Abstract
The immune system varies in cell types, states, and locations. The complex networks, interactions and responses of immune cells produce diverse cellular ecosystems composed of multiple cell types, accompanied by genetic diversity in antigen receptors. Within this ecosystem, innate and adaptive immune cells maintain and protect tissue function, integrity and homeostasis upon changes in functional demands and diverse insults. Characterizing this inherent complexity requires studies at single-cell resolution. Recent advances such as, massively-parallel single cell RNA-Seq and sophisticated computational methods are catalysing a revolution in our understanding of immunology. Here, we provide an overview of the state of single cell genomics methods and an outlook on the use of single-cell techniques to decipher the adaptive and innate components of immunity.
Introduction
The immune system is composed of different cell lineages that reside in primary lymphoid organs; in secondary lymphoid organs, in tissues throughout the body; and that transit through the peripheral blood and lymphatic systems. The cells in these lineages are primary responders to changes in the environment, eliciting a complex network of intracellular circuits and intercellular interactions that result in transient responses within and between cells and cell states, more permanent differentiation choices, and flexible adaptation to their tissue of residence. Thus, the cells of each lymphoid and non-lymphoid tissue are key members of diverse “cellular ecosystems” composed of multiple immune and non-immune cell types, which together maintain and protect tissue function, integrity and homeostasis upon changes in functional demands, including insults and injuries. Hence, immunity involves innate and adaptive immune cells interacting with additional cells to form dynamic cellular communities in tissues.
In seminal studies, immunologists have developed an extensive taxonomy of the cells of the immune system, integrating and unifying their functional characteristics, cell fate, and lineage relations with molecular markers. This effort was enabled by tools ranging from microscopy and flow cytometry to functional assays, animal models and, most recently, genomics.
However, the immune cell census remains incomplete. The immune system harbors a breadth of cell types and states, each of which can be at different stages of differentiation or response to environmental cues such as pathogens. In addition, because of the immune system’s distributed nature, the “same” cell types and states are present in locations throughout the body, but are modified by adaptations that reflect the unique niche and functional demand of their tissue of residence (reviewed in 1). Immune cells pose a further challenge: lymphocytes with particular antigen receptor sequences (such as classical T and B cells, but also iNKT cells, gamma/delta T cells and other populations) are clonal in nature, which introduces subtle yet important genetic diversity into these cell populations. Recent advances in single-cell genomics technologies are beginning to allow us to fill in these gaps by inspecting the immune system one cell at a time.
Technologies for characterisation of the cells of the immune system
Over the years, three major techniques have established themselves for the categorization of immune cells. The most prominent is immunophenotyping through flow cytometry, which can identify cells of the immune system (whilst in suspension) by the single cell expression of both cell-surface and intracellular proteins, including cytokines, and their post-translational modifications (reviewed in 2). In addition, staining, sorting and enrichment or depletion of specific viable cell subsets, including rare cell types, can then be used for downstream experiments. Advances in instrumentation, expansion of the number of parameters measured, and standardization of assays has increased the power, resolution and impact of flow cytometry.
These assays of immune cell suspensions have been complemented by histological assays in situ, for both RNA and proteins, including in situ hybridization (ISH) and single-molecule RNA-fluorescence in situ hybridisation (smRNA-FISH) (reviewed in Lein, Science, this issue) for RNA and immunohistochemistry (IHC) for proteins. Microscopy methods provide high-definition spatial representation of cell types, cell boundaries, neighbors or interacting cells, niches, and tissue contexts, and have been used to characterize immune cells (reviewed in 3). More recently, comprehensive profiling of selected bulk populations of large numbers of cells, including of entire transcriptomes and proteomes, helped discover additional markers (4).
While each of these approaches provided invaluable insights, they suffer from complementary limitations. Single-cell approaches, such as flow cytometry and fluorescence activated cell sorting (FACS), or immunofluorescence and in situ hybridization, have been limited to probing a few selected RNAs or proteins, hindering our ability to study comprehensive profiles and to uncover novel factors due to a bias towards pre-characterized genes. Conversely, genomic analyses have either relied on profiling heterogeneous mixtures, whose ensemble average obscures the diversity of cells in the sample, or, have relied on first sorting sub-populations and then profiling them. The latter sorting strategy is limited to known sub-populations and sorting panels, and can be difficult to implement for small samples, while still masking variation within the sub-population.
Recent advances in single-cell genomics and spatial profiling methods
Over the past few years, the revolution in single-cell genomics has enabled an unbiased genome-wide quantification of molecules in thousands of individual cells, as well as multiplex spatial analysis of proteins and RNA in situ.
Among the single cell profiling approaches, the most mature and widely disseminated method is single-cell RNA-sequencing (scRNA-seq), which aims to measure the expression levels of genes in cells in a comprehensive way. scRNA-seq can be both sensitive and accurate (5, 6) despite the minute amounts of starting material in an individual cell. scRNA-seq has grown over time in both accuracy and scale, from a handful of individual cells to hundreds of thousands in a single experiment (reviewed in 7). Importantly, despite the relative sparsity of the measured profile from any individual cell, many single cell profiles can be analysed with statistical methods that can quantify even subtle changes in gene expression between individual cells (cell-to-cell variation), and also any dynamic expression changes during a response. This has been particularly important in the analysis of profiles of the genomes (reviewed in 8) and epigenomes as either single or simultaneous ‘multi-omics’ modalities (reviewed by Kelsey et al., Science, this issue) of single cells.
Whilst extremely useful, the measurement of RNA expression does not provide information about protein abundance nor post-translational modifications; in a key parallel advance, mass cytometry (CyTOF) (9) allows the detection of dozens of proteins at once in each of millions of individual cells, by mass spectrometric (MS) detection of metal-labelled antibodies. These can also be applied by staining in situ, for spatial measurements (reviewed in 10). Protein and RNA measurements can also be combined, either through index sorting followed by scRNA-seq (11), or through mass cytometry (e.g., PLAYR (12)), or by first staining with DNA-barcoded antibodies and then performing scRNA-seq, to detect both the DNA barcode and profile the transcriptome (CITE-Seq, (13)).
We expect spatially-resolved methods (reviewed in Lein, Science, this issue) to dramatically enhance our understanding of the composition and function of the immune system in future studies but we do not discuss them further here. Instead, we focus on the insights provided by scRNAseq-based approaches.
The scale, unique characteristics and throughput of single-cell genomics and spatial techniques are rapidly leading to the accumulation of complex data, which presents analytical challenges and exciting opportunities. These are an active and exciting area of research, reviewed elsewhere (14, 15).
Insights from single-cell analysis of the immune system
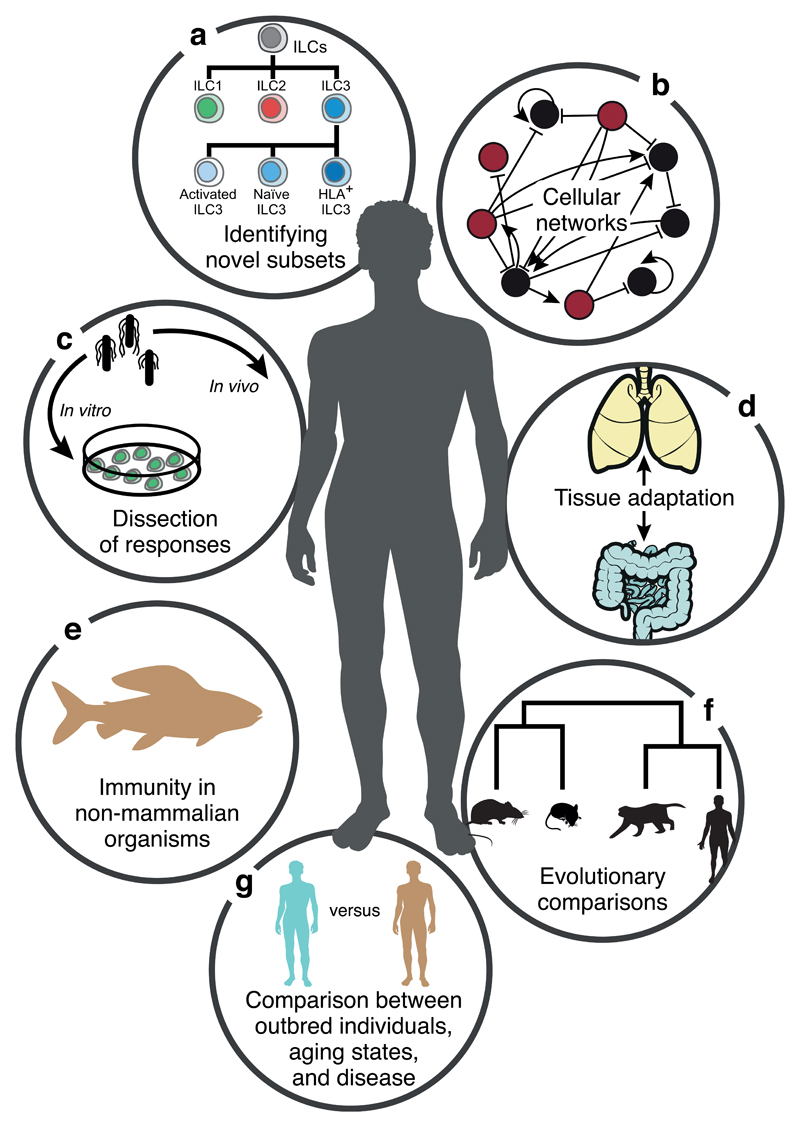
Single cell genomics studies can inform on many key aspects of the immune system (Fig. 1). As the cost of performing single-cell experiments has decreased, it has allowed profiling of a larger number of immune cells across different conditions. Studies have explored both the innate and adaptive branches of the vertebrate immune system with single-cell genomics techniques, providing important insights. These will guide biomedical research, and inspire the next generation of therapies directed towards inflammation, autoimmunity and cancer.
Figure 1. Single cell genomics in immunology.
The immune system is well-suited to studies at single-cell resolution. Immune responses involve a wide variety of cell types that can be further subdivided into more fine-grained subtypes and distinct cell states that can be followed throughout the duration of a response. Furthermore, single-cell analyses provide insights into intercellular networks and allow us to compare immune responses across individuals and species.
Innate immunity
Innate immune responses represent the first barrier of fast-acting defense against pathogens. These intrinsic, cell-autonomous responses are mounted upon stimulation with various pathogen-associated molecules (e.g., viral elements such as dsRNA, or bacterial components such as LPS). Single-cell approaches have been used to study these responses’ stochasticity, for example in dendritic cells (16, 17) and to determine their evolutionary architecture in fibroblasts (18). Further studies of innate immune cells that mediate a rapid, but non-adaptive immune response have illustrated how single-cell genomics can identify novel subpopulations and increase our insight into immune mechanisms.
Dendritic cells, first responders in the immune system
Dendritic cells (DCs) are sentinels of the immune system and are present in the blood, lymphoid organs and across many tissues. These cells engulf antigens, then migrate to lymph nodes to present the antigens to T cells. Early scRNA-seq experiments in immune cells were performed on just eighteen bone marrow-derived mouse dendritic cells (BMDCs) after 4h of stimulation with lipopolysaccharide (LPS), a component of gram-negative bacteria (16). This revealed cellular heterogeneity in cell subsets, co-expressed gene programs and splicing variants. In particular, hundreds of key immune genes were bimodally expressed across cells at four hours after stimulation: whilst some cells expressed them at very high levels, there was little, if any, expression in other cells. A following study profiled 1,700 mouse BMDCs by scRNA-seq along a time course of stimulation with each of three pathogen-associated molecular patterns (PAMPs) (17). This charted how heterogeneity across cells represents temporal changes (asynchronicity) in the response, and that cells are more or less synchronous in their activation and shut-off of different gene modules on different time scales. In particular, a ‘core’ module of antiviral genes is expressed very early by a few ‘precocious’ cells in response to uniform LPS stimulation, but is later activated in all cells. By stimulating and then profiling individual cells in complete isolation, this response was shown to be coordinated by interferon-mediated paracrine signalling from the precocious cells to other immune cells via cell-cell interactions.
Perturb-Seq experiments combine large-scale CRISPR-based perturbations in cells with single-cell RNA-seq and have been used to study this response in the context of dozens of genetic perturbations, especially in transcription factors (19, 20). This clarified the genetic programs controlling two different subsets of cells (19), controlled by different TFs. They also identified new functions for regulators of differentiation, the anti-viral response, and mitochondrial function during immune activation (19),_and identified gene interactions between TFs. Another study (20) further identified interactions and redundancies between developmental and signalling-dependent factors in other mononuclear phagocyte populations.
Single cell genomics can also help dissect the subsets of human innate immune cells. scRNA-seq studies using human DCs have revealed previously unreported DC subsets and shed light on the complexity of the lineage of these cells (21–23). For example, analyzing human tonsils and synovial fluid from rheumatoid arthritis patients showed cells from one novel DC subset are in direct proximity to T cells, supporting their role in mounting an immune response and in autoimmunity (22). Such initial profiling studies should lead to immunophenotyping of newly discovered markers and subsequent functional assays.
Innate Lymphoid Cells (ILCs) and their development
Innate lymphoid cells (ILCs) reside at mucosal barriers and are important for host defense and tissue homeostasis. ILCs are the innate immune system’s functional equivalent of T cell subsets of the adaptive immune system, composed of cytotoxic and non-cytotoxic cells, and resembling T lymphocytes in cytotoxicity and cytokine production but lacking antigen-specific receptors (24). The complete spectrum of immune tasks performed by ILCs is still unknown and, whilst there is evidence that they are important in the processes of inflammation and tissue homeostasis (reviewed in 25), they may be at least partly dispensable in the context of functional T and B cells (26).
The initial discovery of non-cytotoxic ILCs coincided with the era of single-cell RNA-seq, and these methods have helped reveal the heterogeneity, developmental pathways, regulatory circuits and responses to microbiota of ILCs. An analysis of 648 ILCs from human tonsils used gene expression profiles to separate the cells into the expected four distinct populations (ILC1, ILC2, ILC3 and NK cells) with unbiased clustering without relying on any a priori knowledge (27). The transcriptomes of the cells in the four known types, helped identify new marker genes; they also revealed three new subsets of ILC3 cells that are predicted to have distinct functions, on the basis of their cytokine responses and cytokine secretion profiles. Another study analyzed scRNA-Seq profiles of ILC subsets in the colon of mice under different microbiotic conditions (specific pathogen-free facility, antibiotic-treated and germ-free facility) (28). This revealed subsets within the ILC1, 2 and 3 populations, as well as a hybrid set of cell states that may represent plasticity between the ILC1 and ILC3 states.
Single-cell RNA-seq has also been instrumental in resolving details of the trajectory and regulation of the developmental pathway of ILCs (29) (Fig. 2A, B). Analysis of mouse bone marrow progenitors revealed precursor subsets, and delineated distinct ILC development stages and pathways. This study highlighted markers, such as the PD1 receptor on both progenitor cells and cells stimulated after influenza infection and lung inflammation, and regulation by the transcription factor Bcl11b. As the PD1 receptor’s ligand is targeted in tumour immunotherapy, these findings may have implications for disease and therapy.
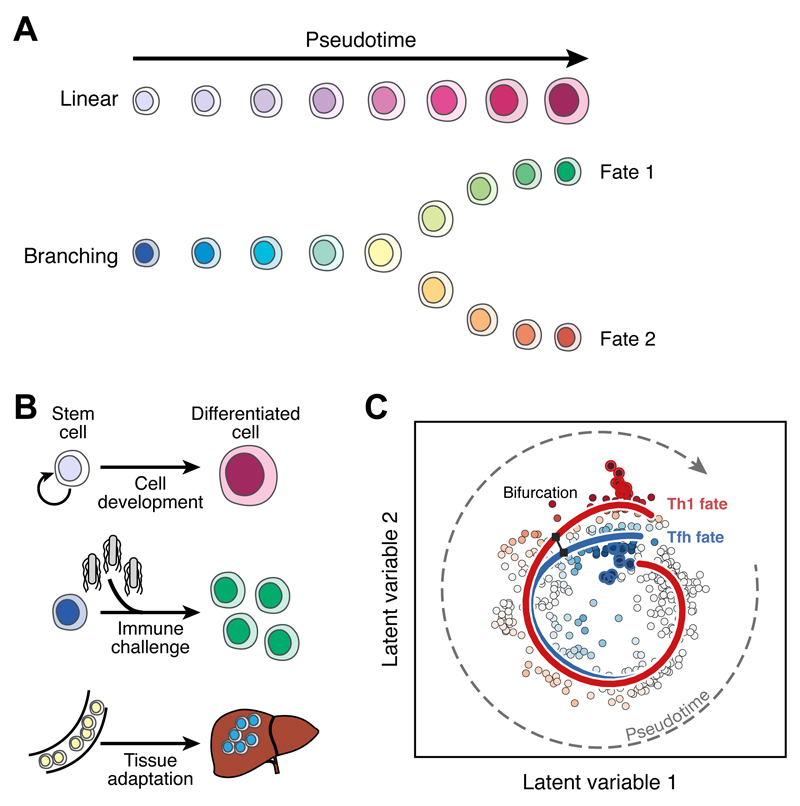
Figure 2. Inferring cellular trajectories from single-cell data.
(A) During a differentiation process, individual cells can be aligned along ‘pseudotime,’ which represents their progression within the differentiation pathway. The processes described in this way can be linear or can involve branches to multiple eventual fates. (B) Examples of biological processes analysed in terms of cellular trajectories include the progression of stem cells to terminally differentiated fates, the response of naive immune cells to infection, and the adaptation of circulating immune cells to the tissues where they ultimately reside. (C) A bifurcating pseudotime trajectory inferred from scRNAseq data generated from a mouse malaria infection model (adapted from (51), reprinted with permission from AAAS and modified with permission of the authors). Each point represents an individual cell following dimensionality reduction using a Bayesian Gaussian Process Latent Variable Model. These cells are then ordered in pseudotime and two simultaneous and bifurcating developmental trajectories (red and blue lines) are inferred using Overlapping Mixtures of Gaussian Processes. The colour of each point indicates the probability that a cell belongs to either the red or the blue trend.
The monocyte-macrophage lineage and its dynamics in health and disease
Macrophages, a broad family of phagocytic cells, not only play a role in encounters with pathogens, but also have diverse roles in tissue homeostasis (e.g., microglia in the brain, osteoclasts in the bone) (30–32). Single cell profiling has shed light on both types of roles, as well as tissue adaptation.
Profiling mouse macrophages infected with fluorescently labelled Salmonella enterica serovar Typhimurium revealed diverse macrophage states correlating with bacterial survival and proliferation, and linking pathogen state to host signalling pathways (33). A similar approach, focused on pathogen proliferation and macrophage response, identified an M2-like state in macrophages infected with actively proliferating bacteria (34). Developing scRNA-seq methods that measure the transcriptional changes in both the immune cell and the pathogen will help inform on the function and mechanisms employed by these cells in defense.
Microglia are a particular lineage of cells within the macrophage family; the tissue resident macrophages in the brain. Surveys with scRNA-seq of the developmental progression of microglia, identified their activation in a mouse model of Alzheimer’s disease (20, 35). A novel microglia subset was associated with neurodegenerative diseases, and linked with relevant markers, spatial localization, and the activity of specific cellular pathways. From the specific sequence of disease progression in animal models, this unique microglia-type showed potential to restrict neurodegeneration.
Comparative studies of innate immune cells and cell-intrinsic responses
One of the challenges in immunology has been to understand how the immune system has evolved across different animal models relative to humans. This was exacerbated by the fact that both compositional and cell intrinsic changes affect such comparisons based on bulk profiles, that subsets defined by cell surface markers may be challenging to match, and that antibodies and markers may be lacking in non-model organisms.
scRNA-seq helps circumvent these challenges. For example, T-cells, NK cells and myeloid-like cell transcriptomes were characterized in zebrafish, a vertebrate model organism (36). In another example, scRNA-seq profiled the cell intrinsic innate immune response of fibroblasts across four mammalian species stimulated with dsRNA (18). This highlights the relationship among divergent gene expression across species, individual cells, and protein sequence divergence.
Adaptive immunity
Novel lymphocyte subsets
Immunologists have gained an understanding of the diversity of lymphocyte subpopulations and their roles in immunity, characterizing populations of lymphocytes such as B cells, CD4+ and CD8+ T cells, NK cells and iNKT cells. Leveraging these advances, scRNA-seq is making further important discoveries of new lymphocyte subpopulations and states, their molecular underpinnings and their relationship to physiology and disease.
For example, the plasticity and complexity of the CD4+ T cell compartment makes it difficult to understand with bulk techniques but particularly amenable to investigation by scRNA-seq methods. scRNAseq analysis of a mouse CD4+ T cell population enabled correlation of a steroidogenic enzyme with a cell surface marker that was used to enrich this subpopulation for further functional characterization (37).
Other studies have focused on the characterization of variation within an immune subset, which can be continuous rather than involving further discrete subsets. For example, scRNA-seq helped characterize the variation within Th17 populations, which can span a continuous spectrum correlated with distinct levels of “pathogenicity”, or the ability to induce an autoimmune disease in an animal model upon adoptive transfer (38, 39). scRNA-seq of Th17 cells from in vivo autoimmune models and from in vitro polarization experiments helped characterize the genetic programs that underlie this diversity, and shed light on the mechanisms that control them.
These strategies are proving beneficial in the context of immunooncology, in both mouse models and patient tumors. Studies of human melanoma (40) and hepatocellular carcinoma (41), and a B16 melanoma mouse model (42), characterized diverse T cell states in the tumor ecosystem (see also below). These studies explored the relationship between cytotoxicity and cell exhaustion, and the roles of particular proteins in suppressive T cell states.
Lymphocyte differentiation
The high-resolution mapping of cell states by scRNA-seq can infer developmental processes in detail. In bulk transcriptomics methods, development and differentiation are studied with time series. In contrast, the single-cell resolution of scRNAseq allows the inference of time series from static snapshots, and reveals the continuum of cell states across timepoints (Figure 2A & B). Known as “pseudotime,” this unobserved dimension measures the progress of cells through a transition. For example, such an approach was used to resolve developmental progressions from single cell qPCR data (43, 44), and later applied to scRNA-seq data in zebrafish hematopoiesis, revealing a continuous spectrum of differentiation in hematopoietic cells (45). An early example of pseudotime inference in the immune system focused on B cells undergoing VDJ recombination. In this work the Wanderlust method inferred trajectories on the basis of similarity of cells according to CyTOF profiles (46).
Notably, when dynamic processes occur concurrently with cell activation or differentiation, for example the induction of cell proliferation in many immune processes, this specific contribution can be either identified and studied (47, 48), or isolated to highlight other phenomena (43, 49). This latter approach may be challenging when cell proliferation is coupled to other immune responses (15).
Layered on top of the progression of differentiation are cell fate decisions. These can be single bifurcations, such as CD4+ T cell fate choices, or complex hierarchies and other relations, as in the whole of haematopoiesis. With appropriate computational analyses, these cell fate decisions can be explored from both snapshot (when differentiation is asynchronous and ongoing) and time course data. Such approaches were applied to study both T cell development (50) and T helper cell differentiation into Th1 and Tfh fates (51) (Figure 2C).
Immune repertoire analyses
The adaptive immune response requires interactions between non-self antigens and antigen-specific receptor molecules expressed by T and B lymphocytes. T cell receptors (TCRs) detect antigenic peptides presented as complexes with major histocompatibility complex (MHC) proteins. B cell receptors (BCRs) bind to proteins or peptides without presentation and are also secreted from B cells as antibodies. The immune system can specifically recognize the necessarily large range of antigenic molecules thanks to the enormous potential diversity of antigen receptor (AgR) amino acid sequences. This diversity is generated by DNA rearrangement in each developing lymphocyte that combines randomly chosen gene segments, and introduces additional variability at the junctions between segments. The vast diversity of AgR molecules allows us to assume that cells with identical receptor DNA sequences arose from the same original developing lymphocyte.
The uniqueness and diversity of AgRs means that single-cell techniques are ideal to study antigen receptor repertoires, as well as their relation to the cells’ subtype and state. Furthermore, AgRs are heterodimeric proteins that comprise two independently encoded protein chains that both typically determine each receptor’s antigen specificity. Single-cell measurements identify the paired receptor chains in each cell.
One line of work focuses on the repertoire of TCR sequences at the single-cell level and provides insights into clonal fates and dynamics. Targeted sequencing of TCR sequences in individual cells combined with immunophenotyping of selected marker genes related fate and clonality in tumor infiltrating lymphocytes (TILs) in colorectal cancer (52). Cells from two populations of IL17-expressing cells (FOXP3-RORC+ and FOXP3+RORC+), often shared TCR sequences, implying a common ancestry. Furthermore, the use of droplet-based encapsulation and targeted AgR amplification detected paired sequences from hundreds of thousands of lymphocytes (53), along with two phenotyping markers.
More recently, AgR sequences have been studied in the context of scRNA-seq, which can simultaneously report on the receptor of a cell and its expression profile (40, 51, 54–56). TCR sequences assembled from scRNA-seq reads (54) (Fig. 3A) were used to demonstrate the breadth of transcriptional states within a single expanded T cell clonotype during Salmonella infection in mice. In another example, TCR analysis was combined with pseudotime and branching inference during the immune response to malaria to show that clonally related sibling cells can be found in both the Th1 and Tfh fates (51) (Fig. 3B). In human melanoma samples (40), similar analysis allowed the relation of the exhaustion state of the cell to the extent of a clone’s expansion.
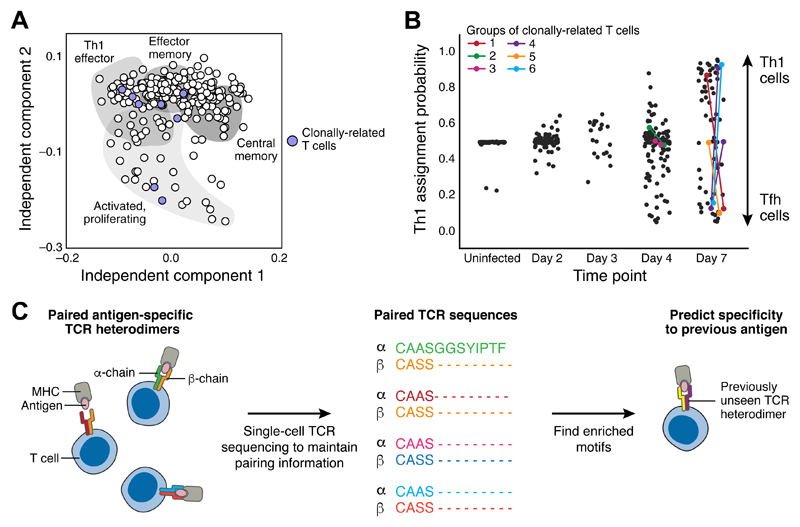
Figure 3. Single-cell analysis of antigen receptor sequences reveals clone distributions between transcriptional states.
(A) Independent component analysis of scRNAseq data from mouse splenic CD4+ T cells during salmonella infection. Each point represents an individual cell. Shaded areas indicate likely functional identities associated with each region of reduced dimensionality space. Purple-filled points indicate cells that are clonally related and share one particular set of TCR sequences. Clonally related cells are distributed throughout the gene expression space (adapted from (54)). (B) Cell type assignment of cells with scRNAseq data from mouse splenic CD4+ T cells at varying timepoints during malaria infection. Each point represents an individual cell with y-axis position indicating the likelihood that it is a Th1 cell rather than a Tfh cell (high values imply a Th1 identity, low values a Tfh identity). Colored points indicate pairs of cells inferred to be clonally related due to shared TCR sequences. Sibling cells can be found such that one is a Th1 cell whilst the other is a Tfh (adapted from (51), reprinted with permission from AAAS and modified with permission of the authors) (C) Datasets (57, 58) of linked alpha and beta TCR chains, provide enough power to allow machine learning inference of common sequence motifs from diverse T cells recognising the same antigen. This raises the possibility that it might be possible in future to more systematically associate TCR sequences with their cognate pMHC sequences. Figure adapted by the authors from (64).
The ideal next step would be to incorporate AgR detection into massively parallel droplet-based whole-transcriptome scRNA-seq. This would allow clonality to be linked to phenotypic state in thousands of cells, at least at a coarse-grained level, and hence illuminate the development of lineages of lymphocyte cell states at unprecedented resolution.
Finally, the systematic and large-scale single cell analysis of AgRs has recently provided evidence that it may be possible to predict a receptor’s cognate antigen using only the receptor sequence, potentially leading to the development of a clinical diagnostic tool. Paired single cell TCR sequences in a variety of antigen-specific T cell populations from multiple donors were determined along with enriched sequence motifs associated with binding to each particular antigen (57, 58). The presence of these enriched motifs in previously unseen TCRs enabled prediction of binding specificity of these receptors (Fig. 3C). Although these studies still require large training sets of TCR sequences with pre-existing knowledge of specificity, they are likely to be an early step on the road to generalizable prediction of TCR specificity from sequence.
Cellular ecosystems: an integrative view of cell-cell interactions in immunity
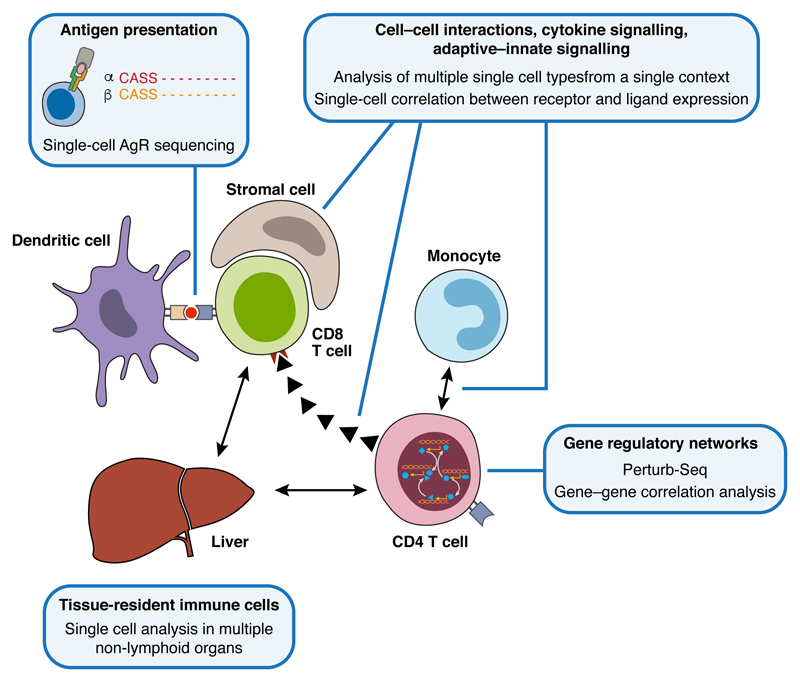
The immune system requires a complex network of interactions to perform its functions (Fig. 4). An integrated view of individual cells, cell types, their spatial organization and functional interaction is needed to achieve a systems-level view of immune function. Single-cell approaches are already accelerating our integrated understanding of immunity and we anticipate much future work in this area.
Figure 4. Network interactions within the immune system.
Immune responses involve networks at multiple scales, ranging from intracellular gene regulatory networks through to long-distance intercellular communication mediated by cytokines or chemokines. A systems approach to understanding these networks will be crucial if we are to fully understand immune biology, and will be accelerated by the application of multiple, different single-cell analysis methods.
Existing single cell studies have begun to demonstrate the power of addressing multiple parts of the immune response at the same time. An analysis of 4,645 individual cells from melanoma patient samples revealed the complexity of the cellular ecosystem present in the tumors (40), and the extent to which it can be compared across individual patients. While malignant cell types exhibit substantial diversity between patients, non-malignant cells (including T cells, B cells, macrophages, endothelial cells, cancer-associated fibroblasts (CAFs), and NK cells) each grouped by type rather than tumor origin. Similar phenomena were observed in other tumor types, for example glioma (59). Moreover, when the single cell profiles are combined with bulk profiles from hundreds of patients, collected by The Cancer Genome Atlas (60, http://cancergenome.nih.gov/), they reveal dependencies between different cell types and the molecules that may mediate them. For example, the correlation between high levels of CD8+ T cells in a tumor and the expression of Complement proteins by its CAFs (40).
Another study analyzed splenic CD4+ T cells, monocytes and DCs in the mouse response to malaria (51), comparing uninfected mice to mice at day 3 post infection. Integrated analyses of the changes in expression of chemokine ligands and receptors across these populations predicted that monocytes would support the differentiation of activated CD4+ T cells towards a Th1 fate rather than the alternative Tfh fate. This was confirmed in an experiment where monocytes were depleted during T cell activation and prior to fate choice.
Both studies inferred processes of intercellular communication from the expression of cognate receptors, co-receptors and/or ligands in scRNA-seq data. Systematic and generalizable approaches for such connections, and relating them to co-variation in cell proportions and states will be of great value. Furthermore, it is clear that methods that analyze single-cell gene expression in a spatially resolved context will be very important to understanding the interactions between cells of the immune system. Indeed, imaging mass cytometry has identified the presence of immune cells within the spatial context of breast cancer tissues (61).
Future applications in immunity
The immune system is comprised of numerous cell types that work in concert to sense and appropriately respond to foreign challenges and physiological changes in order to monitor and maintain health. If the carefully orchestrated functioning of the immune system is perturbed, diseases such as infectious disease, autoimmune disease and cancer can arise.
The rich taxonomies and cell fate maps generated in immunology over the past several decades relate cells by cellular function, differentiation potential and expression of marker proteins. However, to date there is still no complete reference map of immune cells. The additional comprehensive profiles provided by single cell genomic approaches provide an important new tool in this endeavor, helping to address some of the fundamental questions in immunology - from the taxonomy of cells, histological structure in tissues, recruitment to tissues, developmental biology and cell fate and lineage, to physiology and homeostasis and their underlying molecular mechanisms.
Moreover, in order to deeply understand the full scope and function of immune cells, it is most informative to study immune cells in a challenged state—that is, as manifested during disease, infection, development, aging and environmental changes. Studying those in humans often requires handling minute samples; single cell genomic approaches are highly compatible with such limitations in input material.
Finally, genetic perturbations (natural or engineered) can also elicit changes in gene expression that may differ in response to response to environmental cues or changes such as aging (16, 17, 62). As the cost of single-cell experiments goes down, profiling more immune cells across conditions will be possible, through pooled genetic screens, and by economically multiplexing a large number of individuals, using their sequence variations as a natural genetic barcode (63).
Combining single-cell genomics, emerging spatial approaches, immune repertoire analysis, multiplex immunophenotyping, together with established approaches for functional analysis, could also impact the next generation of diagnostics and therapies. For diagnostics, the white cell blood count (WBC) might be reimagined from a tally of major cell populations to an assay that identifies cell signatures (defined by single-cell genomics) of cell types and states and their proportions. For therapy, comparing the role and mechanisms of immune cells in cellular ecosystems between health and disease tissues can help identify new therapeutic targets, as well as better assess the impact of current therapies in the context of clinical trials.
To help usher in this future, as part of the international Human Cell Atlas initiative (www.humancellatlas.org), an effort to generate an Immune Cell Atlas (ICA) is emerging. The ICA will assess the immune system at different stages of differentiation, across different tissues, and in the context of a wide range of diseases. To properly survey the spectrum of immune cells, even the initial pilot effort will include samples from small numbers of patients with a diversity of diseases. Such a systematic characterisation of immunity requires an international collaboration between clinicians, immunologists, genomics experts and computational biologists.
Overall, these types of approaches and projects stand to radically transform our knowledge of immune function and dysfunction in infection, autoimmunity, allergy, inflammatory disorders and cancer and impact therapeutic developments.
Acknowledgments
We thank Jennifer E. Rood, Robert Majovski, Valentine Svensson, Leslie Gaffney and Kerstin Meyer for help with preparation of this manuscript. AR was supported by funds from the Howard Hughes Medicine Institute, NIAID grants U24AI118672 and R24AI072073, the Manton Foundation, and the Klarman Cell Observatory. MJTS and SAT were supported by the Wellcome Trust Grant 206194. AR is a scientific advisory board member for ThermoFisher Scientific and Syros Pharmaceuticals and a consultant for Driver Group; the other authors have no conflicts of interest.
References
- 1.Immunity in the tissues. Nat Immunol. 2013;14:977. doi: 10.1038/ni.2722. [DOI] [PubMed] [Google Scholar]
- 2.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4:648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 3.Bajénoff M, Germain RN. Seeing is believing: a focus on the contribution of microscopic imaging to our understanding of immune system function. Eur J Immunol. 2007;37(Suppl 1):S18–33. doi: 10.1002/eji.200737663. [DOI] [PubMed] [Google Scholar]
- 4.Shay T, Kang J. Immunological Genome Project and systems immunology. Trends Immunol. 2013;34:602–609. doi: 10.1016/j.it.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svensson V, et al. Power Analysis of Single Cell RNA- Sequencing Experiments. bioRxiv. 2016:073692. doi: 10.1038/nmeth.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Svensson V, Vento-Tormo R, Teichmann SA. Moore’s Law in Single Cell Transcriptomics. arXiv [q-bio.GN] 2017 available at http://arxiv.org/abs/1704.01379. [Google Scholar]
- 8.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 9.Bandura DR, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 10.Bodenmiller B. Multiplexed Epitope-Based Tissue Imaging for Discovery and Healthcare Applications. Cell Syst. 2016;2:225–238. doi: 10.1016/j.cels.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Wilson NK, et al. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell. 2015;16:712–724. doi: 10.1016/j.stem.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frei AP, et al. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat Methods. 2016;13:269–275. doi: 10.1038/nmeth.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoeckius M, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017 doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rostom R, Svensson V, Teichmann SA, Kar G. Computational approaches for interpreting scRNA-seq data. FEBS Lett. 2017 doi: 10.1002/1873-3468.12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner A, Regev A, Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol. 2016;34:1145–1160. doi: 10.1038/nbt.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shalek AK, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagai T, Chen X, Miragaia RJ, Gomes T, Rostom R. A Balance Between Regulatory Constraints And Pathogen Pressure Shapes The Evolution Of Innate Immunity. bioRxiv. 2017 (available at http://biorxiv.org/content/early/2017/05/15/137992.abstract) [Google Scholar]
- 19.Dixit A, et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaitin DA, et al. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell. 2016;167:1883–1896.e15. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 21.Breton G, et al. Circulating precursors of human CD1c+ and CD141+ dendritic cells. J Exp Med. 2015;212:401–413. doi: 10.1084/jem.20141441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villani A-C, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356 doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.See P, et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 2017;356 doi: 10.1126/science.aag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eberl G, Colonna M, Di Santo JP, McKenzie ANJ. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348:aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 26.Vély F, et al. Evidence of innate lymphoid cell redundancy in humans. Nat Immunol. 2016;17:1291–1299. doi: 10.1038/ni.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Björklund ÅK, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol. 2016;17:451–460. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- 28.Gury-BenAri M, et al. The Spectrum and Regulatory Landscape of Intestinal Innate Lymphoid Cells Are Shaped by the Microbiome. Cell. 2016;166:1231–1246.e13. doi: 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 29.Yu Y, et al. Single-cell RNA-seq identifies a PD-1(hi) ILC progenitor and defines its development pathway. Nature. 2016;539:102–106. doi: 10.1038/nature20105. [DOI] [PubMed] [Google Scholar]
- 30.Okabe Y, Medzhitov R. Tissue biology perspective on macrophages. Nat Immunol. 2016;17:9–17. doi: 10.1038/ni.3320. [DOI] [PubMed] [Google Scholar]
- 31.Lavin Y, Mortha A, Rahman A, Merad M. Regulation of macrophage development and function in peripheral tissues. Nat Rev Immunol. 2015;15:731–744. doi: 10.1038/nri3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amit I, Winter DR, Jung S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol. 2016;17:18–25. doi: 10.1038/ni.3325. [DOI] [PubMed] [Google Scholar]
- 33.Avraham R, et al. Pathogen Cell-to-Cell Variability Drives Heterogeneity in Host Immune Responses. Cell. 2015;162:1309–1321. doi: 10.1016/j.cell.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saliba A-E, et al. Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella. Nat Microbiol. 2016;2:16206. doi: 10.1038/nmicrobiol.2016.206. [DOI] [PubMed] [Google Scholar]
- 35.Keren-Shaul H, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 2017;169:1276–1290.e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 36.Carmona SJ, et al. Single-cell transcriptome analysis of fish immune cells provides insight into the evolution of vertebrate immune cell types. Genome Res. 2017;27:451–461. doi: 10.1101/gr.207704.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahata B, et al. Single-cell RNA sequencing reveals T helper cells synthesizing steroids de novo to contribute to immune homeostasis. Cell Rep. 2014;7:1130–1142. doi: 10.1016/j.celrep.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaublomme JT, et al. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, et al. CD5L/AIM Regulates Lipid Biosynthesis and Restrains Th17 Cell Pathogenicity. Cell. 2015;163:1413–1427. doi: 10.1016/j.cell.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng C, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 2017;169:1342–1356.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 42.Singer M, et al. A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell. 2016;166:1500–1511.e9. doi: 10.1016/j.cell.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsang JCH, et al. Single-cell transcriptomic reconstruction reveals cell cycle and multi-lineage differentiation defects in Bcl11a-deficient hematopoietic stem cells. Genome Biol. 2015;16:178. doi: 10.1186/s13059-015-0739-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buettner F, Theis FJ. A novel approach for resolving differences in single-cell gene expression patterns from zygote to blastocyst. Bioinformatics. 2012;28:i626–i632. doi: 10.1093/bioinformatics/bts385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macaulay IC, et al. Single-Cell RNA-Sequencing Reveals a Continuous Spectrum of Differentiation in Hematopoietic Cells. Cell Rep. 2016;14:966–977. doi: 10.1016/j.celrep.2015.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bendall SC, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scialdone A, et al. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 48.Leng N, et al. Oscope identifies oscillatory genes in unsynchronized single-cell RNA-seq experiments. Nat Methods. 2015;12:947–950. doi: 10.1038/nmeth.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buettner F, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 50.Setty M, et al. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol. 2016;34:637–645. doi: 10.1038/nbt.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lönnberg T, et al. Single-cell RNA-seq and computational analysis using temporal mixture modeling resolves TH1/TFH fate bifurcation in malaria. Science Immunology. 2017;2:eaal2192. doi: 10.1126/sciimmunol.aal2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Briggs AW, et al. Tumor-infiltrating immune repertoires captured by single-cell barcoding in emulsion. bioRxiv. 2017:134841. [Google Scholar]
- 54.Stubbington MJT, et al. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods. 2016;13:329–332. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Canzar S, Neu KE, Tang Q, Wilson PC, Khan AA. BASIC: BCR assembly from single cells. Bioinformatics. 2017;33:425–427. doi: 10.1093/bioinformatics/btw631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eltahla AA, et al. Linking the T cell receptor to the single cell transcriptome in antigen-specific human T cells. Immunol Cell Biol. 2016;94:604–611. doi: 10.1038/icb.2016.16. [DOI] [PubMed] [Google Scholar]
- 57.Dash P, et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017;547:89–93. doi: 10.1038/nature22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glanville J, et al. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547:94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venteicher AS, et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science. 2017;355 doi: 10.1126/science.aai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol. 2015;19:A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 62.Martinez-Jimenez CP, et al. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science. 2017;355:1433–1436. doi: 10.1126/science.aah4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang HM, et al. Multiplexing droplet-based single cell RNA-sequencing using natural genetic barcodes. bioRxiv. 2017:118778. [Google Scholar]
- 64.Reddy ST. Immunology: The patterns of T-cell target recognition. Nature. 2017 doi: 10.1038/nature23091. [DOI] [PubMed] [Google Scholar]