Abstract
Computed tomography (CT) is critical for various clinical applications, e.g., radiotherapy treatment planning and also PET attenuation correction. However, CT exposes radiation during CT imaging, which may cause side effects to patients. Compared to CT, magnetic resonance imaging (MRI) is much safer and does not involve any radiation. Therefore, recently researchers are greatly motivated to estimate CT image from its corresponding MR image of the same subject for the case of radiotherapy planning. In this paper, we propose a 3D deep learning based method to address this challenging problem. Specifically, a 3D fully convolutional neural network (FCN) is adopted to learn an end-to-end nonlinear mapping from MR image to CT image. Compared to the conventional convolutional neural network (CNN), FCN generates structured output and can better preserve the neighborhood information in the predicted CT image. We have validated our method in a real pelvic CT/MRI dataset. Experimental results show that our method is accurate and robust for predicting CT image from MRI image, and also outperforms three state-of-the-art methods under comparison. In addition, the parameters, such as network depth and activation function, are extensively studied to give an insight for deep learning based regression tasks in our application.
1 Introduction
Computed tomography (CT) imaging is widely used for both diagnostic and therapeutic purposes in various clinical applications. In the cancer radiation therapy, CT image provides Hounsfield units, which is essential for dose calculation in treatment planning. Besides, CT image is also of great importance for attenuation correction of positron emission tomography (PET) in the popular PET-CT scanner [8].
However, patients are exposed to radiation during CT imaging, which can damage normal body cells and further increase potential risks of cancer. Brenner and Hall [1] reported that 0.4 % of cancers were due to CT scanning performed in the past, and this rate will increase to as high as 1.5 to 2 % in the future. Therefore, the use of CT scan should be with great caution. Magnetic resonance imaging (MRI) is a safe imaging protocol. It also provides more anatomical details than CT for diagnostic purpose, but unfortunately cannot be used for either dose calculation or attenuation correction. To reduce unnecessary imaging dose for patients, it is clinically desired to estimate CT image from MR image in many applications.
It is technically difficult to directly estimate CT image from MR image. As shown in Fig. 1, CT and MR images have very different appearances. For example, in CT image, the intensity difference between the prostate and bladder is much smaller than that in MR image. Besides, MR image contains richer texture information than CT image. Therefore, it is challenging to directly estimate a mapping from MRI intensity to CT intensity.
Fig. 1.
A pair of corresponding pelvic MR (left) and CT (right) images from the same subject.
Recently, many researches focus on estimating one modal image from another modality image, e.g., estimating CT image using MRI data. (a) The first category is atlas-based method. These methods first register an atlas (with the attenuation map) to the new subject MR image, and then warp the corresponding attenuation map of atlas to the new MR image as its estimated attenuation map of atlas to the new MR image as its estimated attenuation map [2,3]. However, the performance of these atlas-based methods highly depends on the registration accuracy. (b) The second category is learning-based method, in which non-linear model is learnt from MRI to CT image. Huynh et al. [5] presented an approach to predict CT image from MRI using structured random forest. Such methods often have to first represent the input MR image by features and then map features to output CT image. Thus, the performance of these methods is bound to the quality of feature extraction.
On the other hand, recently the convolutional neural network (CNN) [9] becomes popular in both computer vision and medical imaging fields. As a multilayer and fully trainable model, CNN is able to capture the complex non-linear mapping from the input space to the output space. For the case of 2D image, 2D CNN has been widely used in many applications. However, it is unreasonable to directly apply 2D CNN to process 3D medical images because 2D CNN considers the image appearance slice by slice, thus potentially causing discontinuous prediction results across slices. To address this issue, 3D CNNs have been proposed. Ji et al. [6] presented a 3D CNN model for action recognition in an uncontrolled environment. Tran et al. [13] used 3D CNN to effectively learn spatio-temporal features on a large-scale video dataset. Li et al. [11] applied deep learning models to estimate the missing PET image from the MR image of the same subject.
In this paper, we propose to learn the non-linear mapping from MR to CT images through a 3D fully convolutional neural network (FCN), which is a variation of the conventional CNN. Compared to CNN, FCN generates the structured output, which can better preserve the neighborhood information in the predicted CT image [12]. Specifically, an input MR image is first partitioned into overlapping patches. For each patch, FCN is used to predict the corresponding CT patch. Finally, all predicted CT patches are merged into a single CT image by averaging the intensities of overlapping CT regions. The proposed method is evaluated on a real pelvic CT/MR dataset. Experimental results demonstrate that our method can effectively predict CT image from MR image, and also outperforms three state-of-the-art methods under the comparison. Besides, extensive experiments have been conducted to validate the choice of several key parameters in the FCN, such as network depth and activation function. These parameter evaluation results provide good insight for other regression applications using deep learning.
2 Methods
Deep learning model can learn a hierarchy of features, i.e., high-level features built upon low-level features. CNN [4,9] is one popular type of deep learning models, in which trainable filters and local neighborhood pooling operations are applied in an alternating sequence starting with the raw input images. When trained with appropriate regularization, CNN can achieve superior performance on visual object recognition and image classification tasks [10]. However, most of CNNs are designed for 2D natural images. They are not well suited for medical image analysis, since most of medical images are 3D volumetric images, such as MRI, CT and PET. Compared to 2D CNN, 3D CNN can better model the 3D spatial information due to the use of 3D convolution operations. 3D convolution preserves the spatial neighborhood of 3D image. As a result, 3D CNN can solve the discontinuity problem across slices, which are suffered by 2D CNN. Mathematically, the 3D convolution operation is given by Eq. 1
| (1) |
where x, y, z denotes the 3D voxel position. W is a 3D filter. a is a 3D feature map from the previous (i − 1)-th layer. Initially, a is the input MRI patch. c and C is the index and number of feature maps in the previous layer. i and j are the layer index and filter index, respectively. Pi, Qi and Ri are the dimensions of the i-th filter in 3D space, respectively. f is an activation function that encodes the non-linearity in the CNN.
Structured CNN - FCN
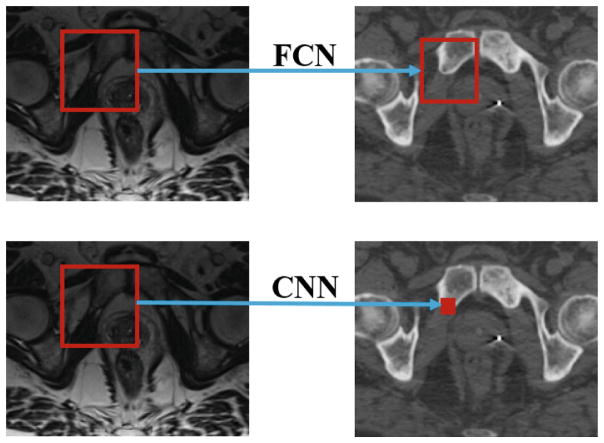
The output of conventional CNN is a single target value, which is unable to preserve neighborhood information in the output space. In this paper, we propose to use FCN to produce the structured output. Instead of predicting the CT intensity voxel by voxel, we use FCN to estimate the CT image in a patch-by-patch manner, as shown in Fig. 2. Compared to using CNN for voxel-wise CT prediction, using FCN for patch-wise CT prediction has several obvious advantages. First, the neighborhood information can be preserved in each predicted CT patch. Second, the prediction efficiency can be greatly improved since a entire CT patch can be predicted by a single pass of forward propagation in the neural network.
Fig. 2.
Illustration of difference between FCN and CNN. The left column shows MR slices, and the right one shows corresponding CT slices.
In the following paragraphs, we will describe the network architecture of FCN used in the MRI-to-CT prediction. Compared to the conventional CNN, the pooling layers are not used in this application. This is because the pooling layers often reduce the spatial resolution of feature maps. Although this property is desirable for tasks, such as image classification, since the pooling over local neighborhood could enhance invariance to certain image distortions, it is not desired in the task of image prediction, where subtle image distortions need to be precisely captured in the prediction process.
3D FCN for Estimating CT Images from MRI Data
Based on the 3D convolution described above, a variety of FCN architectures can be devised. In the following, we describe a 3D FCN architecture, as shown in Fig. 3, for estimating CT patch from MR patch. The training data for this CNN model consists of patches extracted from subjects with both MR and CT images. The size of input MRI patch is 32 × 32 × 16 and the size of output CT patch is 24 × 24 × 12. The input and output patches are in correspondence, which means that they share the same center position in their aligned image space. To generate training samples for FCN, we randomly extracted a large number of patches from 3D MRI volume as inputs, and the corresponding CT image patches as outputs. Patches that cross the boundaries or locate completely in the background were removed. The total number of patches extracted from each volume was 6000, which was sufficient to cover majority of the 3D image volume.
Fig. 3.
The 3D FCN architecture for estimating CT image from MRI image.
In the FCN architecture, we first apply 3-D convolution with a filter size of 7 × 7 × 3 on the input MRI patch to construct 32 feature maps in the first hidden layer. One voxel is padded along the first two dimensions. In the second layer, the outputs of the first layer are fed into another convolutional layer with 64 filters of size 5 × 5 × 3. The third convolutional layer contains 32 feature maps. Each of the feature maps is connected to all the input feature maps through filters of size 3 × 3 × 3. The output layer contains only one feature map generated by 1 filter of size 3 × 3 × 3, and it corresponds to the predicted CT image patch. To keep the same image size, one voxel is padded along three dimensions in the last two layers. In all layers, we set stride as one voxel. The latent nonlinear relationship between MR and CT images is encoded by the large number of parameters in the network.
Caffe [7] is modified to implement the architecture shown in Fig. 3. The network parameters of FCN are updated by back-propagation using stochastic gradient descent algorithm. To train the network, the model hyper-parameters need to be appropriately determined. Specifically, the network weights are initialized by xavier algorithm [4], which can automatically determine the scale of initialization based on the number of input and output neurons. For the network bias, we initialize it to be 0. The initial learning rate and weight decay parameter are determined by conducting a coarse line search, followed by decreasing the learning rate during training. In particular, we have approximately 200,000 training patches. A Titan X GPU is utilized to train the network.
3 Experiments and Results
Data Acquisition and Preprocessing
Our pelvic dataset consists of 22 subjects, each with MR and CT images. The spacings of CT and MR images are 1.172 × 1.172 × 1 mm3 and 1 × 1 × 1 mm3, respectively. In the training stage, CT image is manually aligned to its corresponding MR image to build voxel-wise correspondences. After alignment, CT and MR images of the same subjects have the same image size and spacing. Since only pelvic regions are concerned in this study, we further crop the aligned CT and MR images to reduce the computational burden. Finally, each preprocessed image has a size of 153 × 193 × 50 and a spacing of 1 × 1 × 1 mm3. A typical example of preprocessed CT and MR images is given in Fig. 1.
Parameter Selection of FCN
We evaluate the proposed method on 22 subjects in a leave-one-out cross validation. In our implementation, we adopt FCN to learn the nonlinear mapping from MR image to CT image. There are several factors that could affect the learning process, such as activation function, network depth, the number of filters in each layer and so on. In this paper, we evaluate the effect of different network depths and activation functions in learning this nonlinear mapping. In particular, three popular nonlinear functions are explored, which are the rectified linear units (Relu), sigmoid and tanh functions, respectively. The performance of FCN under different activation functions is shown in Fig. 4. In addition, we also analyze the impact of FCN under different network depths in Fig. 4. The experimental results show that with the Relu activation function, the performance of FCN gradually increases as the increase of network depth. However, for both sigmoid and tanh activation function, PSNR decreases with a deeper network. The best results are obtained with a shallow network (2-layer/3-layer). A simple interpretation is that sigmoid/tanh may suffer from gradient vanishing problem in a deep network although layer-wise training is used, while the Relu does not suffer such problem. The bad performance of Relu is due to the limited nonlinearity of Relu compared to two other activation functions. By increasing the network depth, the nonlinearity can be effectively increased, which renders the improved performance of Relu as the depth increases.
Fig. 4.
Sensitivity analysis of 3 activation functions with respect to network depth.
Experimental Results
To demonstrate the advantage of the proposed method in terms of prediction accuracy, we compare our method with three widely used approaches:
Atlas-based method (Atlas): Here, the MR image of each atlas is first aligned [15] onto the target MR image, and the resulting deformation field is used to warp the CT image of each atlas. The final prediction is obtained by averaging all warped CT images of all atlases.
Structured Random forest based method (SRF): Structured random forest [5] was used to learn a nonlinear mapping between MR image and its corresponding CT image.
Structured random forest and auto-context model (SRF+) based method: Besides the structured random forest, auto-context model (ACM) [14] is further used to iteratively refine the prediction of CT images.
The prediction results by different methods on two typical MR images are shown in Fig. 5. It can be clearly seen that our results are consistent with the ground-truth CT. To quantitatively evaluate our method, we use both mean absolute error (MAE) and peak signal-to-noise ratio (PSNR) to measure the prediction accuracy, as shown in Table 1.
Fig. 5.
Visual comparison of original MR images, the estimated CT images by our method and the ground truth CT images on 2 subjects.
Table 1.
Average of MAE and PSNR on 22 subjects by 4 different methods: Atlas, SRF, SRF+, and FCN (Proposed).
| MAE | Mean(std.) | Med. |
|---|---|---|
| Atlas | 66.1(6.9) | 66.7 |
| SRF | 51.2(3.8) | 51.5 |
| SRF+ | 48.1(4.6) | 48.3 |
| FCN | 42.4(5.1) | 42.6 |
| PSNR | Mean(std.) | Med. |
|---|---|---|
| Atlas | 29.0(2.1) | 29.6 |
| SRF | 31.7(0.9) | 31.8 |
| SRF+ | 32.1(0.9) | 31.8 |
| FCN | 33.4(1.1) | 33.2 |
Quantitative results in Table 1 show that our method outperforms other 3 methods in terms of both MAE and PSNR. Specifically, our method gives an average PSNR of 33.4, which is obviously better than 32.1 obtained by the state-of-the-art SRF+ method.
4 Conclusions
We have developed a 3D FCN model for estimating CT images from MRI images by directly taking MR image patches as input and CT patches as output. The nonlinear relationship between two imaging modalities is captured by a large number of trainable mapping and parameters in the network. We have applied this model to predict CT images from their corresponding MR images. Experiments demonstrate that our proposed method can significantly outperform the three state-of-the-art methods. We also conduct a simple exploration for the important factors in the deep learning regression, which gives an insight of parameter selection in other related regression tasks. Although we considered the FCN model for CT image prediction, this model can also be applied to other related tasks. In our future works, we will explore ways of expediting the computation and designing more effective deep learning models to improve the prediction speed and accuracy.
References
- 1.Brenner DJ, Hall EJ. Computed tomographyałan increasing source of radiation exposure. N Engl J Med. 2007;357(22):2277–2284. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- 2.Burgos N, et al. Robust CT synthesis for radiotherapy planning: application to the head and neck region. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. MICCAI 2015. LNCS. Vol. 9350. Springer; Heidelberg: 2015. pp. 476–484. [Google Scholar]
- 3.Catana C, et al. Toward implementing an MRI-based PET attenuation-correction method for neurologic studies on the MR-PET brain prototype. J Nucl Med. 2010;51(9):1431–1438. doi: 10.2967/jnumed.109.069112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks. International Conference on Artificial Intelligence and Statistics; 2010. pp. 249–256. [Google Scholar]
- 5.Huynh T, et al. Estimating CT image from MRI data using structured random forest and auto-context model. 2015 doi: 10.1109/TMI.2015.2461533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji S, et al. 3D convolutional neural networks for human action recognition. IEEE Trans Pattern Anal Mach Intell. 2013;35(1):221–231. doi: 10.1109/TPAMI.2012.59. [DOI] [PubMed] [Google Scholar]
- 7.Jia Y, et al. Caffe: convolutional architecture for fast feature embedding. Proceedings of the ACM International Conference on Multimedia; ACM; 2014. pp. 675–678. [Google Scholar]
- 8.Kinahan PE, et al. Attenuation correction for a combined 3D PET/CT scanner. Med Phys. 1998;25(10):2046–2053. doi: 10.1118/1.598392. [DOI] [PubMed] [Google Scholar]
- 9.LeCun Y, et al. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 10.LeCun Y, et al. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86(11):2278–2324. [Google Scholar]
- 11.Li R, Zhang W, Suk H-I, Wang L, Li J, Shen D, Ji S. Deep learning based imaging data completion for improved brain disease diagnosis. In: Golland P, Hata N, Barillot C, Hornegger J, Howe R, editors. MICCAI 2014, Part III. LNCS. Vol. 8675. Springer; Heidelberg: 2014. pp. 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nie D, Wang L, Gao Y, Sken D. Fully convolutional networks for multimodality isointense infant brain image segmentation. IEEE 13th International Symposium on Biomedical Imaging (ISBI); IEEE; 2016. pp. 1342–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran D, et al. Learning spatiotemporal features with 3D convolutional networks. 2014 arXiv preprint arXiv:1412.0767. [Google Scholar]
- 14.Zhuowen T, et al. Auto-context and its application to high-level vision tasks and 3D brain image segmentation. IEEE Trans Pattern Anal Mach Intell. 2010;32(10):1744–1757. doi: 10.1109/TPAMI.2009.186. [DOI] [PubMed] [Google Scholar]
- 15.Vercauteren T, et al. Diffeomorphic demons: efficient non-parametric image registration. NeuroImage. 2009;45(1):S61–S72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]