Abstract
The ability to detect biomarkers with ultrahigh sensitivity radically transformed biology and disease diagnosis. However, owing to incompatibilities with infrastructure in current biological and medical laboratories, recent innovations in analytical technology have not received broad adoption. Here, we report a simple, universal ‘add-on’ technology (dubbed EASE) that can be directly plugged into the routine practices of current research and clinical laboratories and that converts the ordinary sensitivities of common bioassays to extraordinary ones. The assay relies on the bioconjugation capabilities and ultrafast and localized deposition of polydopamine at the target site, which permit a large number of reporter molecules to be captured and lead to detection-sensitivity enhancements exceeding 3 orders of magnitude. The application of EASE in the enzyme-linked-immunosorbent-assay-based detection of the HIV antigen in blood from patients leads to a sensitivity lower than 3 fg ml−1. We also show that EASE allows for the direct visualization, in tissues, of the Zika virus and of low-abundance biomarkers related to neurological diseases and cancer immunotherapy.
INTRODUCTION
As recent advances in medicine rapidly unravel the genomic and proteomic signatures of disease development, progression, and response to therapy, sensitive and quantitative analysis of disease biomarkers (e.g., DNA, RNA, and proteins) has become increasingly important in the era of precision medicine where diagnostic and therapeutic decisions are tailored towards individual patients. In parallel, to address the challenge in sensitive and multiplexed biomarker analysis, a large variety of exquisitely designed imaging and detection technologies have also been developed in the past decade1–11. These enabling technologies often leveraging on the unique properties of colloidal nanostructures (e.g., quantum dots, magnetic nanoparticles, and plasmonic nanoparticles) and precisely engineered sensor devices (e.g., nanowire sensors, cantilevers, and microfluidic channels) are so sensitive that their detection limits are commonly seen in the single-molecule level where low-abundance targets such as circulating oligonucleotides, proteins, viruses, and cells can be enumerated with polymerase chain reaction (PCR)-like sensitivity12–18. Despite these remarkable achievements in biotechnology laboratories, broad adoption of these technological innovations by biology and clinical laboratories has been limited, and consequently the impact19. The resistance is a result of multiple factors including complex protocols, specialized reagents and equipment, and most importantly the requirement of different infrastructures, a disconnect that not only elevates the upfront cost but also reduces persistent output and cross-laboratory cross-platform consistency.
Here we present a universal ‘add-on’ technology that can be plugged into virtually all common biodetection and bioimaging techniques, and enhance their sensitivities by approximately three orders of magnitude. This enhancement is achieved by combining horseradish peroxidase (HRP), one of the most popular reporter enzymes in biology, with polydopamine (PDA), arguably the most versatile coating material in surface treatment. Previously, a simple dip-coating protocol has been demonstrated for spontaneous formation of a thin self-adherent PDA film onto a wide range of surfaces20. Interestingly, we found that when dopamine is used in place of the typical HRP substrates, its polymerization rate is increased by 100s of times. In addition, PDA is well known for its outstanding reactivity to the amine, sulfhydryl, and phenol groups in proteins21, enabling site-specific deposition in the vicinity of HRP and the subsequent sensitive detection based on absorption or fluorescence (Fig. 1a). It is worth mentioning that although we discovered the remarkable performance of HRP-PDA combination in biotechnology, similar mechanism has been utilized in nature for perhaps thousands of years. For example, it has been found that invertebrates take advantage of oxidase-catalyzed rapid melanin deposition to treat mechanical injuries and isolate foreign organisms such as parasites22, 23. The chemical reaction is very similar to HRP-catalyzed PDA deposition.
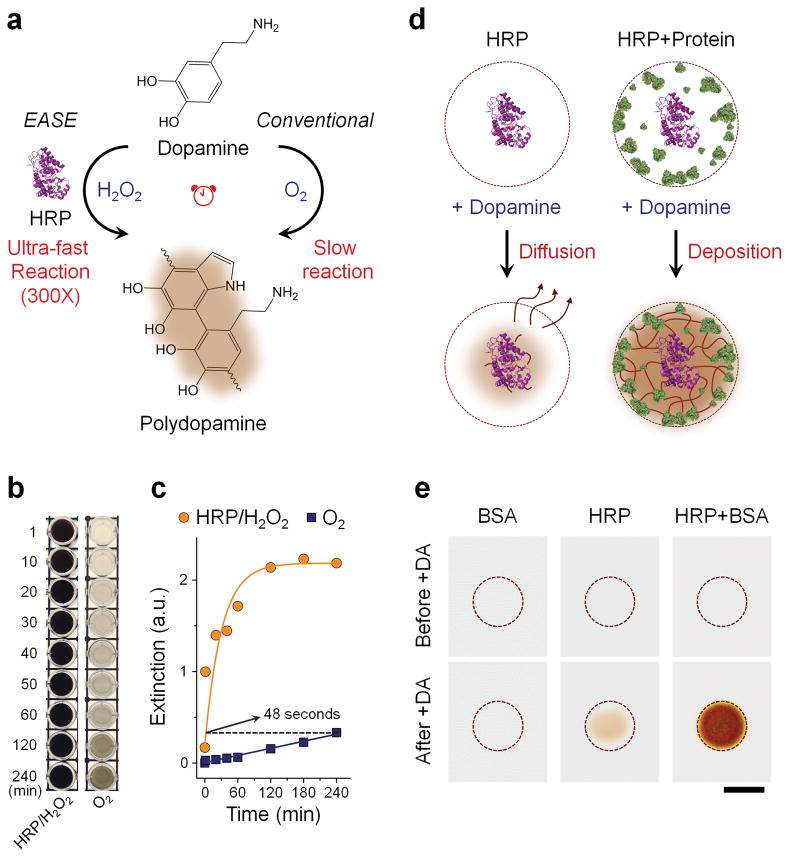
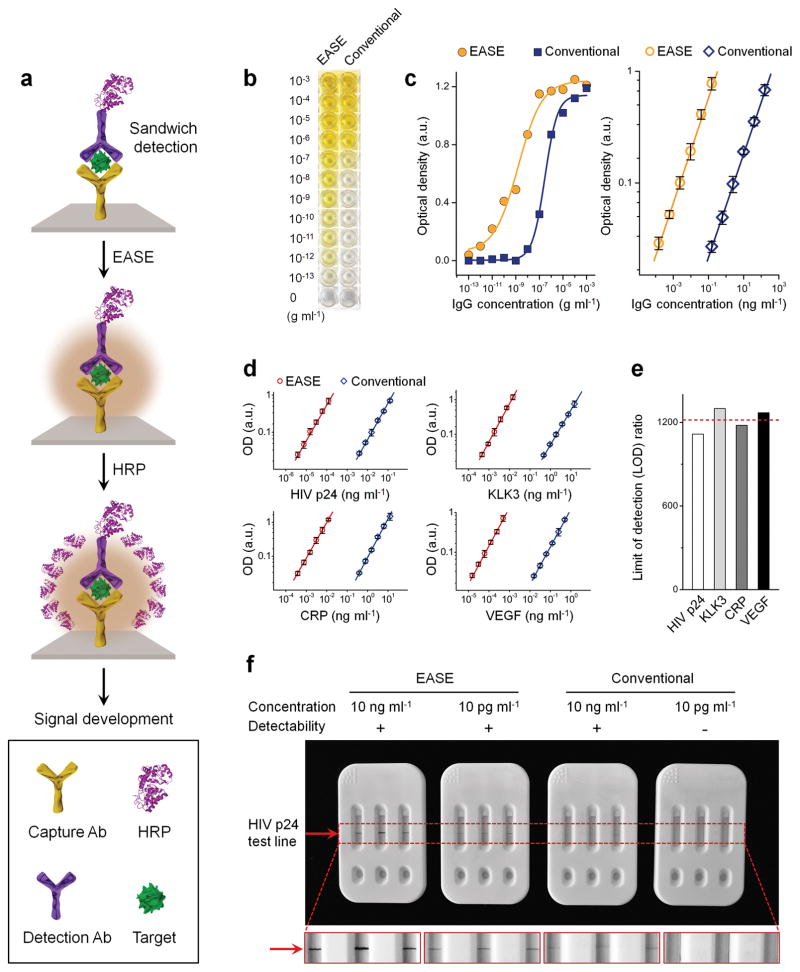
Figure 1. HRP-accelerated dopamine polymerization and deposition.
(a) Schematic illustration of the EASE technology. Dopamine (colorless) slowly oxidizes in the presence of air (O2 as oxidant) and produces brown-black PDA. This polymerization process can be sped up by approximately 300 times under HRP catalysis (H2O2 as oxidant). (b) Visual observation of dopamine polymerization under conventional and HRP catalyzed conditions at various time points. (c) Extinction measured at 700 nm for the samples showing in (b). EASE produces the same optical extinction (700 nm) in 48 s compared to 4 h under conventional conditions. (d) HRP-catalyzed PDA deposition on solid supports. When protein density on the solid support is low (for example only HRP is present), majority of the PDA molecules diffuse away. For solid supports (e.g., flat surface and membrane) with high protein density (e.g., in cells and surfaces blocked with protein molecules for reduced nonspecific binding), rapid and localized deposition of PDA occurs due to the reactivity of PDA to nearby amines (rich in proteins), leading to formation of a dark spot. (e) Images showing membranes immobilized with BSA alone, HRP alone, or HRP/BSA before and after exposing to dopamine. Scale bar, 5 mm.
RESULTS
Enzyme-accelerated ultrafast PDA deposition
To quantify the effect of HRP on PDA polymerization rate, the enzyme-accelerated signal enhancement (EASE) process is compared to the reaction conditions in the conventional dip-coating polymerization procedure where HRP is not present and O2 is the oxidant. As shown in Figure 1b, the dopamine solution slowly changes color from colorless to light grey over a period of four hours, indicating slow PDA formation. In contrast, when HRP and H2O2 of low concentration (typical reaction condition for HRP-catalyzed substrate conversion) were added, the dopamine solution of the same concentration instantly turns to brown-black, showing significantly increased PDA polymerization rate. Quantitative comparison of the reaction kinetics was plotted by measuring the solution light extinction at 700 nm where dopamine has negligible absorption compared to PDA (Supplementary Fig. S1). Under the dip-coating reaction condition, PDA slowly builds up and is not near completion after 4 h of reaction time; whereas under the EASE condition, the PDA solution reaches the same level of light extinction in 48 seconds (plateaus within 1 h), indicating an approximately 300-fold increase in polymerization rate (Fig. 1c).
Next, we characterized whether the EASE process can be confined to the vicinity of HRP molecules (Fig. 1d), a key factor determining the scope of downstream applications. If PDA molecules quickly diffuse away from HRP, the EASE technology will only be useful for improving the enzyme-linked immunosorbent assay (ELISA) by measuring chromogens in solution. If the PDA molecules are confined near HRP, the EASE technology will be broadly applicable to various bioassays beyond ELISA, such as immunohistochemistry (IHC), immunofluorescence (IF), fluorescence in situ hybridization (FISH), and immunoblotting, because the spatial information is preserved. To probe it, HRP was immobilized inside a circle on a nitrocellulose membrane, which was also blocked with bovine serum albumin (BSA). Note that BSA, as a standard blocking agent that helps reduce non-specific binding, serves an additional function here. It also provides reactive chemical groups as the PDA deposition anchor sites. As shown in Figure 1e, when the membrane was exposed to dopamine/H2O2 solution, essentially no PDA was found on the membrane with BSA only (free of background). In contrast, when HRP is present on the membrane, PDA development becomes clearly visible because HRP not only catalyzes the PDA polymerization, but also, as a natural protein molecule, serves as the PDA deposition anchor point. For the membrane incubated with HRP and blocked with BSA, PDA deposition is significantly enhanced due to the high-density reactive sites on the membrane (provided by the BSA molecules) that quickly capture PDA molecules before they diffuse away from the surface. More importantly, the color development is completely confined inside the HRP spot, demonstrating retention of the spatial resolution that makes EASE suited for the aforementioned immuno and hybridization assays.
EASE for immunohistochemistry and immunofluorescence
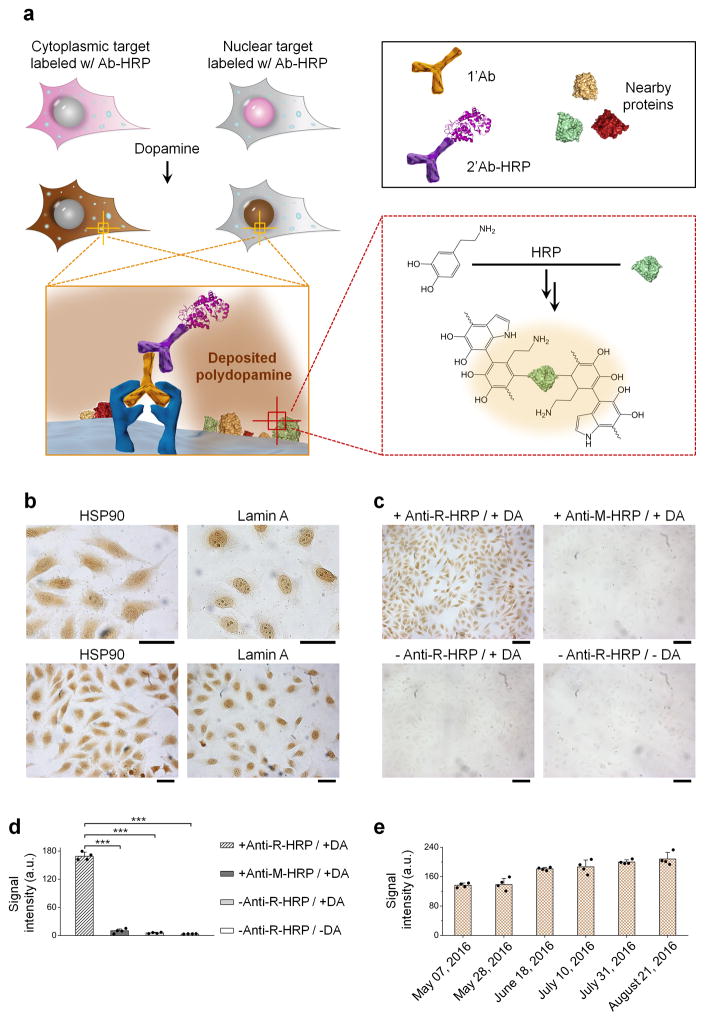
The EASE technology was first applied to IHC and IF, robust technologies capable of interrogating gene expressions in single cells and resolving the heterogeneity issues of complex tissue samples, with well-preserved cell and tissue morphology. IHC and IF work well for high-abundance target molecules, but lacks the sensitivity to detect antigens of low abundance24, 25, in particular in clinical tissue specimens where autofluoresecence can be overwhelmingly high. To test the suitability of EASE, two model antigens, Lamin A (nuclear envelope) and HSP-90 (cytoplasm) were stained in formalin-fixed HeLa cells because these two antigens represent targets in different cell compartments (Fig. 2a). Conventional two-step staining procedure was carried out by incubating cells with the primary antibody (1’Ab) and secondary antibody-HRP (2’Ab-HRP) sequentially, except that dopamine was used as the HRP substrate. Owing to the chromogenic feature of PDA, the staining can be directly visualized. As shown in Figure 2b and Supplementary Figure S3, the staining patterns for both antigens are the same as those obtained with conventional IHC (using 3,3’-diaminobenzidine (DAB) as the substrate) and IF (using quantum dot (QD) labeled secondary antibody) (Supplementary Fig. S4), demonstrating the staining specificity and confirming the confined PDA deposition on the microscopic scale. The specificity is further confirmed by a series of control experiments where either one of the key agents (1’Ab and 2’Ab-HRP) is missing or a mismatched 1’Ab-2’Ab pair is used (Fig. 2c, d and Supplementary Fig. S5). It is also worth mentioning that the PDA chromogens are highly stable after cell staining. As shown in the same group of cells in Figure 2e and Supplementary Figure S6, no obvious signal decay was detected after 4-month, allowing samples to be reexamined after long storage.
Figure 2. IHC-EASE single-cell staining.
(a) Schematic illustration of IHC-EASE. Cells are labeled with 1’Ab and 2’Ab-HRP complex sequentially, and exposed to dopamine. Localized PDA deposition (dark brown) indicates the spatial and abundance information of the target. (b) Bright-field imaging of IHC-EASE stained cells with different magnifications showing cytoplasmic and nuclear staining of HSP90 and Lamin A, respectively. Scale bar, 50 μm. (c) Bright-field imaging of HSP90 staining showing the specificity of IHC-EASE compared to negative controls. Mismatched anti-mouse (M)-HRP, absence of 2’Ab-HRP or dopamine produces negligible signals. Scale bar, 100 μm. (d) Quantitative measurements of the staining intensities of samples shown in (c). Statistical analysis of cells in four random field-of-views shows significant differences between the experiment and control groups. ***P < 0.001 by two-tailed t-test, error bars indicating s.d. (e) Quantitative evaluation of the staining stability upon storage. The staining signal does not decay over a period of three and half months. In fact, the signal slightly increases, likely due to aging of the rapid-formed PDA. Error bars, s.d. over four different images.
To probe the sensitivity enhancement of EASE, fluorescence probes were brought into the assay after PDA deposition, taking advantage of PDA’s remarkable reactivity to any fluorophores with primary amines and the convenience of quantifying fluorescence signals21. Pegylated QDs with terminal amines were used as the fluorophore because of their photostability so that fluorescence intensity can be accurately measured26, 27. As shown in Figure 3a, the fluorescent staining pattern matches that of the PDA, confirming that QD-NH2 immobilization is confined to the PDA network. The specificity is further demonstrated by the control experiments where either one of the key agents (Ab or dopamine) is missing, an isotype 1’Ab is utilized, or un-functionalized QDs are used. As shown in Figure 3b, c and Supplementary Figure S7, the control experiments did not produce detectable signals.
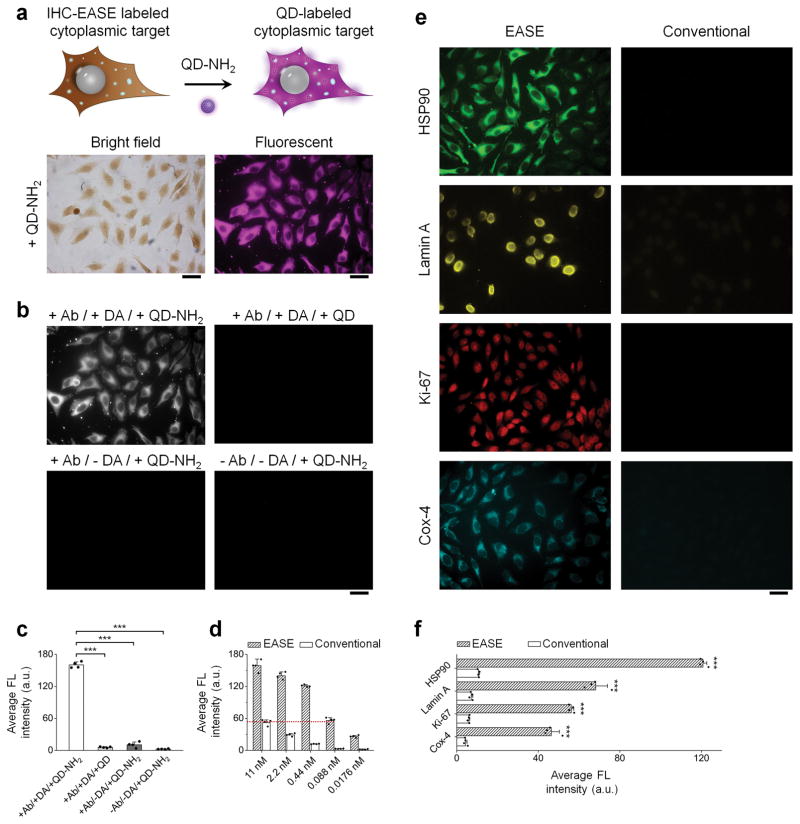
Figure 3. IF-EASE cell staining.
(a) IHC-EASE labeled cells further labeled with QD-PEG-NH2 (NH2-QDs). Fluorescence imaging of HSP90 shows consistent staining pattern after QD adsorption (compared with the bright-field images before QD labeling). Scale bar, 50 μm. (b & c) Verification of staining specificity by comparing the experiment group with various controls (antibody and/or dopamine is missing). Scale bar, 50 μm. The intensity differences between the experiment and controls are highly significant. ***P < 0.001 by two-tailed t-test. Error bars, s.d. over four different images. (d) Quantitative evaluation of IF staining intensity with or without EASE. Signal intensity obtained using IF-EASE at 88 pM 1’Ab is roughly the same as the intensity obtained with conventional IF at 11 nM 1’Ab. Error bar, s.d. over four different images. (e) Fluorescence imaging of four targets (HSP90, Lamin A, Ki-67, and Cox-4) stained with or without EASE at a 1’Ab dilution of 1:25,000. Scale bar, 50 μm. (f) Quantitative measurements of the cell fluorescence intensity showing in (e). The differences are statistically significant. ***P < 0.001 by two-tailed t-test. Error bars, s.d. over four different images.
To evaluate the sensitivity quantitatively, staining was first performed on HSP-90. Unlike ELISA assays where target molecules can be easily immobilized at various densities, engineering cells with a variety of precisely controlled antigen expression levels is extremely difficult. Instead, we reduced the concentration of the 1’Ab in a serial fashion to bring down the signal intensity. As shown in Figure 3d and Supplementary Figure S8, at 1’Ab concentration of 88 pM, IF-EASE achieves the same signal strength compared to conventional IF using 1’Ab of 11 nM, yielding a 125 fold reduction in 1’Ab concentration, which not only indirectly demonstrates the enhancement in imaging sensitivity, but also shows the possibility to reduce the cost of expensive biological agents such as antibodies. The signal enhancement is a result of amplifying a limited number of target molecules (as well as bound HRP) to a polymer network that captures a large number of QDs. Indeed, when we applied four tumor biomarkers (HSP90, Lamin A, Ki-67, and Cox-4) covering various intracellular locations, a 1:25,000 dilution of the primary antibodies (typical IF dilution factor ~ 1:100) produced bright and specific staining similar to those from conventional IF assay using high concentration of 1’Ab (Fig. 3e & f). In contrast, without EASE, 1:25,000 dilutions of the primary antibodies did not produce detectable signals.
Next, to directly evaluate IF-EASE in imaging low-abundance targets, we silenced the expression of GAPDH in HeLa cells using RNA interference (RNAi)-mediated gene knockdown. As shown in Supplementary Figure S9, 36 h post RNAi, the characteristic cytoplasmic distribution of GAPDH can be clearly visualized using IF-EASE, and it is only barely detectable using IF alone. Similarly, at 60 h post RNAi, trace amount of GAPDH can still be detected using IF-EASE, but not with IF alone. This result clearly shows the power of EASE in detected low-abundance targets in cells.
EASE for suspension microarrays
Suspension microarrays are highly multiplexed genotyping and phenotyping platforms used in molecular biology, drug screening, and disease diagnosis29–31. Compared to planar microarrays that are spatially addressable, suspension microarrays are often fabricated by doping microspheres with combinations of luminescent materials and are decoded with flow cytometers (e.g., Luminex microbeads)32, 33. To determine whether an unknown analyte is present or not, conventional methodologies such as direct or sandwich hybridization and immuno-recognition are applied. The suspension microarrays offer advantages such as faster binding kinetics, but their detection sensitivities are essentially the same as the planar counterparts34.
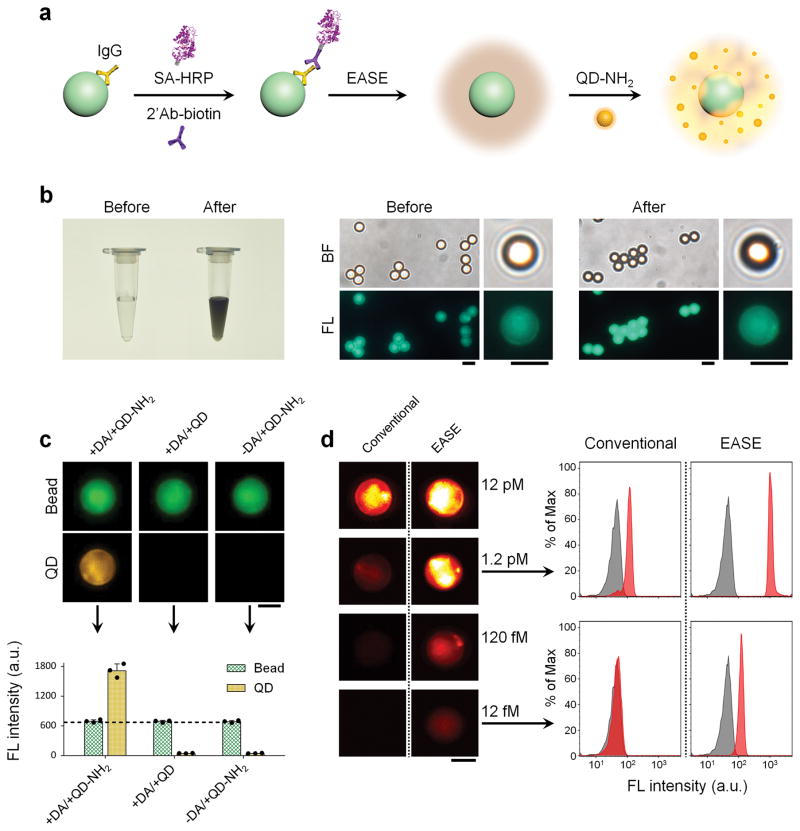
To demonstrate the compatibility of EASE with suspension microarrays, fluorescent microspheres were coated with immunoglobulin G (IgG) to detect a model target, biotinylated 2’Ab. Presence or absence of the analyte was detected with either streptavidin-QD conjugates (conventional sandwich method) or the EASE technology (streptavidin-HRP, PDA, and QD-NH2) (Fig. 4a). Before comparing their sensitivities, we first tested whether PDA deposition on microsphere surface reduces the microsphere fluorescence (would interfere with fluorescence barcodes if multiple colors were doped inside). PDA coating on microsphere is easy to monitor since the solution quickly turns dark brown because of the chromogenic PDA (Fig. 4b), yet microscopy images reveal virtually no change of the microsphere fluorescence before and after PDA coating. Additional quantitative evaluation of the microspheres using fluorometer unambiguously confirms the microscopy result (Supplementary Fig. S10). QDs were used as the fluorescent reporter because of their tunable fluorescence emission and the large Stokes shift (to avoid spectral overlap with the microsphere fluorescence)26, 27. For QDs with various surface chemistries, only aminated QDs bind with the microspheres, showing their interaction is due to the chemical reactions between amines and PDA rather than physical adsorption (Fig. 4c and Supplementary Fig. S11)21. Next, we proceeded with the sensitivity comparison. Flow cytometry and fluorescence microscopy both reveal that the target IgG can be detected at a concentration of 1.2 pM using conventional sandwich assay (streptavidin-QD as the reporter), whereas addition of EASE can push the detection limit down by 2 orders of magnitude (fM range) (Fig. 4d).
Figure 4. Ultrasensitive suspension microarray enabled by EASE.
(a) Schematic illustration showing signal enhancement by EASE. Fluorescent microspheres coated with Abs (IgG) capture 2’Ab-biotin (a model target) in solution. The target molecule is detected by streptavidin (SA)-HRP complex, which catalyzes PDA deposition followed by QD adsorption. (b) Effect of PDA coating on microsphere fluorescence (1×109 beads ml−1, 12 nM 2’Ab-biotin). The dark microsphere suspension shows successful PDA coating, while the microscopy images show no obvious fluorescence change before and after the coating. Scale bar, 5 μm. (c) Representative fluorescence images of the microspheres and corresponding quantitative flow cytometry data showing strong QD signals only when both QD-PEG-NH2 and dopamine are present (1×106 beads ml−1, 12 pM 2’Ab-biotin). Scale bar, 3 μm. Error bars, s.d. over three replicates. (d) Quantitative evaluation of detection sensitivity enhancement by EASE. Representative single-bead fluorescence images and flow cytometry histograms (1×106 beads ml−1) show that the detection sensitivity of the microsphere immunoassay is improved by 100 folds (from 12 pM to 1.2 fM). Scale bar, 3 μm.
To assess the specificity of this ultrasensitive detection assay, two control experiments were conducted. In the first experiment where the target molecule was missing, no significant signals were detected with or without the EASE process, confirming the antibody-antigen binding specificity (Supplementary Fig. S12). Second, we also evaluated the potential crosstalk in a dual-color setup. Two types of microspheres were mixed together, green microsphere with mouse IgG on the surface and yellow microsphere with rabbit IgG. When anti-rabbit IgG was added as the target, strong fluorescence signal from the EASE assay was only detected on the yellow microspheres, free of crosstalk (Supplementary Fig. S13). This remarkable detection specificity lays the foundation for massive parallel screening applications when additional optical barcodes are used35.
EASE for ELISA and lateral flow strips
To demonstrate the versatility of EASE, it was further applied to ELISA and immuno strip tests, robust and poplar biochemical assays. These assays using antibodies for molecular recognition and enzyme-catalyzed chromogen development for target identification are easy to perform, gaining broad applications in both research and clinical laboratories. On the other hand, their mediocre detection sensitivities are also well acknowledged36. Compared to the suspension assays discussed above, a technical feature of these assays is that they are performed on solid supports (flat surfaces or porous membranes), rendering the sample washing steps quick and easy (dip in and out of washing buffer without the need of a centrifuge). This seemingly insignificant feature combined with the unique bioconjugation capability of PDA allows EASE to be carried over for more than one time21. For example, in the first round of amplification, HRP molecules bound to the target can catalyze localized deposition of PDA. The PDA layer can in turn capture a large number of HRP molecules that are capable of catalyzing the conversion of chromogenic substrates (Fig. 5a).
Figure 5. ELISA and lateral flow strips with EASE.
(a) Schematic illustration of the signal enhancement process. Upon target detection, a layer of PDA is coated around the target molecule, which allows a large number of HRP to adsorb. These HRP molecules, in turn, catalyze the substrate (TMB) conversion at a significantly enhanced rate. (b) Visual assessment of the detection sensitivity of ELISA-EASE using mouse IgG as a model target in comparison with conventional ELISA. Colored solutions are visualized in ELISA-EASE wells at target concentrations as low as 10−13 g ml−1, while the conventional assay only produces detectable colors at 10−8 to 10−9 g ml−1 concentration range. (c) Quantitative comparison using values obtained from a plate reader shows the assay response over the full target concentration range (left) and a zoomed-in range close to the essays’ LODs (right). Improvement of approximately 3 orders of magnitude is seen. Error bars, s.d. over three replicates. (d) ELISA working curves for four targets, HIV p24, KLK3, CRP, VEGF, with or without EASE. Error bars, s.d. over three replicates. (e) The average of LOD improvements for all four targets is around 1,200 fold. (f) Strip tests of HIV p24 with or without EASE. Positive signals (indicated by the red arrow) are observed at 10 ng ml−1 and 10 pg ml−1 for the EASE-strips, but the conventional strips can only detect down to 10 ng ml−1. Each strip represents three replicates.
To probe the sensitivity and specificity of ELISA with or without EASE, a standard sandwich ELISA assay was established to detect mouse IgG (model target). Serial dilution of the target molecule resulted in gradients of color development that can be easily visualized by naked eye (substrate: tetramethylbenzidine or TMB). As shown in Figure 5b, without EASE, color development in the ELISA assay is visible at a target concentration between 10−7 and 10−8 g ml−1; with EASE as an add-on step, the color development becomes clearly visible at 10−12 g ml−1. This significantly improved limit of detection (LOD) was further quantified on a plate reader. The standard curve relating signal strength and target concentration is shown in Figure 5c (left panel), with a zoomed-in low-concentration range plotted in the right panel. The plate-reader readouts reveal the ELISA LODs (3 s.d. from the background) were 85.3 fg ml−1 (with EASE) and 108 pg ml−1 (without EASE), a 1,266 fold improvement. The specificity of the ELISA assays was demonstrated by control experiments where the target molecule was missing (Supplementary Fig. S14) or high-concentration non-target analytes were introduced (Supplementary Fig. S15). The robustness of the EASE-aided ELISA was further demonstrated in another four disease biomarkers: HIV capsid antigen p24 (HIV p24), kallikrein 3 (KLK3), c-reactive protein (CRP), and vascular endothelial growth factor (VEGF). Similarly, their calculated values of LODs of ELISA-EASE are 2.87 fg ml−1, 0.31 pg ml−1, 0.24 pg ml−1, and 11.5 fg ml−1 (Fig. 5d and Supplementary Fig. S16), respectively, representing an average 1,217 fold improvement over the conventional ELISA (Fig. 5e).
Building on the remarkable sensitivity enhancement achieved on ELISA plate, the HIV biomarker p24 was further tested using lateral flow strips (Supplementary Fig. S17), a simple and low-cost bioassay, sharing similar detection mechanism as ELISA (conducted in porous membranes rather than on flat surfaces), but is better suited for point-of-care diagnosis37. As shown in Figure 5f and Supplementary Figure S18, the strip test detects p24 at a concentration of 10 ng ml−1 (spiked HIV p24 antigen in phosphate-buffered saline (PBS)) under conventional conditions (using DAB as the substrate), whereas EASE can improve it by at least 1,000 times (10 pg ml−1), enabling ultrasensitive detection of HIV antigens with naked-eye.
With the EASE platform validated in the above bioassays, we moved on to real biological problems that require much improved detection sensitivity to resolve. We demonstrate the power of EASE assay in detection of four biologically significant low-abundance targets, HIV in blood, in situ protein detection in brain samples, Zika virus (ZIKV) imaging in the placenta, and programmed death-ligand 1 (PD-L1) in tumor.
Early diagnosis of HIV using ELISA-EASE
Early diagnosis of HIV provides timely access to treatments, thus improving patient outcomes and quality of life38. A study of ~16,000 patients on antiretroviral (ARV) treatment between shows substantial numbers beginning ARV later than recommended due to late diagnosis39. For adults, early knowledge of infection also leads to behavioral changes that can reduce 30% new infections per year40. For children and infants, earlier diagnosis is even more important. At this time, over 200,000 children acquire HIV worldwide every year, with most cases due to transmission to infants from their mothers during pregnancy, birth, or breastfeeding. HIV progresses rapidly in infants – without treatment they can die within months – but early treatment by ART greatly improves outcomes. Large-scale programs (e.g., President’s Emergency Plan for AIDS Relief (PEPFAR)) have made ART available, but early diagnosis remains a barrier to treatment.
HIV can be detected in blood or plasma by 1) nucleic acid amplification tests (NAAT), 2) lab based immunoassays (ELISA), or 3) rapid tests (similar to pregnancy tests). In general, NAAT is sensitive, but very expensive, and rapid test is of low performance and cannot be used in infants (false positive due to antibodies from mom). For decades, ELISA has been the workhorse laboratory HIV test and is the first test in the Centers for Disease Control and Prevention (CDC) testing algorithm. The sensitivity of ELISA, however, has been a major limitation (even for the most recent generation, detections are made around two weeks after infection). How to push the detection to an earlier time has been a major unmet clinical need.
We used the ELISA assay to detect p24 antigen in real patient samples, the key protein that makes up most of the viral capsid, in patient sera. Quantitative measurement of its presence in serum is highly valuable to blood screening, diagnosis of infection, and monitoring treatment responses41. As recommended by the CDC, HIV p24 antigen detection using ELISA offers a number of advantages such as reduced cost, fast assay times, and applicability in low-resource settings. On the other hand, it is commonly acknowledged that p24 ELISA is an insensitive assay with a LOD of approximately 10 pg ml−1,42 limiting its use to samples with high viral loads. Layering the EASE technology on top, however, can convert the ordinary detection sensitivity of ELISA to extraordinary, as shown in the above ELISA studies conducted in buffers.
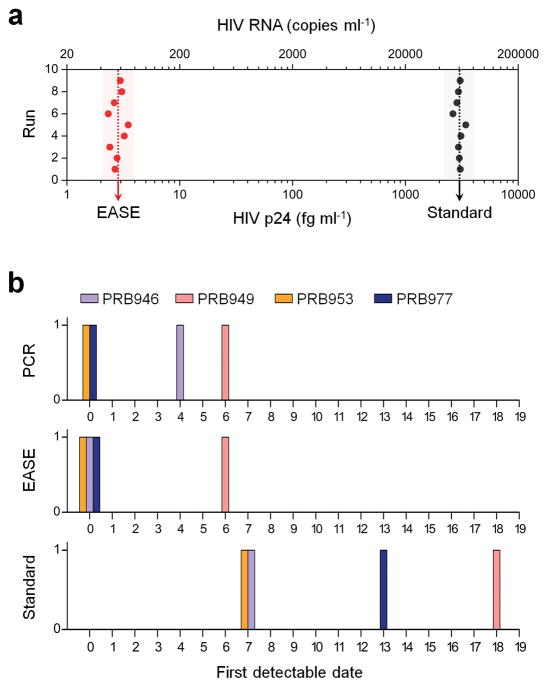
To demonstrate its ability in clinical diagnosis, sera from 24 donors (obtained from SeraCare, Milford, MA and Discovery Life Sciences, Los Osos, CA) were assayed with either standard ELISA or ELISA with EASE. Among these samples, four were obtained from HIV-infected patients (PRB 946, PRB 949, PRB 953, and PRB 977) whose viral loads had been determined using PCR (data from SeraCare); and 20 HIV-negative donors were included to exclude biased results due to nonspecific interactions (Supplementary Fig. S19). The analytical LOD was determined by spiking HIV p24 antigen of various concentrations into plasma. Results from 9 repeated runs performed on 9 consecutive days show a highly consistent value (Fig. 6a and Supplementary Fig. S20) of 2.84 fg ml−1 for ELISA-EASE, representing a 1,060 fold improvement over standard ELISA. Theoretical calculation indicates that this level of protein detection corresponds to samples contain approximately 56 copies ml−1 of RNA or 28 ml−1 viral particles43, on par with the sensitivity of PCR, which requires sophisticated instruments and long assay time. Indeed, when ELISA-EASE was applied to the HIV infected patient samples (multiple bleeds obtained on different dates), it can pick up the viral infection on average 10 days earlier (similar to PCR) than the standard ELISA assay (Table 1, Fig. 6b and Supplementary Fig. S21). This remarkable sensitivity potentially can provide a precious time window for treating other time-sensitive infections (e.g., viral and bacterial infections) and diseases (e.g., heart diseases) as well.
Figure 6. Early diagnosis of HIV in patient blood samples using ELISA-EASE.
(a) LOD values (obtained in 9 runs performed on different days) of ELISA with or without EASE for detection of HIV p24 spiked in plasma. The average LOD of ELISA with EASE is 2.84 fg ml−1, 1,060 fold lower than that of the standard ELISA. (b) First date when HIV infection becomes detectable since the first bleed for the four patients. Positive detection is made within the first week for ELISA-EASE and PCR, whereas the conventional ELISA detects infection 2–3 weeks later when the viral load is high.
Table 1. Viral load assessment using ELISA, ELISA-EASE, and PCR in four HIV-infected patients’ plasma samples.
Serial bleeds were collected from individual patients over a course of 18 days during the development of HIV infection (first bleed as day 0). ELISA-EASE detects HIV p24 as early as PCR does.
| Patient ID | Phlebotomy date | EASE (pg ml−1) | Standard (pg ml−1) |
|---|---|---|---|
| PRB 946 | 0 | 0.006* | ↓ |
| 4† | 0.807 | ↓ | |
| 7 | 26.86 | 19.22* | |
| 11 | 39.70 | 50.63 | |
| PRB 949 | 0 | ↓ | ↓ |
| 6† | 0.029* | ↓ | |
| 9 | 0.561 | ↓ | |
| 18 | 22.05 | 17.22* | |
| PRB 953 | 0† | 0.043* | ↓ |
| 3 | 1.320 | ↓ | |
| 7 | 23.36 | 16.01* | |
| 10 | 39.99 | 50.97 | |
| PRB 977 | 0† | 0.009* | ↓ |
| 2 | 0.121 | ↓ | |
| 13 | >100 | >100* | |
| 15 | >100 | >100 |
↓ blow the quantitation range;
first detectable date using PCR;
first detectable date using ELISA.
Resolving corticotrophin releasing factor (CRF) distribution in the brain using IF-EASE
CRF and its canonical G-protein coupled receptors, corticotrophin releasing factor receptor type 1 (CRFR1) and CRFR2 play an essential role in stress responsiveness regulated by the central nervous system44. Alterations in the function of the CRF system and changes in CRF receptor signaling are broadly linked to neuropsychiatric disorders including addiction and depression45. The ability to resolve the spatial distribution of CRF receptors in the brain will transform our understanding of how these receptors influence neural circuit function and how alterations in the expression and distribution of these receptors contribute to the disease states. Detection of CRF receptors has been largely limited to in situ hybridization detection on the mRNA level and radio-ligand binding assays 46, 47, which provide poor spatial resolution. High-resolution localization of these receptors using conventional immunostaining techniques has been limited by the low levels of receptor expression. To test the effectiveness of EASE technology to enhance CRFR1 detection using antibody staining, we performed immunostaining for CRFR1 using conventional methods and EASE. Analysis of CRFR1 detection revealed only a small number of CRFR1-positive cells in the cerebral cortex of the mouse brain using conventional immunostaining (Supplementary Fig. S22). In contrast, EASE amplification revealed numerous CRFR1-positive cells including both small diameter and large diameter cells, indicative of expression in both interneurons and pyramidal neurons, respectively (Supplementary Fig. S22b). Additionally, EASE detection of CRFR1 localized the protein to the cell bodies of both cell types, as well as the apical dendrites of pyramidal neurons.
Direct imaging of ZIKV infection in the placenta using IF-EASE
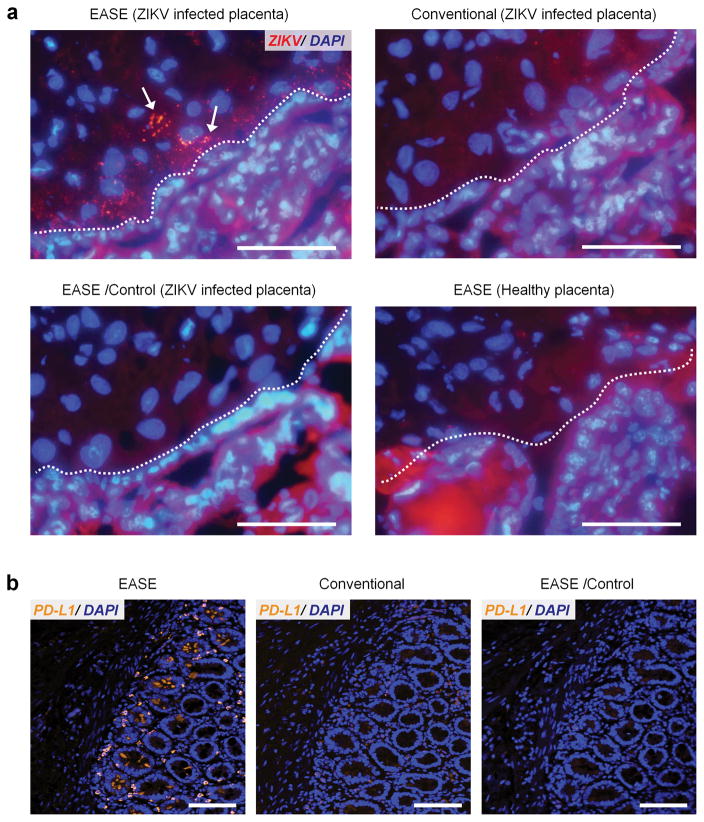
Zika is a mosquito-borne flavivirus initially identified in the 1950s’ in monkeys. Its recent outbreak in Brazil has been correlated with cases of fetal microcephaly as well as Guillian-Barré, raising major global concerns. While there is now scientific consensus, including our own work48, that ZIKV indeed causes fatal brain injury, the mechanism of how it occurs is largely unknown. qPCR and deep sequencing are capable of identifying ZIKV in the placenta, but cannot elucidate the means by which ZIKV crossed the placental barrier due to their inability to track ZIKV through conventional immunohistologic analysis. The EASE technology enabled direct visualization of ZIKV infected cells within the placental chorionic villus core of pregnant nonhuman primates. As shown in Figure 7a and Supplementary Figure S23, the infected cells appear in the mesenchymal core in close proximity to the cytotrophoblast cell layer. The EASE technology opens a new avenue to understand fetal brain injury and microcephaly caused by ZIKV and potentially to prevent mother-to-child transmission.
Figure 7. Sensitive imaging of ZIKV in placenta and PD-L1 in FFPE pancreatic tumor specimens.
(a) Representative fluorescent images of ZIKV in placental chorionic villi (nuclei counter-stained with DAPI). Scale bar, 100 μm. ZIKV infected cells indicated by arrows can only be observed through IF-EASE, not with conventional IF. Staining specificity is verified using the controls (without primary Ab, or IF-EASE in non-infected placentas). Dashed lines, cytotrophoblast cell layer (identified by morphology). Infected cells appear within the chorionic villus core and villi beneath in close proximity to the cytotrophoblast cell layer. The red background signal is due to tissue autofluorescence, which can be reduced under confocal imaging where the excitation source is a laser (narrow band). (b) Representative fluorescence micrographs of PD-L1 expression in pancreatic specimens from the patient (SU-09-21157), samples counter-stained with DAPI. Scale bar, 100 μm. PD-L1 staining can be easily observed through the IF-EASE technology, but very difficult using the conventional IF. The control experiment (without primary Ab) did not show detectable signals either.
PD-L1 imaging in patient tumor specimens using IF-EASE
PD-L1, also known as CD-274 or B7-H1 is a cell surface ligand, which binds and triggers PD-1, a potent immune-inhibitory receptor on T cells49. Monoclonal antibodies which block this interaction, by binding either PD-L1 or PD-1 have proven to be efficacious immune-oncology agents in a variety of tumor types49–54. Immunohistochemical assays for detecting PD-L1+ cells within tumors have also been approved as companion diagnostic tests for patient selection in limited therapeutic indications, but broader application of anti-PDL1 IHC is limited by both biologic and technical factors. PD-L1 expression vary broadly across a wide range and levels below the detection thresholds of current IHC assays still have biologic significance55. Therefore, we set out to test whether EASE can be used to detect low-level PD-L1 signals while preserving good signal-to-noise ratios, an unmet clinical need for immunotherapy. Clinical formalin-fixed paraffin-embedded (FFPE) pancreatic tumor specimens with low PD-L1 expression were used to test the performance of IF-EASE with the conventional IF. As shown in Figure 7b and Supplementary Figure S24, specific detection of PD-L1 is readily achieved with IF-EASE, whereas the signals detected by conventional IF technology are at extremely low levels. These exciting results address the unmet clinical need of detecting low abundance targets in FFPE tissues (high autofluorescence background), proving the feasibility for future large-scale studies.
DISCUSSION
Since the invention of a simple immersion procedure for slow coating of a PDA layer onto virtually any surface20, this mussel-inspired surface chemistry has intrigued the scientists to explore a wide variety of applications such as sensing, tissue engineering, and bioimaging56, 57. Similar to the oxidase-catalyzed rapid melanin deposition found in invertebrates22, 23, we found that HRP can speed up PDA polymerization by approximately 300 times. More importantly, due to the excellent reactivity of PDA to primary amines, the polymer chains quickly crosslink with nearby biomolecules (rich in many reactive chemical groups including NH2), forming a localized network for immobilization of a large number of reporter molecules and nanoparticles (as long as they have accessible amine groups) for signal enhancement while preserving the spatial information. This technology, dubbed EASE, has been studied in various contexts including immunohistochemistry and immunofluorescence for single cell imaging, ELISA, lateral flow strips, and suspension microarrays. Consistently, it improves bio-imaging and -detection sensitivity by at least 2–3 orders of magnitude regardless of the assay format. Most significantly, EASE achieves this remarkable sensitivity without changing the design of common assay formats, or requiring specialized equipment and reagents, in contrast to most ultrasensitive detection technologies invented in the past 10–20 years. Therefore, EASE can be directly incorporated into the current biology and clinical infrastructure for immediate impact.
The flexibility of this general technology has allowed us to go beyond technology development and tackle a number of real biological problems that cannot be solved (at least extremely difficult to solve) using conventional bioassays. We applied EASE to ELISA-based detection of HIV infection in patient blood samples. For comparison, the measurements were benchmarked against the gold-standard assays, standard ELISA and PCR. Our results show that the EASE-enabled ELISA outperforms the standard ELISA by >1,000 times in sensitivity, which translates into detection of 2–3 viruses per 100 μl of blood. This sensitivity is similar to that of PCR-based approaches allowing HIV detection 1–2 weeks earlier, yet ELISA is faster and cheaper to perform, and compatible with point-of-care (POC) applications (costly thermocycler not needed). Furthermore, EASE is a robust process that can be applied to a variety of real biological and clinical problems, such as brain biology, in situ virus imaging in placenta, and PD-L1 imaging for immunotherapy. We envision that no or only minor developments are needed for broad technology adoption and application, such as early detection of viruses and bacteria without culture, counting disease biomarkers of low abundance, and monitoring treatment responses.
METHODS
Preparation of dopamine solution for EASE
Dopamine hydrochloride powder (15 mg) was dissolved rapidly in tris buffer (10 mM, 3 ml) at pH8.5, followed by quick addition of H2O2 (1M, 60 μl). The mixture solution was used fresh.
Polydopamine deposition
Small droplets of HRP (0.1 μg) in PBS buffer and/or BSA (15 μg) in PBS buffer were placed on nitrocellulose membrane and air-dried for 1 hour at room temperature. The membranes were further exposed to the EASE assay for 1 minute and washed with PBS for 30 seconds.
Cell culture and fixation
HeLa cells were cultured in MEM medium with L-glutamine, 10% fetal bovine serum, and antibiotics (60 μg ml−1 streptomycin and 60 U ml−1 penicillin) in glass-bottom 24-well plates to 60–80% confluency. Before IF staining, cells were rinsed with 1X tris-buffered saline (TBS), fixed with 4% formaldehyde in TBS for 30 minutes, permeabilized with 2% DTAC (dodecyltrimethylammonium chloride)/TBS for 30 minutes followed by 0.25% TritonX-100/TBS for 5 minutes and washed five times with TBS (each time 3 minutes). The fixed cells were stored in 1X PBS at 4 °C.
Cell imaging and signal analysis
An Olympus IX-71 inverted fluorescence microscope equipped with a true-color charge-coupled device (QColor5, Olympus), a LSM 510 Meta confocal microscpe (Zeiss, Dublin, CA) and a hyper-spectral imaging camera (Nuance, 420–720 nm spectral range, CRI, now Advanced Molecular Vision) were used for cell imaging. Low-magnification images were obtained with a 20X objective (NA 0.75, Olympus) and high-magnification with 40X and 100X oil-immersion objectives (NA 1.40, Olympus). Wide UV filter cube (330–385 nm band-pass excitation, 420 nm long-pass emission, Olympus) was used for imaging of all QD probes. All images were acquired with cells attached to the coverslip bottom of the well and immersed in PBS without anti-fading reagents. For quantitative comparisons, the same exposure time and gain were applied during imaging. Nuance image analysis software and ImageJ were used to identify regions of interest that included stained cells and excluded ‘blank’ cell-free areas. Average fluorescence intensity throughout all regions of interest within a single image was recorded. Identical analysis was performed on 4 images (containing ~40 cells per field of view) taken from different areas of the sample to obtain an overall average staining intensity and assess signal variation.
IHC/IF-EASE single cell imaging
Prior to staining, the endogenous peroxidase activity of cells was quenched by 3% H2O2 solution. Cells were first blocked with 2% BSA/0.1% casein in 1X PBS for 30 minutes. Rabbit anti-Lamin A IgG (LOT: L1293, Sigma-Aldrich) or anti-HSP90 IgG (LOT: SAB4300541, Sigma-Aldrich) diluted in PBS buffer containing 6% BSA was added to the cells. After 1-hour incubation, cells were washed three times (5 minutes each) with PBS containing 2% BSA, followed by another 1-hour incubation with goat anti-rabbit IgG (H+L) HRP-2’Ab (LOT: RA230590, ThermoFisher). Unbound antibodies were washed away with PBS with 2% BSA (5 min X 3), and fresh enzyme substrate (dopamine or DAB) was added to cells for 15 minutes incubation. The ideal staining result is strong chromogen signal of interested target locations with low nonspecific signals in background. To characterize the staining stability after storage, the stained cells were stored in 1X PBS at 4 °C, and washed with fresh PBS every four days. Images were captured every three weeks on the same cell subset with the same exposure and gain. For immunofluorescence imaging with QDs, after the PDA development step, amine-functionalized PEG-coated QDs (10 nM) were incubated with cells for 1 hour.
Preparation of antigen-coated fluorescent beads
IgG purified from mouse and rabbit serum were covalently linked to the surface of green and yellow fluorescent beads, via 2-step carbodiimide-mediated cross-linking between the carboxylic groups on bead surface and the primary amines on IgG. Briefly, fluorescent beads were first washed and suspended in MES buffer (pH 4.8) with 0.01% Tween-20 at 0.1 w/v% (~107 beads ml−1) and activated for 15 minutes upon addition of 10 mg ml−1 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and 10 mg ml−1 N-hydroxysulfosuccinimide (sulfo-NHS). The activated beads were washed by centrifugation (5,000 g X 2 min) twice using 50mM borate buffer (pH 8.5) with 0.01% Tween-20 to remove excess crosslinkers and then incubated with IgG (2.5 mg ml−1) in borate buffer with 0.01% Tween-20 for 6 hours. The resulting IgG-coated beads were washed 3 times to remove excess IgG, resuspended in PBS (with 0.5% BSA), and stored at 4 °C.
Suspension microarray with EASE
Biotinylated goat anti-mouse and goat anti-rabbit IgGs were detected by the antibody-coated green and yellow beads. PBS containing 0.5% BSA was used as incubation and blocking buffer throughout the experiment. All incubation steps were carried out at room temperature under gentle rotation. All washing steps were done by centrifuging down the microbeads at 3,000 g for 2 minutes. Each microbead type was resuspended in 100 μl PBS at a final concentration of 1×106 beads ml−1. The beads were first incubated in the blocking buffer for 30 minutes. Biotinylated anti-mouse or -rabbit IgGs were added to the bead solution, incubated for 30 minutes, washed 3 times with PBS (0.5% BSA), and resuspended in 100 μl buffer. Then HRP-streptavidin probes (1:3000 dilution) were added to the bead solution, incubated for 30 minutes, washed 3 times with PBS (0.5% BSA), resuspended in 100 μl dopamine solution for EASE, followed by 15 min incubation. The microbeads were washed another 3 times in BSA-free PBS, and mixed with amine-functionalized PEG-coated QDs (1 nM final concentration) for 1 hour incubation. At the end of QD incubation, the beads were washed 5 times with DI water and concentrated in 10 μl water for microscopy examination. Hyper-spectral imaging camera (Nuance, 420–720 nm spectral range, CRI, now Advanced Molecular Vision) and software were used to unmix and quantify fluorescence signal components. False-color composite images were obtained by merging individual channels. For quantitative analysis, Nuance image analysis software was used to automatically identify regions of interest that included QD labelling. Identical analysis was performed on 5 images (containing at least 20 beads per field of view). High-throughput quantitative analysis was achieved on a LSR-II flow cytometer (BD Biosciences). For each sample, at least 5,000 beads were counted. The flow cytometry data was analyzed with FlowJo 9.3.3 (TreeStar).
ELISA-EASE
Mouse IgG, HIV p24, KLK3, CRP and VEGF (commercial kits purchased from Abcam (REF: ab151276, Cambridge, MA) or R&D Systems (LOT: DHP240; DKK300; DCRP00; DVE00)) were used as model targets for the ELISA experiments. 96-well plastic plates coated with capture antibodies were first blocked with PBS containing 2% BSA. 200 μl samples with serial dilutions and control samples were added into different wells. The wells were covered with adhesive strips and incubated for 2 hours at room temperature, washed 4 times, incubated with Ab-HRP conjugates for 2 hours at room temperature, washed 4 times with PBS (6% BSA), incubated with dopamine solution for 15 minutes, washed 3 times with PBS, incubated with HRP (1 nM) in PBS for 1 hour, and washed 4 times with PBS (6% BSA). 200 μl of the substrate solution was added to each well and the reaction was quenched after 20 min incubation in dark. Absorbance at 450 nm (optical density) was measured using an Infinite M 200 plate reader (Tecan). The results were compared with those obtained with conventional ELISA assays.
The sensitivity of ELISA-EASE in detecting HIV p24 in plasma was probed by spiking HIV p24 of known concentrations into the plasma from healthy donors. For plasma samples from both HIV infected patients and healthy donors, immune complex disruption and neutralization procedures were applied to treat the samples. 20 μl 5% Triton X-100, 90 μl plasma samples, 90 μl glycine reagent (1.5 M) were mixed and incubated for 1 hour at 37 °C. 90 μl tris buffer (1.5 M) was then added into the mixed solution and incubated for 10 minutes at room temperature. The plasma samples from HIV positive groups with high HIV p24 concentration were diluted (10× and 100×) to fit the ELISA working ranges for measurement.
Lateral flow test-EASE
The striper unit, BioDot ZX1010 (BioDot), is equipped with 4 frontline dispensers. Reagents (capture antibody) to be striped were aspirated through the end of the frontline dispenser. The nitrocellulose membrane (Sartorius CN95) was placed on the stage of the striper and secured, and then the frontline dispensers were adjusted to the appropriate position above the nitrocellulose membrane. The striper was programed to release the reagents at a rate of 1 μl cm−1. The membrane was placed in a forced air oven at 37 °C for 30 minutes before cooling in a desiccated environment. Once cooled, the membrane was placed on a backing card (DCN MIBA-020), and then the wick (GE Healthcare, CF5) was laid over the nitrocellulose with a 2 mm overlap. The completed card was placed in the staging area of the guillotine strip cutter (Kinbio ZQ200), and cut into 4 mm wide strips before being stored in Mylar bags that are sealed shut after including desiccant packets until use.
HIV p24 was used as a model target for the lateral flow test. Capturing antibodies (HIV p24 antibody) were immobilized onto nitrocellulose membrane. The membrane was blocked with 0.5% tween-20/2% BSA in PBS for 30 minutes. The membrane was then exposed to HIV p24 sample solutions (10 min). After washing (3X), the strips were treated with HIV p24 antibody-HRP conjugates for 30 minutes and washed 3 times again. DAB was used as the enzyme substrate for 10 min color development.
RNA interference
GAPDH expression knock-down was done by transfecting siRNA targeting GAPDH into HeLa cells. Annealed siRNA with 3′-TT overhangs was purchased from IDT (Coralville, IA). The sense strand sequence is 5′-CAUCAUCCCUGCCUCUACUTT-3′ 28. HeLa cells were grown in a 10 cm TC-treated dish, trypsinized, and mixed in suspension with culture medium containing 25 nM GAPDH siRNA, together with 0.5 μl per well DharmaFECT-2 transfection reagent (Dharmacon). The cells (500 μl cell suspension per well) were then seeded into a glass-bottom 24-well plate, and incubated for 36 or 60 hours. Following RNAi, the cells were processed for staining using IF-EASE. Primary antibody used is: anti-GAPDH (rabbit, LOT: G9545, Sigma-Aldrich).
Histology preparation of brain tissues for CRFR1 staining
Mice were deeply anesthetized with 50 mg/kg of Beuthanasia-D and transcardially perfused with phosphate-buffered saline (PBS), followed by 4% paraformaldehyde. Whole brain tissue was dissected, fixed overnight in 4% paraformaldehyde, and cryoprotected by soaking in a 30% sucrose solution for 48 hours. The brains were flash frozen in OCT and stored at −80 °C. The frozen brains were then cryosectioned to 30 μm-thick sections and stored in 1× PBS with 0.1% NaAz prior to immunostaining.
CRFR1 IF staining in brain sections
Coronal 30 μm sections were selected based on a reference atlas (Franklin and Paxinos) and analyzed for protein expression. Primary antibody against CRFR1 (Novus Biologicals, cat. No. NLS1778) was diluted 1:100. Cy3- or HRP-labeled secondary antibodies (donkey anti-rabbit, Jackson Immunolabs, and goat anti-rabbit) were diluted 1:250 58. Sections were incubated in 3% hydrogen peroxide 1× TBS buffer (10 min) to quench the intrinsic peroxide in tissue, washed with 1× TBS for 10 minutes, and blocked with 1× TBST (TBS + 0.3% TritonX 100) with 3% donkey serum for 60 minutes. The blocked sections were stained with the primary antibody diluted in the blocking buffer overnight, washed three times in 1× TBS for 10 minutes, and incubated in Cy3- or HRP-conjugated secondary antibodies for 1 hour at room temperature. IF-EASE were applied as described above (amine-Cy3 was used as the reactive fluorophore). The sections were washed three more times in 1× TBS and mounted.
Immunostaining of ZIKV-infected placenta
Placental samples were collected from pregnant pigtail macaques (Macaca nemestrina), who were inoculated with ZIKV (strain FSS13025, Cambodia 2010) or from a normal pregnancy 48. Formaldehyde-fixed sections of frozen placental chorionic villi were stained using both conventional IF and IF-EASE. The primary antibody (ZIKV E-protein Clone ZV-13, Diamond lab) was diluted 1:200 59. Other reagents such as the labeled secondary antibodies as well as the staining protocol were the same as the ones described in the CRFR1 experiments. Healthy control was used for studying the specificity of IF-EASE. Adjacent tissue slides were used for all staining conditions.
PD-L1 immunostaining of pancreatic tumor specimens
The FFPE pancreatic tumor tissue specimens from two patients (SU-09-21157; SU-10-26808) were deparaffinized by washing the slides with xylene (7 min, 3 times), 100% ethanol (2 min, twice), 95% ethanol (2 min, twice), 70% ethanol (2 min, twice) and DI water (2 min). The sections were then incubated in 3% hydrogen peroxide in 1× TBS buffer (30 min) to quench the intrinsic peroxide. Antigen retrieval was performed by incubating the sections with the Trilogy antigen retrieval buffer under high pressure (15 min), cooling down (20 min), and washing with 1× TBS (5 min, 2 times). The sections were subsequently stained using both conventional IF and IF-EASE. The protocols are the same as the ones described immediately above except the primary antibody is mouse anti-PD-L1 (1:150 dilution, Cell signaling Technology, REF: 29122S). Adjacent tissue slides were used for all staining conditions.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information.
Supplementary Material
Acknowledgments
This work was supported in part by the NIH (R21CA192985, R01AI100989, AI083019, AI104002, and AI060389) and the Department of Bioengineering at the University of Washington. J.L. thanks the Howard Hughes Medical Institute for a student fellowship. We are also grateful to Prof. Barry Lutz and Mr. Daniel Leon for help with the lateral flow test, and Dr. Pavel Zrazhevskiy for discussions on immunostaining.
Footnotes
AUTHOR CONTRIBUTIONS
J.L. and X.H.G. conceived the idea and designed the project. J.L. and W.T. performed majority of the experiments, with helps from M.A.B. and L.S.Z. for CRF imaging in the brain, from M.A.D., K.M.A.W, M.G, and L.R. for ZIKA imaging, and R.H.P. for PD-L1 imaging. All authors were involved in data analysis. J.L., L.S.Z., K.M.A.W, M.G, L.R., R.H.P. and X.H.G. wrote the paper.
COMPETING FINACIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Howes PD, Chandrawati R, Stevens MM. Colloidal nanoparticles as advanced biological sensors. Science. 2014;346:1247390. doi: 10.1126/science.1247390. [DOI] [PubMed] [Google Scholar]
- 2.Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 3.Kelley SO, et al. Advancing the speed, sensitivity and accuracy of biomolecular detection using multi-length-scale engineering. Nature nanotechnology. 2014;9:969–980. doi: 10.1038/nnano.2014.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 5.Kosaka PM, et al. Detection of cancer biomarkers in serum using a hybrid mechanical and optoplasmonic nanosensor. Nature nanotechnology. 2014;9:1047–1053. doi: 10.1038/nnano.2014.250. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Lorenzo L, de La Rica R, Alvarez-Puebla RA, Liz-Marzan LM, Stevens MM. Plasmonic nanosensors with inverse sensitivity by means of enzyme-guided crystal growth. Nature materials. 2012;11:604–607. doi: 10.1038/nmat3337. [DOI] [PubMed] [Google Scholar]
- 7.He L, Ozdemir SK, Zhu J, Kim W, Yang L. Detecting single viruses and nanoparticles using whispering gallery microlasers. Nature nanotechnology. 2011;6:428–432. doi: 10.1038/nnano.2011.99. [DOI] [PubMed] [Google Scholar]
- 8.Thomas RK, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nature Medicine. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 9.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nature Biotechnology. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 10.Wu G, et al. Bioassay of prostate-specific antigen (PSA) using microcantilevers. Nature Biotechnology. 2001;19:856–860. doi: 10.1038/nbt0901-856. [DOI] [PubMed] [Google Scholar]
- 11.Schallmeiner E, et al. Sensitive protein detection via triple-binder proximity ligation assays. Nature Methods. 2007;4:135–137. doi: 10.1038/nmeth974. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe R, et al. Arrayed lipid bilayer chambers allow single-molecule analysis of membrane transporter activity. Nature communications. 2014;5 doi: 10.1038/ncomms5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma W, et al. Attomolar DNA detection with chiral nanorod assemblies. Nature communications. 2013;4 doi: 10.1038/ncomms3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haun JB, Devaraj NK, Hilderbrand SA, Lee H, Weissleder R. Bioorthogonal chemistry amplifies nanoparticle binding and enhances the sensitivity of cell detection. Nature nanotechnology. 2010;5:660–665. doi: 10.1038/nnano.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vollmer F, Arnold S. Whispering-gallery-mode biosensing: label-free detection down to single molecules. Nature Methods. 2008;5:591–596. doi: 10.1038/nmeth.1221. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Tang HX, Roukes ML. Ultra-sensitive NEMS-based cantilevers for sensing, scanned probe and very high-frequency applications. Nature nanotechnology. 2007;2:114–120. doi: 10.1038/nnano.2006.208. [DOI] [PubMed] [Google Scholar]
- 17.Cooper MA, et al. Direct and sensitive detection of a human virus by rupture event scanning. Nature Biotechnology. 2001;19:833–837. doi: 10.1038/nbt0901-833. [DOI] [PubMed] [Google Scholar]
- 18.Rissin DM, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nature Biotechnology. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burst bubbles. Nature. 2015;526:609–610. doi: 10.1038/526609b. [DOI] [PubMed] [Google Scholar]
- 20.Lee H, Dellatore SM, Miller WM, Messersmith PB. Mussel-inspired surface chemistry for multifunctional coatings. Science. 2007;318:426–430. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H, Rho J, Messersmith PB. Facile conjugation of biomolecules onto surfaces via mussel adhesive protein inspired coatings. Advanced Materials. 2009;21:431–434. doi: 10.1002/adma.200801222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soderhall K, Cerenius L. Role of the prophenoloxidase-activating system in invertebrate immunity. Current Opinion in Immunology. 1998;10:23–28. doi: 10.1016/s0952-7915(98)80026-5. [DOI] [PubMed] [Google Scholar]
- 23.Cerenius L, Soderhall K. The prophenoloxidase-activating system in invertebrates. Immunological Reviews. 2004;198:116–126. doi: 10.1111/j.0105-2896.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- 24.Weber R, et al. Threshold of detection of Cryptosporidium oocysts in human stool specimens: evidence for low sensitivity of current diagnostic methods. Journal of Clinical Microbiology. 1991;29:1323–1327. doi: 10.1128/jcm.29.7.1323-1327.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahler M, Ngo JT, Schulte-Pelkum J, Luettich T, Fritzler MJ. Limited reliability of the indirect immunofluorescence technique for the detection of anti-Rib-P antibodies. Arthritis Research & Therapy. 2008;10:1. doi: 10.1186/ar2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zrazhevskiy P, Sena M, Gao X. Designing multifunctional quantum dots for bioimaging, detection, and drug delivery. Chemical Society Reviews. 2010;39:4326–4354. doi: 10.1039/b915139g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nature materials. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 28.Zrazhevskiy P, et al. Cross-platform DNA encoding for single-cell imaging of gene expression. Angewandte Chemie International Edition. 2016;55:8975–8978. doi: 10.1002/anie.201603945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Battersby BJ, Lawrie GA, Trau M. Optical encoding of microbeads for gene screening: alternatives to microarrays. Drug Discovery Today. 2001;6:19–26. [Google Scholar]
- 30.Braeckmans K, De Smedt SC, Leblans M, Pauwels R, Demeester J. Encoding microcarriers: present and future technologies. Nature Reviews Drug Discovery. 2002;1:447–456. doi: 10.1038/nrd817. [DOI] [PubMed] [Google Scholar]
- 31.Nolan JP, Sklar LA. Suspension array technology: evolution of the flat-array paradigm. TRENDS in Biotechnology. 2002;20:9–12. doi: 10.1016/s0167-7799(01)01844-3. [DOI] [PubMed] [Google Scholar]
- 32.Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman JR. Advanced multiplexed analysis with the FlowMetrixTM system. Clinical chemistry. 1997;43:1749–1756. [PubMed] [Google Scholar]
- 33.Dunbar SA. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clinica Chimica Acta. 2006;363:71–82. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan WC, et al. Luminescent quantum dots for multiplexed biological detection and imaging. Current Opinion in Biotechnology. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 35.Han M, Gao X, Su JZ, Nie S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nature Biotechnology. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 36.Klein D, Hurley LB, Merrill D, Quesenberry CP., Jr Review of medical encounters in the 5 years before a diagnosis of HIV-1 infection: implications for early detection. JAIDS Journal of Acquired Immune Deficiency Syndromes. 2003;32:143–152. doi: 10.1097/00126334-200302010-00005. [DOI] [PubMed] [Google Scholar]
- 37.Fu E, et al. Enhanced sensitivity of lateral flow tests using a two-dimensional paper network format. Analytical chemistry. 2011;83:7941–7946. doi: 10.1021/ac201950g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palella FJ, et al. Survival benefit of initiating antiretroviral therapy in HIV-infected persons in different CD4+ cell strata. Annals of internal medicine. 2003;138:620–626. doi: 10.7326/0003-4819-138-8-200304150-00007. [DOI] [PubMed] [Google Scholar]
- 39.Holodniy M, et al. Relationship between antiretroviral prescribing patterns and treatment guidelines in treatment-naive HIV-1-infected US veterans (1992–2004) Journal of Acquired Immune Deficiency Syndromes. 2007;44:20–29. doi: 10.1097/01.qai.0000248354.63748.54. [DOI] [PubMed] [Google Scholar]
- 40.Marks G, Crepaz N, Janssen RS. Estimating sexual transmission of HIV from persons aware and unaware that they are infected with the virus in the USA. Aids. 2006;20:1447–1450. doi: 10.1097/01.aids.0000233579.79714.8d. [DOI] [PubMed] [Google Scholar]
- 41.Miles SA, et al. Rapid serologic testing with immune-complex-dissociated HIV p24 antigen for early detection of HIV infection in neonates. New England Journal of Medicine. 1993;328:297–302. doi: 10.1056/NEJM199302043280501. [DOI] [PubMed] [Google Scholar]
- 42.Nishanian P, Huskins KR, Stehn S, Detels R, Fahey JL. A simple method for improved assay demonstrates that HIV p24 antigen is present as immune complexes in most sera from HIV-infected individuals. Journal of Infectious Diseases. 1990;162:21–28. doi: 10.1093/infdis/162.1.21. [DOI] [PubMed] [Google Scholar]
- 43.Marozsan AJ, et al. Relationships between infectious titer, capsid protein levels, and reverse transcriptase activities of diverse human immunodeficiency virus type 1 isolates. Journal of Virology. 2004;78:11130–11141. doi: 10.1128/JVI.78.20.11130-11141.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bale TL, Vale WW. CRF and CRF receptors: role in stress responsivity and other behaviors. Annual Review of Pharmacology and Toxicology. 2004;44:525–557. doi: 10.1146/annurev.pharmtox.44.101802.121410. [DOI] [PubMed] [Google Scholar]
- 45.Zorrilla EP, Logrip ML, Koob G. Corticotropin releasing factor: a key role in the neurobiology of addiction. Frontiers in neuroendocrinology. 2014;35:234–244. doi: 10.1016/j.yfrne.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Pett K, et al. Distribution of mRNAs encoding CRF receptors in brain and pituitary of rat and mouse. Journal of Comparative Neurology. 2000;428:191–212. doi: 10.1002/1096-9861(20001211)428:2<191::aid-cne1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 47.Weathington JM, Cooke BM. Corticotropin-releasing factor receptor binding in the amygdala changes across puberty in a sex-specific manner. Endocrinology. 2012;153:5701–5705. doi: 10.1210/en.2012-1815. [DOI] [PubMed] [Google Scholar]
- 48.Waldorf KMA, et al. Fetal brain lesions after subcutaneous inoculation of Zika virus in a pregnant nonhuman primate. Nature Medicine. 2016;22:1256–1259. doi: 10.1038/nm.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keir ME, et al. PD-1 and its ligands in tolerance and immunity. Annual Review of Immunology. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brahmer JR, et al. Phase I study of single-agent anti–programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. Journal of Clinical Oncology. 2010;28:3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamid O, et al. Safety and tumor responses with lambrolizumab (anti–PD-1) in melanoma. New England Journal of Medicine. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garon EB, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. New England Journal of Medicine. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 53.Rizvi NA, et al. Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): a phase 2, single-arm trial. The Lancet Oncology. 2015;16:257–265. doi: 10.1016/S1470-2045(15)70054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nghiem PT, et al. PD-1 blockade with pembrolizumab in advanced Merkel-cell carcinoma. New England Journal of Medicine. 2016;374:2542–2552. doi: 10.1056/NEJMoa1603702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, et al. PD-L1 expression in human cancers and its association with clinical outcomes. OncoTargets and Therapy. 2016;9:5023–5039. doi: 10.2147/OTT.S105862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, Ai K, Lu L. Polydopamine and its derivative materials: synthesis and promising applications in energy, environmental, and biomedical fields. Chemical Reviews. 2014;114:5057–5115. doi: 10.1021/cr400407a. [DOI] [PubMed] [Google Scholar]
- 57.Li J, et al. Stably doped conducting polymer nanoshells by surface initiated polymerization. Nano Letters. 2015;15:8217–8222. doi: 10.1021/acs.nanolett.5b03728. [DOI] [PubMed] [Google Scholar]
- 58.Sanford CA, et al. A central amygdala CRF circuit facilitates learning about weak threats. Neuron. 2017;93:164–178. doi: 10.1016/j.neuron.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao H, et al. Structural basis of Zika virus-specific antibody protection. Cell. 2016;166:1016–1027. doi: 10.1016/j.cell.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information.