Abstract
Rice (Oryza sativa L.) is one of the most important staple food in the world. To meet the increasing demand for food, a strategy for improving rice yield is needed. Alleles of wild relatives are useful because they confer adaptation to plants under diverse harsh environments and have the potential to improve rice. O. barthii is a wild rice species endemic to Africa and the known progenitor of the African cultivated rice, O. glaberrima. To explore the genetic potential of the O. barthii as a genetic resource, 40 chromosome segment substitution lines (CSSL) of O. barthii in the background of the elite japonica cultivar Koshihikari were developed and evaluated to identify QTLs associated with 10 traits related to flag leaf morphology, grain yield and other agronomic traits. More than 90% of the entire genome of the donor parent was represented in contiguous or overlapping chromosome segments in the CSSLs. Evaluation of the CSSLs for several agriculturally important traits identified candidate chromosome segments that harbors QTLs associated with yield and yield-related traits. These results suggest that alleles from O. barthii might be used as a novel genetic resource for improving the yield-related traits in cultivars of O. sativa.
Keywords: Oryza barthii, chromosome segment substitution lines (CSSLs)
Introduction
The wild species of the genus Oryza serve as a virtually untapped reservoir of genetic diversity that can be used to improve rice production. The genus is composed of two cultivated (O. sativa and O. glaberrima) and 22 wild species, that represents between 15 to 25 million years of evolutionary diversification (Vaughan 1994). Evidence suggests that wild rice relatives may contain genes that are capable of improving both the yield and quality of the rice seeds (Tanksley et al. 1996). However, the expressions of these favorable genes are often masked by the effects of other deleterious genes (Xiao et al. 1996). The establishment of molecular markers has greatly advanced the development of genetic maps, which in turn aided in the exploitation of the genetic potential of these wild species for the improvement of yield and quality of modern rice cultivars (Ren et al. 2003). Using genetic maps, the introduction of genes from wild rice relatives with the AA genome into Asian cultivated rice has been possible, contributing in expanding rice genetic diversity. Similarly, the genome sequences of many wild rice relatives have become valuable resources for a more extensive exploitation of wild Oryza germplasm to enhance rice breeding programs (Zhang et al. 2014b). These approaches have led to the discovery of “hidden” genes from wild rice relatives, in particular, from O. rufipogon, which is the progenitor of Asian cultivated rice O. sativa (Tanksley and McCouch 1997).
Chromosome segment substitution lines (CSSLs) from interspecific hybridization represent a powerful and useful genetic resource for genome research, especially quantitative trait loci (QTL) mapping and gene cloning (Ali et al. 2010). CSSLs are generated by crossing a donor parent (wild rice or popular rice cultivars) with a recipient, usually an elite line, followed by several backcrossing to the recurrent parent. The resulting progenies possess one or few homozygous chromosome fragments of the donor genotype in the genetic background of the recurrent parent. A CSSL set covers the whole donor genome in small, contiguous or overlapping chromosome fragments (Gutierrez et al. 2010, Yano 2001). This allows the evaluation of the effect of each chromosome segments on useful agronomic traits and use of the target segments in breeding. At least 10 sets of CSSLs have been constructed in rice (Bian et al. 2010, Doi et al. 1997, Ebitani et al. 2005, Furuta et al. 2014, 2016, Kubo et al. 2002, Shim et al. 2010, Takai et al. 2007), and many agronomic QTLs have been identified and some were cloned using map-based methods (Shen and Xing 2014, Tan et al. 2007, Ujiie and Ishimaru. 2013, Yang et al. 2014, Zhang et al. 2011). However, most of these CSSLs used Asian wild and cultivated species and there has been no report of the use of African wild relatives such as O. barthii.
O. barthii is annual, wild African rice that is known to be the progenitor of the African cultivated rice species, O. glaberrima. This species have tolerance to biotic and abiotic stresses (Khush 1997). The edible seeds of this species are sometimes harvested for local use but harvests are often very limited because of its opened-shape panicle, high shattering rate and low grain number. Recently, the genome of O. barthii has been sequenced, and several genetic comparisons between O. barthii and O. glaberrima have been conducted to identify the domestication pathway of rice in Africa (Orjuela et al. 2014, Zhang et al. 2014b). However, only a few studies about the genetic diversity of these species have been performed. Based on past studies on Asian wild rice, O. barthii is also a potential resource of genetic diversity that can be tapped to increase yield of cultivated rice. Here, we report the development and evaluation of CSSLs harboring O. barthii genomic segments in the genetic background of O. sativa cv. Koshihikari. Chromosomal segments from the entire genome of O. barthii were successfully introduced into the genomic background of Koshihikari as contiguous, overlapping segments. The CSSLs exhibited significantly different phenotypes for panicle length, number of primary branches, number of grains per panicle, grain length, grain width and 100-grain weight.
Materials and Methods
Development of the BSLs
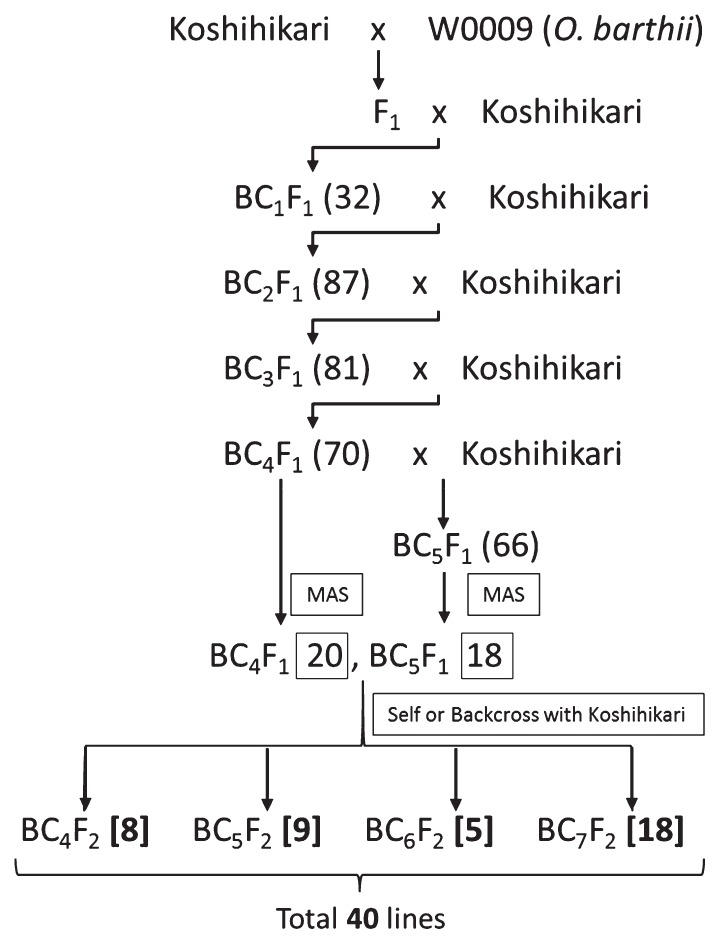
The donor parent, O. barthii Acc. W0009 and the recurrent parent O. sativa cv. Koshihikari were provided by the National Institute of Genetics (NIG), Japan. The breeding scheme used to develop the CSSLs, hereafter referred to as BSLs, is presented in Fig. 1. The BSLs were developed by first crossing cv. Koshihikari with O. barthii to produce the F1 hybrid. The resulting F1 was then backcrossed to Koshihikari to produce 32 BC1F1 plants. Successive backcrossing with the recurrent parent produced 87 BC2F1, 81 BC3F1, 70 BC4F1 and 66 BC5F1 plants. Genotyping using 129 single nucleotide polymorphisms via the AcycloPrime-FP Detection System and Fluorescence Polarization Analyzer (Perkin Elmer Life Science, Boston, MA, USA) was carried out for 70 BC4F1 and 66 BC5F1 to identify backcross lines having 1–2 long, contiguous chromosome segments at a few target chromosomes. The SNP markers, which were developed using the Build 2 pseudomolecules of cv. Nipponbare, were evenly distributed across the 12 rice chromosomes at an average marker interval of 3.0 Mb. A total of 20 BC4F1 and 18 BC5F1 lines were identified to have the target wild introgression. The selected backcross lines were then subjected to additional backcrossing with Koshihikari and subsequent self-pollination to fix the genotypes to homozygotes. Finally, a BSL set composed of 40 individuals with overlapping substituted chromosome segments from the donor were developed. All of the backcrossed lines that were derived from the successive backcrossing of the F1 with Koshihikari were cultivated at the experimental field in Kisarazu, Chiba, Japan.
Fig. 1.
Breeding scheme for developing BSLs carrying O. barthii chromosome segments in O. sativa cv. Koshihikari genetic background. Numbers in parenthesis indicate the number of lines produced for each backcross generation whereas boxed numbers show the number of candidate lines for BSLs selected by marker assisted selection (MAS). Numbers in brackets present the number of BSLs finally selected from the resulting CSSLs.
Agronomic evaluation of the BSLs
The BSLs were evaluated for several agriculturally important traits in the experimental field of Nagoya University in Togo, Aichi, Japan in 2016. Seedlings of all plant materials were first raised in the greenhouse and then transplanted in the field 30 days after sowing. Ten plants of each BSL and Koshihikari were planted in single-row plots at a spacing of 30 cm between rows and 15 cm between hills. The main shoot of five plants at the 3rd–7th hills of each row were measured for flag leaf length, flag leaf width, panicle length, number of grains per panicle, number of primary branches per panicle, 100-grain weight and spikelet fertility. The panicle number in five plants in the same position was counted. Data on the flag leaf length was based on measurements of the full length of the leaf blade of the flag leaf, whereas data on flag leaf width was obtained as a measure of the widest point of the leaf blade of flag leaf. Data on grain length and grain width were obtained from 20 seeds that were air-dried for 2 months in the glasshouse. Grain length and grain width were measured using a scanned image analyzing software, SmartGrain (Tanabata et al. 2012). Dunnett’s multiple comparison test at 95% confidence interval (P < 0.05) was used to evaluate significance of difference in the BSLs compared to Koshihikari. The data on the other qualitative traits such as seed pericarp color, awn formation and shattering, we did not measure quantitatively but characterized by its outstanding phenotype.
Results
Characteristics of the BSLs
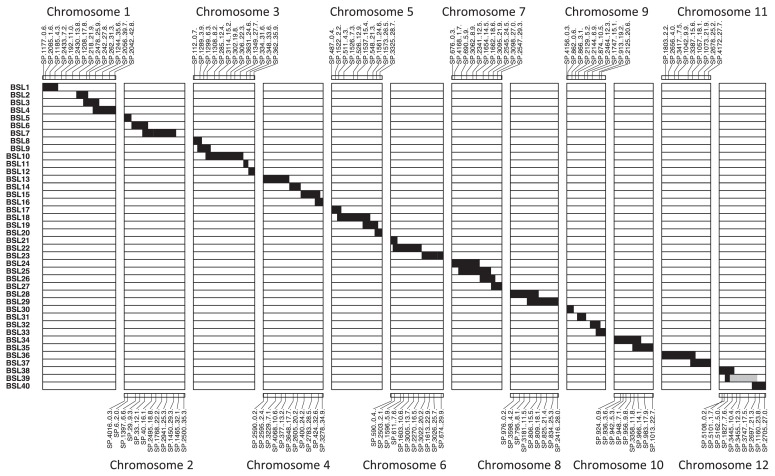
All BSLs carry homozygous chromosome segments except for one line, which still carry a heterozygous O. barthii segment on chromosome 12. This fragment represents around 2% of the donor parent’s genome, measures approximately 12.5 Mb and is flanked by the SNP markers SP-5162 and SP-2697. Chromosomes 1, 3, 4, 5, 7 and 9 of O. barthii were each covered by 4–5 BSLs, whereas chromosomes 2, 6, 8, 10, 11 and 12 were each represented by 2–3 BSLs. The whole genome of O. barthii was represented as contiguous or overlapping chromosome segments in the BSL set (Fig. 2). More than 94% of the donor parent’s genome was represented in contiguous or overlapping chromosome segments in the BSL set. Approximately 4% of the donor genome was not successfully represented as indicated by the small gaps in the genetic map of the BSL set. Those unrepresented regions were in chromosomes 1 (defined by the SNP marker SP-192 to SP-1208), 2 (defined by SP-1465 to SP-2500) and 9 (defined by SP-2129 and SP-2464).
Fig. 2.
Graphical representation of the genotypes of the 40 BSLs. White bars and black bars indicate homozygous chromosomal segments derived from Koshihikari (O. sativa) and W0009 (O. barthii), respectively. Gray bars represent heterozygous regions. The SNP markers used for MAS are indicated with their physical positions (Mb) for each chromosome.
Putative chromosome segments controlling important agronomic traits in the BSLs
Data on the agronomic traits of the BSLs under experimental field conditions are presented in Table 1. A total of 66 significant changes in the tested traits were identified in the BSLs, 28 of which had positive and 38 had negative changes on the phenotypic values compared to Koshihikari.
Table 1.
Measurements of agronomic traits in 40 BSLs
| Chr. No. | Line name | Flag leaf length (cm) | Flag leaf width (cm) | Number of panicles/plant | Panicle length (cm) | Number of primary branches/panicle | Grain character | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Number of grains/panicle | Grain length (mm) | Grain width (mm) | 100-grain weight (g) | Fertility (%) | |||||||
| Koshihikari | 29.43 | 0.81 | 15.8 | 18.66 | 9.60 | 113.80 | 6.49 | 3.61 | 2.04 | 95.10 | |
|
| |||||||||||
| 1 | BSL1 | 30.46 | 0.96a | 11.6 | 19.46 | 10.80 | 153.20a | 6.79 | 3.21 | 1.92b | 93.16 |
| 1 | BSL2 | 25.73 | 0.70 | 10.8 | 20.32 | 9.60 | 120.40 | 6.81 | 3.21 | 1.91 | 95.25 |
| 1 | BSL3 | 25.58 | 0.72 | 12.3 | 20.80 | 9.60 | 105.00 | 6.83 | 2.84b | 2.08 | 94.93 |
| 1 | BSL4 | 30.71 | 0.82 | 12.0 | 20.48 | 9.75 | 119.60 | 6.46 | 3.36 | 2.04 | 91.57 |
| 2 | BSL5 | 28.90 | 0.78 | 14.0 | 21.26 | 9.40 | 116.40 | 6.35 | 2.94b | 1.95 | 90.39 |
| 2 | BSL6 | 29.06 | 0.84 | 13.0 | 21.22 | 11.00 | 130.40 | 7.32a | 3.36 | 2.18 | 90.14 |
| 2 | BSL7 | 26.09 | 0.77 | 12.4 | 21.70 | 10.00 | 126.00 | 6.76 | 3.09b | 1.98 | 73.75b |
| 3 | BSL8 | 27.75 | 0.88 | 11.6 | 20.12 | 9.80 | 127.20 | 6.91 | 3.33 | 2.30a | 93.53 |
| 3 | BSL9 | 28.09 | 0.75 | 11.4 | 20.62 | 9.20 | 117.60 | 6.81 | 3.09b | 2.56a | 92.45 |
| 3 | BSL10 | 30.67 | 0.82 | 11.4 | 20.68 | 12.00a | 156.60a | 6.98 | 3.11b | 2.07 | 90.98 |
| 3 | BSL11 | 34.51 | 0.77 | 12.3 | 20.46 | 12.00a | 163.40a | 6.67 | 3.15b | 1.87 | 89.77 |
| 3 | BSL12 | 27.73 | 0.82 | 10.9 | 18.92 | 10.40 | 119.00 | 6.64 | 3.14b | 1.94 | 92.24 |
| 4 | BSL13 | 28.29 | 0.85 | 13.2 | 20.18 | 9.40 | 110.80 | 6.52 | 3.18 | 2.29a | 93.93 |
| 4 | BSL14 | 26.80 | 0.82 | 12.8 | 18.52 | 8.20 | 93.60 | 6.98 | 3.17 | 2.24 | 94.48 |
| 4 | BSL15 | 25.93 | 0.52b | 12.8 | 18.52 | 8.60 | 91.40 | 6.12 | 3.10b | 2.05 | 91.90 |
| 4 | BSL16 | 26.33 | 0.60 | 15.4 | 21.52 | 11.40 | 145.00 | 6.15 | 3.19b | 1.94 | 93.02 |
| 5 | BSL17 | 26.88 | 0.68 | 11.4 | 17.38 | 8.40 | 96.40 | 6.92 | 2.68b | 1.61b | 89.57 |
| 5 | BSL18 | 27.82 | 0.68 | 18.5 | 21.50 | 10.00 | 124.20 | 6.72 | 2.80b | 1.79b | 92.96 |
| 5 | BSL19 | 26.05 | 0.72 | 15.2 | 18.90 | 9.80 | 124.60 | 6.85 | 3.18 | 2.18 | 93.45 |
| 5 | BSL20 | 27.79 | 0.71 | 16.8 | 21.65 | 11.20 | 161.80a | 6.74 | 3.29 | 2.09 | 94.21 |
| 6 | BSL21 | 31.44 | 0.79 | 14.5 | 19.45 | 10.40 | 122.80 | 6.27 | 3.04b | 2.10 | 93.70 |
| 6 | BSL22 | 32.37 | 0.82 | 13.6 | 20.95 | 12.80a | 171.20a | 6.29 | 2.86b | 2.12 | 75.67b |
| 6 | BSL23 | 27.13 | 0.67 | 14.6 | 22.23a | 10.00 | 131.60 | 6.88 | 3.38 | 2.14 | 90.39 |
| 7 | BSL24 | 30.66 | 0.78 | 10.4 | 22.80a | 9.80 | 127.80 | 6.32 | 2.99b | 2.06 | 89.78 |
| 7 | BSL25 | 27.13 | 0.86 | 12.4 | 19.90 | 13.00a | 156.00a | 6.43 | 3.02b | 1.84b | 86.52b |
| 7 | BSL26 | 31.20 | 0.56 | 11.4 | 17.61 | 10.00 | 136.60 | 6.70 | 3.03b | 1.90b | 94.15 |
| 7 | BSL27 | 37.25a | 0.94a | 14.6 | 23.54a | 12.00a | 166.40a | 6.47 | 3.05b | 1.97 | 91.53 |
| 8 | BSL28 | 27.86 | 0.79 | 17.2 | 20.40 | 11.40 | 162.80a | 6.95 | 3.20 | 1.93 | 80.81b |
| 8 | BSL29 | 22.68 | 0.81 | 16.1 | 19.52 | 11.75a | 167.50a | 6.88 | 3.27 | 2.17 | 92.56 |
| 9 | BSL30 | 30.29 | 0.87 | 15.3 | 18.81 | 8.60 | 106.60 | 6.59 | 3.15b | 2.02 | 93.37 |
| 9 | BSL31 | 25.14 | 0.84 | 12.8 | 19.40 | 9.60 | 130.00 | 6.62 | 3.16b | 2.03 | 94.45 |
| 9 | BSL32 | 29.63 | 0.84 | 14.4 | 20.94 | 10.40 | 129.60 | 6.31 | 2.88b | 1.87b | 93.21 |
| 9 | BSL33 | 28.20 | 0.80 | 14.6 | 18.78 | 9.60 | 109.00 | 6.46 | 3.17b | 1.95 | 93.53 |
| 10 | BSL34 | 34.14 | 0.78 | 10.8 | 20.62 | 11.00 | 131.40 | 6.83 | 3.20 | 2.16 | 94.10 |
| 10 | BSL35 | 31.65 | 0.83 | 11.6 | 22.02a | 10.20 | 123.40 | 6.80 | 3.30 | 2.13 | 92.26 |
| 11 | BSL36 | 30.18 | 0.88 | 18.2 | 20.42 | 9.80 | 113.40 | 6.64 | 3.16b | 1.87b | 92.59 |
| 11 | BSL37 | 31.68 | 0.86 | 17.5 | 20.36 | 9.60 | 118.80 | 6.83 | 3.03b | 2.09 | 92.58 |
| 12 | BSL38 | 29.86 | 0.90a | 15.0 | 21.43 | 11.00 | 128.20 | 6.59 | 3.15b | 1.99 | 92.56 |
| 12 | BSL39 | 35.09 | 0.85 | 14.7 | 19.16 | 9.20 | 112.20 | 7.21a | 3.37 | 2.10 | 94.53 |
| 12 | BSL40 | 32.38 | 0.79 | 12.5 | 21.45 | 8.80 | 111.80 | 7.03 | 3.07b | 1.85b | 90.57 |
significantly higher than Koshihikari,
significantly lower than Koshihikari.
Significance was calculated at P<0.05, values shown are means of 5 samples.
Flag leaf length and width
Among the BSLs tested, only BSL27 showed longer average flag leaf, which was 26.6% longer than that of Koshihikari. BSL15 had the narrowest flag leaf (0.52 cm), which was 35.8% narrower than that of Koshihikari. BSL27 on the other hand, showed a wider phenotype for flag leaf, suggesting the higher potential for photosynthesis in the flag leaf of BSL27 than Koshihikari.
Panicle numbers per plant and panicle length
There are no significant differences in the number of panicles per plant. On the other hand, significant differences in panicle length were detected in 4 BSLs with substituted segments from chromosomes 6, 7 and 10 of O. barthii. All 4 BSLs recorded a significant increase in panicle length (18–26%), with BSL27 recording the longest panicles (23.5 cm). This represents a 26.1% increase in average panicle length compared to Koshihikari.
Number of primary branches and grains per panicle
There were only positive effects in the number of primary branches and grains per panicle detected in the BSLs. BSL10, 11, 22, 25, 27 and 29 exhibited significantly more primary branches per panicle compared to Koshihikari. A significant decrease in the number of primary branches per panicle was not observed in any line. The number of grains per panicle is one of the most important components of yield and it has been one of the main targets for improvement of rice breeding. Nine lines showed a 24–50% increase in the number of grains per panicle relative to Koshihikari, with BSL22 recording more than 1.5 times the number of grains than Koshihikari. Even though many of the BSLs did not show significant differences in the number of primary branches, a moderate positive correlation between the number of primary branches and grains per panicle was detected (correlation coefficient = 0.7269). These results indicate that O. barthii has several superior alleles controlling the numbers of primary branches per panicle, which led to the increase in the number of grains per panicle in the BSLs.
Grain length and width
Only BSL6 and 39 showed 11–12.8% longer grains than Koshihikari. Both lines harbored chromosome 2 and 12 from O. barthii. On the other hand, most of the BSLs recorded decreases in grain width compared to Koshihikari, which showed 3.04 mm average grain width. These lines have introgressed segments from chromosomes 1, 2, 3, 4, 5, 6, 7, 9, 11 and 12 of the donor parent and an 11–25% decrease in grain width. BSL17 and 18 with segments from chromosomes 5 of O. barthii significantly reduced the grain width of Koshihikari by 22–25%.
100-grain weight
Three lines (BSL8, 9 and13) showed significantly higher 100-grain weight compared to Koshihikari, which recorded an average of 2.38 g. These BSLs carry a fragment of chromosome 3 and 4 of the donor genome and increased 100-grain weight by around 20%. A reduction in 100-grain weight was observed in 8 lines having fragments from chromosome 1, 5, 7, 9, 11 and 12 of O. barthii.
Spikelet fertility
Koshihikari had a spikelet fertility of around 95%. All the BSLs showed lower spikelet fertility compared to Koshihikari, with BSL7 recording the lowest fertility (73.75%). This line has a fragment of chromosome 2 from O. barthii. Another 3 lines, BSL22, BSL25 and BSL28 that including the region of chromosome 6, 7 and 8 respectively also registered significant reductions ranging from 9 to 20%.
Other traits
Other traits that were clearly expressed in few of the BSLs were also identified. Although these were not quantitatively measured and subjected to QTL analysis, the traits can be clearly associated with major gene(s) or QTLs. For instance, red pericarp color in grain was observed in BSL24 and 25, which carries approximately 8.6 Mb and 16.0 Mb of chromosome 7 of O. barthii. The overlapped region in these two lines possessed the Rc gene (Os07g0211g00), which encodes the bHLH transcription factor known to regulate pericarp color (Furukawa et al. 2007, Wang et al. 2014). Another trait identified was the long awns in BSL14 and 29, which carried segments of chromosome 4 and 8, respectively, of O. barthii. This coincides with the location of RAE1 and RAE2, which have been reported to regulate awn elongation in rice (Bessho-Uehara et al. 2016, Furuta et al. 2015). BSL4 and 19 having segments from chromosome 1 and 5 of O. barthii exhibited grain shattering. This segments coincided with the location of known QTLs included the qSh1 and SH5 genes (Konishi et al. 2006, Yoon et al. 2014).
Discussion
Rice was domesticated from its wild relatives by humans (Wang et al. 1992). However, only a small portion of the genetic variation found in nature was captured in the domestication process, and this has limited the genetic base that forms the foundation of modern cultivars (Ladizinsky 1985). Although wild rice species are valued as a source of genetic variation, they usually have low yields. Nevertheless, our results indicate that the African wild relatives have genes that can increase rice yield despite its overall inferior appearance.
In this study, we developed a set of rice chromosome segment substitution lines harboring O. barthii genome in the background of the japonica cultivar, Koshihikari. The donor parent, W0009, has been characterized as having the characteristic morphology of the African wild rice, as well as tolerance to multiple biotic/abiotic stresses according to Oryzabase (https://shigen.nig.ac.jp/rice/oryzabase). The primary objective of this study was to develop a wide population containing O. barthii genetic materials for rice breeding and genomic research. Total 40 lines that composed the BSL set covered the whole genome of O. barthii in the Koshihikari genetic background. A lot of significant differences in the traits examined were detected between the BSLs and Koshihikari (Table 1).
Grain shape and 100-grain weight were the major determinants of yield from O. barthii. For grain length, only 2 lines having segments from chromosome 2 and 12 from O. barthii recorded an increase in grain length. Based on the rice QTL database Q-TARO, the previously reported QTLs in the substituted chromosomal regions were determined (Yonemaru et al. 2010). The region on chromosome 2 was close to HDA703, which regulates grain size (Hu et al. 2009), whereas the region on chromosome 12 has not been reported to be associated with QTLs related to grain length or size. Most of the BSLs showed narrower grain width than Koshihikari, especially all the lines harboring a segment of chromosome 7 of O. barthii. In a previous study of recombinant inbred lines derived from the cross between O. sativa and O. rufipogon, a QTL cluster controlling several traits related to plant architecture has been reported at the distal end of the short arm of chromosome 7 (Onishi et al. 2007). The Q-TARO database identifies the genes in chromosome 7 for grain width named grb7 or qSGW7 (Amarawathi et al. 2008, Zhou et al. 2015). For 100-grain weight, we identified 3 lines that were associated with a significant increase in the value of this trait (P < 0.05). These lines have segments of chromosome 3 and 4 from O. barthii, which has not only been associated with genes regulating 100-grain weight, but has been reported to harbor QTLs such as OsPT2, qGY-3 and gwt4a (Lin et al. 1995, Mao et al. 2003, Zhang et al. 2014a). These lines, however, had no detectable correlation with flag leaf length or width. It is suggested that photosynthesis rate in the flag leaf do not contribute in 100-grain weight and vice versa. This is evident in BSL27, which showed the largest flag leaf area but did not record any increase in 100-grain weight. A reduction in 100-grain weight was detected in 8 lines, 2 of which are carrying segments from chromosome 5 of O. barthii. This region coincides with qGW5 which regulates 100-grain weight in rice (Wan et al. 2008). BSL32, which harbors a segment of chromosome 9 of O. barthii showed an 8.3% decrease in 100-grain weight compared to Koshihikari. This segment coincides with the location of a novel QTL for 1000-grain weight (qTGW9) that has been reported by Qiao et al. (2016).
The number of grains per panicle and its correlated trait, number of primary branches per panicle are also important traits for the improvement of yield. Nine lines out of 40 BSLs showed significantly different number of grains per panicle compared to Koshihikari. BSL22 recorded the highest number of grains per panicle (171.2). Some QTLs and genes related to these 2 traits were detected. For example, we could find the QTL on chromosome 8, named WFP to increase the panicle branching number according to Q-TARO database (Miura et al. 2010). Furthermore, there are several genes detected on the long arm of chromosome 7. Photoperiod-related genes such as GHD7 and PRR37 have been reported to contribute in increasing grain number (Weng et al. 2014, Yang et al. 2014). However, no QTLs or candidate genes controlling grain number per panicle have been reported on chromosome 6 included in BSL22. This suggests that a novel QTL regulating grain number per panicle exists on this chromosome region from O. barthii. The results also indicate that the BSLs in this study will prove an efficient population for QTL identification, and that the use of different wild rice accessions in the development of such genetic resource can lead to the discovery of novel genes. As for spikelet fertility, other CSSLs having chromosome segments from other wild rice species or relatives such as GLSL (O. glaberrima, Shim et al. 2010) and RSL (O. rufipogon, Furuta et al. 2014) also showed lower spikelet fertility than Koshihikari. The chromosome 6 region in BSL22 overlaps with the reported location of GSL5, a candidate gene regulating spikelet fertility (Shi et al. 2015).
Plant breeding requires the combination of design and science to improve the genetic basis of new crop varieties with better agronomic traits. Approaches in plant breeding have advanced to the use of molecular markers and high-density single nucleotide polymorphism (SNP) arrays to combine phenotype and genotype. The development of wild rice CSSLs has provided a broad platform for both wild rice genomic research and QTL mapping. Novel genes found in wild rice using these CSSLs could provide a new genetic resource for breakthroughs in rice breeding. In the present study, BSLs were mainly evaluated for yield and yield related traits. However, O. barthii has also been reported to harbor genes for disease and insect resistance (Cheema et al. 2008, Fujita et al. 2004, Vaughan 1994). Evaluation of the performance of the 40 BSLs under different biotic stresses would facilitate the identification of genes controlling resistance to diseases and pests. Thus, BSLs could serve as useful genetic resource not only for improvement of agricultural traits, but also for understanding physiological and ecological differences between O. barthii and O. sativa. These CSSL populations could also be used for mapping and cloning novel QTL/genes. The identified QTL regions present in the CSSLs would allow a regional candidate-gene association mapping approach in natural populations to reveal natural allelic variations that could explain the expression of desirable traits. Moreover, this set of CSSLs will also allow us to proceed with field trials in different environments to identify novel QTL for other complex quantitative traits.
Acknowledgments
We thank the National Institute of Genetics for the seeds of O. barthii Acc. W0009. This research was supported by a Japan Society for the Promotion of Science fellowship (Grant 15J03740), the Core Research for Evolutional Science and Technology by JST, a MEXT Grant-in-Aid for Scientific Research on Innovative Areas (Grant 16H01464), and the Canon foundation. This project was also supported by SATREPS by JICA and JST.
Literature Cited
- Ali, M.L., Sanchez, L.P., Yu, S.B., Lorieux, M. and Eizenga, G.C. (2010) Chromosome segment substitution lines: a powerful tool for the introgression of valuable genes from Oryza wild species into cultivated rice (O. sativa). Rice 3: 218–234. [Google Scholar]
- Amarawathi, Y., Singh, R., Singh, K.A., Singh, P.V., Mohapatra, T., Sharma, R.T. and Singh, K.N. (2008) Mapping of quantitative trait loci for basmati quality traits in rice (Oryza sativa L.). Mol. Breed. 21: 49–65. [Google Scholar]
- Bessho-Uehara, K., Wang, D.R., Furuta, T., Minami, A., Nagai, K., Gamuyao, R., Asano, K., Angeles-Shim, R.B., Shimizu, Y., Ayano, M.et al. (2016) Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proc. Natl. Acad. Sci. USA 113: 8969–8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, J.M., Jiang, L., Liu, L.L., Wei, X.J., Xiao, Y.H., Zhang, L.J., Zhao, Z.G., Zhai, H.Q. and Wan, J.M. (2010) Construction of a new set of rice chromosome segment substitution lines and identification of grain weight and related traits QTLs. Breed. Sci. 60: 305–313. [Google Scholar]
- Cheema, K.K., Grewal, N.K., Vikal, Y., Sharma, R., Lore, J.S., Das, A., Bhatia, D., Mahajan, R., Gupta, V., Bharaj, T.S.et al. (2008) A novel bacterial blight resistance gene from Oryza nivara mapped to 38 Kb region on chromosome 4L and transferred to Oryza sativa L. Genet. Res. 90: 397–407. [DOI] [PubMed] [Google Scholar]
- Doi, K., N. Iwata and A. Yoshimura (1997) The construction of chromosome substitution lines of African rice (Oryza glaberrima Steud.) in the background of Japonica rice (O. sativa L.). Rice Genet. Newsl. 14: 39–41. [Google Scholar]
- Ebitani, T., Takeuchi, Y., Nonoue, Y., Yamamoto, T., Takeuchi, K. and Yano, M. (2005) Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar ‘Kasalath’ in a genetic background of japonica elite cultivar ‘Koshihikari’. Breed.Sci. 55: 65–73. [Google Scholar]
- Fujita, D., Doi, K., Yoshimura, A. and Yasui, H. (2004) Introgression of a resistance gene for green leafhopper from Oryza barthii into cultivated rice, Oryza sativa L. Rice. Genet. Newsl. 21: 64. [Google Scholar]
- Furukawa, T., Maekawa, M., Oki, T., Suda, I., Iida, S., Shimada, H., Takamure, I. and Kadowaki, K. (2007) The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J. 49: 91–102. [DOI] [PubMed] [Google Scholar]
- Furuta, T., Uehara, K., Angeles-Shim, R.B., Shim, J., Ashikari, M. and Takashi, T. (2014) Development and evaluation of chromosome segment substitution lines (CSSLs) carrying chromosome segments derived from Oryza rufipogon in the genetic background of Oryza sativa L. Breed. Sci. 63: 468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta, T., Komeda, N., Asano, K., Uehara, K., Gamuyao, R., Angeles-Shim, R.B., Nagai, K., Doi, K., Wang, D.R., Yasui, H.et al. (2015) Convergent loss of awn in two cultivated rice species Oryza sativa and Oryza glaberrima is caused by mutations in different loci. G3 (Bethesda) 5: 2267–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta, T., Uehara, K., Angeles-Shim, R.B., Shim, J., Nagai, K., Ashikari, M. and Takashi, T. (2016) Development of chromosome segment substitution lines harboring Oryza nivara genomic segments in Koshihikari and evaluation of yield-related traits. Breed. Sci. 66: 845–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, A.G., Carabali, S.J., Giraldo, O.X., Martinez, C.P., Correa, F., Prado, G., Tohme, J. and Lorieux, M. (2010) Identification of a rice stripe necrosis virus resistance locus and yield component QTLs using Oryza sativa × O. glaberrima introgression lines. BMC Plant Biol. 10: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., Qin, F., Huang, L., Sun, Q., Li, C., Zhao, Y. and Zhou, D.X. (2009) Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem. Biophys. Res. Commun. 388: 266–271. [DOI] [PubMed] [Google Scholar]
- Khush, G.S. (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35: 25–34. [PubMed] [Google Scholar]
- Konishi, S., Izawa, T., Lin, S.Y., Ebana, K., Fukuta, Y., Sasaki, T. and Yano, M. (2006) An SNP caused loss of seed shattering during rice domestication. Science 312: 1392–1396. [DOI] [PubMed] [Google Scholar]
- Kubo, T., Aida, Y., Nakamura, K., Tsunematsu, H., Doi, K. and Yoshimura, A. (2002) Reciprocal chromosome segment substitution series derived from Japonica and Indica cross of rice (Oryza sativa L.). Breed. Sci. 52: 319–325. [Google Scholar]
- Ladizinsky, G. (1985) Founder effect in crop-plant evolution. Econ. Bot. 39: 191–199. [Google Scholar]
- Lin, H.X., Qian, H.R., Zhuang, J.Y., Lu, J., Min, S.K., Xiong, Z., Huang, N. and Zheng, K.L. (1995) Interval mapping of QTLs for yield and other related characters in rice. Rice Genet. Newsl. 12: 251–253. [DOI] [PubMed] [Google Scholar]
- Mao, B.B., Cai, W.J., Zhang, Z.H., Hu, Z.L., Li, P., Zhu, L.H. and Zhu, Y.G. (2003) Characterization of QTLs for harvest index and source-sink characters in a DH population of rice (Oryza sativa L.). Acta Genetica Sinica 30: 1118–1126. [PubMed] [Google Scholar]
- Miura, K., Ikeda, M., Matsubara, A., Song, X.J., Ito, M., Asano, K., Matsuoka, M., Kitano, H. and Ashikari, M. (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42: 545–549. [DOI] [PubMed] [Google Scholar]
- Onishi, K., Horiuchi, Y., Ishigoh-Oka, N., Takagi,, K., Ichikawa, N., Maruoka, M. and Sano, Y. (2007) A QTL cluster for plant architecture and its ecological significance in Asian wild rice. Breed. Sci. 57: 7–16. [Google Scholar]
- Orjuela, J., Sabot, F., Chéron, S., Vigouroux, Y., Adam, H., Chrestin, H., Sanni, K., Lorieux, M. and Ghesquière, A. (2014) An extensive analysis of the African rice genetic diversity through a global genotyping. Theor. Appl. Genet. 127: 2211–2223. [DOI] [PubMed] [Google Scholar]
- Qiao, W., Qi, L., Cheng, Z., Su, L., Li, J., Sun, Y., Ren, J., Zheng, X. and Yang, Q. (2016) Development and characterization of chromosome segment substitution lines derived from Oryza rufipogon in the genetic background of O. sativa spp. indica cultivar 9311. BMC Genomics 17: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, F., Lu, B.R., Li, S., Huang, J. and Zhu, Y. (2003) A comparative study of genetic relationships among the AA-genome Oryza species using RAPD and SSR markers. Theor. Appl. Genet. 108: 113–120. [DOI] [PubMed] [Google Scholar]
- Shen, G.J. and Xing, Y.Z. (2014) Two novel QTLs for heading date are identified using a set of chromosome segment substitution lines in rice (Oryza sativa L.). J. Genet. Genomics 41: 659–662. [DOI] [PubMed] [Google Scholar]
- Shi, X., Sun, X., Zhang, Z., Feng, D., Zhang, Q., Han, L., Wu, J. and Lu, T. (2015) GLUCAN SYNTHASE-LIKE 5 (GSL5) plays an essential role in male fertility by regulating callose metabolism during microsporogenesis in rice. Plant Cell Physiol. 56: 497–509. [DOI] [PubMed] [Google Scholar]
- Shim, R.A., Angeles, E.R., Ashikari, M. and Takashi, T. (2010) Development and evaluation of Oryza glaberrima Steud. chromosome segment substitution lines (CSSLs) in the background of O. sativa L. cv. Koshihikari. Breed. Sci. 60: 613–619. [Google Scholar]
- Takai, T., Nonoue, Y., Yamamoto, S., Yamanouchi, U., Matsubara, K., Liang, Z.W., Lin, H.X., Ono, N., Uga, Y. and Yano, M. (2007) Development of chromosome segment substitution lines derived from backcross between indica donor rice cultivar ‘Nona bokra’ and japonica recipient cultivar ‘Koshihikari’. Breed. Sci. 57: 257–261. [Google Scholar]
- Tan, L., Liu, F., Xue, W., Wang, G., Ye, S., Zhu, Z., Fu, Y., Wang, X. and Sun, C. (2007) Development of Oryza rufipogon and O. sativa introgression lines and assessment for yield-related quantitative trait loci. J. Integr. Plant Biol. 49: 871–884. [Google Scholar]
- Tanabata, T., Shibaya, T., Hori, K., Ebana, K. and Yano, M. (2012) SmartGrain: high-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol. 160: 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S.D., Grandillo, S., Fulton, T.M., Zamir, D., Eshed, Y., Petiard, V., Lopez, J. and Beck-Bunn, T. (1996) Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor. Appl. Genet. 92: 213–224. [DOI] [PubMed] [Google Scholar]
- Tanksley, S.D. and McCouch, S.R. (1997) Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 277: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Ujiie, K. and Ishimaru, K. (2013) Identification of chromosome regions affecting leaf area with rice chromosome segment substitution lines. Plant Prod. Sci. 16: 31–36. [Google Scholar]
- Vaughan, D.A. (1994) The wild relatives of rice: a genetic resources handbook. International Rice Research Institute, Manila, p. 137. [Google Scholar]
- Wan, X., Weng, J., Zhai, H., Wang, J., Lei, C., Liu, X., Guo, T., Jiang, L., Su, N. and Wan, J. (2008) Quantitative trait loci (QTL) analysis for rice grain width and fine mapping of an identified QTL allele gw-5 in a recombination hotspot region on chromosome 5. Genetics 179: 2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Yu, Y., Haberer, G., Marri, P.R., Fan, C., Goicoechea, J.L., Zuccolo, A., Song, X., Kudrna, D., Ammiraju, J.S.et al. (2014) The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 46: 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., Second, G. and Tanksley, S.D. (1992) Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theor. Appl. Genet. 83: 565–581. [DOI] [PubMed] [Google Scholar]
- Weng, X., Wang, L., Wang, J., Hu, Y., Du, H., Xu, C., Xing, Y., Li, X., Xiao, J. and Zhang, Q. (2014) Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol. 164: 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, J., Grandillo, S., Ahn, S., McCouch, S.R., Tanksley, S., Li, J. and Yuan, L. (1996) Genes from wild rice improve yield. Nature 384: 223–224. [Google Scholar]
- Yang, S., Weers, B.D., Morishige, D.T. and Mullet, J.E. (2014) CONSTANS is a photoperiod regulated activator of flowering in sorghum. BMC Plant Biol. 14: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M. (2001) Genetic and molecular dissection of naturally occurring variation. Curr. Opin. Plant Biol. 4: 130–135. [DOI] [PubMed] [Google Scholar]
- Yonemaru, J., Yamamoto, T., Fukuoka, S., Uga, Y., Hori, K. and Yano, M. (2010) Q-TARO: QTL Annotation Rice Online Database. Rice 3: 194. [Google Scholar]
- Yoon, J., Cho, L.H., Kim, S.L., Choi, H., Koh, H.J. and An, G. (2014) The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 79: 717–728. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Hu, B., Li, W., Che, R., Deng, K., Li, H., Yu, F., Ling, H., Li, Y. and Chu, C. (2014a) OsPT2, a phosphate transporter, is involved in the active uptake of selenite in rice. New Phytol. 201: 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Zhu, T., Xia, E., Shi, C., Liu, Y., Zhang, Y., Liu, Y., Jiang, W., Zhao, Y., Mao, S.et al. (2014b) Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc. Natl. Acad. Sci. USA 111: E4954–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Wang, Q., Jiang, L., Liu, L., Wang, B., Shen, Y., Cheng, X. and Wan, J. (2011) Fine mapping of qSTV11KAS, a major QTL for rice stripe disease resistance. Theor. Appl. Genet. 122: 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Miao, J., Gu, H., Peng, X., Leburu, M., Yuan, F., Gu, H., Gao, Y., Tao, Y., Zhu, J.et al. (2015) Natural variations in SLG7 regulate grain shape in rice. Genetics 201: 1591–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]