Abstract
Only 3% of the transcribed human genome is translated into protein, and small non-coding RNAs from these untranslated regions have demonstrated critical roles in transcriptional and translational regulation of proteins. Here, we provide a resource that will facilitate cell line selection for gene expression studies involving sncRNAs in cancer research. As the most accessible and tractable models of tumours, cancer cell lines are widely used to study cancer development and progression. The NCI-60 panel of 59 cancer cell lines was curated to provide common models for drug screening in 9 tissue types; however, its prominence has extended to use in gene regulation, xenograft models, and beyond. Here, we present the complete small non-coding RNA (sncRNA) transcriptomes of these 59 cancer cell lines. Additionally, we examine the abundance and unique sequences of annotated microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs), and reveal novel unannotated microRNA sequences.
Subject terms: Cancer genomics, RNA sequencing, Non-coding RNAs
Background & Summary
The NCI-60 Human Tumour Cell Lines Screen is an initiative started by the National Institutes of Health (NIH) in the late 1980s, focusing on the development of 59 human tumour cell lines for use as an in vitro drug screen model1–3 (Table 1 (available online only)). These cell lines, derived from nine solid and blood malignancies, have shown great utility both in its original purpose for therapeutic screening as well as in basic cancer research (reviewed by Shoemaker et al.4). They have since been extensively characterized for various molecular features, including karyotypic complexity1, DNA fingerprinting2, gene expression microarray profiling5,6, and human leukocyte antigen typing3. However, the small non-coding RNA (sncRNA) transcriptomes of the NCI-60 cell lines have yet to have been reported at the sequencing level.
Table 1. NCI-60 cell line characteristics.
| Tissue | Cell line | Subtype | Cell type of origin | Doubling time (hrs) | Year of origin | Culture conditions |
|---|---|---|---|---|---|---|
| BREAST | BT-549 | Ductal Carcinoma | Epithelial | 53.9 | 1978 | RPMI-1640+10% FBS |
| HS-578T | Carcinoma | Epithelial | 53.8 | 1977 | DMEM+10% FBS | |
| MCF-7 | Adenocarcinoma | Epithelial | 25.4 | 1973 | MEM+10% FBS | |
| MDA-MB-231 | Adenocarcinoma | Epithelial | 41.9 | 1974 | L-15 Medium+10% FBS | |
| T-47D | Ductal Carcinoma | Epithelial | 45.5 | 1981 | DMEM+10% FBS | |
| CNS | SF-539 | Gliblastoma multiforme | Right Temporal Lobe | 35.4 | 1986 | MEM+10% FBS |
| SF-295 | Glioblastoma | Left Temporal Lobe | 29.5 | 1986 | RPMI-1640+10% FBS | |
| SF-268 | Highly Anaplastic Astrocytoma | Right Parietal Lobe | 33.1 | 1987 | RPMI-1640+10% FBS | |
| U251 | Glioblastoma | Non-Epithelial (CNS) | 23.8 | 1978 | EMEM+10% FBS | |
| SNB-75 | Glioblastoma | Non-Epithelial (CNS) | 62.8 | 1988 | RPMI-1640+10% FBS | |
| SNB-19 | Astrocytoma | Left parieto-occipital | 34.6 | 1980 | Ham's F10+10% FBS | |
| COLON | COLO 205 | Dukes' Type D, colorectal adenocarcinoma | Epithelial | 23.8 | 1975 | RPMI-1640+10% FBS |
| HCT-15 | Colon adenocarcinoma | Epithelial | 20.6 | 1979 | RPMI-1640+10% FBS | |
| HCC2998 | Colon adenocarcinoma | Epithelial | 31.5 | 1988 | RPMI-1640+10% FBS | |
| HCT-116 | Colon adenocarcinoma | Epithelial | 17.4 | 1981 | McCoy's 5a+10% FBS | |
| HT-29 | Colon adenocarcinoma | Epithelial | 19.5 | 1964 | RPMI-1640+10% FBS | |
| KM12 | Colon adenocarcinoma | Epithelial | 23.7 | 1988 | DMEM+10% FBS | |
| SW-620 | Dukes' Type D, colorectal adenocarcinoma | Epithelial | 20.4 | 1977 | L15+10% FBS | |
| LEUKEMIA | CCRF-CEM | Acute lymphoblastic leukemia | T lymphoblast | 26.7 | 1964 | RPMI-1640+10% FBS |
| HL-60(TB) | Acutre promyelocytic leukemia | Promyeloblast | 28.6 | 1977 | IMDM+20% FBS | |
| K-562 | Chronic myelogenous leukemia | Lymphoblast | 19.6 | 1975 | IMDM+20% FBS | |
| MOLT-4 | Acute lymphoblastic leukemia | Lymphoblast | 27.9 | 1980 | RPMI-1640+10% FBS | |
| RPMI 8226 | Plasmacytoma; myeloma | B Lymphocyte | 33.5 | 1966 | RPMI-1640+10% FBS | |
| SR | Large cell immunoblastic lymphoma | Lymphoblast | 28.7 | 1983 | RPMI-1640+10% FBS | |
| MELANOMA | MALME-3M | Malignant melanoma | Fibroblast | 46.2 | 1975 | IMDM+20% FBS |
| LOX-IMVI | Amelanotic melanoma | Skin | 20.5 | 1988 | RPMI-1640+10% FBS | |
| M14 | Malignant melanoma | Skin | 26.3 | 1976 | RPMI-1640+10% FBS | |
| MDA-MB-435 | Previously described as ductal carcinoma | Melanocyte | 25.8 | 1976 | L15+10% FBS | |
| SK-MEL-28 | Cutaneous melanoma | Skin | 35.1 | 1976 | DMEM+10% FBS | |
| SK-MEL-5 | Malignant melanoma | Stellate | 25.2 | 1977 | MEM+10% FBS | |
| SK-MEL-2 | Malignant melanoma | Skin | 45.5 | 1975 | MEM+10% FBS | |
| UACC-257 | Malignant melanoma | Skin | 38.5 | 1991 | RPMI-1640+10% FBS | |
| UACC-62 | Melanotic melanoma | Skin | 31.3 | 1991 | RPMI-1640+10% FBS | |
| NSCLC | A549 | Adenocarcinoma | Epithelial | 22.9 | 1972 | RPMI-1640+10% FBS |
| EKVX | Adenocarcinoma | Epithelial | 43.6 | 1988 | RPMI-1640+10% FBS | |
| HOP 92 | Large cell carcinoma | Epithelial | 79.5 | 1991 | RPMI-1640+10% FBS | |
| HOP 62 | Adenocarcinoma | Epithelial | 39 | 1989 | RPMI-1640+10% FBS | |
| NCI-H23 | Adenocarcinoma | Epithelial | 33.4 | 1980 | RPMI-1640+10% FBS | |
| NCI-H322M | Small cell bronchioloalveolar carcinoma | Epithelial | 35.3 | 1991 | RPMI-1640+10% FBS | |
| NCI-H226 | Squamous cell carcinoma; mesothelioma | Epithelial | 61 | 1980 | RPMI-1640+10% FBS | |
| NCI-H460 | Carcinoma; large cell lung cancer | Epithelial | 17.8 | 1982 | RPMI-1640+10% FBS | |
| NCI-H522 | Adenocarcinoma | Epithelial | 38.2 | 1985 | RPMI-1640+10% FBS | |
| OVARIAN | IGR-OV1 | Ovarian endometrioid adenocarcinoma | Endometreoid | 31 | 1985 | RPMI-1640+10% FBS |
| NCI/ADR-RES | High Grade Ovarian Serous Adenocarcinoma | Epithelial | 34 | 1986 | RPMI-1640+20% FBS | |
| OVCAR-3 | Adenocarcinoma | Epithelial | 34.7 | 1982 | RPMI-1640+10% FBS | |
| OVCAR-8 | High Grade Ovarian Serous Adenocarcinoma | Epithelial | 26.4 | 1984 | RPMI-1640+10% FBS | |
| OVCAR-4 | High Grade Ovarian Serous Adenocarcinoma | Epithelial | 41.4 | 1984 | RPMI-1640+10% FBS | |
| OVCAR-5 | High Grade Ovarian Serous Adenocarcinoma | Epithelial | 48.8 | 1984 | RPMI-1640+10% FBS | |
| SK-OV-3 | Adenocarcinoma | Epithelial | 48.7 | 1973 | RPMI-1640+10% FBS | |
| PROSTATE | DU-145 | Prostate carcinoma | Epithelial | 32.3 | 1978 | MEM+10% FBS |
| PC-3 | Adenocarcinoma | Epithelial | 27.1 | 1980 | F-12K+10% FBS | |
| RENAL | A498 | Carcinoma | Epithelial | 66.8 | 1977 | MEM+10% FBS |
| CAKI-1 | Clear cell renal cell carcinoma | Epithelial | 39 | 1975 | McCoy's 5a+10% FBS | |
| 786-0 | Renal cell adenocarcinoma | Epithelial | 22.4 | 1976 | RPMI-1640+10% FBS | |
| ACHN | Renal cell adenocarcinoma | Epithelial | 27.5 | 1979 | MEM+10% FBS | |
| RXF393 | Renal cell carcinoma | Epithelial | 62.9 | 1991 | RPMI-1640+10% FBS | |
| SN12C | Renal cell carcinoma | Epithelial | 29.5 | 1986 | DMEM+10% FBS | |
| TK-10 | Clear cell renal cell carcinoma | Epithelial | 51.3 | 1987 | RPMI-1640+10% FBS | |
| UO-31 | Renal cell carcinoma | Epithelial | 41.7 | 1991 | DMEM+10% FBS |
The advent of next-generation sequencing has revealed the large proportion of non-coding genes in the human genome, and the relevance of these non-coding species in regulating the expression of both neighbouring and distant protein-coding genes. In the context of cancer, microRNAs (miRNAs) remain the best-studied non-coding RNA species, and have been implicated in all stages of cancer: initiation, progression, and response to therapy (reviewed by Hayes et al.7). Recent advances in the bioinformatic tools used for the discovery of small non-coding RNA have considerably expanded the number of known miRNA sequences8. Other types of sncRNA, including PIWI-interacting RNAs (piRNAs), small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs) are emerging topics in cancer biology (reviewed by Ng et al. and Mannoor et al.9,10). Beyond their functions in gene regulation, sncRNAs are attractive prognostic biomarkers due to their abundance and stability in various biofluids11.
We sequenced the sncRNA transcriptomes of the 59 cell lines in the panel (Fig. 1). SncRNA profiles were generated using the OASIS analysis platform v2.0 (ref. 12). For known sncRNA species (miRNAs, piRNAs, snoRNA, snRNA, and rRNA), high quality reads were mapped to the hg38 build of the human genome and quantified based on annotations containing their specific chromosomal locations. Detection of novel miRNAs was performed using well-established prediction algorithms that assess reads for miRNA folding characteristics, among other factors that indicate the probability that the tested sequence belongs to the miRNA family of sncRNAs13. In total, the genomic loci of 49,961 sncRNAs were examined. Using a detection threshold of greater than or equal to 5 reads across all tissues, we detected a total of 24,621 unique sncRNAs [Data Citation 1].
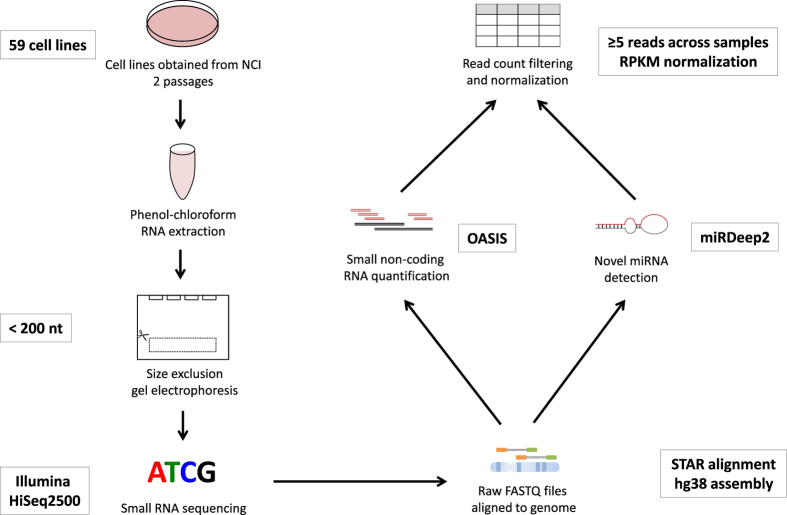
Figure 1. Experimental workflow.
Graphical representation of experimental procedure used to extract, process, and analyze RNA from cell lines.
We then examined the genomic distribution of the detected sncRNAs across all tissue types (Table 2,Fig. 2). Notably, sncRNAs are expressed across all chromosomes in every tissue type assessed. SncRNA loci commonly expressed among all tissues may indicate their involvement in preserved biological or cancer-relevant processes, whereas differences in expression may denote tissue specificity.
Table 2. Average number of sncRNA species detected and sequencing coverage per tissue type.
|
Number of sncRNA |
Sequencing details |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | miRNA | novel miRNA | piRNA | snoRNA | snRNA | Other | Average number of reads per sample | Average contig length | Avg. quality | %GC | Average coverage | |
| Tissue type | 24,794 | 2,509 | 288 | 19,018 | 259 | 1,602 | 412 | 23,457,233 | 22.34 | 33.25 | 46.26% | 28.94 |
| Breast | 11,079 | 1,905 | 183 | 6,909 | 239 | 1,248 | 403 | 25,310,156 | 22.32 | 33.23 | 45.81% | 25.36 |
| CNS | 10,120 | 1,793 | 180 | 6,150 | 236 | 1,175 | 397 | 25,633,608 | 22.35 | 33.34 | 44.57% | 45.08 |
| Colon | 13,985 | 1,977 | 211 | 8,050 | 232 | 1,276 | 392 | 25,850,349 | 22.22 | 33.2 | 46.15% | 22.97 |
| Leukemia | 10,728 | 1,841 | 185 | 5,604 | 228 | 1,112 | 387 | 18,921,562 | 22.41 | 33.25 | 48.77% | 20.23 |
| Melanoma | 9,536 | 1,830 | 179 | 4,694 | 224 | 1,020 | 383 | 16,116,671 | 22.36 | 32.64 | 46.56% | 28.45 |
| NSCLC | 15,707 | 2,051 | 232 | 9,385 | 236 | 1,293 | 398 | 23,407,812 | 22.34 | 33.3 | 46.18% | 22.8 |
| Ovarian | 10,422 | 1,916 | 188 | 6,312 | 238 | 1,167 | 398 | 27,221,870 | 22.4 | 34.12 | 46.56% | 41.49 |
| Prostate | 6,167 | 1,393 | 121 | 2,589 | 216 | 771 | 344 | 35,637,053 | 22.3 | 33.54 | 44.44% | 30.11 |
| Renal | 14,532 | 1,943 | 209 | 8,549 | 234 | 1,288 | 396 | 23,949,373 | 22.3 | 33 | 45.99% | 27.05 |
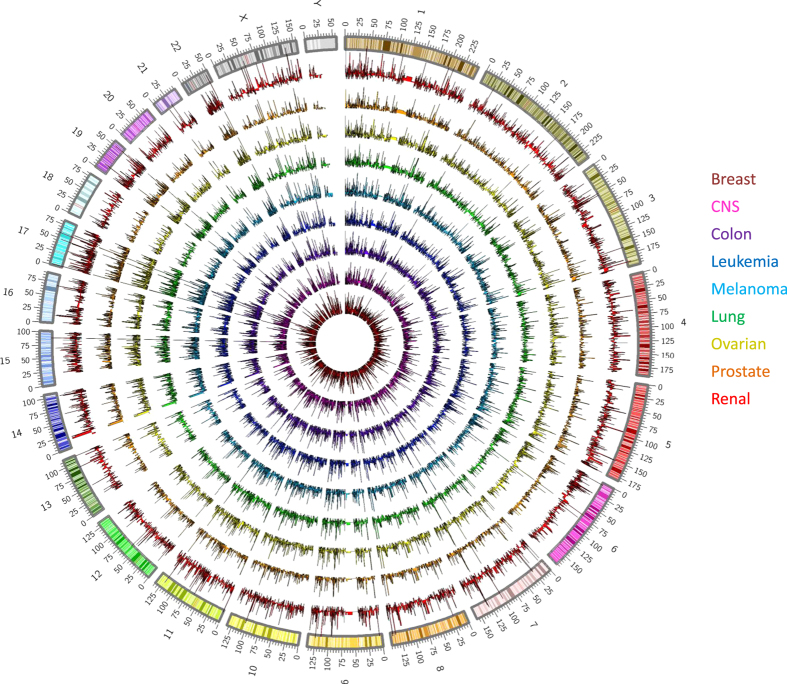
Figure 2. Genome-wide distribution of expressed small non-coding RNA by tissue type.
Genomic position of sncRNAs detected (reads≥5) in each tissue type in reference to the hg38 chromosome build karyotype. From inner-most ring to outer: breast (red), CNS (magenta), colon (purple), leukemia (blue), melanoma (teal), lung (green), ovarian (yellow), prostate (orange), and renal (red).
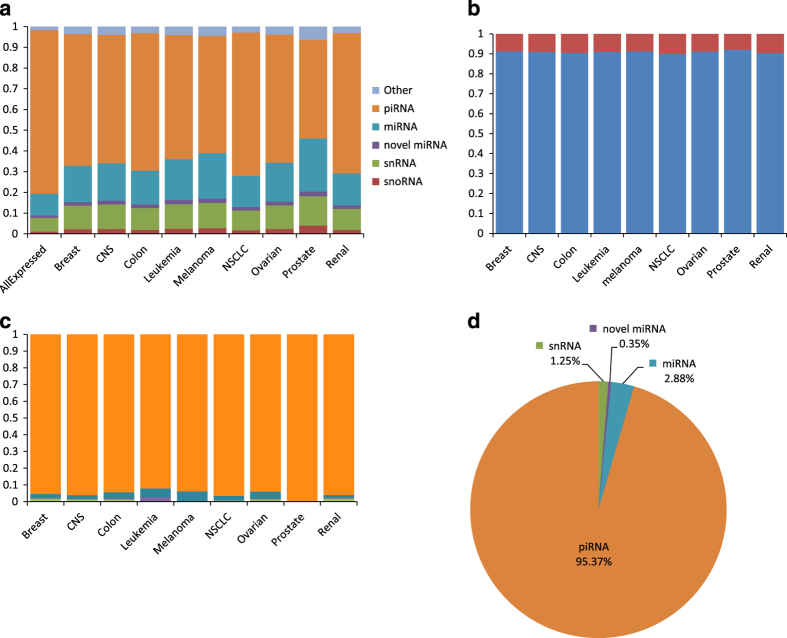
We also examined the relative frequency of detection for each sncRNA species, both in the entire NCI-60 cell line panel and in lines grouped by organ type (Fig. 3a). Beyond those annotated in miRBase (v.21), novel unannotated miRNAs were determined by integrating secondary structure formation potential with free energy scoring14. These novel miRNAs represent an increase of approximately 10% of total miRNAs expressed across all tissue types (Fig. 3b), highlighting the constant expansion of the known non-coding transcriptome as sequencing technologies and bioinformatic tools advance. Consistent with the number of annotated loci in the human genome, piRNAs represent the largest proportion of sncRNA species expressed, followed by miRNA and snRNA (Fig. 3a). Of note, an appreciable number of tissue-specific piRNA sequences across all tissues analyzed increased the relative fraction of piRNAs for all tissues expressed (Fig. 3c,d). Thus, as parts of the small non-coding RNA transcriptome are significantly understudied, we provide this resource to the research community for studying sncRNA-related genetic and epigenetic regulation in cancer using the NCI-60 cell models.
Figure 3. sncRNA distribution by tissue type.
(a) Relative fraction of sncRNA species detected per tissue type. (b) Average fraction of currently annotated (blue) and novel unannotated (red) miRNA per tissue type. (c) Relative fraction of tissue-specific unique sncRNA sequences detected per tissue type. (d) Fraction of tissue-specific unique sncRNA species.
Methods
Cell line and sequencing information
Cell line doubling times were obtained directly from the National Institutes of Health NCI (https://dtp.cancer.gov/discovery_development/nci-60/cell_list.htm), and year-of-origin information refers to data of first publication containing the cell line (Table 1 (available online only)). Cell lines were obtained directly from the National Cancer Institute (NCI), were thawed and passaged twice precisely before total RNA was manually extracted using phenol-chloroform protocols from all cell lines using Trizol reagent (Invitrogen, CA, USA). 5,000 ng of extracted RNA per sample was used for sequencing input. Sequencing was performed in accordance with The Cancer Genome Atlas miRNA sequencing protocol (described by Chu et al.15). Briefly, after ligation to adaptors, 15 cycles of PCR was performed for amplification (98 °C-15 s, 62 °C-30 s and 72 °C-15 s), followed by 5 min at 72 °C. Small RNA exclusion was performed using gel extraction on a 3% MetaPhor Agarose gel (Lonza Inc., Basel, Switzerland), selecting species shorter that 200 nucleotides in order to enrich for targets optimized at 22 nucleotides in length, and was subsequently ethanol-precipitated. Library quality was confirmed by analysis on the Agilent Bioanalyzer DNA1000 chip (Agilent Technologies). Small non-coding RNA sequencing was performed on the Illumina HiSeq2500 platform at the Michael Smith Genome Sciences Centre at the BC Cancer Research Centre, with 8 multiplexed libraries per sequencing lane (Table 3 (available online only), Fig. 1)15,16. Data resulting from small non-coding RNA sequencing can be found on the Sequence Read Archive [Data Citation 2].
Table 3. Sequencing quality metrics for sequenced cell line.
| Tissue | External ID | Sample name (.fastq) | Total reads | Total alignments | Aligned | Total unaligned | Unaligned | Total unique | Unique | Total non-unique | Non-unique | Coverage | Avg. coverage depth | Avg. length | Avg. quality | %GC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Breast | BT-549 | MX1381-C9C07ANXX-1-CACTGT | 26,779,757 | 20,087,593 | 74.84% | 6,738,932 | 25.16% | 19,994,122 | 74.66% | 46,703 | 0.17% | 0.62% | 23.36 | 22.33 | 33.02 | 47.70% |
| HS-578T | MX1383-C9C07ANXX-3-GGGGTT | 24,961,809 | 20,875,168 | 83.42% | 4,137,590 | 16.58% | 20,773,358 | 83.22% | 50,861 | 0.20% | 0.35% | 41.95 | 22.13 | 34.19 | 45.13% | |
| MCF-7 | MX1384-C9C07ANXX-4-CTGGGT | 43,741,142 | 32,170,069 | 73.27% | 11,690,617 | 26.73% | 31,931,263 | 73.00% | 119,262 | 0.27% | 0.81% | 28.46 | 22.27 | 33.55 | 42.37% | |
| MDA-MB-231 | MX1384-C9C07ANXX-4-GCCGGT | 16,798,227 | 11,552,251 | 68.68% | 5,261,551 | 31.32% | 11,521,123 | 68.59% | 15,553 | 0.09% | 0.43% | 19.43 | 22.45 | 33.25 | 46.01% | |
| T-47D | MX1387-C9C07ANXX-7-CTAAGG | 14,269,847 | 9,004,918 | 63.04% | 5,274,372 | 36.96% | 8,986,041 | 62.97% | 9,434 | 0.07% | 0.48% | 13.58 | 22.4 | 32.16 | 47.83% | |
| CNS | SF-268 | MX1386-C9C07ANXX-6-TAGTTG | 35,537,775 | 32,273,096 | 90.47% | 3,385,641 | 9.53% | 32,031,463 | 90.13% | 120,671 | 0.34% | 0.28% | 84.29 | 22.35 | 34.68 | 46.13% |
| SF-295 | MX1386-C9C07ANXX-6-CCGGTG | 26,519,319 | 23,694,543 | 89.10% | 2,890,215 | 10.90% | 23,563,806 | 88.86% | 65,298 | 0.25% | 0.24% | 70.66 | 22.39 | 34.43 | 42.31% | |
| SF-539 | MX1386-C9C07ANXX-6-ATCGTG | 25,500,558 | 23,012,418 | 90.00% | 2,549,670 | 10.00% | 22,889,448 | 89.76% | 61,440 | 0.24% | 0.25% | 65.79 | 22.25 | 34.49 | 46.43% | |
| SNB-19 | MX1387-C9C07ANXX-7-GCGTGG | 32,717,872 | 21,567,743 | 65.75% | 11,204,285 | 34.25% | 21,459,531 | 65.59% | 54,056 | 0.17% | 0.85% | 18.3 | 22.44 | 32.24 | 43.60% | |
| SNB-75 | MX1387-C9C07ANXX-7-CATGGG | 12,680,815 | 7,882,308 | 62.10% | 4,805,816 | 37.90% | 7,867,693 | 62.04% | 7,306 | 0.06% | 0.37% | 15.13 | 22.2 | 32.05 | 45.20% | |
| U251 | MX1387-C9C07ANXX-7-ATTCCG | 20,845,308 | 13,006,365 | 62.30% | 7,858,915 | 37.70% | 12,966,435 | 62.20% | 19,958 | 0.10% | 0.58% | 16.29 | 22.47 | 32.17 | 43.76% | |
| Colon | COLO 205 | MX1381-C9C07ANXX-1-TCAAGT | 18,351,752 | 13,907,298 | 75.66% | 4,466,918 | 24.34% | 13,862,401 | 75.54% | 22,433 | 0.12% | 0.44% | 22.77 | 22.23 | 33.19 | 45.58% |
| HCC2998 | MX1382-C9C07ANXX-2-GTAGCC | 22,418,943 | 16,741,250 | 74.53% | 5,710,380 | 25.47% | 16,675,921 | 74.38% | 32,642 | 0.15% | 0.88% | 13.4 | 21.83 | 32.83 | 47.12% | |
| HCT-116 | MX1382-C9C07ANXX-2-TACAAG | 19,321,624 | 13,822,479 | 71.42% | 5,521,561 | 28.58% | 13,777,662 | 71.31% | 22,401 | 0.12% | 0.54% | 18.43 | 22.23 | 33.06 | 46.02% | |
| HCT-15 | MX1382-C9C07ANXX-2-ATGTTT | 22,372,577 | 15,816,346 | 70.57% | 6,585,278 | 29.43% | 15,758,281 | 70.44% | 29,018 | 0.13% | 0.62% | 18.15 | 22.24 | 32.92 | 46.00% | |
| HT-29 | MX1383-C9C07ANXX-3-CAAGTT | 43,295,013 | 36,704,351 | 84.42% | 6,746,834 | 15.58% | 36,392,459 | 84.06% | 155,720 | 0.36% | 0.64% | 40.79 | 22.23 | 34.19 | 45.58% | |
| KM12 | MX1383-C9C07ANXX-3-GTCCTT | 21,097,776 | 17,022,531 | 80.52% | 4,108,873 | 19.48% | 16,955,321 | 80.37% | 33,582 | 0.16% | 0.41% | 29.68 | 22.35 | 34.03 | 46.33% | |
| SW-620 | MX1387-C9C07ANXX-7-TTGCGG | 34,094,755 | 22,349,352 | 65.38% | 11,803,916 | 34.62% | 22,232,420 | 65.21% | 58,419 | 0.17% | 0.92% | 17.57 | 22.46 | 32.21 | 46.44% | |
| Leukemia | CCRF-CEM | MX1381-C9C07ANXX-1-GATCTG | 16,675,728 | 12,545,480 | 75.12% | 4,148,470 | 24.88% | 12,509,054 | 75.01% | 18,204 | 0.11% | 0.40% | 22.55 | 22.48 | 33.4 | 49.05% |
| HL-60(TB) | MX1382-C9C07ANXX-2-TGCTTT | 14,448,374 | 9,588,877 | 66.29% | 4,870,207 | 33.71% | 9,567,467 | 66.22% | 10,700 | 0.07% | 0.51% | 13.5 | 22.12 | 32.72 | 49.74% | |
| K-562 | MX1383-C9C07ANXX-3-TCGCTT | 35,495,593 | 28,500,403 | 80.03% | 7,089,711 | 19.97% | 28,311,603 | 79.76% | 94,279 | 0.27% | 0.60% | 34.46 | 22.43 | 34.08 | 48.92% | |
| MOLT-4 | MX1384-C9C07ANXX-4-GAGAGT | 17,384,261 | 12,413,016 | 71.30% | 4,989,141 | 28.70% | 12,377,237 | 71.20% | 17,883 | 0.10% | 0.49% | 18.25 | 22.49 | 33.61 | 48.27% | |
| RPMI 8226 | MX1385-C9C07ANXX-5-AGGAAT | 17,460,565 | 14,241,842 | 81.43% | 3,242,340 | 18.57% | 14,194,637 | 81.30% | 23,588 | 0.14% | 0.46% | 22.37 | 22.47 | 34.06 | 48.91% | |
| SR | MX1387-C9C07ANXX-7-CCACTC | 12,064,850 | 6,751,739 | 55.92% | 5,318,416 | 44.08% | 6,741,131 | 55.87% | 5,303 | 0.04% | 0.48% | 10.23 | 22.48 | 31.62 | 47.70% | |
| Melanoma | LOX-IMVI | MX1383-C9C07ANXX-3-CCTATT | 31,657,831 | 25,924,121 | 81.64% | 5,812,011 | 18.36% | 25,767,653 | 81.39% | 78,167 | 0.25% | 0.47% | 39.9 | 22.38 | 34.1 | 45.79% |
| M14 | MX1383-C9C07ANXX-3-GTTTGT | 16,456,810 | 13,115,289 | 79.57% | 3,361,434 | 20.43% | 13,075,479 | 79.45% | 19,897 | 0.12% | 0.32% | 29.6 | 22.45 | 34.08 | 47.29% | |
| MALME-3M | MX1383-C9C07ANXX-3-AGATGT | 97,640 | 42,454 | 43.48% | 55,187 | 56.52% | 42,452 | 43.48% | 1 | 0% | 0.01% | 4.85 | 22.33 | 29.76 | 46.02% | |
| MDA-MB-435 | MX1384-C9C07ANXX-4-TATCGT | 16,462,112 | 11,741,084 | 71.22% | 4,736,983 | 28.78% | 11,709,183 | 71.13% | 15,946 | 0.10% | 0.50% | 16.9 | 22.36 | 33.4 | 47.73% | |
| SK-MEL-2 | MX1386-C9C07ANXX-6-TGAGTG | 30,951,870 | 27,402,647 | 88.25% | 3,636,213 | 11.75% | 27,228,840 | 87.97% | 86,817 | 0.28% | 0.32% | 61.56 | 22.51 | 34.52 | 46.50% | |
| SK-MEL-28 | MX1386-C9C07ANXX-6-CGCCTG | 14,712,878 | 12,743,244 | 86.48% | 1,988,450 | 13.52% | 12,705,638 | 86.36% | 18,790 | 0.13% | 0.22% | 41.57 | 22.26 | 34.24 | 46.57% | |
| SK-MEL-5 | MX1386-C9C07ANXX-6-GCCATG | 12,394,333 | 10,855,860 | 87.48% | 1,552,173 | 12.52% | 10,828,474 | 87.37% | 13,686 | 0.11% | 0.19% | 41.26 | 22.5 | 34.4 | 46.38% | |
| UACC-257 | MX1388-C9C07ANXX-8-AGCTAG | 46,062 | 9,213 | 20.00% | 36,849 | 80.00% | 9,213 | 20.00% | 0 | 0% | 0% | 2.13 | 22.17 | 26.55 | 46.63% | |
| UACC-62 | MX1388-C9C07ANXX-8-GTATAG | 22,270,501 | 15,834,593 | 70.97% | 6,465,114 | 29.03% | 15,776,225 | 70.84% | 29,162 | 0.13% | 0.62% | 18.26 | 22.24 | 32.7 | 46.13% | |
| NSCLC | A549 | MX1381-C9C07ANXX-1-GCCTAA | 24,542,337 | 18,533,593 | 75.35% | 6,048,721 | 24.65% | 18,453,696 | 75.19% | 39,920 | 0.16% | 0.53% | 25.18 | 22.39 | 33.18 | 45.04% |
| EKVX | MX1382-C9C07ANXX-2-AAGCTA | 18,358,000 | 13,056,340 | 71.01% | 5,321,501 | 28.99% | 13,016,669 | 70.90% | 19,830 | 0.11% | 0.46% | 20.08 | 22.13 | 32.99 | 45.71% | |
| HOP 62 | MX1382-C9C07ANXX-2-GCATTT | 22,874,592 | 16,227,434 | 70.81% | 6,678,209 | 29.19% | 16,165,375 | 70.67% | 31,008 | 0.14% | 0.58% | 19.83 | 22.18 | 32.86 | 45.15% | |
| HOP 92 | MX1382-C9C07ANXX-2-CGTACG | 16,671,005 | 11,307,213 | 67.74% | 5,378,661 | 32.26% | 11,277,492 | 67.65% | 14,852 | 0.09% | 0.49% | 16.63 | 22.16 | 32.7 | 45.80% | |
| NCI-H226 | MX1384-C9C07ANXX-4-TCTTCT | 18,745,702 | 12,999,215 | 69.24% | 5,766,399 | 30.76% | 12,959,420 | 69.13% | 19,883 | 0.11% | 0.52% | 17.92 | 22.38 | 33.28 | 47.25% | |
| NCI-H23 | MX1384-C9C07ANXX-4-CTATCT | 19,234,429 | 13,677,426 | 71.00% | 5,578,694 | 29.00% | 13,634,066 | 70.88% | 21,669 | 0.11% | 0.51% | 19.47 | 22.44 | 33.36 | 48.52% | |
| NCI-H322M | MX1384-C9C07ANXX-4-GATGCT | 40,017,514 | 30,184,128 | 75.16% | 9,938,560 | 24.84% | 29,974,031 | 74.90% | 104,923 | 0.26% | 0.95% | 22.98 | 22.47 | 33.58 | 45.76% | |
| NCI-H460 | MX1385-C9C07ANXX-5-AGCGCT | 12,712,247 | 9,871,928 | 77.57% | 2,851,889 | 22.43% | 9,848,794 | 77.47% | 11,564 | 0.09% | 0.32% | 22.39 | 22.38 | 33.72 | 47.50% | |
| NCI-H522 | MX1385-C9C07ANXX-5-CGGCCT | 37,514,479 | 30,982,049 | 82.29% | 6,643,642 | 17.71% | 30,759,900 | 81.99% | 110,937 | 0.30% | 0.55% | 40.76 | 22.53 | 34.02 | 44.85% | |
| Ovarian | IGR-OV1 | MX1383-C9C07ANXX-3-AGTCTT | 28,718,340 | 23,886,771 | 82.95% | 4,897,725 | 17.05% | 23,754,573 | 82.72% | 66,042 | 0.23% | 0.42% | 41.25 | 22.41 | 34.16 | 46.10% |
| NCI/ADR-RES | MX1384-C9C07ANXX-4-ATCAGT | 22,291,994 | 16,348,364 | 73.20% | 5,974,630 | 26.80% | 16,286,408 | 73.06% | 30,956 | 0.14% | 0.54% | 21.99 | 22.37 | 33.49 | 47.02% | |
| OVCAR-3 | MX1385-C9C07ANXX-5-AATTAT | 29,981,152 | 25,036,208 | 83.26% | 5,017,926 | 16.74% | 24,890,371 | 83.02% | 72,855 | 0.24% | 0.46% | 38.94 | 22.41 | 34.16 | 47.68% | |
| OVCAR-4 | MX1385-C9C07ANXX-5-CCGTAT | 28,168,179 | 22,957,608 | 81.29% | 5,271,643 | 18.71% | 22,835,575 | 81.07% | 60,961 | 0.22% | 0.43% | 38.31 | 22.39 | 34.02 | 47.21% | |
| OVCAR-5 | MX1385-C9C07ANXX-5-TAGGAT | 24,989,662 | 20,898,933 | 83.43% | 4,141,256 | 16.57% | 20,797,961 | 83.23% | 50,445 | 0.20% | 0.40% | 37.13 | 22.3 | 34.22 | 45.64% | |
| OVCAR-8 | MX1385-C9C07ANXX-5-ATAGAT | 21,340,411 | 17,484,019 | 81.76% | 3,891,733 | 18.24% | 17,413,389 | 81.60% | 35,289 | 0.17% | 0.35% | 36.62 | 22.44 | 34.22 | 46.25% | |
| SK-OV-3 | MX1386-C9C07ANXX-6-AAAATG | 35,063,352 | 31,836,638 | 90.46% | 3,344,843 | 9.54% | 31,600,666 | 90.12% | 117,843 | 0.34% | 0.30% | 76.21 | 22.49 | 34.54 | 46.04% | |
| Prostate | DU-145 | MX1382-C9C07ANXX-2-CTGATC | 42,719,250 | 30,543,575 | 71.24% | 12,284,018 | 28.76% | 30,327,141 | 70.99% | 108,091 | 0.25% | 1.01% | 21.68 | 22.17 | 32.93 | 42.80% |
| PC-3 | MX1385-C9C07ANXX-5-GCTCAT | 28,554,855 | 23,674,991 | 82.68% | 4,944,659 | 17.32% | 23,545,544 | 82.46% | 64,652 | 0.23% | 0.44% | 38.54 | 22.42 | 34.15 | 46.08% | |
| Renal | 786-0 | MX1381-C9C07ANXX-1-CGTGAT | 28,239,458 | 21,417,620 | 75.65% | 6,875,229 | 24.35% | 21,310,920 | 75.47% | 53,309 | 0.19% | 0.53% | 28.79 | 22.32 | 33.21 | 45.18% |
| A498 | MX1381-C9C07ANXX-1-ACATCG | 23,983,341 | 18,160,244 | 75.56% | 5,861,378 | 24.44% | 18,083,724 | 75.40% | 38,239 | 0.16% | 0.49% | 26.57 | 22.26 | 33.14 | 44.19% | |
| ACHN | MX1381-C9C07ANXX-1-TGGTCA | 26,253,335 | 20,447,816 | 77.70% | 5,854,225 | 22.30% | 20,350,495 | 77.52% | 48,615 | 0.19% | 0.50% | 29.38 | 22.42 | 33.4 | 44.66% | |
| CAKI-1 | MX1381-C9C07ANXX-1-ATTGGC | 20,365,416 | 15,568,695 | 76.31% | 4,825,051 | 23.69% | 15,512,066 | 76.17% | 28,299 | 0.14% | 0.38% | 29.39 | 22.37 | 33.42 | 44.53% | |
| RXF393 | MX1386-C9C07ANXX-6-CTTTTG | 29,863,988 | 26,677,423 | 89.05% | 3,269,280 | 10.95% | 26,512,156 | 88.78% | 82,552 | 0.28% | 0.34% | 57.33 | 22.42 | 34.42 | 48.85% | |
| SN12C | MX1387-C9C07ANXX-7-TGTTGG | 23,377,833 | 15,428,844 | 65.88% | 7,976,649 | 34.12% | 15,373,563 | 65.76% | 27,621 | 0.12% | 0.67% | 16.45 | 22.28 | 32.21 | 45.57% | |
| TK-10 | MX1387-C9C07ANXX-7-TTCTCG | 26,774,556 | 17,534,744 | 65.36% | 9,275,498 | 34.64% | 17,463,407 | 65.22% | 35,651 | 0.13% | 1.16% | 10.66 | 21.92 | 31.69 | 49.55% | |
| UO-31 | MX1388-C9C07ANXX-8-TCTGAG | 12,737,055 | 8,368,476 | 65.64% | 4,376,740 | 34.36% | 8,352,163 | 65.57% | 8,152 | 0.06% | 0.34% | 17.82 | 22.44 | 32.53 | 45.42% | |
|
MEAN |
23,457,233 | 18,073,055 | 74.34% | 5,429,370 | 25.66% | 17,982,756 | 74.18% | 45,107 | 0.16% | 0.49% | 29 | 22.34 | 33.25 | 46.26% | ||
| Standard Deviation | 9,294,225 | 7,957,399 | 11.69% | 2,519,478 | 11.69% | 7,888,112 | 11.63% | 35,871 | 0.08% | 0.22% | 17 | 0.14 | 1.28 | 1.64% | ||
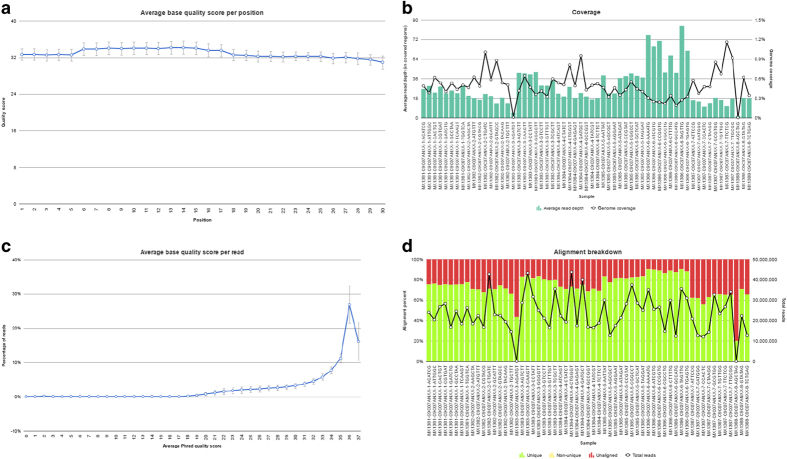
Pre-processing and small non-coding RNA species detection
Small-RNA sequencing data was analyzed according to published protocols17. In order to extract information for the sncRNA species of interest, unaligned reads (in FASTQ format) were trimmed for adaptors (Cutadapt v1.7.1) and based on sequencing quality (‘trim bases’ from Partek Flow v6.0.17.0614) to reach a Phred quality score ≥20 (Fig. 4a–d). FASTQ files were then aligned using the Spliced Transcripts Alignment to a Reference (STAR v2.4.1d) aligner to the human genome (hg38)18. Quantification algorithms (featureCounts v1.4.6 (ref. 19) were applied using chromosomal location annotations for known miRNA (Mirbase v.21 (ref. 20), piRNA (piRNAbank v.2 (ref. 21), snoRNA (Ensembl v.84 (ref. 22), and snRNA (Ensembl v.84 (ref. 22) locations12. Detection of novel miRNA is performed using the miRDeep2 algorithm (v2.0.0.5), which considers the relative free energy of miRNAs and their random folding P-values13. Chromosomal position of expressed small RNAs was plotted against and hg38 karyotype obtained from UCSC Genome Browser (Fig. 2). According to OASIS sncRNA software recommendation (v2.0), sncRNA species were considered expressed if the total reads across all samples considered summed to ≥5 reads12. Data resulting from species quantification can be found in Data Citation 1.
Figure 4. Sequencing and mapping quality.
(a) Phred quality score per sncRNA base position. (b) Genome-wide read depth (column) and genome coverage (line) per sample. (c) Fraction of sequencing reads per Phred score. (d) Percentage of total reads aligned (unique: green, unaligned: red).
Normalization and quantification
Raw reads were scaled/normalized using reads per kilobase exon per million mapped reads (RPKM) method23, and expression correlation matrices were created using Pearson scores with unsupervised hierarchal clustering performed using one-minus-Pearson correlation scores (Fig. 5). For validation of sncRNA expression, we then correlated miRNA species present in two published microarray cohorts of the NCI-60 cell lines. For the 50 (of the 59) cell lines also present in the Sanger Cell Line Database24 (http://www.cancerrxgene.org/translation/CellLine), raw reads from each unique sequence were correlated with expression of the sequence previously detected by microarray by rank-normalized Spearman`s correlations (Table 4 (available online only)), and performed a similar analysis against all cell lines present in the cohort described by Sokilde et al.5.
Figure 5.
Data Records
Raw unaligned sequencing reads (in FASTQ file format) are available through the Sequence Read Archive (Data Citation 2). Raw sequencing file names (in FASTQ format) are listed in Table 3 (available online only). A summary of raw sequencing reads for each detected small RNA species are available at through Figshare (Data Citation 1).
Technical Validation
High-throughput sequencing allows for direct in-depth analyses of the human genome, recently revealing a critical role for the expression of the non-coding transcriptome in both genetic and epigenetic regulatory processes.
Sequencing quality control
We examined only high-confidence reads from miRNA sequencing. Samples were sequenced to an average depth of 22.34±0.14 (mean±s.d.; Table 3 (available online only), Fig. 4b). In order to assure only the calling of high-quality sequencing reads, we filtered detected reads to only to include Phred scores ≥20. On average, samples had a Phred score of 33.24±1.28 (Table 3 (available online only), Fig. 4c). Additionally, reads for each sample had an average percent GC content of 46.26±1.6% (Table 3) (available online only). Unsupervised hierarchical clustering and similarity (one-minus-Spearman correlation) of normalized reads revealed relative similarity of sncRNA expression profiles across all cell lines and tissue types analyzed (Fig. 5).
miRNA detection validation
In order to validate the detection of the sncRNA species in these cell lines, we correlated the raw reads per miRNA detected with corresponding miRNA detected by microarray24,25. This analysis was performed for the 50 NCI-60 cell lines present in the Sanger Cell Line miRNA Normalized Data from the Broad Institute (http://www.broadinstitute.org/cgi-bin/cancer/datasets.cgi; File name: Sanger_miR_data1.pn.cn.matlab2.res). Using Spearman’s Rank-Order correlation, we analyzed the correlation of this RMA-normalized miRNA expression to reads obtained from sequencing this cell line panel. Expression of miRNAs in all lines analyzed correlated significantly between sequencing and microarray analysis (Table 4 (available online only); P-values <0.0001, rmean=0.67). Similarly, we correlated sequencing-detected miRNA expression against a complete NCI-60 microarray cohort described by Sokilde et al.5. In this study, profiling was performed on the LNA-enhanced mercury Dx 9.2 microarray platform, and data was log2-normalized after pre-processing (Table 4; P-value range <0.0001–0.0647, rmean=0.28). Microarray data from multiple platforms was compared to sequencing data presented here in order to de-emphasize platform bias and illustrate the need for comprehensive profiling when considering small RNA expression26.
Table 4. Spearman correlation of sequenced NCI-60 cell lines to published miRNA microarray expression levels.
|
Broad |
Sokilde
et al. |
|||||
|---|---|---|---|---|---|---|
| Cell line | r | 95% confidence interval | p (two-tailed) | r | 95% confidence interval | p (two-tailed) |
| 786-0 | 0.7064 | 0.651–0.7543 | <0.0001 | 0.3252 | 0.1976–0.4421 | <0.0001 |
| A598 | n/a | n/a | n/a | 0.2644 | 0.1327–0.3869 | <0.0001 |
| A549 | 0.6809 | 0.6218–0.7324 | <0.0001 | 0.1815 | 0.04619–0.3103 | 0.0071 |
| ACHN | 0.6959 | 0.6389–0.7453 | <0.0001 | 0.27 | 0.1387–0.392 | <0.0001 |
| BT-549 | n/a | n/a | n/a | 0.3202 | 0.1922–0.4376 | <0.0001 |
| CAKI-1 | 0.7004 | 0.6441–0.7492 | <0.0001 | 0.1604 | 0.02453–0.2905 | 0.0175 |
| CCRF-CEM | n/a | n/a | n/a | 0.1251 | -0.01157–0.2571 | 0.0647 |
| COLO-205 | 0.6766 | 0.6169–0.7287 | <0.0001 | 0.2974 | 0.1677–0.4169 | <0.0001 |
| DU-145 | 0.726 | 0.6735–0.7711 | <0.0001 | 0.3293 | 0.2019–0.4457 | <0.0001 |
| EKVX | 0.6248 | 0.558–0.6836 | <0.0001 | 0.2634 | 0.1317–0.386 | <0.0001 |
| HCC2998 | 0.6512 | 0.5879–0.7066 | <0.0001 | 0.3391 | 0.2125–0.4545 | <0.0001 |
| HCT-116 | 0.7019 | 0.6458–0.7505 | <0.0001 | 0.3393 | 0.2127–0.4547 | <0.0001 |
| HCT-15 | 0.7193 | 0.6658–0.7654 | <0.0001 | 0.2704 | 0.1391–0.3924 | <0.0001 |
| HL-60 | 0.6358 | 0.5704–0.6932 | <0.0001 | 0.2533 | 0.1211–0.3767 | 0.0002 |
| HOP-62 | 0.6899 | 0.632–0.7401 | <0.0001 | 0.2566 | 0.1245–0.3798 | 0.0001 |
| HOP-92 | 0.6576 | 0.5951–0.7121 | <0.0001 | 0.2288 | 0.09537–0.3542 | 0.0006 |
| HS-578-T | 0.6283 | 0.5619–0.6867 | <0.0001 | 0.2857 | 0.1554–0.4063 | <0.0001 |
| HT-29 | 0.6956 | 0.6386–0.7451 | <0.0001 | 0.2875 | 0.1572–0.4079 | <0.0001 |
| IGR-OV1 | 0.6986 | 0.642–0.7476 | <0.0001 | 0.3065 | 0.1774–0.4251 | <0.0001 |
| K-562 | n/a | n/a | n/a | 0.2657 | 0.1341–0.3881 | <0.0001 |
| KM12 | n/a | n/a | n/a | 0.3265 | 0.1989–0.4432 | <0.0001 |
| LOX-IMVI | 0.6699 | 0.6091–0.7228 | <0.0001 | 0.2637 | 0.132–0.3862 | <0.0001 |
| M14 | 0.6938 | 0.6365–0.7435 | <0.0001 | 0.2585 | 0.1265–0.3815 | 0.0001 |
| MALME-3M | 0.5848 | 0.513–0.6485 | <0.0001 | 0.3069 | 0.1779–0.4256 | <0.0001 |
| MCF-7 | 0.7235 | 0.6707–0.769 | <0.0001 | 0.3271 | 0.1996–0.4438 | <0.0001 |
| MDA-MB-231 | 0.7125 | 0.658–0.7596 | <0.0001 | 0.1837 | 0.04847–0.3123 | 0.0064 |
| MDA-MB-435 | 0.7081 | 0.653–0.7558 | <0.0001 | 0.2907 | 0.1607–0.4109 | <0.0001 |
| MOLT-4 | 0.7034 | 0.6476–0.7518 | <0.0001 | 0.2809 | 0.1502–0.402 | <0.0001 |
| NCI/ADR-RES | n/a | n/a | n/a | 0.3046 | 0.1754–0.4234 | <0.0001 |
| NCI-H226 | 0.6901 | 0.6323–0.7403 | <0.0001 | 0.3613 | 0.2365–0.4743 | <0.0001 |
| NCI-H23 | 0.631 | 0.565–0.689 | <0.0001 | 0.2649 | 0.1332–0.3873 | <0.0001 |
| NCI-H322M | n/a | n/a | n/a | 0.3624 | 0.2378–0.4754 | <0.0001 |
| NCI-H460 | 0.6395 | 0.5746–0.6964 | <0.0001 | 0.2794 | 0.1486–0.4006 | <0.0001 |
| NCI-H522 | 0.7047 | 0.649–0.7529 | <0.0001 | 0.2856 | 0.1552–0.4062 | <0.0001 |
| OVCAR-3 | 0.6951 | 0.638–0.7446 | <0.0001 | 0.3629 | 0.2383–0.4758 | <0.0001 |
| OVCAR-4 | 0.6731 | 0.6128–0.7256 | <0.0001 | 0.3422 | 0.2158–0.4573 | <0.0001 |
| OVCAR-5 | 0.7101 | 0.6552–0.7575 | <0.0001 | 0.2952 | 0.1655–0.415 | <0.0001 |
| OVCAR-8 | 0.6613 | 0.5994–0.7154 | <0.0001 | 0.3407 | 0.2142–0.456 | <0.0001 |
| PC-3 | 0.6373 | 0.5721–0.6945 | <0.0001 | 0.2893 | 0.1592–0.4096 | <0.0001 |
| RPMI-8226 | 0.6704 | 0.6098–0.7233 | <0.0001 | 0.2641 | 0.1325–0.3866 | <0.0001 |
| RXF393 | 0.5656 | 0.4915–0.6316 | <0.0001 | 0.3085 | 0.1797–0.427 | <0.0001 |
| SF-268 | 0.6788 | 0.6193–0.7305 | <0.0001 | 0.2812 | 0.1505–0.4022 | <0.0001 |
| SF-295 | 0.6831 | 0.6243–0.7343 | <0.0001 | 0.2986 | 0.1691–0.418 | <0.0001 |
| SF-539 | 0.6623 | 0.6005–0.7162 | <0.0001 | 0.3037 | 0.1745–0.4227 | <0.0001 |
| SK-MEL-2 | 0.6294 | 0.5632–0.6876 | <0.0001 | 0.2851 | 0.1546–0.4057 | <0.0001 |
| SK-MEL-28 | 0.6765 | 0.6167–0.7286 | <0.0001 | 0.2867 | 0.1564–0.4072 | <0.0001 |
| SK-MEL-5 | 0.6095 | 0.5408–0.6702 | <0.0001 | 0.3681 | 0.2439–0.4804 | <0.0001 |
| SK-OV-3 | 0.658 | 0.5956–0.7125 | <0.0001 | 0.2613 | 0.1295–0.384 | <0.0001 |
| SN12C | 0.7182 | 0.6645–0.7644 | <0.0001 | 0.321 | 0.193–0.4382 | <0.0001 |
| SNB-19 | 0.6126 | 0.5441–0.6729 | <0.0001 | 0.2332 | 0.09998–0.3583 | 0.0005 |
| SNB-75 | n/a | n/a | n/a | 0.294 | 0.1642–0.4139 | <0.0001 |
| SR | n/a | n/a | n/a | 0.1599 | 0.02397–0.29 | 0.0179 |
| SW-620 | 0.7124 | 0.6578–0.7595 | <0.0001 | 0.3805 | 0.2575–0.4915 | <0.0001 |
| T-47D | 0.6827 | 0.6238–0.7339 | <0.0001 | 0.3928 | 0.2709–0.5023 | <0.0001 |
| TK-10 | 0.7137 | 0.6593–0.7606 | <0.0001 | 0.1981 | 0.06341–0.3258 | 0.0032 |
| U-251 | 0.6242 | 0.5573–0.6831 | <0.0001 | 0.2862 | 0.1558–0.4067 | <0.0001 |
| UACC-257 | 0.4873 | 0.4049–0.5618 | <0.0001 | 0.2332 | 0.09994–0.3583 | 0.0005 |
| UACC-62 | 0.676 | 0.6161–0.7281 | <0.0001 | 0.3074 | 0.1785–0.426 | <0.0001 |
| UO-31 | n/a | n/a | n/a | 0.2975 | 0.1672–0.4175 | <0.0001 |
Additional information
How to cite this article: Marshall, E. A. et al. Small non-coding RNA transcriptome of the NCI-60 cell line panel. Sci. Data 4:170157 doi: 10.1038/sdata.2017.157 (2017).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by grants from the Canadian Institutes for Health Research (CIHR FRN-143345), the National Institute of Health (NIH-1R01HD089713-01) and scholarships from CIHR and the University of British Columbia. The authors would like to thank May Zhang, Miwa Suzuki and Emma Conway for assistance with cell culture and RNA harvesting. The authors would also like to thank Monica Fuss for assistance in data analysis.
Footnotes
The authors declare no competing financial interests.
Data Citations
- Marshall E. A. 2017. figshare. https://doi.org/10.6084/m9.figshare.c.3811156
- 2017. NCBI Sequence Read Archive. SRP109305
References
- Lorenzi P. L. et al. DNA fingerprinting of the NCI-60 cell line panel. Molecular cancer therapeutics 8, 713–724 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschke A. V. et al. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer research 63, 8634–8647 (2003). [PubMed] [Google Scholar]
- Adams S. et al. HLA class I and II genotype of the NCI-60 cell lines. Journal of translational medicine 3, 11 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R. H. The NCI60 human tumour cell line anticancer drug screen. Nature reviews Cancer 6, 813–823 (2006). [DOI] [PubMed] [Google Scholar]
- Sokilde R. et al. Global microRNA analysis of the NCI-60 cancer cell panel. Molecular cancer therapeutics 10, 375–384 (2011). [DOI] [PubMed] [Google Scholar]
- Kohn K. W. et al. Gene expression profiles of the NCI-60 human tumor cell lines define molecular interaction networks governing cell migration processes. PLoS ONE 7, e35716 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J., Peruzzi P. P. & Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends in molecular medicine 20, 460–469 (2014). [DOI] [PubMed] [Google Scholar]
- Londin E. et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proceedings of the National Academy of Sciences of the United States of America 112, E1106–E1115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K. W. et al. Piwi-interacting RNAs in cancer: emerging functions and clinical utility. Molecular cancer 15, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannoor K., Liao J. & Jiang F. Small nucleolar RNAs in cancer. Biochimica et biophysica acta 1826, 121–128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majem B., Rigau M., Reventos J. & Wong D. T. Non-coding RNAs in saliva: emerging biomarkers for molecular diagnostics. International journal of molecular sciences 16, 8676–8698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capece V. et al. Oasis: online analysis of small RNA deep sequencing data. Bioinformatics 31, 2205–2207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander M. R. et al. Discovering microRNAs from deep sequencing data using miRDeep. Nature biotechnology 26, 407–415 (2008). [DOI] [PubMed] [Google Scholar]
- Friedlander M. R., Mackowiak S. D., Li N., Chen W. & Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic acids research 40, 37–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu A. et al. Large-scale profiling of microRNAs for The Cancer Genome Atlas. Nucleic acids research 44, e3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker-Santos D. D. et al. Developmental transcription factor NFIB is a putative target of oncofetal miRNAs and is associated with tumour aggressiveness in lung adenocarcinoma. The Journal of pathology 240, 161–172 (2016). [DOI] [PubMed] [Google Scholar]
- Martinez V. D. et al. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Scientific reports 5, 10423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K. & Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- Kozomara A. & Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic acids research 42, D68–D73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai Lakshmi S. & Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic acids research 36, D173–D177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aken B. L. et al. The Ensembl gene annotation system. Database 2016, baw093–baw093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Yang W. et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic acids research 41, D955–D961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enfield K. S. et al. MicroRNA gene dosage alterations and drug response in lung cancer. Journal of biomedicine & biotechnology 2011, 474632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik S. K. et al. Expression of microRNAs in the NCI-60 cancer cell-lines. PLoS ONE 7, e49918 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Marshall E. A. 2017. figshare. https://doi.org/10.6084/m9.figshare.c.3811156
- 2017. NCBI Sequence Read Archive. SRP109305