics (1975-present)
1.1. History
It was believed for a long time that cancer of the pancreas was not existent, and it was not until 1882 when a solid tumor was successfully excised for the first time from this tissue. The patient, however, died within hours of being discharged (Busnardo et al., 1983). Since then, many surgical procedures have been proposed for resection of malignant pancreatic tissue. Walter Kausch and Allan Whipple started performing pancreatic surgery around 1910 and developed a method known as pancreaticoduodenectomy (Kausch-Whipple procedure). Early on, this difficult surgical procedure led to high morbidity and mortality and a poor long-term outcome, which prompted many to wonder if the surgery should be performed at all. Nonetheless, significant advances have been made to date and pancreaticoduodenectomy is currently the standard operative procedure in patients diagnosed with pancreatic cancer (Bachmann et al., 2006).
Despite the advances, surgical resection still provides the patient with the highest probability for survival, but is usually only be performed when the tumor is localized to the pancreas (Vincent et al., 2011). Currently, patients’ median survival is 14–20 months with up to a 25% 5-year survival rate following surgical resection (Bachmann et al., 2006; Yachida et al., 2010). Although substantial progress has been made in understanding this disease, mortality rates in this malignancy have been progressively rising, while the incidence and mortality rates of other common cancers have been declining (Vincent et al., 2011).
1.2. Incidence
Pancreatic ductal adenocarcinoma (PDAC) is currently the fourth leading cause of cancer-related deaths in the United States and its incident rate has been steadily rising. In the current year, 48,960 new cases of PDAC are expected to arise in the United States and of these cases, 51% and 49% are projected to affect males and females, respectively. The estimated number of deaths is 20,710 in males and 19,850 in females. 83% of all PDAC diagnoses will result in patient death (Siegel et al., 2015). While the probability of developing PDAC is 30% greater in males, age and racial/ethnic backgrounds play important roles in the incidence rates. Of all diagnosed cases, about 27% are diagnosed between ages of 75 and 84, whereas about 9% are diagnosed between the ages of 45 and 54. African-Americans (15.7 out of 100,000) have the highest incidence rate of developing PDAC amongst the different racial/ethnic groups while Asian-Americans/Pacific Islanders (9.8 out of 100,000) have the lowest (Howlader et al., 2013). Nonetheless, this malignancy is deadly irrespective of gender, age or race.
1.3. Survival
Detecting the tumor early and when it is confined to the pancreas only (localized) is the most promising means by which to significantly increase patient survival; however, only 9% of PDAC diagnoses are made at this early stage (Howlader et al., 2013). For other common carcinomas, identifying the tumor at the localized stage results in 5-year survival rates of 99% (breast and prostate), 92% (kidney), 90% (colon/rectum), 83% (oral cavity/pharynx), 64% (stomach) and 54% (lung/bronchus). Still the 5-year survival rate for PDAC that is detected at this early stage is still only 26% (Bachmann et al., 2006; Howlader et al., 2013). Ultimately, researchers and clinicians alike are striving to discover ways to detect PDAC as early as possible and improve treatment responses.
As PDAC progresses into the regional stage, in which the malignant cancer invades surrounding organs/tissues and the lymph nodes, the 5-year survival rate for patients decreases to only 10%. Detection of the tumor, however, is more likely (28%). As with all cancers, once the primary tumor metastasizes to distant organs via the circulatory system, the probability of survival diminishes significantly. In the case of PDAC, the 5-year survival rate for this disease once it has reached distant organs is only 2%. Detection at this stage is 53% (Howlader et al., 2013), but unfortunately few patients respond to the available interventions.
Over the past 4 decades, 5-year survival rates have increased across several different cancers. The 5-year survival rate for prostate cancer increased from 68% in 1975 to >99% in 2010. Patients diagnosed with leukemia between 2004 and 2010 had a 5-year survival rate of 60%, whereas between 1975 and 1977, it was only 34%. Pancreatic cancer, on the other hand, has not seen such drastic improvements. Between 1975 and 1977, the 5-year survival rate was 3%. It increased to 4% between 1987 and 1989 and currently, the 5-year survival rate is 7% (Siegel et al., 2015). Although there is a statistical significance (p < 0.05) between 1975 and the present, the 4% difference over the last forty years is not impressive. We need to better understand the reasons for low survival and develop methods for identifying potential therapeutic targets to create more sufficient drugs against this deadly disease. This review focuses on the advances that have been made over the last few decades in understanding the biology of the low survival rates in PDAC patients and to introduce potential novel biomarkers and therapeutic targets in PDAC.
2. Factors contributing to poor survival
There are several reasons why PDAC patients have such low survival. First, there are few viable methods for detecting this malignancy early. Second, PDAC is a highly aggressive cancer and can metastasize early. Finally, few targeted therapies exist for treating PDAC, and those therapies that are used remain relatively ineffective.
2.1. Limited diagnostic methods—symptoms and biomarkers
Pancreatic cancer remains one of the most difficult cancers to detect during its pre-invasive stages. Once it has metastasized, only 6% of patients survive beyond 5 years and thus it is virtually incurable. Thus, detecting the tumor while it is still localized to the pancreas provides the best possibility for survival. However, because patients rarely have physical symptoms during these pre-invasive stages—the pancreas is a visceral organ and imaging modalities are costly—it will require validation of diagnostic biomarkers before this cancer can be detected during these early stages. Currently, the most reliable and frequently used biomarker of PDAC is carbohydrate antigen 19-9 (CA19-9), also known as sialylated Lewis (a) antigen (Maitra and Hruban, 2008; Wingren and Sandstrom, 2012). CA19-9’s sensitivity ranges from 69% to 98% and its specificity from 46% to 98% (Wingren and Sandstrom, 2012). Consequently, CA19-9 is non-PDAC specific and can provide false positive results as it is found in both benign and malignant gastrointestinal tumors. Moreover, in 10% of the population, CA19-9 can be deficient due to persons who genotypically cannot produce the protein (Wingren and Sandstrom, 2012). Several recent efforts have been made to identify novel serum biomarkers of PDAC (Harsha et al., 2009; Nolen et al., 2014; Shaw et al., 2014), but to date, no clinical benefit of these studies has been realized. Thus, it is crucial for future efforts to identify novel PDAC-specific biomarkers that could be used to diagnose this cancer earlier.
2.2. PDAC is an aggressive malignancy
As a consequence of PDAC developing in a moderately symptom-free manner, metastasis has usually already occurred in the majority of the cases at the time of diagnosis. (Deer et al., 2010; Wolfgang et al., 2013).
2.2.1. Development of PDAC
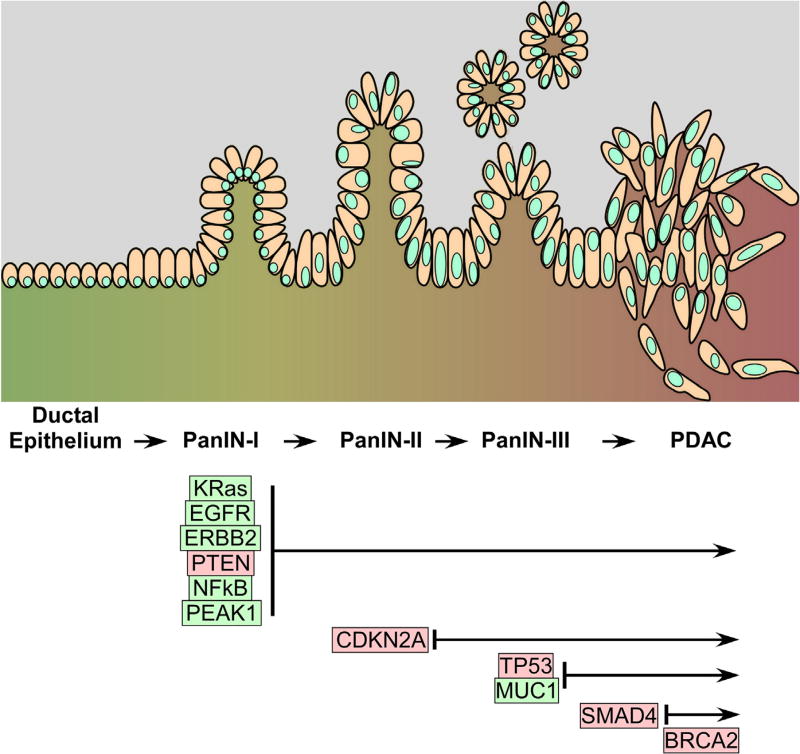
The development of malignant pancreatic tumors from normal pancreatic tissue occurs through non-invasive precursor lesions known as pancreatic intraepithelial neoplasms (PanINs), intraductal papillary mucinous neoplasms (IPMN) or mucinous cystic neoplasms (MCN) (Maitra and Hruban, 2008). Invasive pancreatic ductal cancer cells most commonly arise from PanINs while acquiring specific gene mutations as they progress from PanIN-1 to PanIN-3 (Vincent et al., 2011). PanINs differ from one another based on the degree of cytological and tissue architecture atypia (Maitra and Hruban, 2008). Genetic mutations occur as early as PanIN-1, where the activation of the KRas oncogene is present in 95% of PDAC cases. As these early neoplasms transition to PanIN-2 and PanIN-3, tumor suppressor genes such as CDKN2A (p16/Ink4A), TP53 (p53), and Smad4 (DPC4) are often times inactivated or deleted (Vincent et al., 2011). Fig. 1 illustrates PanIN-to-PDAC progression along with genetic alterations in specific genes as the disease progresses.
Fig. 1.
Top—schematic of the morphological changes that occur at the cellular and tissue levels during PanIN → PDAC progression. PanIN-Is are characterized by hyperproliferative cells with some papillary structures. PanIN-IIs are characterized by loss of basal localization of the cell nucleus and nuclear atypia. PanIN-IIIs are characterized by more pronounced nuclear atypia and intraluminal (in situ) cell clusters. PDAC is characterized by loss of basement membrane, local cellular invasion and systemic metastasis. Bottom—most commonly associated genetic alterations associated with the various stages of PanIN → PDAC progression. Red indicates loss-of-function changes. Green indicates gain-of-function changes. Additional genetic alterations are discussed in the text and in Table 1.
IPMNs are mucin-producing neoplasms that arise within the pancreatic duct or one of its branches (Maitra and Hruban, 2008). They have many genetic mutations as seen in PanINs with one major difference: IPMNs rarely show Smad4 deletion (Maitra and Hruban, 2008; Vincent et al., 2011). Alternatively, MCNs typically arise in females and contain an ovarian-like stromal cells and proteins. Unlike IPMNs, MCNs do not communicate with large pancreatic ducts. Interestingly, IPMNs and MCNs are found in patients who are on average 3–5 years younger than those with PanIN-derived invasive lesions (Maitra and Hruban, 2008).
2.2.2. Invasive carcinomas
From the appearance of the tumor tissue, it is evident that pancreatic cancers are extremely infiltrative neoplasms. Surgically resected tumors contain vascular and perineural invasions, which leads to metastasis to regional lymph nodes, the liver and other distant sites (Maitra and Hruban, 2008).
2.2.2.1. Early vs. late dissemination and metastasis
Two recent reports have addressed the timing of metastasis in PDAC using unique, but complementary methods (Rhim et al., 2012; Yachida et al., 2010). Nonetheless, on the surface these reports may seem to contradict, but a careful analysis reveals that they point to distinct mechanisms and timing of local and systemic PDAC dissemination/metastasis. Rhim et al. (2012) demonstrated that epithelial-to-mesenchymal transition (EMT) and dissemination of pancreatic cancer cells can occur early—even before a malignant tumor can be detected. Using Pdx1-Cre mouse models, they generated PDA-specific mutations in the KRas and p53 genes (see Sections 2.2.1 and 2.2.3 for more details). They fluorescently tagged epithelial pancreatic cancer cells with RosaYFP and tracked them and monitored both epithelial (E-Cadherin) and mesenchymal markers (Zeb1 and Fsp1). They noticed that in in mice harboring PDA-causing mutations, E-cadherin expression decreased while Zeb1 and Fsp1 expression significantly increased in the pre-invasive PanIN lesions. This suggested that EMT was being induced in these tissues and could lead to local invasion and dissemination of these cells at early stages during disease progression. Thus, they hypothesized that mesenchymal PanIN cells may have invasive properties. In support of this, they noticed YFP+ cells had crossed the basement membrane and dissociated from any apparent pancreatic epithelial structure. This raised the question of whether circulating pancreatic cells (CPCs) from PanINs seeded distant organs. Usage of YFP lineage labeling, 4/11 8- to 10-week-old mice revealed YFP+ cells seeded within the liver, suggesting that prior to tumor formation, cells found in PanIN 2 and 3 lesions are present in the circulation and are able to seed distant organs. These data taken together propose that metastasis is an early event, precedes primary tumor formation in PDAC and may lead to cells lying dormant in distant tissues—only detectable after identification of the primary tumor.
On the other hand, work from Yachida et al. (2010) suggested that pancreatic cancer can remain localized at the primary tumor for over a decade suggesting that early diagnosis may drastically improve the overall patient survival. They generated data by sequencing the genomes from human patient pancreatic cancer tissue (n = 7). In all patients, metastasis was found in two or more sites, most commonly in the liver, lung and peritoneum. Comparing the genomes from each patient’s tissue, they were able to identify mutations in two categories. ‘Founder’ mutations referred to mutations that were found in all samples (pancreas, liver, lung and peritoneum) from a given patient whereas ‘progressor’ mutations were all other mutations that were not present in each sample from a given patient. ‘Founder’ mutations had a mean frequency across patients of 64% while ‘progressor’ mutations had a mean frequency of 36%. This suggested that these mutations arose prior to metastatic lesion development. These data also aided the discovery of one parental clone per patient. Using single nucleotide polymorphism (SNP) chip data and the sequencing data, it was found that the majority of the homozygous mutations (89%) were present in the parental clones and they carried the typical mutations found in PDAC (KRas, TP52 and Smad4). Each parental clone gave rise to many subclones, which eventually led to metastasis. Using their mathematical model, they conservatively predict that the time it takes for an initiated tumor cell to give rise to a parental clone is 11.7 ± 3.1 years. And the time a parental clone gives rise to subclones with metastatic capacity takes an additional 6.8 ±3.4 years. Finally, they suggest the time it takes between metastasis and patient death to be 2.7 ± 1.2 years. Taken together, these data propose that the progression of pancreatic cancer from initiation to metastasis could be well over a decade, thus implying a much longer time period for early detection.
2.2.3. Highly resistant to current available therapies
2.2.3.1. Desmoplasia and vascularity
Desmoplasia, often referred to as deposition of stromal cells and extracellular matrix (ECM), plays a major role in aiding tumor progression by inducing therapy resistance (Feig et al., 2012), (Whatcott et al., 2015). The accumulation of ECM components such as collagen, fibronectin, proteoglycans, and non-tumor cells distorts the normal arrangement of pancreatic tissue; therefore, causing compression on blood vessels, which ultimately leads to failed drug delivery (Feig et al., 2012). Additionally, stromal cells can be co-opted by the tumor cells to promote the progression of the disease. Depletion of the desmoplastic stroma may be a therapeutic option in PDAC patients in the near future. Section 4.4 below discusses this tumor compartment in more detail as it relates to cancer progression.
Because accumulation of ECM components and stromal cells compresses against blood vessels, the vasculature within pancreatic tissue is also affected. Deficiency in vasculature explains the poor delivery of drugs to tumor tissue (Neesse et al., 2011). This can also be problematic to tumor cells for growth. With limited access to vasculature, pancreatic cancer cells must rely on their ability to reprogram metabolic pathways to provide the necessary ingredients to survive and proliferate (Le et al., 2012). This presents two problems: (1) drug delivery is inefficient due to limited access to the vasculature and (2) tumor cells are able to survive and proliferate despite nutrient and resource deprivation.
2.2.3.2. Current anti-cancer therapies
2.2.3.2.1. Gemcitabine
Currently, there are two FDA approved drugs normally administered together to PDAC patients. However, there is minimal increase in survival. Gemcitabine is administered to PDAC patients as chemotherapy at all stages of the disease and it inhibits DNA replication and repair (Kobayashi et al., 2014). Pancreatic cancer cells build resistance to gemcitabine. One mechanism of resistance is autophagy in response to the drug, which prevents the cells from entering the apoptotic pathway (Papademetrio et al., 2014).
2.2.3.2.2. Erlotinib
Unlike gemcitabine, erlotinib is a targeted therapy and tyrosine kinase inhibitor designed to block the epidermal growth factor receptor (EGFR) and is commonly given to PDAC patients along with gemcitabine. Erlotinib can also potentiate gemcitabine-induced apoptosis (Ng et al., 2002). Since not all PDAC patients carry the EGFR mutation, erlotinib is helpful for a small percentage of PDAC patients and even then, the cancer cells do not respond to the drug. Even with erlotinib and gemcitabine administered together, the overall survival is prolonged very minimally. Details regarding resistance mechanisms are explained in Sections 4.1 (Oncogene addiction) and 4.2 (Synthetic lethality). Thus, it is of great importance to discover novel therapeutic targets to fight against this disease.
2.2.3.2.3. Radiation therapy
Radiation therapy (RT) is a common treatment option for cancer patients where high-energy particles or waves directly target proliferating tumor cells. In this regard, RT is often times given to patients with localized tumors; however, RT can also be administered for locally advanced tumors. For PDAC patients, RT may be a treatment option for the small population of those diagnosed at the earliest stage. Using RT for PDAC patients has been rather controversial though, as results have not shown much improvement in overall survival (Goodman and Hajj, 2013). Patients with resectable tumors can receive RT before (neoadjuvant) or after (adjuvant) surgical resection. Neoadjuvant RT is the preferred route as physicians would like to see shrinkage of the tumor before surgically removing it. Often times, an invasive surgery, such as the Whipple procedure, may lead to increased mortality and morbidity rates; therefore, post-operative complications may be greater with the addition of RT. For that reason, neoadjuvant RT can minimize the degree of resection, risk of survival and rarely, eliminate the tumor. On the contrary, patients may not respond to RT and this can postpone surgery. By delaying surgical resection, this provides an opportunity for the localized tumor cells to spread. Adjuvant RT bypasses this risk because the localized tumor has been surgically removed. Adjuvant RT is applied mainly to prevent recurrence, an event where >80% of surgically curable PDAC patients suffer from within the first year after surgery (Goodman and Hajj, 2013). Recurrence is the main reason for the low 5-year survival rate of patients who have had their tumor surgically removed from their pancreas (Hishinuma et al., 2006). Future efforts should focus on neoadjuvant RT to minimize tumor size in surgically curable PDAC patients.
3. Common genetic mutations or gene expression alterations
3.1. Genetically engineered mouse models (GEMMs)
Genetically engineered mouse models (GEMMs) for PDAC research were created to recapitulate both genetic and morphologic alterations that often lead to the development of the disease. Experiments conducted in mouse models have shown identical results to human PDAC. Over the past decade, pancreatic cancer GEMMs have garnered significant attention since they show promise for assisting in the development of new diagnostic and therapeutic approaches. Here, we discuss genes alterations that have been genetically engineered into mice and summarize what we have learned from these experiments.
3.1.1. KRas
Kirsten rat sarcoma (KRas) is the most commonly mutated gene in PDAC (95%) (Maitra and Hruban, 2008; Tanaka, 2015; Vincent et al., 2011); therefore, GEMMs carrying the KRas mutation are heavily studied models. Although KRas would be an ideal gene to therapeutically target, research over the last 30 years has shown disappointing results in targeting KRas, and the gene has been deemed “undruggable.” The activating point-mutation of KRas is predominately found on codon G12 (98%), and less commonly found on codons G13 and Q61 (Eser et al., 2014). KRas mutations (or KRasG12D) impair essential GTPase activity of the KRas protein by blocking its interaction with GTPase-activating proteins (GAPs) needed to hydrolyze GTP. Thus, KRas remains constitutively active and stimulates downstream pathways to induce proliferation, metabolic reprogramming, anti-apoptosis, invasion, migration, metastasis, etc, all hallmarks of cancer progression (Eser et al., 2014). Therefore, studying KRasG12D in GEMMs is crucial for the overall understanding of tumor development and progression of PDAC. It has been shown that oncogenic KRas not only initiates PDAC, but is also necessary for tumor progression and maintenance (Collins et al., 2012a; Hingorani et al., 2003; Ying et al., 2012) as well as growth of metastatic lesions (Collins et al., 2012b). As a result, it has been shown that GEMMS containing activating mutations in KRas undergo tumor initiation and progression, as seen in humans.
3.1.2. p53
p53 is a tumor suppressing transcription factor which is inactivated in 75% of PDAC cases (Maitra and Hruban, 2008; Tanaka, 2015). p53 can promote cell cycle arrest by regulating the G1/S checkpoint, maintain G2/M arrest and initiate apoptosis (Maitra and Hruban, 2008). Therefore, loss of function of this key cell cycle regulator can lead to uncontrollable cell division and tumor progression. Hingorani et al. (2005) demonstrated in GEMMs that initiated pre-invasive disease by KRasG12D develops into an invasive and metastatic disease by the additional inactivating point-mutation in TP53, Trp53R172H, which mirrors human PDAC. Wagner et al. (1998) created a model in which mice spontaneously develop pancreatic cancer by the over-expression of TGFα. When the p53 mutant is introduced to these mice, tumor development is accelerated and mice develop invasive carcinomas (Wagner et al., 2001). Mouse models carrying the p53 mutation have illustrated the importance of its loss of function in PDAC progression such that upon mutation of the gene, the disease becomes invasive and metastatic.
3.1.3. p16
CDKN2A (p16 or Ink4) is the most commonly inactivated (via mutation) tumor suppressor in PDAC and normally arises during PanIN-2 (Maitra and Hruban, 2008; Tanaka, 2015). Like p53, p16 has important functions in regulating the cell cycle as it inhibits cell cycle progression through the G1-S checkpoint (Maitra and Hruban, 2008). Loss of function of this gene is found in over 90% of PDAC cases (Maitra and Hruban, 2008; Tanaka, 2015); therefore, generating a mouse model carrying this mutation was important in the overall understanding of pancreatic cancer progression. Aguirre et al. (2003) generated the mouse model Ink4a/Arflox to sustain Cre-mediated removal of exons 2 and 3 to exclude p16INK4A and p19ARF at the protein level. They noticed that when Ink4a inactivation was combined with KRasG12D, there was a rapid progression in local invasion and growth; whereas when either gene was mutated alone, this phenotype was not observed. They also demonstrated similarities between murine pancreatic tumors to human pancreatic adenocarcinoma on the tissue level; thus, suggesting a parallel progression model between murine models and human disease.
3.1.4. SMAD4 and TβSRII
SMAD4 (DPC4), another tumor suppressor and transcription factor, mediates TGFβ signaling, which has important growth-inhibitory effects by regulating specific target genes (Bardeesy et al., 2006); thus, loss of this signaling pathway may cause cells to grow inappropriately due to deregulation (Ijichi et al., 2006). SMAD4 is deleted in 55% of PDAC cases normally during late PanIN-3 lesions (Maitra and Hruban, 2008; Tanaka, 2015). SMAD4 deletion has also been suggest to not occur until PDAC (Chiao et al., 2011; Chiao and Ling, 2011). Thus, SMAD4-dependent TGFβ signaling can provide tumor cells an advantage in growth in the absence of SMAD4. SMAD4 deletion alone does not impair normal pancreatic growth, development or function. It has also been shown that SMAD4 deletion alone does not initiate pancreatic cancer (Bardeesy et al., 2006). Because we know KRasG12D initiates PanIN formation, and development of PDAC requires additional mutations, SMAD4 deletion has been shown to contribute to pancreatic cancer progression when combined with activating KRas mutations in the murine pancreas. Bardeesy et al. (2006) demonstrated that Pdx1-Cre Smad4lox/lox along with Pdx1-Cre LSL-KRasG12D rapidly developed into intraductal papillary mucinous neoplasias (IPMN), identical to IPMNs found in humans. They also showed that PDAC development was accelerated upon Smad4 deletion in KRasG12D INK4A/ARF heterozygous mice. At the same time, they observed that cells from these animals responded to TGFβ treatment by increasing their proliferation. In related work, deletion of the TGFβ Type-II receptor (T(βRII)—a mutation that is found in a subset of PDAC patients—promotes tumor progression in GEMMs. Ptf1a-Cre; Tgfbr2flox/flox mice demonstrated that blockage of TGFβ-signaling was insufficient for pancreatic development and did not initiate pancreatic tumor formation. However, when combined with the KRas-activating mutation there was a rapid progression to PDAC that resulted with a median survival of 59 days. Symptoms such as weight loss, bloody ascites and jaundice were observed in these mice, which recapitulate symptoms found in human PDAC patients (Ijichi et al., 2006). We have learned that the canonical TGFβ-signaling pathway is essential for normal pancreatic tissue growth regulation such that deletion of either SMAD4 or TβRII contributes significantly to PDAC progression.
3.1.5. MUC1
MUC1 is a glycoprotein expressed on the apical surfaces of normal glandular epithelia and functions to bind pathogens and is involved in cell signaling. In PDAC, MUC1 is unusually glycosylated and is found to be overexpressed in over 60% of PDAC cases, normally in high-grade PanINs (Tinder et al., 2008). Therefore, MUC1 can play an important role in tumor progression. Using a KRasG12D mouse model to express MUC1 in levels similar to that in human tumors of the pancreas, Tinder and colleagues observed PDAC progression at accelerated rates relative to KRas and MUC1 mutations alone. The weight of the tumor, levels of metastatic lesions and cell proliferation were greater in the double mutant mice, suggesting that overexpression of MUC1 in the context of KRasG12D leads to accelerated tumor progression (Tinder et al., 2008). In another study conducted by this same group, it was shown that MUC1 overexpression induced EMT, which led to an increase in invasion and metastasis. Genetic deletion of MUC1 also significantly reduced EMT was significantly reduced (Roy et al., 2011). They also demonstrated that MUC1-null mice had slower tumor progression and metastatic rates along with less tumorigenic capacity (Besmer et al., 2011). MUC1 has shown clear implications of its oncogenic capabilities in mouse models. Mechanistically, more recent work has implicated MUC1 in the regulation metabolism and hypoxic response via its regulation of HIF1α expression/stability and activity. Notably, this role for MUC1 connects the cellular microenvironment to metabolism within the epithelial tumor cell compartment (Chaika et al., 2012a).
3.1.6. LKB1
Germ-line mutation in the LKB1 gene have been shown to induce hereditary intestinal polyposis syndrome, more commonly known as Peutz-Jeghers syndrome, which increases the risk of developing pancreatic cancer by more than 100-fold (Giardiello et al., 2000; Mehenni et al., 2006). LKB1 can regulate cell growth and apoptosis and can activate adenosine monophosphate-activated protein kinase (AMPK) to negatively regulate rapamycin kinase (Shaw et al., 2004). Therefore, generating a mouse model for this tumor suppressor gene was essential for our general understanding of PDAC. Hezel et al. (2008) demonstrated that Pdx1-Cre; Lkb1flox/flox mice developed pancreatic serous cystadenomas. Also, mice lacking Lkb1 had impaired acinar cell structure that often led to acinar-ductal metaplasia (ADM). Although PDAC was never observed in these models, precursor lesions were seen with high frequency. In the context of KRas-activating mutations, the additional mutation of Lkb1 (Pdx1-Cre; KRasG12D; Lkb1flox/+) resulted in a median survival rate that was dramatically reduced relative to the respective single-mutants. Additionally, the number of PanIN lesions were significantly greater in the double-mutants. It was also shown that homozygous loss of Lkb1 (Pdx1-Cre; Lkb1flox/flox) led to pancreatic tumor development in 100% of the mice and further decreased median survival. Although homozygous loss of Lkb1 led to PDAC, it was demonstrated that loss of a single allele was sufficient in PDAC progression (Morton et al., 2010). To conclude, LKB1 is a critical tumor suppressor gene such that a single deletion of an allele can lead to PDAC precursor lesions and eventually to PDAC in the background of activated KRas.
3.1.7. SRC
The tyrosine kinase SRC has been fundamentally linked with cellular transformation and changes in its activity can influence proliferation, invasion and migration (Bromann et al., 2004). Although SRC is usually not mutated in PDAC, its overexpression is seen in 70% of the cases (Lutz et al., 1998).Therefore, SRC’s increased activity in PDAC suggests that its regulation is disrupted during tumor progression. One way that SRC can be activated is through reduced abundance or activity of C-terminal Src Kinase (CSK). CSK’s phosphorylation of SRC at Y527 enables SRC to be continuously active (Irby and Yeatman, 2000). Shields et al. (2011) generated a mouse model with depleted CSK in the context of mutated KRas: Pdx1-Cre; LSL-KRasG12D; CSKf/f. Precursor lesions were detectable as early as 3 weeks in these mice and PDAC was seen in 5–8 weeks with a median survival rate of just 6 weeks. Also, mice harboring deletion of CSK alone did not develop precursor lesions. Thus, the data suggests that Src accelerates pancreatic tumor progression in a KRas-dependent manner and that activation of Src alone is not sufficient to initiate pancreatic cancer.
3.1.8. EGFR
The epidermal growth factor receptor (EGFR) family contains 4 transmembrane receptors that have specific roles in the cell. EGFR (ERBB1 or HER1) can either homodimerize or heterodimerize with other ERBB receptors when it binds one of its seven ligands, most commonly epidermal growth factor (EGF) or TGFα. Upon ligand binding and dimerization, many signaling pathways are activated intracellularly (Roskoski, 2014). Currently, the EGFR antagonist erlotinib is the only FDA approved targeted therapy for pancreatic cancer but has shown nominal effects on increasing patient survival because of acquired resistance to the drug. Although EGFR mutations are found in less than 3% of PDAC cases (Maitra and Hruban, 2008; Navas et al., 2012), its presence has been demonstrated to be extremely crucial for pancreatic cancer progression. IHC staining revealed no staining of EGFR in normal pancreatic tissue in mice; however, there was significantly more staining in metaplasia, pancreatitis, PanINs (Shen et al., 2013) and PDAC (Navas et al., 2012). Navas et al. (2012) showed that EGFR signaling is required for acinar-ductal metaplasia (ADM) in the presence of KRas activating mutations. In Elas-KRasG12V; Egfrlox/lox mice, PanIN lesions or PDAC were not observed; in contrast, mice harboring the KRas-activating mutation with a wild-type EGFR or a single depleted allele displayed many PanIN lesions that eventually led to PDAC. To further understand the necessity of EGFR, it was also shown that additional mutation of p16, which is a commonly mutated tumor suppressor in PDAC, did not progress in PanIN formation in KRas-activating and EGFR depleted mice. However, this effect was not reproduced when p53 was inactivated demonstrating that EGFR functions independent of p53 in most pancreatic cancers.
3.2. Pre-clinical xenograft studies
3.2.1. ERBB2
EGFR can also dimerize with ErbB2 (HER2). This heterodimerization leads to tumor cell proliferation, adhesion, migration (Komoto et al., 2009) and suppress apoptosis (Chou et al., 2013); therefore, if not regulated properly, ERBB2 can lead to uncontrollable cell growth and metastasis in pancreatic cancer. Unlike other malignancies in which ERBB2 is known to contribute to cancer initiation and progression, it is amplified in only 2% of PDAC patients (Chou et al., 2013). Yet, its expression is upregulated in over 50% of patients suggesting that this receptor may be an important target for therapeutic intervention in PDAC (Roskoski, 2014). Like EGFR, ERBB2 has shown increased staining in PanIN lesions compared to normal pancreatic tissue (Shen et al., 2013). Since drugs targeting both EGFR (e.g., erlotinib) and ERBB2 (e.g., lapatinib and trastuzumab) already exist, the idea of dual inhibition of these EGFR kinases was investigated. Studies have shown promise of dual inhibition of EGFR and ERBB2, yet the extent of this benefit and how this regimen may be combined with gemcitabine-based therapy is not clear (Larbouret et al., 2007; Walsh et al., 2013). Interestingly, our group previously reported on the effect of ERBB2 knockdown in PDAC cell growth and metastasis in vivo. Surprisingly, we discovered that when ERBB2 was silenced or suppressed with the drug trastuzumab, cellular levels of the novel kinase PEAK1 and Src activity were increased and the cells were more aggressive—decreasing the time to pre-morbidity by almost three weeks (Kelber et al., 2012). Notably, this effect could be reversed by blocking PEAK1 kinase.
3.2.2. PEAK1
Pseudopodium-enriched atypical kinase one (PEAK1), or SKG269, has been recently identified as a potential biomarker and therapeutic target for PDAC (Wang et al., 2010). This catalytically active non-receptor tyrosine kinase can control cell migration by localizing to integrin-mediated focal adhesions and actin stress fibers by regulating the cytoskeleton and altering cell shape (Kelber et al., 2012). PEAK1 can also receive signals via growth factors, which can promote proliferation, survival and motility, all important characteristics of tumor progression. Of importance, PEAK1 is seen to be overexpressed in PDAC, along with other malignancies, and its upregulation has been shown to be driven by oncogenic KRas via Src-dependent transcriptional regulation (Kelber et al., 2012). In vivo, mice carrying tumor cells with PEAK1 knocked down (shPEAK1) displayed smaller tumors, less metastasis to many distant organs and longer survival relative to parental and shControl mice (Kelber et al., 2012). Since targeting KRas has shown little-to-no success, targeting downstream effectors maybe the next option. Since PEAK1 upregulation is driven by oncogenic KRas, PEAK1 can be a therapeutic target since its expression has been shown to be necessary for pancreatic tumor progression. In this regard, we’ve also recently reported that PEAK1 translation is dependent upon the post-translational hypusination of eIF5A Furthermore, targeting the hypusination enzymes DOHH and DHPS with CPX or GC7, respectively, can block PEAK1 protein production and tumorigenic properties in pancreatic cancer cells (Fujimura et al., 2014).
3.2.3. HIF1α
Hypoxia-inducible factor 1-α (HIF1α) has been shown to be overexpressed in pancreatic cancer and functions to promote tumor progression (Zhong et al., 1999). Once cells are deficient of oxygen, HIF1α induces transcription of important genes involved in angiogenesis and erythropoiesis. This increases oxygen delivery in areas deprived of oxygen (Semenza, 2003). Thus, HIF1α is necessary for tumor cells to proliferate and survive by increasing oxygen delivery and is therefore overexpressed in many types of cancers. At the same time, overexpression of HIF1α also increases drug resistance. With that said, targeting HIF1α for therapy can be promising. Zhao et al. (2015a,b) showed that inhibiting HIF1α and with simultaneous gemcitabine treatment significantly enhanced anti-tumor effects in PDAC both in vitro and in vivo. Table 1 summarizes the most commonly studied genes in PanIN-to-PDAC progression.
Table 1.
Common genetic alterations and their roles in PDAC.
| Gene | Genetic alteration | Role in PDAC |
|---|---|---|
| KRas | Activating mutation | An oncogene that promotes proliferation, metabolic reprogramming, anti-apoptosis, invasion, migration (Eser et al., 2014) |
| p53 | Inactivation | Deregulation of the G1/S checkpoint/G2/M arrest and apoptosis because of its inactivation (Maitra and Hruban, 2008) |
| p16 | Inactivation | Deregulation of the G1/S checkpoint because of its inactivation (Maitra and Hruban, 2008) |
| SMAD4/ TβRII | Deletion/deletion | Growth-inhibitory effects are deregulated because of the absence of either gene (Bardeesy et al., 2006; Ijichi et al., 2006) |
| BRCA2/BRCA1 | Inactivation/inactivation | Repair of damaged DNA and disposal of irreparable DNA is deregulated due to the inactivation of either gene (Maitra and Hruban, 2008) |
| Notch2/Notch1 | Overexpression/deletion | Notch2 regulates PanIN progression by increasing MYC signaling (Mazur et al., 2010). Notch1 depletion increases cell proliferation (Hanlon et al., 2010) |
| β-catenin | Activating mutation | Promotes SPN formation (Heiser et al., 2008) by mediating Wnt-targeted gene expression (Wideltz, 2005) |
| MUC1 | Overexpression | Increases tumor weight, induces proliferation, metastatic rates and EMT (Besmer et al., 2011) |
| RAC1 | Overexpression | Induces ADM formation and PDAC progression (Heid et al., 2011) |
| LKB1 | Inactivation | Deregulation of cell growth and apoptosis which leads to ADM formation and PDAC progression (Hezel et al., 2008; Shaw et al., 2004) |
| Ikk2/ PTEN | Upregulation/deactivation | Ikk2 regulates PanIN progression by inducing the Notch and NF-κB pathways (Hagemann et al., 2009; Maniati et al., 2011). PTEN depletion activates the NF-κB and PI3K/AKT pathways (Ying et al., 2011) |
| SRC | Overexpression | Induces proliferation, invasion and migration that leads to PanIN formation and PDAC progression (Bromann et al., 2004; Shields et al., 2011) |
| EGFR | Overexpression | Activates the PI3K/AKT pathway (Roskoski, 2014) and induces ADM and PanIN formation (Navas et al., 2012) |
| ERBB2 | Amplification/upregulation | Heterodimerization with EGFR increases proliferation, adhesion, migration (Komoto et al., 2009) and suppresses apoptosis (Chou et al., 2013) |
| PEAK1 | Overexpression | Increases cell migration by regulating the cytoskeleton and alters cell shape. Increases proliferation, survival and motility (Kelber et al., 2012) |
| HIF1α | Overexpression | Induces angiogenesis and erythropoiesis by regulating target genes to increase oxygen delivery, leads to increased proliferation and survival (Semenza, 2003) |
4. Oncogene addiction, synthetic lethality, stem cells and stroma in PDAC
4.1. Oncogene addiction
Cancer cells often times depend on specific oncogenes for their growth and survival. This concept is known as “oncogene addiction” (Weinstein and Joe, 2008). Targeting a single oncogene that drives tumor cell survival would be the most ideal approach for defeating malignancies; however, there has been little success with this approach. Because these tumor cells are exceedingly reliant on specific oncogenes, it has been predicted that these genes or cells carry second-site mutations prior to drug treatment that ultimately lead to drug resistance (McCormick, 2011). Erlotinib, a tyrosine kinase inhibitor, targets oncogenic epidermal growth factor receptor (EGFR) and is commonly administered with gemcitabine to PDAC patients. Overall, survival minimally increases as patients acquire resistance. One potential mechanism for this acquired resistance may be the “addiction” pancreatic cancer cells have for EGFR such that a second-site mutation substitutes for EGFR’s oncogenic functions. In the case of lung cancer, a second-site mutation of T790M in EGFR was found in 63% of patients with acquired resistance (Yu et al., 2013). It has also been shown that patients with colorectal cancer acquire resistance towards cetuximab, a common EGFR inhibitor for metastatic colorectal cancers patients, due to an acquired S492R mutation on the extracellular domain of EGFR (Montagut et al., 2012). Alternatively, oncogene addiction can also lead to activation of specific tumorigenic pathways, which ultimately lead to tumor cell survival and drug resistance. EGFR can activate the PI3K/AKT pathway, which induces proliferation and inhibits apoptosis (Roskoski, 2014). It has been shown that this pathway remains active despite EGFR being inhibited. Guix et al. (2008) illustrated in squamous cell cancer that the insulin-like growth factor 1 receptor (IFG1R) pathway had increased activity and was responsible for the increased PI3K/Akt activity during EGFR inhibition. Inhibiting both EGFR with gefitinib and IFG1R with an antibody in a mouse model inhibited PI3K/Akt signaling, reduced cell growth and tumor size. It is evident that tumor cells acquire oncogene addiction and developing multi-targeted therapies may be required for treatment.
4.2. Synthetic lethality
Synthetic lethality refers to cell death when two or more genes are mutated at the same time; however, if either gene is mutated alone, the cells remain viable (Ferrari et al., 2010). Because of oncogene addiction, we have learned that pancreatic cancer cells depend on EGFR function for growth and survival such that upon inhibition of this oncogene using erlotinib, second site mutations arise and pathways regulated by EGFR remain active. Therefore, targeting EGFR and downstream effectors simultaneously may be required to induce synthetic lethality. Combination therapy of gemcitabine and erlotinib has been well studied and is currently the most common therapeutic intervention for pancreatic cancer. Although the prolonged survival is statistically significant compared to gemcitabine alone, combination therapy of gemcitabine and erlotinib increases the survival rate minimally. Miyabayashi et al. (2013) demonstrated that gemcitabine induces MAPK signaling and this activity increases expression of EGFR in PDAC cells grown in a mouse model harboring KRasG12D and Tgfbr2KO mutations. When introducing erlotinib in this context, EGFR activity was inhibited; however, gemcitabine continued to induce MAPK signaling which led to elevated EGFR levels. Therefore, a MAPK kinase (MEK) inhibitor was added to inhibit MAPK activity and gemcitabine-induced MAPK signaling and EGFR expression was reduced. Thus, targeting downstream effectors, such as MAPK, along with targeting EGFR may improve overall survival. Similarly, Diep et al. (2011) showed that combination treatment of erlotinib and two MEK inhibitors displayed significant synergistic effects and increased antitumorigenic activity. Another potential gene that is synergistically lethal with EGFR inhibition is the S6 kinase ribosomal protein S6 kinase 2 (RPS6KA2). Activated RPS6KA2 has been observed to be downstream of EGFR/RAS/MAPK/ERK signaling and has been shown to be activated by EGF, independent of KRas mutations. Knockdown of RPS6KA2 led to apoptosis only in the presence of erlotinib and increased erlotinib’s effect on tumor cell survival (Milosevic et al., 2013). Identifying additional genes for targeted therapy along with erlotinib is essential for increasing overall survival of PDAC patients. By inducing synthetic lethality, patient survival is likely to increase.
4.3. Stem cells
Cancer stem cells (CSCs) have recently been reported as potential therapeutic targets; however, they are difficult to target because of the small percentage of the tumor that they occupy. Typically, CSCs inhabit less than 5% of total tumor cells, innately have enormous proliferative potential (Li et al., 2007) and can be resistant to standard therapies. Such drugs given to induce cell death can temporarily result in tumor reduction; however, this approach can easily neglect the limited number of CSCs—consequently, the tumor builds resistance against these drugs and can relapse (Dean et al., 2005). As a result, CSCs survive and can regenerate the tumor in true stem cell fashion. In an immunocompromised mouse model, human pancreatic cancer cells were xenografted and a highly tumorigenic subpopulation of pancreatic CSCs were identified using stem cell markers CD44, CD24 and epithelial-specific antigen (ESA). CD44+CD24+ESA+ cells displayed a 100-fold increased tumorigenic potential compared to nontumorigenic cells, stem cell properties such as self-renewal and differentiation, as well as increased expression of the developmental signaling molecule sonic hedgehog, which is commonly upregulated in pancreatic cancer. Interestingly, mice injected with as little as 100CD44+CD24+ESA+ cells led to pancreatic cancers indistinguishable from those observed in humans (Li et al., 2007). Similarly, Hermann et al. (2007) demonstrated that CD133+ pancreatic CSCs displayed highly tumorigenic characteristics in xenografted mouse models. As few as 500CD133+ CSCs injected into mice were able to induce orthotopic tumor formation, whereas 1 million CD133− CSCs did not display tumor presence. Importantly, the group was able to show that CSCs do not represent a homogeneous population of tumor-initiating cells; rather, specific migrating CSCs were critically involved in metastasis. Of these cells, CXCR4 was seen to be expressed and depletion of this gene abolished metastatic activity of pancreatic cancer cells. Because CSCs are limited in number, perhaps targeting stem cell regulators may be more beneficial. Nodal and Activin, members of the TGFβ family, are essential regulators of human embryonic stem cell fate (Vallier et al., 2005). Inhibiting/targeting their receptors ALK4/7 reversed CSC self-renewal and tumorigenic capacity. However, targeting this receptor is challenging because of the extensive stroma that is often present in pancreatic cancer, as well as the pleiotropic effects that these receptors play in other tissues. As mentioned above, the sonic hedgehog pathway has been observed to be upregulated in pancreatic cancer, especially in stromal cells. Therefore, inhibiting stroma-specific molecules can increase drug delivery to tumor cells. With that said, inhibition of both the hedgehog pathway and ALK4/7 has been suggested as a therapeutic strategy for targeting pancreatic CSCs (Lonardo et al., 2011).
4.4. Stroma
Inhibiting stroma-specific molecules; however, remains an area of intense interest. The surrounding stroma of PDAC tumors constitutes the majority of the tumor mass and PDAC is one of the most stroma-rich cancers. PDAC stroma consists of many extracellular matrix components, fibroblasts, myofibroblasts, pancreatic stellate cells, immune cells, blood vessels, cytokines and growth factors (Feig et al., 2012). In theory, targeting PDAC stromal components should reduce tumor growth and potentially aid in drug delivery. Olive et al. (2009) showed that inhibiting hedgehog signaling enhanced gemcitabine delivery in a mouse model that led to increased survival. They inhibited a downstream hedgehog effector Smoothened (Smo), using IPI-926, and found that mice responded better to gemcitabine leading to increased survival reduced metastatic burden in the liver. On the contrary, targeting stromal components does not always lead to tumor suppression. Ozdemir et al. (2014) demonstrated that depletion of myofibroblasts in GEMMs with activated KRas and deleted Tgfbr2 led to increased tumor invasion and decreased overall survival. Myofibroblast depletion also illustrated suppression of angiogenesis, enhanced tumor hypoxia, induction of EMT, CSC-like phenotypes and an overall alteration in the ECM. Mice with depleted myofibroblasts did not show increased survival upon treatment with gemcitabine relative to control mice; rather, there were no statistical differences. The group suggested that gemcitabine may target myofibroblasts, which leads to tumor progression as previously described, and may explain a potential reason for drug resistance in PDAC patients. Finally, recent evidence suggests that both the tumor epithelial and stromal compartments are enriched for metabolic activity, emphasizing that regulators of metabolism maybe new and novel targets to simultaneously reduce both tumor and stromal cell contributions to PDAC (Chaika et al., 2012b). In agreement with this, a recent report has demonstrated that vitamin C can target mutant KRas cancer cells by GLUT1-/ROS-mediated inactivation of GAPDH (Yun et al., 2015). This offers promise in PDAC, as most tumors are positive for activating mutations in KRas. It is evident that stromal targeting is a promising approach for PDAC treatment, but identifying exactly what to target requires more research.
5. Identifying new diagnostic biomarkers and therapeutic targets
5.1. Proteomics
Over the past decade, proteomics has quickly found its place as a useful technique for identifying potential biomarkers or therapeutic targets since this method can detect the protein and post-translationally modified protein levels (Anderson and Anderson, 1998). Identification of such biomolecules may help in predict disease prognosis and identification of new targets; thus, in the case of PDAC, proteomics is very practical. One application of this method is to compare normal and tumor tissue from PDAC patients for global protein level differences. An increase or decrease in a protein’s level in the tumor may suggest whether a particular protein is involved in tumor progression or suppression. A potential hurdle, however, arises when isolating PDAC tissue from its microenvironment. Because PDAC is highly desmoplastic, containing all types of cells, it becomes difficult to dissect the tumor cell versus stromal cell proteomes (Cowan and Maitra, 2014; Erkan et al., 2010). Another similar application is to compare global protein levels between primary and metastatic PDAC tumors in order to identify the proteins that are involved in the process of metastasis (Aebersold and Mann, 2003). McKinney and Kee (2012) examined protein levels from cells derived from primary pancreatic tumors and compared them to the protein levels from cells derived from metastatic sites. By using standard proteomic methods, they analyzed their data through bioinformatics and were able to identify 540 specific proteins uniquely expressed in the primary tumor cells and 487 in metastatic cells. Of these proteins, the differential levels of 134 of them were statistically significant between the two populations (McKinney and Kee, 2012). If we further look into these 134 proteins, we may be able to better understand which proteins are responsible for metastasis and potentially develop drugs targeting these proteins. For a disease like PDAC, which has a dire need for novel biomarkers and therapeutic targets, identifying proteins via proteomics holds great promise for addressing many unanswered questions.
5.2. Secretomics
A branch of proteomics is secretomics, which analyzes all of the secreted proteins from cells. Secreted proteins play important roles in cell signaling and are pivotal for priming tumor cells to disseminate during metastasis (Pavlov and Diamandis, 2010). Analysis of secretome samples from pancreatic cancer cells or pancreatic juice are most accessible; therefore, studies should be conducted using these samples. One quantitative method for identifying proteins via mass spectrometry is known as stable isotope labeling with amino acids in cell culture (SILAC). Gronborg et al. (2006) used this technique to identify 145 differentially secreted proteins with greater than a 1.5 fold change from pancreatic cancer-derived cells compared to normal pancreatic epithelial cells. Of these proteins, many were indicated to have altered expression prior to their work; however, new proteins were discovered with altered expression. These novel proteins could serve as biomarkers as they are secreted at different levels in PDAC cells. Studying these proteins, among others, could provide valuable information at the different steps of PDAC progression. Currently, the best available biomarker of pancreatic cancer is CA 19-9, a secreted protein that is used to screen for in pancreatic cancer in patients (Gronborg et al., 2006; Pavlov and Diamandis, 2010). Another approach in identifying proteins from the secretome is by investigating exosomes from tumor tissue. Exosomes are membrane-enclosed vesicles that carry proteins and nucleic acids and are secreted into the circulation by all cells (Keller et al., 2006). Analyzing the contents within cancer-specific exosomes can provide insight on proteins involved in metastasis. At the same time, these proteins may also serve as biomarkers for early detection of cancer. Using mass spectrometry analyses, glypican-1 (GPC1) was recently discovered to be an enriched cell-surface protein on pancreatic cancer cell-derived exosomes. GPC1+ circulating exosomes were significantly higher in patients with PDAC relative to healthy individuals. Interestingly, wild-type KRas was present in both GPC1+ and GPC1− circulating exosomes whereas mutant KRas was detected only in GPC1+ circulating exosomes; thus, further validating the specificity of GPC1+ circulating exosomes to PDAC. When comparing the specificity and sensitivity of the standard PDAC biomarker CA 19-9 to GPC1+ circulating exosomes, it was shown that CA 19-9 levels were high in the sera of patients with benign pancreatic disease and patients with PDAC relative to healthy individuals. On the other hand, GPC1+ circulating exosomes were elevated in patients with PDAC only and that GPC1+ circulating exosome levels were similar in healthy individuals compared to patients with benign pancreatic disease. At the same time, CA 19-9 expression was indistinguishable between sera from healthy individuals and patients with pancreatic cancer precursor lesions while GPC1+ circulating exosomes were significantly higher in patients with precursor lesions relative to healthy donors (Melo et al., 2015).Thus, GPC1+ circulating exosomes provides more specificity and sensitivity as a biomarker for patients with PDAC relative to CA 19-9. It is evident that the secretome can offer useful information in regards to identifying novel proteins that may serve as biomarkers or therapeutic targets and further research should focus in this area.
5.3. Sequencing
Determining the order of nucleotide bases across the genome or exome is known as sequencing. Sequencing can be done on DNA or RNA. Similar to proteomics, these methods may be used to compare genomic or transcriptomic differences between PDAC tissue and normal pancreatic tissue. These methods enable researchers to identify gene mutations and isoform preferences, in addition to overall expression patterns. If an oncogenic mutation is identified, drugs may be developed to target the site of mutation. Since not all of the cells in the organism will carry this mutation, the specificity of the drug to target the tumor cells will increase while limiting off-target effects. Like proteomics, sequencing becomes difficult when isolating PDAC tissue from its microenvironment because of the vast amounts of stromal cells and inflammatory cells (Cowan and Maitra, 2014; Erkan et al., 2010). In a recent publication, Witkiewicz et al. (2015) used microdissection techniques to separate pancreatic tumor cells from its microenvironment in 109 surgically resected pancreatic cancer patients. These cells were subjected to whole-exome sequencing and new mutations were discovered using multiple sequencing techniques. Mutation Significance of covariance (MutSigCV) is a technique used to identify mutations from patient tumor issue (Lawrence et al., 2013). MutSigCV analysis of these tumor cells resulted in 24 significantly mutated genes that occurred in >3.5% of the cases. Some of these mutations have been previously described, such as KRas, TP53, CDKN2A and Smad4; however, novel genes such as BCLAF1, IRF6, FLG, AXIN1, GLI3 and PIK3CA were identified to be significantly mutated using this sequencing approach. Interestingly, the group was able to discover the amplification of MYC, which correlated with poor outcome. Sequencing techniques can provide valuable information for discovering novel therapeutic targets.
5.4. Circulating tumor cells
Circulating tumor cells (CTCs) are cells derived from a primary tumor that have entered the vasculature and circulate within the blood stream looking to seed in distant organs (Gupta and Massague, 2006). CTCs appear in extremely low frequencies in the blood; about 1–10CTCs are found within 1 mL of blood (Miller et al., 2010). From the total amount of CTCs that escape the primary tumor, only 0.01% develops into metastases (Zhe et al., 2011). This minute amount of CTCs found in the blood makes it difficult to detect their presence; however, if identified, CTCs can be essential biomarkers for cancer initiation and progression. A common method used for detecting CTCs is by using antibodies against antigens found on the cell surface of these cells (Cen et al., 2012; Yu et al., 2011). Epithelial cell adhesion molecule (EpCAM) is the marker that is used to detect CTCs as expression of EpCAM in blood cells is absent. CellSearch (Veridex) is an FDA approved method that uses ferrofluids enriched with EpCAM antibody-coated magnetic beads to capture CTCs. Once fixed, the cells are visualized by staining for antibodies for epithelial cytokeratins 8, 18 and 19. Staining for CD45 is also required as it is a marker for leukocytes and is used as a negative control. Cells that positively stain for the cytokeratins and do not stain for CD45 are classified as CTCs (Cen et al., 2012), (Yu et al., 2011). A study to detect CTCs in multiple cancers was conducted using this method. CTCs found in prostate and breast cancers were significantly greater than those found in pancreatic cancer. Albeit CTCs from pancreatic cancer were detected, their levels were comparable to CTCs found in nonmalignant diseases (Allard et al., 2004). Thus, a drawback using this method for CTC detection in pancreatic cancer is its low sensitivity and it is EpCAM dependent. Aside from detecting CTCs, fluorescently tagging these cells and tracking them in a mouse model could provide valuable information on their role in cancer progression. Rhim et al. (2012) traced CTCs in mice by tagging yellow fluorescent protein (YFP) to PanIN and PDAC cells. They also tagged EMT markers and concluded that CTCs from PanIN and PDAC animals were phenotypically similar and largely maintained a mesenchymal phenotype. This was confirmed by qPCR from isolated YFP+ cells. Although the group was able to track CTCs in a mouse model, the issue arises in clinical settings. If we can improve antigen-specificity to PDAC, we can use antibodies specific to the CTCs to detect them; therefore, newer technologies are required for better detection of the limited amount of CTCs present in the blood with higher specificity to pancreatic cancer. Table 2 summarizes the advantages, disadvantages and potential therapeutic options for the methods of identifying novel diagnostic markers and/or therapeutic targets.
Table 2.
Methods for identification of new diagnostic biomarkers and therapeutic targets.
| Method | Advantages | Disadvantages | Potential uses |
|---|---|---|---|
| Proteomics | Insight on proteins required for tumor progression/metastasis (Aebersold and Mann, 2003; Erkan et al., 2010) | Isolating cells from microenvironment is difficult (Anderson and Anderson, 1998; Cowan and Maitra, 2014) |
|
| Secretomics |
|
Most secretome studies are performed in vitro (Gronborg et al., 2006) | Biomarkers |
| Sequencing |
|
Isolating cells from microenvironment is difficult (Anderson and Anderson, 1998; Cowan and Maitra, 2014) | Therapeutic targets |
| Circulating tumor cells (CTCs) | Provides useful information for metastasis (Rhim et al., 2012) |
|
Biomarkers |
6. Concluding remarks
Research on pancreatic cancer has vastly increased over the past few decades, yet the survival rate has remained rather low. Advancements in technologies have provided us a “blue-print” of PDAC initiation, development and progression. This has given us the opportunity to identify potential biomarkers and therapeutic targets for this malignancy; however, more studies are required for definitive treatment options. Due to the lack of noticeable symptoms while the tumor is localized to the pancreas, a “trace-back-footsteps” approach ought to be implemented such that future studies should focus on metastatic mechanisms that come with pancreatic cancer since the majority of PDAC patients are diagnosed at this stage. By identifying genes required for metastasis, working backwards towards regulators of these genes can explain what triggers cells to metastasize from the lymph nodes. Once this is accomplished, we will have a better understanding of what genes are required for tumor cells to spread from the pancreas. And this “moving-backwards” method may eventually identify the cell-of-origin for this lethal disease.
Acknowledgments
We thank the members of the Kelber Lab at California State University Northridge. Funding that supported this and related work came from the CSUN College of Science and Math, Medtronic/Minimed, CSUPERB and the Sidney Stern Memorial Trust.
References
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Aguirre AJ, Bardeesy N, Sinha M. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard W, Matera J, Miller M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- Anderson N, Anderson N. Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis. 1998;19:1853–1861. doi: 10.1002/elps.1150191103. [DOI] [PubMed] [Google Scholar]
- Bachmann J, Michalski C, Martignoni M. Pancreatic resection for pancreatic cancer. HPB (Oxford) 2006;8:346–351. doi: 10.1080/13651820600803981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N, Cheng KH, Berger JH. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besmer D, Curry J, Roy L. Pancreatic ductal adenocarcinoma mice lacking mucin 1 have a profound defect in tumor growth and metastasis. Cancer Res. 2011;71:4432–4442. doi: 10.1158/0008-5472.CAN-10-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromann P, Korkaya H, Courtneidge SA. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene. 2004;23:7957–7968. doi: 10.1038/sj.onc.1208079. [DOI] [PubMed] [Google Scholar]
- Busnardo A, DiDio L, Tidrick R. History of the pancreas. Am. J. Soc. 1983;146:539–550. doi: 10.1016/0002-9610(83)90286-6. [DOI] [PubMed] [Google Scholar]
- Cen P, Ni X, Yang J. Circulating tumor cells in the diagnosis and management of pancreatic cancer. Biochem. Biophys. Acta. 2012;1826:350–356. doi: 10.1016/j.bbcan.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaika NV, Gebregiworgis T, Lewallen ME. MUC1 mucin stabilizes and activates hypoxia-inducible factor 1 alpha to regulate metabolism in pancreatic cancer. Proc. Natl. Acad. Sci. U. S. A. 2012a;109:13787–13792. doi: 10.1073/pnas.1203339109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaika NV, Yu F, Purohit V. Differential expression of metabolic genes in tumor and stromal components of primary and metastatic loci in pancreatic adenocarcinoma. PLoS One. 2012b;7:e32996. doi: 10.1371/journal.pone.0032996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiao PJ, Ling J. Kras Pten, NF-kappaB, and inflammation: dangerous liaisons. Cancer Discov. 2011;1:103–105. doi: 10.1158/2159-8290.CD-11-0115. [DOI] [PubMed] [Google Scholar]
- Chou A, Waddell N, Cowley M. Clinical and molecular characterization of HER2 amplified-pancreatic cancer. Genome Med. 2013;5:78. doi: 10.1186/gm482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M, Bednar F, Zhang Y. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J. Clin. Investig. 2012a;122:639–653. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M, Brisset J, Zhang Y. Metastatic pancreatic cancer is dependent on oncogenic Kras in mice. PLoS One. 2012b:7. doi: 10.1371/journal.pone.0049707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan R, Maitra A. Genetic progression of pancreatic cancer. Cancer J. 2014;20:80–84. doi: 10.1097/PPO.0000000000000011. [DOI] [PubMed] [Google Scholar]
- Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat. Rev. Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- Deer E, Gonzalez-Hernandez J, Coursen J. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep C, Munoz R, Choudhary A. Synergistic effect between erlotinib and MEK inhibitors in KRAS wild-type human pancreatic cancer cells. Clin. Cancer Res. 2011;17:2744–2756. doi: 10.1158/1078-0432.CCR-10-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkan M, Reiser-Erkan C, Michalski C. Tumor microenvironment and progression of pancreatic cancer. Exp. Oncol. 2010;32:128–131. [PubMed] [Google Scholar]
- Eser S, Schnieke A, Schneider G. Oncogenic KRAS signaling in pancreatic cancer. Br. J. Cancer. 2014;18:4266–4276. doi: 10.1038/bjc.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig C, Gopinathan A, Neesse A. The pancreas cancer microenvironment. Clin. Cancer Res. 2012;18:4266–4276. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari E, Lucca C, Foiani M. A lethal combination for cancer cells: synthetic lethality screenings for drug discovery. Eur. J. Cancer. 2010;46:2889–2895. doi: 10.1016/j.ejca.2010.07.031. [DOI] [PubMed] [Google Scholar]
- Fujimura K, Wright T, Strnadel J, et al. A hypusine-eIF5A-PEAK1 switch regulates the pathogenesis of pancreatic cancer. Cancer Res. 2014;74(22):6671–6681. doi: 10.1158/0008-5472.CAN-14-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardiello F, Brensinger J, Tersmette A. Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology. 2000;119:1447–1453. doi: 10.1053/gast.2000.20228. [DOI] [PubMed] [Google Scholar]
- Goodman KA, Hajj C. Role of radiation therapy in the management of pancreatic cancer. J. Surg. Oncol. 2013;107:86–96. doi: 10.1002/jso.23137. [DOI] [PubMed] [Google Scholar]
- Gronborg M, Kristiansen T, Hruban R. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol. Cell. Proteom. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- Guix M, Faber AC, Wang SE. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J. Clin. Investig. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Hagemann T, Biswas S, Lawrence T. Regulation of macrophage function in tumors: the multifaceted role of NF-kappaB. Blood. 2009;113:3139–3146. doi: 10.1182/blood-2008-12-172825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon L, Avila J, Demarest R. Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res. 2010;70:4280–4286. doi: 10.1158/0008-5472.CAN-09-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsha HC, Kandasamy K, Ranganathan P. A compendium of potential biomarkers of pancreatic cancer. PLoS Med. 2009;6:e1000046. doi: 10.1371/journal.pmed.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid I, Lubeseder-Martellato C, Sipos B. Early requirement of Rac1 in a mouse model of pancreatic cancer. Gastroenterology. 2011;2011:2. doi: 10.1053/j.gastro.2011.04.043. [DOI] [PubMed] [Google Scholar]
- Heiser P, Cano D, Landsman L. Stabilization of β-catenin induces pancreas tumor formation. Gastroenterology. 2008;135:1288–1300. doi: 10.1053/j.gastro.2008.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Hezel AF, Gurumurthy S, Granot Z. Pancreatic LKB1 deletion leads to acinar polarity defects and cystic neoplasms. Mol. Cell. Biol. 2008;28:2414–2425. doi: 10.1128/MCB.01621-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Hishinuma S, Ogata Y, Tomikawa M. Patterns of recurrence after curative resection of pancreatic cancer, based on autopsy findings. J. Gastrointest. Surg. 2006;10:511–518. doi: 10.1016/j.gassur.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Howlader NNA, Krapcho M, Garshell J, Neyman N, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA. SEER Cancer Stat. Rev 2013 [Google Scholar]
- Ijichi G, Chytil A, Gorska AE. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006;20:3147–3160. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irby R, Yeatman T. Role of Src expression and activation in human cancer. Oncogene. 2000;19:5636–5642. doi: 10.1038/sj.onc.1203912. [DOI] [PubMed] [Google Scholar]
- Kelber JA, Reno T, Kaushal S. KRas induces a Src/PEAK1/ErbB2 kinase amplification loop that drives metastatic growth and therapy resistance in pancreatic cancer. Cancer Res. 2012;72:2554–2564. doi: 10.1158/0008-5472.CAN-11-3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S, Sanderson M, Stoeck A. Exosomes: from biogenesis and secretion to biological function. Immunol. Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Mizuno S, Murata Y. Gemcitabine-based chemoradiotherapy followed by surgery for borderline resectable and locally unresectable pancreatic ductal adenocarcinoma: significance of the CA19-9 reduction rate and intratumoral human equilibrative nucleoside transporter 1 expression. Pancreas. 2014;43:350–360. doi: 10.1097/MPA.0000000000000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoto M, Nakata B, Amano R. HER2 overexpression correlates with survival after curative resection of pancreatic cancer. Cancer Sci. 2009;100:1243–1247. doi: 10.1111/j.1349-7006.2009.01176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larbouret C, Robert B, Teulon I. Combination of anti-EGFR and anti-HER2 antibodies: hope in pancreatic cancer treatment. Bull. Cancer. 2007;94:860–862. [PubMed] [Google Scholar]
- Lawrence M, Stojanov P, Polak P. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Rajeshkumar N, Maitra A. Conceptual framework for cutting the pancreatic cancer fuel supply. Clin. Cancer Res. 2012;18:4285–4290. doi: 10.1158/1078-0432.CCR-12-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Lonardo E, Hermann P, Mueller M. Nodal/activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell. 2011;9:433–446. doi: 10.1016/j.stem.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Lutz MP, Esser IB, Flossmann-Kast BB. Overexpression and activation of the tyrosine kinase Src in human pancreatic carcinoma. Biochem. Biophys. Res. Commun. 1998;243:503–508. doi: 10.1006/bbrc.1997.8043. [DOI] [PubMed] [Google Scholar]
- Maitra A, Hruban RH. Pancreatic cancer. Annu. Rev. Pathol. 2008;3:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniati E, Bossard M, Cook N. Crosstalk between the canonical NF-κB and Notch signaling pathways inhibits Pparγ expression and promotes pancreatic cancer progression in mice. J. Clin. Investig. 2011;121:4685–4699. doi: 10.1172/JCI45797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur P, Einwachter H, Lee M. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. PNAS. 2010;107:13438–13443. doi: 10.1073/pnas.1002423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F. Cancer therapy based on oncogene addiction. J. Surg. Oncol. 2011;103:464–467. doi: 10.1002/jso.21749. [DOI] [PubMed] [Google Scholar]
- McKinney K, Kee J. Sindram D. Identification of differentially expressed proteins from primary versus metastatic pancreatic cancer cells using subcellular proteomics. Cancer Genom. Proteom. 2012;9:257–263. [PubMed] [Google Scholar]
- Mehenni H, Resta N, Park J. Cancer risks in LKB1 germline mutation carriers. Gut. 2006;55:984–990. doi: 10.1136/gut.2005.082990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo S, Luecke L, Kahlert C. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Doyle G, Terstappen L. Significance of circulating tumor cells detected by the cellsearch system in patients with metastatic breast colorectal and prostate cancer. J. Oncol. 2010 doi: 10.1155/2010/617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milosevic N, Kuhnemuth B, Muhlberg L. Synthetic lethality screen identifies RPS6KA2 as modifier of epidermal growth factor receptor activity in pancreatic cancer. Neoplasia. 2013;15:1354–1362. doi: 10.1593/neo.131660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabayashi K, Iijchi H, Mohri D. Erlotinib prolongs survival in pancreatic cancer by blocking gemcitabine-induced MAPK signals. Cancer Res. 2013;73:2221–2234. doi: 10.1158/0008-5472.CAN-12-1453. [DOI] [PubMed] [Google Scholar]
- Montagut C, Dalmases A, Bellosillo B. Identification of a mutation in the extracellular domain of the epidermal growth factor receptor conferring cetuximab resistance in colorectal cancer. Nat. Med. 2012;18:221–223. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- Morton J, Jamieson N, Karim S. LKB1 haploinsufficiency cooperates with Kras to promote pancreatic cancer through suppression of p21-dependent growth arrest. Gastroenterology. 2010;139:586–597. doi: 10.1053/j.gastro.2010.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas C, Hernandez-Porras I, Schuhmacher AJ. EGF receptor signaling is essential for kras oncogene-driven pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:318–330. doi: 10.1016/j.ccr.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neesse A, Michl P, Frese K. Stromal biology and therapy in pancreatic cancer. Gut. 2011;60:861–868. doi: 10.1136/gut.2010.226092. [DOI] [PubMed] [Google Scholar]
- Ng S, Tsao M, Nicklee T. Effects of the epidermal growth factor receptor inhibitor OSI-774, Tarceva, on downstream signaling pathways and apoptosis in human pancreatic adenocarcinoma. Mol. Cancer Ther. 2002;1:777–783. [PubMed] [Google Scholar]
- Nolen B, Brand R, Prosser D. Prediagnostic serum biomarkers as early detection tools for pancreatic cancer in a large prospective cohort study. PLoS One. 2014:9. doi: 10.1371/journal.pone.0094928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive KP, Jacobetz MA, Davidson CJ. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir B, Pentcheva-Hoang T, Carstens J. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunsuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papademetrio D, Cavaliere V, Simunovich T. Interplay between autophagy and apoptosis in pancreatic tumors in response to gemcitabine. Target Oncol. 2014;9:123–134. doi: 10.1007/s11523-013-0278-5. [DOI] [PubMed] [Google Scholar]
- Pavlov M, Diamandis E. The cancer cell secretome: a good source for discovering biomarkers. J. Proteom. 2010;73:1896–1906. doi: 10.1016/j.jprot.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Rhim AD, Mirek ET, Aiello NM, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Roy L, Sahraei M, Subramani D. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30:1449–1459. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Shaw R, Kosmatka M, Bardeesy N. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. PNAS. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw V, Lane B, Jenkinson C. Serum cytokine biomarker panels for discriminating pancreatic cancer from benign pancreatic disease. Mol. Cancer. 2014:13. doi: 10.1186/1476-4598-13-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Wang Q, Cheng S. The biological features of PanIN initiated from oncogenic KRas mutation in genetically engineered mouse models. Cancer Lett. 2013;339:135–143. doi: 10.1016/j.canlet.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Shields DJ, Murphy EA, Desgrosellier JS. Oncogenic Ras/Src cooperativity in pancreatic neoplasia. Oncogene. 2011 doi: 10.1038/onc.2010.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Miller K, Jemal A. Cancer statistics 2015. CA Cancer J. Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- Tanaka S. Molecular pathogenesis and targeted therapy of pancreatic cancer. Ann. Surg. Oncol. 2016;2:197–205. doi: 10.1245/s10434-015-4463-x. [DOI] [PubMed] [Google Scholar]
- Tinder T, Subramani D, Basu G. MUC1 enhances tumor progression and contributes toward immunosuppression in a mouse model of spontaneous pancreatic adenocarcinoma. J. Immunol. 2008;181:3116–3125. doi: 10.4049/jimmunol.181.5.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L, Alexander M, Pedersen R. Activin/nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 2005;111:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- Vincent A, Herman J, Schulick R. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Luhrs H, Kloppel G. Malignant transformation of duct-like cells originating from acini in transforming growth factor transgenic mice. Gastroenterology. 1998;115:1254–1262. doi: 10.1016/s0016-5085(98)70098-8. [DOI] [PubMed] [Google Scholar]
- Wagner M, Greten F, Weber C. A murine tumor progression model for pancreatic cancer recapitulating the genetic alterations of the human disease. Genes Dev. 2001;15:286–293. doi: 10.1101/gad.184701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh N, Kennedy S, Larkin A. EGFR and HER2 inhibition in pancreatic cancer. Investig. New Drugs. 2013;31:558–566. doi: 10.1007/s10637-012-9891-x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kelber JA, Tran Cao HS. Pseudopodium-enriched atypical kinase 1 regulates the cytoskeleton and cancer progression [corrected] Proc. Natl. Acad. Sci. U. S. A. 2010;107:10920–10925. doi: 10.1073/pnas.0914776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein I, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- Whatcott C, Diep C, Jiang P. Desmoplasia in primary tumors and metastatic lesions of pancreatic cancer. Clin. Cancer Res. 2015;21:3561–3568. doi: 10.1158/1078-0432.CCR-14-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wideltz R. Wnt signaling through canonical and non-canonical pathways: recent progress. Growth Factors. 2005;23:111–116. doi: 10.1080/08977190500125746. [DOI] [PubMed] [Google Scholar]
- Wingren C, Sandstrom A, Segersvard R. Identification of serum biomarker signatures associated with pancreatic cancer. Cancer Res. 2012;72:2481–2490. doi: 10.1158/0008-5472.CAN-11-2883. [DOI] [PubMed] [Google Scholar]
- Witkiewicz A, McMillan E, Balaji U. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang C, Herman J, Laheru D. Recent progress in pancreatic cancer. CA Cancer J. Clin. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Elpek KG, Vinjamoori A. Pten is a major tumor suppressor in pancreatic ductal adenocarcinoma and regulates an NF-kappaB-cytokine network. Cancer Discov. 2011;1:158–169. doi: 10.1158/2159-8290.CD-11-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman A, Lyssiotis C. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Stott S, Toner M. Circulating tumor cells: approaches to isolation and characterization. J. Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Arcila M, Rekhtman N. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin. Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Mullarky E, Lu C. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;350(6266):1391–1396. doi: 10.1126/science.aaa5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Ren H, Jia L. Inhibition of HIF-1α by PX-478 enhances the anti-tumor effect of gemcitabine by inducing immunogenic cell death in pancreatic ductal adenocarcinoma. Oncotarget. 2015a;6:2250–2262. doi: 10.18632/oncotarget.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li F, Li Y. Co-delivery of HIF1α siRNA and gemcitabine via biocompatible lipid-polymer hybrid nanoparticles for effective treatment of pancreatic cancer. Biomaterials. 2015b;46:13–25. doi: 10.1016/j.biomaterials.2014.12.028. [DOI] [PubMed] [Google Scholar]
- Zhe X, Cher M, Bonfil R. Circulating tumor cells: finding the needle in the haystack. Am. J. Cancer Res. 2011;1:740–751. [PMC free article] [PubMed] [Google Scholar]
- Zhong H, De Marzo A, Laughner E. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]