Abstract
Florigen is a mobile flowering signal in plants that has a strong impact on plant reproduction and is considered one of the important targets for crop improvement. At the molecular level, florigen is represented as a protein product encoded by the FLOWERING LOCUS T (FT) gene, which is highly conserved across flowering plants and thus the understanding of this protein is expected to be applied to the improvement of many crops. Recent advances in molecular genetics, cell biology and structural biology in plants revealed the presence of intercellular receptors for florigen, a transcriptional complex essential for florigen to function, and also shed light on the molecular basis of pleiotropic function of florigen beyond flowering. Furthermore, cutting-edge technologies, such as live cell imaging and next generation sequencing revealed the precise distribution of florigen and transcriptional targets of the florigen activation complex (FAC) during early stages of floral transition. These understandings will help future crop improvement through the regulation of flowering and other plant developmental processes.
Keywords: florigen, shoot apical meristem, flowering, FT
Introduction
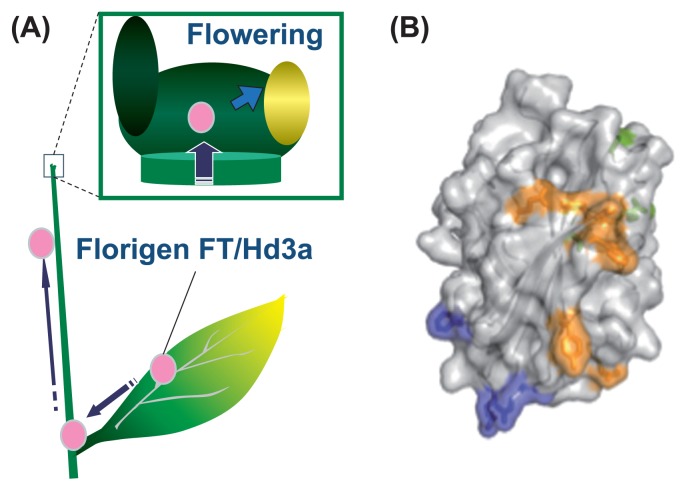
Florigen is a systemic signal that initiates flowering in plants (Chailakhyan 1936). It is synthesized in leaves and transported to the shoot apical meristem (SAM) where it promotes floral transition (Fig. 1A). The molecular function of florigen has long been an important question in the field of flowering studies (Tamaki et al. 2007, Zeevaart 2006). A decade ago the molecular nature of florigen was revealed as the protein product encoded by the FLOWERING LOCUS T (FT) genes in plants (Fig. 1B) (Corbesier et al. 2007, Tamaki et al. 2007). The identification of florigen facilitated the understanding of its molecular mechanism for the perception and function of FT florigen in detail (Tsuji and Taoka 2014). A combination of recent advances in techniques such as next generation sequencing and live cell imaging identified its precise distribution and overall function in the shoot apical meristem (Tamaki et al. 2015).
Fig. 1.
Molecular nature of florigen. (A) Concept of florigen. Florigen is generated in leaf vasculature, transported to the shoot apical meristem and promote flowering. Filled circle in magenta indicate florigen FT/Hd3a protein. Square at the top of the stem indicate the enlargement of the shoot apex. In the enlarged picture, green and yellow ovals indicate leaf or flower primordia, respectively. (B) Cristal structure of Hd3a florigen. Regions colored by blue, yellow and green indicate the regions essential for receptor binding, regions essential for transcriptional activation and other essential residues for florigen function, respectively.
Regulation of flowering time is an important target for plant breeding because the control of flowering to a favorable time provides successful grain production in a given cropping area (Jung and Muller 2009). Flowering at unfavorable seasons causes loss of yield due to insufficient growth of photosynthetic organs or poor fertility due to heat or cold stress during reproduction. Thus, understanding the mechanisms of flowering, and especially of the function of florigen, can contribute to novel breeding techniques in crops to produce cultivars that can start their reproductive stage at optimal seasons (Tsuji et al. 2011). Recent advances in plant genomics help in identifying important regulators of flowering control in many crop species, and also contribute to the understanding of the diversity for future improvements of floral transition. Studies on the control of flowering include many important areas of plant sciences, such as photoperiodic flowering pathway and day length perception, molecular phonology in field-grown plants, and natural variation of flowering time (Hori et al. 2016, Izawa 2015). Understanding the function of florigen can contribute to many of these research fields because florigen is highly conserved across flowering plants (Tsuji et al. 2013b). Here I summarize our current understanding of the molecular function of florigen, focusing mainly on rice (Oryza sativa) florigen Heading date 3a (Hd3a).
Molecular nature of florigen
Recent advances in molecular genetics in plants revealed the molecular nature of florigen as a globular protein named FT, which satisfies the major prerequisites for florigen as a systemic floral signal (Corbesier et al. 2007, Tamaki et al. 2007). These prerequisites were determined from the long history of classical physiological studies, and they are as follows: (1) produced in leaves under a favorable day length, (2) transported from leaves to the SAM through phloem, and (3) conserved across angiosperms (Fig. 1A). Here I will briefly introduce the molecular nature of florigen by describing how FT proteins are recognized.
Photoperiodic flowering has long been considered as a systemic event and the efforts to characterize the genetic factors regulating this process succeeded through molecular genetic analysis of model plants (Andres and Coupland 2012). Mutants for flowering time regulation in Arabidopsis and quantitative trait loci (QTL) analysis of heading date in rice provided rich information for understanding the mechanisms of photoperiodic flowering. FT in Arabidopsis and Hd3a in rice are genes identified through these studies, and they are orthologs that encode proteins similar to the phosphatidylehtanolamine binding protein (PEBP) in animals (Kardailsky et al. 1999, Kobayashi et al. 1999, Kojima et al. 2002). PEPB is a small globular protein with the size of 20 to 25 kD, and shares small pocket on the surface which can interact with the small anions in animal homologs. Both genes are expressed from the phloem of leaves when flowering-promotive day lengths are given—long days for Arabidopsis and short days for rice. After identification of FT/Hd3a genes, the characteristics of these genes were analyzed in detail at the molecular level. In rice, the precise sites for Hd3a promoter activity, Hd3a mRNA accumulation and Hd3a protein distribution were examined using transgenic plants, and results suggested that Hd3a protein is the mobile floral signal in rice (Tamaki et al. 2007). Hd3a promoter is active in the phloem of the leaf blade under short day conditions, and Hd3a mRNA is detected only from the leaf blades. On the other hand, Hd3a protein, which was visualized by a fusion protein with green fluorescent protein (GFP) expressed from Hd3a promoter, accumulates in the SAM. These observations provided evidence to show that translated Hd3a protein moves from leaf phloem tissue to the SAM, where it initiates floral transition. The results obtained in studies using Arabidopsis, which included the grafting of FT-expressing plants, support this conclusion (Notaguchi et al. 2008). Transgenic plants with increased amount of FT/Hd3a flowered earlier and plants with less FT flowered later. In rice, the natural variation of flowering time well correlates with the level of Hd3a and its homolog RICE FLOWERING LOCUS T1 (RFT1) expressions (Takahashi et al. 2009). Altogether, it is now widely accepted that FT/Hd3a is the florigen signal.
Florigen receptor and activation complex
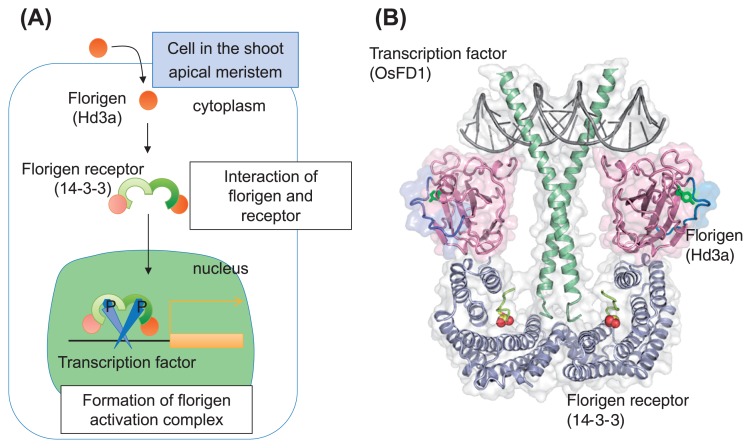
The next important question is how FT protein exerts floral transition in the SAM. A model from Arabidopsis suggested that a bZIP domain containing transcription factor, FD, interacts with FT to form a transcriptional complex that activates the floral identity genes such as APETALA1 (AP1, Abe et al. 2005, Wigge et al. 2005). However, the precise mechanism for the complex formation has been poorly understood. Molecular genetics and biochemical analyses from rice indicated that the interaction between FT and FD was mediated by another class of FT interacting protein, named 14-3-3, and cellular imaging study indicated a dynamic process of complex formation (Fig. 2, Taoka et al. 2011).
Fig. 2.
Formation of florigen activation complex (FAC). (A) Scheme for FAC formation in a shoot apex cell of rice. After florigen Hd3a reaches the cell at the shoot apex, Hd3a interacts with 14-3-3 proteins in the cytoplasm, then translocated to the nucleus to form FAC with FD. The resultant FAC activates downstream gene expression. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. The black bar and orange rectangle in the nucleus indicate a promoter and a downstream gene for FAC, and arrow indicate transcriptional activation. (B) Crystal structure of FAC with the modeled bZIP and DNA. Two molecules of florigen Hd3a (magenta), a dimer of florigen receptor 14-3-3 (blue), and C-terminus region of OsFD1 (green sticks and red balls) essential for direct binding are solved as structure of FAC. DNA at the top (gray) and bZIP region of the transcription factor OsFD1 (green) at the center of the structure are modeled structure.
Several studies have shown a similar complex formation in the rice homologs of Arabidopsis FT and FD, Hd3a and OsFD1, but interestingly, direct interactions in vitro using purified proteins have not been observed. This suggests the presence of additional components that can mediate the interaction of Hd3a and OsFD1 in the cell. This missing link was identified from the assay using another class of Hd3a interacting protein, 14-3-3, because 14-3-3 interacts directly with Hd3a and OsFD1 through different interfaces on the surface of 14-3-3, and addition of 14-3-3 results in the formation of Hd3a-14-3-3-OsFD1 complex in vitro. 14-3-3 is a scaffold protein that can interact with various proteins at the phosphorylated serine or threonine residues in the target proteins. They form dimers to build their W-shaped structure. Mutation analysis revealed that the interaction between Hd3a and OsFD1 depends on the interaction of 14-3-3 with both the proteins and the function of Hd3a depends completely on its ability to form a complex with OsFD1 through 14-3-3. Since the complex formation is essential for Hd3a florigen function, this is named as florigen activation complex (FAC) (Fig. 2B, Taoka et al. 2011).
Cellular imaging of how FAC is formed in the cell revealed that 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Hd3a forms a subcomplex with 14-3-3 in the cytoplasm, where there is an accumulation of 14-3-3. The presence of OsFD1 results in the translocation of Hd3a-14-3-3 subcomplex from the cytoplasm into the nucleus where OsFD1 localizes. In the nucleus Hd3a-14-3-3-OsFD1 tri-protein complex (FAC) is formed and it activates downstream genes such as OsMADS15, a homolog of AP1 in rice. Based on this dynamic mechanism of complex formation, 14-3-3 proteins are considered as the receptor of Hd3a florigen (Fig. 2A, Taoka et al. 2011).
Florigen distribution and downstream gene expression in the SAM
Another question on florigen function is how florigen changes its distribution and coordinates downstream gene expression. Early studies of rice florigen imaging failed to obtain detailed localization during floral transition and subsequent inflorescence development (Tamaki et al. 2007). Recent advances in the imaging of living plant cells and organs enabled the observation of Hd3a accumulation precisely at early stages of the inflorescence development. Hd3a-GFP was not detected in the SAM at the vegetative stage, but its accumulation was clearly observed in the SAM at the early stages of inflorescence development (Tamaki et al. 2015). Hd3a-GFP is less abundant in the newly developed branch primordia in the inflorescences (primary branch meristem), but it accumulates at the late stages of branch primordia. This may contribute to restrict precocious initiation of floral meristem in the inflorescences. In fact, overexpression of Hd3a-GFP from phloem reduced the number of inflorescence branches because of precocious conversion of branch meristem to the floral meristem (Tamaki et al. 2015). In addition, recent analysis of the genetic basis of heterosis identified florigen function in the inflorescence architecture and the yield of crops (Krieger et al. 2010, Park et al. 2014). In tomato, optimal level of expression for genes encoding FAC components contributes fruit yield through the regulation of the timing of floral meristem development (Jiang et al. 2013, Krieger et al. 2010). FT/Hd3a homolog was identified as the cause of heterosis because heterozygous plants for one of the FT homologs showed prolonged development of branches that bear fruits in cultivated tomato. In rice, extensive genome-wide association studies for heterosis identified Hd3a as the candidate gene that contributes to the expression of heterosis (Huang et al. 2016). It is interesting to investigate the molecular mechanism that limits florigen Hd3a distribution or its transport in the SAM to optimize inflorescence development. Molecules that may control florigen transport are suggested from the recent molecular genetic analysis of the mutants in Arabidopsis and rice (Liu et al. 2012, Song et al. 2017, Zhu et al. 2016). The C2-domain containing protein FT-INTERCTING PROTEIN 1 (FTIP1) (Liu et al. 2012, Song et al. 2017) and SODIUM POTASSIUM ROOT DEVECTIVE 1 (NaKR1) (Zhu et al. 2016) are these candidates, but the mutations in these genes dramatically reduce FT expression itself thus it needs more detailed analysis to reach the precise understanding of the mechanisms in FT transport.
The floral transition and subsequent inflorescence development are controlled by the activity of proteins encoded by the downstream genes of florigen FT/Hd3a in the SAM. Thus, identification of these downstream genes is important for understanding florigen function. Transcriptome analysis using microarray and RNA-seq, combined with laser microdissection, a microtechnique to dissect SAM, helped the identification of genes that are regulated by florigen FT/Hd3a (Kobayashi et al. 2012). The major group of downstream genes encodes MADS-box transcription factors in AP1/FRUITFUL clade and SEPALLATA clade (Schmid et al. 2003). In rice and Arabidopsis, mutations in some of these genes delay flowering, and attenuate the effect of FT/Hd3a florigen (Kobayashi et al. 2012, Torti et al. 2012, Wang et al. 2009). Thus these downstream genes consist of a gene expression network essential for flowering induced by florigen FT/Hd3a.
Interestingly, recent RNA-seq experiments identified that the expression of transposable elements (TEs) genes are regulated by florigen Hd3a in rice (Tamaki et al. 2015). Hd3a induces silencing of a subset of TEs at the early stages of floral transition. The silenced TEs are distributed in similar proportion to all the TEs in the rice genome which are concentrated around the centromeres (International Rice Genome Sequencing Project 2005), indicating that the mechanisms by which Hd3a silences TEs may be a generic one that can affect it in a genome-wide manner. The possible mechanisms of this regulation include changes of expressions or activities in DNA methylation enzymes and RNA-dependent DNA methylation pathways because these pathways silences TEs and changes of their activities affect TE expressions in genome-wide manner (reviewed in Matzke and Mosher 2014). The silencing of TEs upon flowering can contribute to the defense of the genome against the activity of TEs and it is possible that the changes in TE expression can act as controlling elements for the genes that play a role during floral transition in rice (Tamaki et al. 2015).
Florigen function beyond flowering
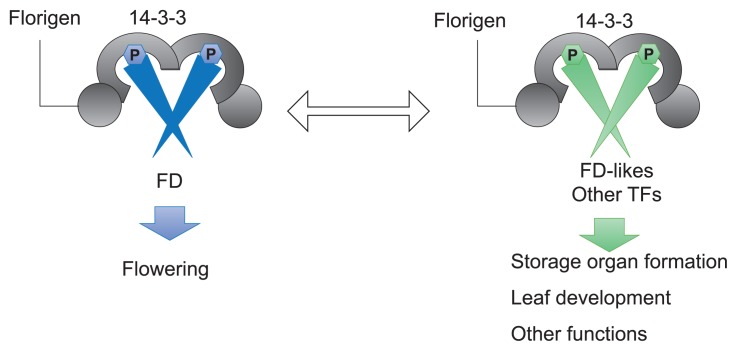
Florigen FT/Hd3a regulates diverse developmental processes beyond flowering, including the timing of reproduction, formation of storage organs and contributes to plant architecture (Tsuji and Taoka 2014). The molecular mechanism of the pleiotropic function of florigen FT/Hd3a can be explained by a model from FAC. In this section I will summarize examples of multiple functions of FT/Hd3a florigen and explain the mechanisms by the FAC model (Fig. 3).
Fig. 3.
Molecular basis of pleiotropic function of florigen. FAC containing FD as a transcription factor subunit promotes flowering (right). On the other hand, FAC with transcription factors (TFs) other than FD, such as FD-like proteins, promotes processes other than flowering such as storage organ formation and leaf development.
Tuber formation in potato is induced by short days, and classical grafting experiments have suggested the involvement of a mobile tuberization signal (Navarro et al. 2011). This signal is generated in leaves upon exposure to short days and transported to the underground stem or stolons where it initiates tuberization (Navarro et al. 2011). This mechanism is quite similar to that of florigen and therefore, the tuber forming signal or tuberigen was considered as a molecule related to florigen. Recently, potato FT homolog Solanum tuberosum SELF PRUNING 6A (StSP6A) has been shown to be the molecular signature of this mobile signal (Navarro et al. 2011). StSP6A forms a FAC-like complex, tuberigen activation complex (TAC), with 14-3-3 and FD-like proteins. Interestingly, the closest FD homolog of potato, StFD, is not involved in tuber formation, but FD-like protein StFDL1a/1b promotes tuber formation (Teo et al. 2017). This suggests that the exchange of transcription factors in FAC can change the function of FT depending on the developmental context. Same conclusions were obtained from molecular genetic analysis of growth cessation in poplar and leaf development and lateral branching in rice (Tsuji et al. 2013a, 2015, Tylewicz et al. 2015). In poplar FT2 promotes vegetative growth and inhibits bud set during summer, and this function is regulated by FAC containing the neo-functionalized FD-like protein PtFDL1 (Tylewicz et al. 2015). In rice, FAC, containing another FD homolog in rice, OsFD2, regulates leaf development (Tsuji et al. 2013a), and FAC with an unknown transcription factor promotes lateral branching in the axillary meristem (Tsuji et al. 2015). From these evidences, it is clear that the functions of florigen at the molecular level beyond flowering are diversification of FD function and the exchange of transcription factor components of FAC at the specific developmental context (Tsuji et al. 2013b).
Conclusion
The molecular nature of the systemic floral signal, florigen, is a protein product encoded by the FT gene, which is highly conserved across flowering plants. FT is expressed in leaves and transported to the SAM. In SAM cells, FT interacts with its receptor 14-3-3 in the cytoplasm and is translocated into the nucleus to interact with the transcription factor FD. The resultant tri-protein complex, FAC, activates downstream genes to initiate floral transition in the SAM. FT distribution in the SAM explains its function during inflorescence development, and its contribution to the yield of some crops, through the regulation of balance between inflorescence branching and floral meristem formation. Besides flowering, florigen shows multiple functions such as formation of storage organs and contributes to plant architecture. The exchange of transcription factors in the FAC is the molecular basis of this pleiotropic function. Understanding the molecular function of florigen by cutting-edge technologies will help future crop improvement programs through the regulation of flowering and other plant developmental processes.
Acknowledgement
I thank Dr. K. Shimamoto (passed away on 28th September 2013, Nara Institute of Science and Technology) for encouraging the florigen studies. This work is supported by grants from Grant-in-Aid for Scientific Research on Innovative Areas (16H06466) to H.T..
Footnotes
This review paper won a Young Scientist Award 2014 of Japanese Society of Breeding.
Literature Cited
- Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K. and Araki, T. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056. [DOI] [PubMed] [Google Scholar]
- Andres, F. and Coupland, G. (2012) The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13: 627–639. [DOI] [PubMed] [Google Scholar]
- Chailakhyan, M.K. (1936) New facts in support of the hormonal theory of plant development. C. R. Acad. Sci. URSS 13: 79–83. [Google Scholar]
- Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., Giakountis, A., Farrona, S., Gissot, L., Turnbull, C.et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Huang, X., Yang, S., Gong, J., Zhao, Q., Feng, Q., Zhan, Q., Zhao, Y., Li, W., Cheng, B., Xia, J.et al. (2016) Genomic architecture of heterosis for yield traits in rice. Nature 537: 629–633. [DOI] [PubMed] [Google Scholar]
- Hori, K., Matsubara, K. and Yano, M. (2016) Genetic control of flowering time in rice: integration of Mendelian genetics and genomics. Theor. Appl. Genet. 129: 2241–2252. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequnce of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- Izawa, T. (2015) Deciphering and prediction of plant dynamics under field conditions. Curr. Opin. Plant Biol. 24: 87–92. [DOI] [PubMed] [Google Scholar]
- Jiang, K., Liberatore, K.L., Park, S.J., Alvarez, J.P. and Lippman, Z.B. (2013) Tomato yield heterosis is triggered by a dosage sensitivity of the florigen pathway that fine-tunes shoot architecture. PLoS Genet. 9: e1004043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, C. and Muller, A.E. (2009) Flowering time control and applications in plant breeding. Trends Plant Sci. 14: 563–573. [DOI] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J. and Weigel, D. (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kobayashi, K., Yasuno, N., Sato, Y., Yoda, M., Yamazaki, R., Kimizu, M., Yoshida, H., Nagamura, Y. and Kyozuka, J. (2012) Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24: 1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M. and Araki, T. (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T. and Yano, M. (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43: 1096–1105. [DOI] [PubMed] [Google Scholar]
- Krieger, U., Lippman, Z.B. and Zamir, D. (2010) The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 42: 459–463. [DOI] [PubMed] [Google Scholar]
- Liu, L., Liu, C., Hou, X., Xi, W., Shen, L., Tao, Z., Wang, Y. and Yu, H. (2012) FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 10: e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, M.A. and Mosher, R.A. (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408. [DOI] [PubMed] [Google Scholar]
- Navarro, C., Abelenda, J.A., Cruz-Oro, E., Cuellar, C.A., Tamaki, S., Silva, J., Shimamoto, K. and Prat, S. (2011) Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478: 119–122. [DOI] [PubMed] [Google Scholar]
- Notaguchi, M., Abe, M., Kimura, T., Daimon, Y., Kobayashi, T., Yamaguchi, A., Tomita, Y., Dohi, K., Mori, M. and Araki, T. (2008) Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 49: 1645–1658. [DOI] [PubMed] [Google Scholar]
- Park, S.J., Jiang, K., Tal, L., Yichie, Y., Gar, O., Zamir, D., Eshed, Y. and Lippman, Z.B. (2014) Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 46: 1337–1342. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Uhlenhaut, N.H., Godard, F., Demar, M., Bressan, R., Weigel, D. and Lohmann, J.U. (2003) Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012. [DOI] [PubMed] [Google Scholar]
- Song, S., Chen, Y., Liu, L., Wang, Y., Bao, S., Zhou, X., Teo, Z.W.N., Mao, C., Gan, Y. and Yu, H. (2017) OsFTIP1-mediated regulation of florigen transport in rice is negatively regulated by the ubiquitin-like domain kinase OsUbDKγ4. Plant Cell 29: 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., Teshima, K.M., Yokoi, S., Innan, H. and Shimamoto, K. (2009) Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc. Natl. Acad. Sci. USA 106: 4555–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki, S., Matsuo, S., Wong, H.L., Yokoi, S. and Shimamoto, K. (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036. [DOI] [PubMed] [Google Scholar]
- Tamaki, S., Tsuji, H., Matsumoto, A., Fujita, A., Shimatani, Z., Terada, R., Sakamoto, T., Kurata, T. and Shimamoto, K. (2015) FT-like proteins induce transposon silencing in the shoot apex during floral induction in rice. Proc. Natl. Acad. Sci. USA 112: E901–E910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka, K., Ohki, I., Tsuji, H., Furuita, K., Hayashi, K., Yanase, T., Yamaguchi, M., Nakashima, C., Purwestri, Y.A., Tamaki, S.et al. (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335. [DOI] [PubMed] [Google Scholar]
- Teo, C.-J., Takahashi, K., Shimizu, K., Shimamoto, K. and Taoka, K. (2017) Potato tuber induction is regulated by interactions between components of a tuberigen complex. Plant Cell Physiol. 58: 365–374. [DOI] [PubMed] [Google Scholar]
- Torti, S., Fornara, F., Vincent, C., Andres, F., Nordstrom, K., Gobel, U., Knoll, D., Schoof, H. and Coupland, G. (2012) Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24: 444–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, H., Taoka, K. and Shimamoto, K. (2011) Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 14: 45–52. [DOI] [PubMed] [Google Scholar]
- Tsuji, H., Nakamura, H., Taoka, K. and Shimamoto, K. (2013a) Functional diversification of FD transcription factors in rice, components of florigen activation complexes. Plant Cell Physiol. 54: 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, H., Taoka, K. and Shimamoto, K. (2013b) Florigen in rice: complex gene network for florigen transcription, florigen activation complex, and multiple functions. Curr. Opin. Plant Biol. 16: 228–235. [DOI] [PubMed] [Google Scholar]
- Tsuji, H. and Taoka, K. (2014) Florigen signaling. Enzymes 35: 113–144. [DOI] [PubMed] [Google Scholar]
- Tsuji, H., Tachibana, C., Tamaki, S., Taoka, K., Kyozuka, J. and Shimamoto, K. (2015) Hd3a promotes lateral branching in rice. Plant J. 82: 256–266. [DOI] [PubMed] [Google Scholar]
- Tylewicz, S., Tsuji, H., Miskolczi, P., Petterle, A., Azeez, A., Jonsson, K., Shimamoto, K. and Bhalerao, R.P. (2015) Dual role of tree florigen activation complex component FD in photoperiodic growth control and adaptive response pathways. Proc. Natl. Acad. Sci. USA 112: 3140–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.W., Czech, B. and Weigel, D. (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749. [DOI] [PubMed] [Google Scholar]
- Wigge, P.A., Kim, M.C., Jaeger, K.E., Busch, W., Schmid, M., Lohmann, J.U. and Weigel, D. (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059. [DOI] [PubMed] [Google Scholar]
- Zeevaart, J.A. (2006) Florigen coming of age after 70 years. Plant Cell 18: 1783–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Liu, L., Shen, L. and Yu, H. (2016) NaKR1 regulates long-distance movement of FLOWRING LOCUS T in Arabidopsis. Nat. Plants 2: 16075. [DOI] [PubMed] [Google Scholar]