Abstract
Purpose of review
The goal of this review is to summarize the current knowledge in the field regarding the non-canonical activation of the NRF2 pathway. Specifically, we address what role p62 plays in mediating this pathway, which pathologies have been linked to the p62-dependent activation of NRF2, as well as what therapeutic strategies could be used to treat diseases associated with the non-canonical pathway.
Recent findings
It has recently been shown that autophagic dysfunction leads to the aggregation or autophagosomal accumulation of p62, which sequesters KEAP1, resulting in prolonged activation of NRF2. The ability of p62 to outcompete NRF2 for KEAP1 binding depends on its abundance, or post-translational modifications to its key domains. Furthermore, the relevance of the p62-dependent activation of NRF2 in disease has been demonstrated in human hepatocellular carcinomas, as well as neurodegenerative diseases.
Summary
These findings indicate that targeting p62, or the enzymes that modify it, could prove to be an advantageous strategy for treating diseases associated with autophagy dysregulation and prolonged activation of NRF2. Other therapeutic possibilities include restoring proper autophagic function, or directly inhibiting NRF2 or its targets, to restore redox and metabolic homeostasis. Future studies will help further clarify the mechanisms, regulation, and relevance of the non-canonical pathway in driving disease pathogenesis.
Keywords: non-canonical pathway, NRF2, KEAP1, autophagy, p62, therapeutics
Introduction
The cellular response to stress consists of an interconnected network of signaling pathways designed to sense the stressor, mitigate the damage, and initiate a response that corrects the insult and restores the cell to homeostasis. Which stress response pathways are activated depends on the timing and nature of the insult, as well as the cell and tissue type in which the insult occurs, with some of the same cellular machinery playing very different roles in chronic versus acute responses in different cell types. One stress response pathway critical in responding to increased xenobiotic or oxidative stress is the NRF2-KEAP1 pathway. Nuclear factor E2-related factor 2 (NRF2) is a nuclear transcription factor that transcribes anti-oxidant response element (ARE)-containing genes, many of which encode anti-oxidant and detoxifying enzymes critical in mitigating oxidative damage and restoring metabolic and redox homeostasis to the cell. Under normal physiological conditions, NRF2 is bound in the cytosol by Kelch-like ECH-associated protein 1 (KEAP1), which recruits the Cullin-3/Ring-Box1 (Cul3/Rbx1) E3 ubiquitin ligase complex, ubiquitylating NRF2, thus targeting it for proteasomal degradation [1]. However, under stress conditions, oxidative or electrophilic modification of key cysteine residues in KEAP1 (i.e. C151) results in a conformational change that prevents ubiquitylation of NRF2, allowing newly synthesized NRF2 to translocate to the nucleus and initiate transcription of its target genes [2]. This regulated mode of NRF2 activation, which occurs as a direct result of increased oxidative or electrophilic stress, is termed “canonical activation”, and many disease treatment strategies harness the protective power of a controlled NRF2 response, including prevention of cancer and diabetes, by utilizing the electrophilic modification of KEAP1 to transiently activate the NRF2 pathway [3–7].
Another pathway that plays a critical role in mediating oxidative stress is the autophagy-lysosome pathway. Autophagy is a tightly regulated cellular degradation pathway responsible for the removal of damaged proteins and organelles, including oxidatively damaged proteins and dysfunctional mitochondria, both of which can propagate further oxidative damage and cellular dysfunction if not properly removed. The autophagic response consists of three main stages, initiation, elongation, and fusion, with each step being mediated by a coordinated set of protein-protein interactions that are critical in ensuring proper flux through the pathway. The importance of autophagy in removing damaged cellular constituents is evidenced by the fact that autophagic dysfunction at any step in the pathway results in the accumulation of pathogenic proteins and organelles, an underlying cause of a number of disease states, including neurodegeneration, cardiovascular disease, liver disease, lung disease, and cancer [8, 9]. Importantly, a link between dysregulation of the autophagy pathway and activation of NRF2 has also been demonstrated. Autophagy blockage, either via genetic ablation of the key autophagy initiation proteins Beclin-1, ATG5, or ATG7, or exposure to the environmental toxicant arsenite, results in the accumulation of the autophagy adapter protein p62/SQSTM1 [10, 11]. The accumulation of p62 is a hallmark of pathologies associated with autophagic dysfunction, and since p62 is a multi-domain protein that can interact with a host of protein targets, its accumulation results in the sequestration and loss of function of a number of its binding partners, including KEAP1 [12].
KEAP1 interacts with the KEAP1-interacting region (KIR) of p62 via a DPSTGE motif [13, 14], which is similar to the ETGE motif involved in NRF2-KEAP1 binding. The sequestration of KEAP1 by p62 allows stabilization of NRF2, which can then translocate to the nucleus and initiate target gene transcription. Since activation of NRF2 occurs independently of cysteine modifications, this mode of p62-dependent regulation of NRF2 is termed “non-canonical activation” [13]. In this review, we will discuss the current understanding of the mechanisms and disease relevance of the non-canonical activation of the NRF2 pathway. Furthermore, we will examine the multi-faceted role of p62 in mediating this response, as well as possible therapeutic approaches that could be utilized to restore proper regulation of both the autophagy and NRF2 pathways to treat diseases where the non-canonical activation of NRF2 is prevalent.
The non-canonical NRF2 pathway
Discovery of the non-canonical activation of NRF2 dates back to 2010, when five separate groups, including ours, discovered that p62 competes with NRF2 for KEAP1 binding [14, 11, 13, 15, 16]. This p62-KEAP1-NRF2 axis was further validated by the discovery that p62 contains an ARE, with the p62-dependent upregulation of NRF2 increasing transcription of p62 itself, creating a positive feedback loop that promotes prolonged activity of NRF2 [14]. Intriguingly, the p62 ARE was recently verified, along with a number of ARE-like sequences in other key autophagy proteins, further demonstrating a crucial link between NRF2 activation and the autophagy pathway [17]. Mentioned briefly above, the non-canonical mechanism of NRF2 activation is driven by autophagic dysfunction, with the autophagosomal accumulation of p62 and sequestration of its target proteins, including KEAP1, leading to prolonged activation of the NRF2 pathway. Autophagic dysfunction can occur as a result of genetic ablation of key autophagy proteins, such as ATG5, ATG7, or Beclin-1, all of which are critical in initiating the formation of the autophagosome, or as a result of exposure to exogenous stressors, such as the environmental toxicant arsenic, which causes the sequestration of KEAP1 into p62 positive autophagosomes that are unable to fuse with the lysosome to complete the autophagic process [10]. The relevance of autophagic dysfunction as it pertains to the non-canonical activation of NRF2 in disease will be discussed in greater detail below.
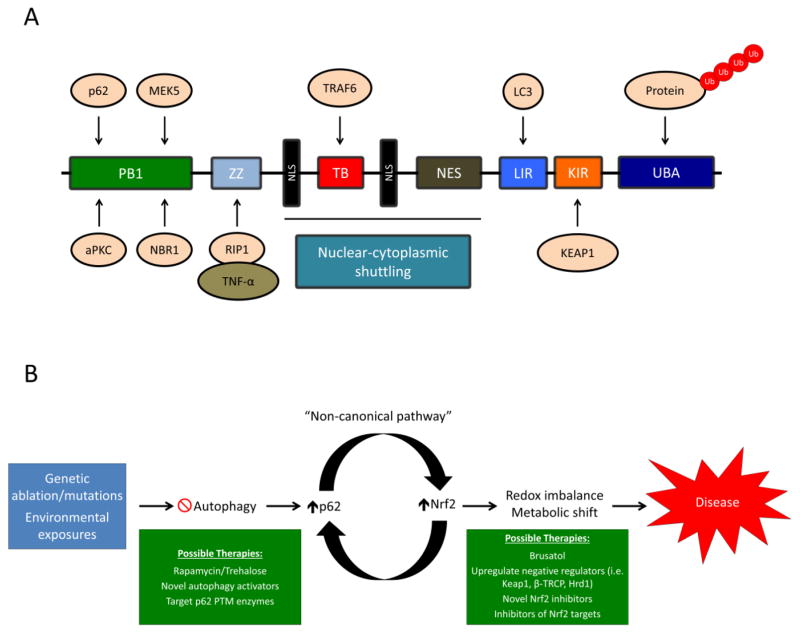
As mentioned above, the accumulation of p62 is a critical driving force behind the non-canonical pathway of NRF2 activation. There are a number of domains in p62 that govern its interacting partners and function, including a: 1) Phem and Box1 (PB1) domain, responsible for dimerization and oligomerization of p62 with other PB1 containing proteins, including p62 itself, atypical protein kinase C (aPKC), dual specificity mitogen-activated protein kinase kinase 5 (MEK5), and next to BRCA1 gene 1 protein (NBR1); 2) ZZ-type zinc finger domain, which interacts with TNF-α via binding to the receptor interacting protein 1 (RIP-1); 3) Tumor necrosis factor receptor-associated factor 6 (TRAF6) binding (TB) domain, which mediates binding to TRAF6, an E3 ubiquitin ligase involved in protein ubiquitylation; 4–6) Two nuclear localization signal (NLS) domains and one nuclear export signal domain (NES), responsible for the nuclear-cytoplasmic shuttling of p62; 7) LC3 interacting region (LIR), which mediates the interaction between p62 and the autophagosomal protein microtubule-associated protein 1A/1B-light chain 3 (LC3); 8) KEAP1-interacting region, the domain that contains the DPSTGE motif responsible for mediating the p62-KEAP1 interaction; and 9) Ubiquitin associated (UBA) domain, which binds to poly-ubiquitylated cargo [18, 19] (Figure 1A). The ability of p62 to self-oligomerize and heterodimerize with other PB1 domain containing proteins, as well as interact with specific binding partners, including KEAP1 and LC3, and bind to poly-ubiquitylated proteins, are critical to its function as an adapter protein and autophagic substrate; however, when p62 is not properly turned over, it’s diversity of binding partners is what also contributes to the sequestration and aggregation of p62-associated proteins.
Figure 1. The domain structure of p62 and possible therapeutic interventions to treat the p62-dependent activation of NRF2.
(A) p62 is a multifunctional protein consisting of the following domains: 1) PB1 domain, responsible for p62 self-oligomerization and binding to other PB1 containing proteins (i.e. aPKC, MEK5, and NBR1, shown above); 2) ZZ domain, interacts with RIP-1, which recruits TNF-α and mediates cell death; 3) TB domain, binds to TRAF6, which mediates protein ubiquitylation; 4–6) NLS and NES domains, control the nuclear-cytoplasmic shuttling of p62; 7) LIR, binds to LC3 in the autophagosomal membrane, recruiting cargo for autophagic degradation; 8) KIR, binds to KEAP1; and 9) UBA domain, binds to poly-ubiquitylated proteins. (B) Possible autophagy and NRF2-based therapeutic interventions for treating diseases associated with non-canonical activation of NRF2. Genetic alterations to key autophagy proteins, or chronic exposure to environmental toxicants (i.e. arsenic), result in autophagy blockage and the p62-dependent upregulation of NRF2. Prolonged activation of NRF2 results in changes to the cellular metabolic and redox state, contributing to the pathogenesis of certain diseases. Restoring autophagic function via autophagy activating compounds, preventing the post-translational modifications of p62, or directly inhibiting the function of NRF2 or its downstream targets, could prove viable therapeutic options in the treatment of diseases associated with increased levels of p62 and NRF2.
Interestingly, a number of post-translational modifications to p62 can also modulate its interaction with its binding partners [20]. For example, phosphorylation of p62 by the mammalian target of rapamycin (mTOR) at S349 in the KIR domain enhances p62-KEAP1 binding and the subsequent accumulation of NRF2 [21]. Furthermore, phosphorylation of S403 in the UBA domain of p62 by Unc-51 Like Autophagy Activating Kinase 1 (ULK1), Casein kinase II (CKII), or Tank-binding kinase 1 (TBK1), which promotes recruitment of ubiquitylated cargo, plays an important role in the degradation of p62 and its substrates, although whether or not this modification also drives KEAP1 degradation is still uncertain [22–24]. Conversely, ubiquitylation of K7 by tripartite motif family 21 (TRIM21) in the PB1 domain of p62 prevents its oligomerization and interaction with KEAP1, keeping the KEAP1-dependent degradation of NRF2 intact [25]. These studies indicate that the accumulation of p62, as well as enzyme-mediated post-translational modifications of relevant amino acids in its critical domains, are important in mediating the non-canonical activation of NRF2. Thus, understanding the mechanism that contributes to the p62-KEAP1-dependent upregulation of NRF2 will play an important role in targeting diseases where autophagic dysfunction and the prolonged activation of NRF2 are observed.
Relevance of the non-canonical pathway in disease
There are a number of diseases where the increased expression or aggregation/autophagosomal accumulation of p62 have been linked to the non-canonical activation of the NRF2 pathway. Interestingly, non-canonical activation of NRF2 can have beneficial or detrimental outcomes depending on the disease context. For example, one of the most notable examples of detrimental non-canonical activation of NRF2 occurs in hepatocellular carcinomas, with human hepatocellular carcinoma cell lines (HCC) exhibiting increased aggregates of p62, including the phosphorylated form, as well as KEAP1, accounting for the elevated levels of NRF2 observed in these cell lines. Furthermore, p62 is necessary for the increased activity of NRF2 in HCC, as ablation of p62 prevented activation of NRF2 and its downstream target genes [26]. Aggregates positive for phosphorylated p62 and KEAP1 are also observed in HCC patient samples [21], indicating this mechanism of NRF2 activation could play a critical role in mediating the progression of hepatocellular carcinomas. Mice with ATG5 specifically deleted in the liver exhibit increased liver inflammation, fibrosis, and tumorigenesis, all of which can be reversed by deletion of NRF2 [27], indicating autophagy deficiency and NRF2 activation play a critical role in numerous liver-based pathologies. The non-canonical pathway may also play a negative role in neurodegenerative disease, as the aggregation of phosphorylated p62 and KEAP1 is observed in neurofibrillary tangles found in postmortem Alzheimer’s disease brain tissue, which also correlates with an increase in NRF2 target gene expression [28–30]. Furthermore, the S349T mutation in the KIR of p62, which prevents phosphorylation of p62 by mTOR, has also been shown to result in suppressed NRF2 signaling, possibly affecting osteoclast and osteoblast differentiation in patients with Paget’s disease of the bone [31].
The non-canonical activation of NRF2 is not always associated with negative outcomes. The p62-dependent activation of NRF2 plays a critical role in preventing the oxidative liver damage that results from increased lipogenesis, which is commonly observed during metabolic disorders such as non-alcoholic fatty liver disease (NAFLD) and type II diabetes, with the stress activated proteins sestrin 1 and sestrin 2 enhancing degradation of the p62-KEAP1 complex to activate NRF2 [32–34]. Also quercetin, a common dietary flavonoid, prevents the cytotoxicity associated with hepatotoxicants such as acetaminophen and carbon tetrachloride by disrupting KEAP1-NRF2 binding, either through direction interaction with KEAP1 cysteines, or by enhancing the p62-dependent sequestration of KEAP1, to upregulate the NRF2 response [35]. Liver specific deletion of ATG5 also alleviates acetaminophen toxicity via prolonged activation of NRF2 and increased hepatocyte proliferation [36]. These studies demonstrate that the non-canonical activation of NRF2, either via genetic or pharmacological means plays an important role in a diverse array of disease pathologies, and can have either beneficial or detrimental outcomes depending on the tissue type and context of the p62-KEAP1 interaction.
Possible therapeutic strategies
While a number of “canonical” NRF2-based therapies exist, with dimethyl fumarate (DMF) currently approved for the treatment of multiple sclerosis [37], therapeutic strategies targeting the non-canonical activation of NRF2 are limited. There are a number of possible strategies that could be employed to prevent the sustained activation of NRF2 that occurs as a result of the autophagosomal accumulation of p62 and KEAP1 or the phospho-p62-dependent autophagic degradation of KEAP1 (Figure 1B). The first tactic would be to restore proper autophagic function using activators of the autophagy pathway. To date, there are few, if any, targeted activators of autophagy. The most promising thus far is the inhibitor of mTOR rapamycin, which has known immunosuppressive effects [38], but has also been shown to activate autophagy in a number of experimental settings. Interestingly, rapamacyin has been shown to prevent degradation of KEAP1 via autophagy by blocking the phosphorylation of p62 [21]. Thus, rapamycin treatment, through the targeted inhibition of p62 phosphorylation, could prevent the autophagic degradation of KEAP1 and subsequent non-canonical upregulation of NRF2, especially in cases linked to environmental exposures such as arsenic. Another promising possibility, at least in a neuronal setting, is the disaccharide trehalose, which has been shown to stimulate the autophagic removal of neuropathic proteins [39]. A number of high-throughput screening studies have also identified other autophagy inducers that could also prove useful once further testing confirms their specificity and mechanism of action [40–42]. Inhibiting ULK1, CKII, or TBKI, the enzymes responsible for phosphorylating S403 in the UBA domain of p62, or enhancing the activity of TRIM21 to prevent p62 oligomerization and heterodimerization, could also prove to be viable therapeutic interventions should a targeted compound that modulates the activity of these enzymes be discovered (Figure 1B).
Another tactic for treating diseases in which there is prolonged activation of NRF2 is to directly inhibit NRF2 itself, or the function of its target proteins, to restore proper redox and metabolic homeostasis (Figure 1B). Our lab discovered one of the only established inhibitors of NRF2, brusatol, which could be used to mitigate some of the negative effects that result from sustained NRF2 activation. Furthermore, direct inhibitors of NRF2 target proteins could also be utilized. Another intriguing possibility would be to re-establish or enhance the activity of the negative regulators of NRF2 (i.e. KEAP1, β-transducin repeat-containing protein (β-TRCP), or HRD1), although currently there are no established compounds capable of achieving this effect. Furthermore, an inhibitor of the phospho-p62-KEAP1 interaction that does not directly affect KEAP1-NRF2 binding has recently been discovered [43], implying that compounds targeting the p62-KEAP1 interaction can be utilized to prevent the non-canonical upregulation of NRF2. Therefore, discovering novel inhibitors of NRF2 or its target proteins, or developing novel compounds that enhance the negative regulation and degradation of NRF2 could prove useful in re-establishing physiological NRF2 signaling (Figure 1B).
Conclusions
Similar to the canonical activation of NRF2, the role of the non-canonical pathway is cell-type and context dependent, and our understanding of the mechanisms underlying this mode of NRF2 regulation is still growing. What is well established is the critical role of the p62-KEAP1 interaction in driving the sustained activation of NRF2. While the initial discovery of the non-canonical activation of NRF2 was based upon autophagic dysfunction and the subsequent autophagosomal accumulation/aggregation of p62, recent work has shown that post-translational modification of key amino acid residues in p62 can also dictate its interaction with KEAP1 and contribute to prolonged activation of the NRF2-mediated response. These findings are critical, not only in contributing to our understanding of the p62-dependent sequestration or degradation of KEAP1, but also in providing novel therapeutic targets that can be utilized to treat diseases where enhanced expression of p62 and increased levels of NRF2 are prevalent. Thus, understanding the mechanism by which NRF2 is activated is of critical importance in developing a therapeutic regimen for NRF2-based disease treatment or prevention.
Acknowledgments
The authors are funded by the following grants from the National Institutes of Health: ES023758 (EC & DDZ), CA154377 (DDZ), ES015010 (DDZ), DK109555 (DDZ), and ES006694 (a center grant).
Footnotes
Conflict of interest
The authors have nothing to disclose.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors
References
Recently published papers of particular interest have been highlighted as:
• Of importance
•• Of major importance
- 1.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24(24):10941–53. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23(22):8137–51. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slocum SL, Kensler TW. Nrf2: control of sensitivity to carcinogens. Arch Toxicol. 2011;85(4):273–84. doi: 10.1007/s00204-011-0675-4. [DOI] [PubMed] [Google Scholar]
- 4.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27(20):2179–91. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng H, Whitman SA, Wu W, Wondrak GT, Wong PK, Fang D, et al. Therapeutic potential of Nrf2 activators in streptozotocin-induced diabetic nephropathy. Diabetes. 2011;60(11):3055–66. doi: 10.2337/db11-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chartoumpekis DV, Kensler TW. New player on an old field; the keap1/Nrf2 pathway as a target for treatment of type 2 diabetes and metabolic syndrome. Curr Diabetes Rev. 2013;9(2):137–45. doi: 10.2174/1573399811309020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su ZY, Shu L, Khor TO, Lee JH, Fuentes F, Kong AN. A perspective on dietary phytochemicals and cancer chemoprevention: oxidative stress, nrf2, and epigenomics. Top Curr Chem. 2013;329:133–62. doi: 10.1007/128_2012_340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(7):651–62. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 9.Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol. 2013;8:105–37. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau A, Zheng Y, Tao S, Wang H, Whitman SA, White E, et al. Arsenic inhibits autophagic flux, activating the Nrf2-Keap1 pathway in a p62-dependent manner. Mol Cell Biol. 2013;33(12):2436–46. doi: 10.1128/MCB.01748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–23. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 12.Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282(24):4672–8. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- 13.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30(13):3275–85. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285(29):22576–91. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan W, Tang Z, Chen D, Moughon D, Ding X, Chen S, et al. Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy. 2010;6(5):614–21. doi: 10.4161/auto.6.5.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Copple IM, Lister A, Obeng AD, Kitteringham NR, Jenkins RE, Layfield R, et al. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J Biol Chem. 2010;285(22):16782–8. doi: 10.1074/jbc.M109.096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •17.Pajares M, Jimenez-Moreno N, Garcia-Yague AJ, Escoll M, de Ceballos ML, Van Leuven F, et al. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12(10):1902–16. doi: 10.1080/15548627.2016.1208889. This study verifies that p62 is a transcriptional target of NRF2, and that a number of other autophagy proteins contain ARE-like sequences, highlighting the link between the autophagy and NRF2 pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nezis IP, Stenmark H. p62 at the interface of autophagy, oxidative stress signaling, and cancer. Antioxid Redox Signal. 2012;17(5):786–93. doi: 10.1089/ars.2011.4394. [DOI] [PubMed] [Google Scholar]
- 19.Puissant A, Fenouille N, Auberger P. When autophagy meets cancer through p62/SQSTM1. Am J Cancer Res. 2012;2(4):397–413. [PMC free article] [PubMed] [Google Scholar]
- 20.Rojo de la Vega MDM, Chapman E, Zhang DD. NRF2-targeted therapeutics: New targets and modes of NRF2 regulation. Curr Opin Toxicol. 2016:1. doi: 10.1016/j.cotox.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••21.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51(5):618–31. doi: 10.1016/j.molcel.2013.08.003. This paper was the first to introduce the enhanced interaction between phosphorylated p62 and KEAP1 as a means of upregulating NRF2 in liver pathologies. Targeting the post-translational modifications that occur on p62, and not just its aggregation, could prove to be an important therapeutic strategy in treating diseases with autophagy dysregulation and/or high levels of NRF2. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44(2):279–89. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 23.Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37(2):223–34. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim J, Lachenmayer ML, Wu S, Liu W, Kundu M, Wang R, et al. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet. 2015;11(2):e1004987. doi: 10.1371/journal.pgen.1004987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••25.Pan JA, Sun Y, Jiang YP, Bott AJ, Jaber N, Dou Z, et al. TRIM21 Ubiquitylates SQSTM1/p62 and Suppresses Protein Sequestration to Regulate Redox Homeostasis. Mol Cell. 2016;62(1):149–51. doi: 10.1016/j.molcel.2016.03.015. This study highlights the importance of TRIM21 as a negative regulator of p62 aggregation and sequestration of its interacting partners. This article also indicates the importance of post-translational modifications to p62 in modulating its function. [DOI] [PubMed] [Google Scholar]
- 26.Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193(2):275–84. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ni HM, Woolbright BL, Williams J, Copple B, Cui W, Luyendyk JP, et al. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61(3):617–25. doi: 10.1016/j.jhep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanji K, Maruyama A, Odagiri S, Mori F, Itoh K, Kakita A, et al. Keap1 is localized in neuronal and glial cytoplasmic inclusions in various neurodegenerative diseases. J Neuropathol Exp Neurol. 2013;72(1):18–28. doi: 10.1097/NEN.0b013e31827b5713. [DOI] [PubMed] [Google Scholar]
- 29.Tanji K, Miki Y, Ozaki T, Maruyama A, Yoshida H, Mimura J, et al. Phosphorylation of serine 349 of p62 in Alzheimer’s disease brain. Acta Neuropathol Commun. 2014;2:50. doi: 10.1186/2051-5960-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamazaki H, Tanji K, Wakabayashi K, Matsuura S, Itoh K. Role of the Keap1/Nrf2 pathway in neurodegenerative diseases. Pathol Int. 2015;65(5):210–9. doi: 10.1111/pin.12261. [DOI] [PubMed] [Google Scholar]
- 31.Wright T, Rea SL, Goode A, Bennett AJ, Ratajczak T, Long JE, et al. The S349T mutation of SQSTM1 links Keap1/Nrf2 signalling to Paget’s disease of bone. Bone. 2013;52(2):699–706. doi: 10.1016/j.bone.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 32.Bae SH, Sung SH, Oh SY, Lim JM, Lee SK, Park YN, et al. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 2013;17(1):73–84. doi: 10.1016/j.cmet.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Rhee SG, Bae SH. The antioxidant function of sestrins is mediated by promotion of autophagic degradation of Keap1 and Nrf2 activation and by inhibition of mTORC1. Free Radic Biol Med. 2015;88(Pt B):205–11. doi: 10.1016/j.freeradbiomed.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Ro SH, Semple IA, Park H, Park H, Park HW, Kim M, et al. Sestrin2 promotes Unc-51-like kinase 1 mediated phosphorylation of p62/sequestosome-1. FEBS J. 2014;281(17):3816–27. doi: 10.1111/febs.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji LL, Sheng YC, Zheng ZY, Shi L, Wang ZT. The involvement of p62-Keap1-Nrf2 antioxidative signaling pathway and JNK in the protection of natural flavonoid quercetin against hepatotoxicity. Free Radic Biol Med. 2015;85:12–23. doi: 10.1016/j.freeradbiomed.2015.03.035. [DOI] [PubMed] [Google Scholar]
- 36.Ni HM, Boggess N, McGill MR, Lebofsky M, Borude P, Apte U, et al. Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicol Sci. 2012;127(2):438–50. doi: 10.1093/toxsci/kfs133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bomprezzi R. Dimethyl fumarate in the treatment of relapsing-remitting multiple sclerosis: an overview. Ther Adv Neurol Disord. 2015;8(1):20–30. doi: 10.1177/1756285614564152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sehgal SN. Rapamune (RAPA, rapamycin, sirolimus): mechanism of action immunosuppressive effect results from blockade of signal transduction and inhibition of cell cycle progression. Clin Biochem. 1998;31(5):335–40. doi: 10.1016/s0009-9120(98)00045-9. [DOI] [PubMed] [Google Scholar]
- 39.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282(8):5641–52. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 40.Levine B, Packer M, Codogno P. Development of autophagy inducers in clinical medicine. J Clin Invest. 2015;125(1):14–24. doi: 10.1172/JCI73938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •41.Li Y, McGreal S, Zhao J, Huang R, Zhou Y, Zhong H, et al. A cell-based quantitative high-throughput image screening identified novel autophagy modulators. Pharmacol Res. 2016;110:35–49. doi: 10.1016/j.phrs.2016.05.004. This article discusses how a novel inhibitor of the phosphorylated-p62-KEAP1 interaction, K67, and its derivatives, modulate the p62-KEAP1-NRF2 pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vakifahmetoglu-Norberg H, Xia HG, Yuan J. Pharmacologic agents targeting autophagy. J Clin Invest. 2015;125(1):5–13. doi: 10.1172/JCI73937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasuda D, Nakajima M, Yuasa A, Obata R, Takahashi K, Ohe T, et al. Synthesis of Keap1-phosphorylated p62 and Keap1-Nrf2 protein-protein interaction inhibitors and their inhibitory activity. Bioorg Med Chem Lett. 2016;26(24):5956–9. doi: 10.1016/j.bmcl.2016.10.083. [DOI] [PubMed] [Google Scholar]