Abstract
Prostate cancer is the second leading cause of male cancer deaths due to disease progression to castration resistant prostate cancer (CRPC). Androgen receptor (AR) splice variants including AR-V7 function as constitutively active transcription factors in CRPC cells, thereby promoting resistance to AR-targeted therapies. To date, there are no AR variant specific treatments for CRPC. Here we report that the splicing of AR variants AR-V7 as well as AR-V1 and AR-V9 is regulated coordinately by a single polyadenylation signal in AR intron 3. Blocking this signal with morpholino technology or silencing of the polyadenylation factor CPSF1 caused a splice switch that inhibited expression of AR variants and blocked androgen-independent growth of CRPC cells. Our findings support the development of new therapies targeting the polyadenylation signal in AR intron 3 as a strategy to prevent expression of a broad array of AR variants in CRPC.
Keywords: prostate cancer, androgen receptor, alternative polyadenylation, antisense oligonucleotides, mRNA processing
Introduction
Patients with advanced prostate cancer are treated with androgen deprivation therapy (ADT), followed by second generation AR-targeted therapies abiraterone and enzalutamide when the disease progresses to castration resistant prostate cancer (CRPC). Persistent AR signaling throughout CRPC progression underlies resistance. One mechanism of persistent AR signaling is synthesis of constitutively active AR variants (AR-Vs) lacking the ligand binding domain (1). This is supported by data showing that patients harboring AR-V7-positive circulating tumor cells display resistance to abiraterone and enzalutamide (2–4).
The best-characterized mechanism underlying AR-V expression is the emergence of structural rearrangements in the AR gene, which have been shown to occur frequently in CRPC (5). AR gene rearrangements underlie overexpression of diverse AR-Vs with structure and function similar to AR-V7 in patient tumors (5–8). Although AR-V7 overexpression has been linked to AR gene rearrangements in prostate cancer cell lines, AR-V7 levels in patient tumors appear to be independent of AR gene rearrangements (5).
In this study, we used genome engineering to develop models of AR-V7-positive CRPC that more accurately reflected AR-V7 expression levels observed in clinical CRPC. Interrogation of these models demonstrated that splicing of AR-V7 occurred coordinately with splicing of additional AR-Vs, and that all of these splicing events were regulated by a single polyadenylation signal residing in AR intron 3. Use of morpholino technology to interfere with this AR alternative polyadenylation mechanism provided proof-of-principle for inhibiting expression and function of AR-Vs in CRPC.
Materials and Methods
Cell culture
22Rv1 cells (CRL-2505) were obtained from ATCC (Manassas, VA, USA) in 2013 and cultured according to ATCC protocol in RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum and 1X penicillin/streptomycin. LNCaP95 cells were a gift from Jun Luo (Johns Hopkins University) received in 2012 and were cultured in phenol red free RMPI 1640 (Invitrogen) supplemented with 10% fetal bovine serum and 1X penicillin/streptomycin. VCaP cells were obtained from ATCC (CRL-2876) in 2011 and were cultured in DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum and 1X penicillin/streptomycin. ATCC authenticates human cell lines using short tandem repeat analysis. LNCaP95 cells were authenticated by multiplex ligation-dependent probe amplification as described (7) to validate genomic copy number of AR and control regions on X and Y chromosomes matched parental LNCaP cells. LNCaP95 were further authenticated by PCR of genomic DNA and Sanger sequencing to validate presence of the AR T878A mutation matching parental LNCaP cells. Authenticated cells were expanded and frozen in liquid nitrogen. Cells were kept in culture no longer than 3 months after resuscitation of these frozen stocks, with the exception of 22Rv1 AR genetic correction experiments as outlined below. Aliquots of supernatants from cells in culture were evaluated quarterly for mycoplasma contamination using a PCR-based method (9).
Genetic correction of AR in 22Rv1
Construction of tAR2 TALENs was carried out as previously described (6). Repeat variable dinucleotide (RVD) modules used to generate tAR2 plasmids are listed in Supplementary Table S1. To generate 22Rv1 duplication-corrected subclones, cells were electroporated with tAR2 vectors and a single strand annealing (SSA) reporter, pJET: mCherry-PGK-EF1a-GFPssa. This SSA reporter was constructed using conventional cloning into the pJET1.2 vector (Clontech, #K1231). The PGK-EF1a is a bidirectional promoter that expresses mCherry from the PGK promoter and a GFPssa construct from the EF1a promoter. The GFPssa consists of the first 445bp of GFP, a SalI-SacI cloning site for annealed TALEN cutsite oligos, and the last 605bp of GFP including a 330 bp overlap region. Cells with TALEN nuclease activity were identified by GFP positivity and sorted by flow cytometry into single wells of 96 well plates. Clones were screened by PCR and restriction enzyme digestion of the PCR products to detect disruption of an Afl III restriction enzyme site in the tAR-2 spacer sequence. Subclones lacking this Afl III site were further screened by multiplex ligation-dependent probe amplification (MLPA) as described (7). Genomic DNA was isolated using Nucleospin tissue kit (Catalog 740952, Machery-Nagel). Genomic DNA was amplified using Phusion-HF (M0530S New England Biolabs, Ipswich, MA) according to the manufacturer protocol and the following primers: AR int2 F1: 5′-CTCCCAACAAGTGATCAGTAGTCAGAAAATGG-3′, AR int2 R1: 5′-CAACAGGGTATCTTATTTTGCAAACCCTAAGTC-3.′ Products were cloned and sequenced as previously described (6). Genomic PCR products were column purified with Qiaquick PCR purification kit (28104 Qiagen, Valencia, CA) and digested with 10 units of Afl III (R0541S New England Biolabs, Ipswich, MA) for 2 hours. Restriction analysis was performed via agarose gel electrophoresis.
siRNA transfections
For siRNA transfections, cells in medium supplemented with 10% charcoal-stripped FBS were mixed with 200 pmol targeted or nontargeting control siRNAs (Dharmacon). siRNAs were obtained from Dharmacon: nontargeting negative control (D-001210-01), CPSF1 smartpool: M-020395-01-0005 CPSF1 individual siRNAs: D-020395-01-0002, D-020395-02-0002, D-020395-03-0002, D-020395-04-0002, CPSF2 smartpool (M-013404-00-0005), CPSF3 smartpool (M-006365-00-0005), CPSF4 smartpool (M-012292-02-0005), WDR33 smartpool (M-017101-02-0005), FIP1L1 smartpool (M-014670-00-0005), U2AF2 (L-012380-02), SRSF1 (L-018672-01), PTBP1 (L-003528-00). siRNA sequences designed to target AR CE3 were as follows: siCE3-01: 5′-GUAGUUGUGAGUAUCAUGA-3′, siCE3-02: SENSE: 5′-AGGACGGGCUGUAGAAGUAUU-3′, ANTISENSE: 5′-UACUUCUACAGCCCGUCCUUU-3′, siCE3-03: SENSE: 5′-CAAAAUGACCAGACCCUGAUU-3′, ANTISENSE: 5′-UCAGGGUCUGGUCAUUUUGUU-3′.
Cell/siRNA mixtures were electroporated in a 4-mm gap-width cuvette (BTX) with a BTX Square Wave Electroporator (22Rv1 and sublines: 350 V, 10 ms pulse, LNCaP95: 305 V, 10 ms pulse). Following a 15 minute recovery, cells were seeded in appropriate culture medium. VCaP cells were transfected with Lipofectamine 2000 according to manufacturer instructions.
Morpholino transfections
GeneTools’ design tool was used to design a 25-mer consisting of the sequence 5′-GTGTATTAATGGCTTTATTAAGGGA-3′ targeted to the CE3 PAS (CE3 pAM). As a control, a standard commercially available nontargeting control morpholino was used (GeneTools). Morpholinos were labeled with 3′ fluorescein to visualize transfection efficiency. For morpholino transfections, cells were mixed with 10 μM CE3 pAM or control morpholino, electroporated, and seeded in culture dishes as described above. For VCaP and LNCaP 95 studies, Vivo-morpholinos (GeneTools) functionalized for cellular uptake were designed as 25-mer sequences corresponding to the same sequences for control and CE3 pAM and added at a final concentration of 1 μM directly to medium supplemented with 5% FBS and 1X penicillin/streptomycin.
Western blots
Cells were lysed in 1X Laemmli buffer (65 mmol/L Tris-HCl (pH 7.0), 2% (w/v) SDS, 5% β-mercaptoethanol, 10% (v/v) glycerol, and 0.5% (w/v) bromophenol blue). Equal volumes of lysates were loaded onto 4–20% or 7.5% PAGE gels (TGX, Biorad) and electrophoresed in 1X SDS PAGE running buffer, followed by transfer to PVDF membrane (Immobilon-P, Millipore). Blots were incubated with antibodies specific to CPSF1 (Bethyl, A301-580, 1:1000), AR (Santa Cruz Santa Cruz Biotechnology N-20, 1:2000), extracellular signal-regulated kinase-2 (ERK-2; Santa Cruz Biotechnology D-2, 1:4000), and AR-V7 (Precision Antibodies, 1:1000). HRP-conjugated goat-anti-mouse (#sc 2005, lot#D1515), and goat-anti-rabbit (#sc 2004, lot#B1315) secondary antibodies were obtained from Santa Cruz and were used at 1:10000 dilution. Blots were incubated in enhanced chemiluminescence substrate (WesternBright, Advansta #K12045-D50 or SuperSignal West Pico, Thermo #1859674) and exposed to X-ray film for chemiluminescence detection.
RNA purification and reverse transcription
RNA was isolated from cells using the Reliaprep RNA Miniprep System (Promega) and purified according to kit manufacturer instructions, including a 30 minute room temperature DNase treatment. RNA was eluted in nuclease free water. RNA concentration and integrity were assessed by denaturing gel electrophoresis or by spectrophotometry with a NanoDrop 2000 (ThermoFisher). Reverse transcription was performed using the GoScript Reverse Transcription kit (Promega) with 0.5–5 μg input RNA and random hexamers according to manufacturer instructions.
Quantitative PCR
Quantitative PCR was performed using Quanta PerfeCTa master mix, 200 nM forward and reverse primers, and 2 μL input cDNA (serially diluted in nuclease free water 1:50 for 18S samples) in 20 μL reactions. Negative controls included no-template and no-RT reactions for each primer set. Cycling conditions were: i. 95°C for 3 minutes, ii. 95°C for 15s, iii. 60°C for 30s. Steps ii-iii were repeated for a total of 40 cycles. All PCR reactions were subjected to thermal melting to confirm that each reaction gave single peaks. All reactions were carried out in technical duplicate, with at least two biological replicates. Quantitative PCR was performed on a BioRad CFX Connect Real-Time PCR Detection System or a BioRad MyiQ Single Color Real-Time PCR Detection System. Cycle thresholds (Ct) were determined using the Biorad CFX manager software. Changes in mRNA expression were calculated by ΔΔCt=ΔCttarget - ΔCtcontrol. Relative changes in mRNA expression levels are represented graphically as fold changes, wherein relative mRNA fold change= 2-ΔΔCt. All statistical tests were performed in GraphPad. Primer sequences are described in the Supplementary Methods.
Mean mRNA fold change for each target gene is shown throughout the manuscript. Error bars for qPCR experiments indicate 95% confidence intervals. Mean mRNA fold changes relative to controls were subjected to unpaired t-tests. All statistical analyses were performed in GraphPad.
Cell growth assays
Electroporated 22Rv1 cells and sublines were seeded at 20,000 cells per well in 24 well dishes in RPMI 1640 supplemented with 10% charcoal-stripped FBS. Twenty-four hours after seeding, medium was exchanged with RPMI 1640 medium containing 10% charcoal stripped FBS supplemented with 1 nM dihydrotestosterone (DHT) or vehicle control (ethanol). Crystal violet staining assays were performed as described (10) and absorbance was measured at 560 nm. Significance was assessed by two tailed t-tests in GraphPad.
AR RNA sequencing
For AR RNA-seq, RNA isolated from 22Rv1 cells was subjected to reverse transcription using the Clontech Advantage RT kit using both oligo-dT and random hexamer primers as per the manufacturer’s recommendations. cDNA samples were submitted to the University of Minnesota Genomics Center for RNA-seq library preparation and hybrid capture with a custom AR-based SureSelect (Agilent) bait library (7) using the SureSelect QXT reagent kit (Agilent) as per manufacturer’s recommendations. Data were analyzed as outlined in the Supplemental Methods section.
AR DNA sequencing
Genomic DNA isolated from isogenic gene corrected sub-lines 22Rv1-undup1 and 22Rv1-undup3 was submitted to the University of Minnesota Genomics Center for DNA-seq library preparation and hybrid capture as described in (7). LUMPY and DELLY structural variant callers were employed as described in (5). Freebayes and Varscan were used to detect mutations in 22Rv1 sub-lines as described in (5).
Analysis of CPSF complex alterations in primary and metastatic prostate cancer
We queried publicly available datasets using cBioPortal (11) to ascertain whether the CPSF complex exhibited changes in mRNA abundance, copy number, and mutation status from both primary (12) and metastatic prostate cancer (13). Mutual exclusivity plots were generated in Excel from scores of mutual exclusivity or co-occurrence obtained from cBioPortal.
TCGA RNA-seq analysis
Raw RNA-seq data from the TCGA prostate cancer dataset was downloaded from the NCBI database of genotypes and phenotypes (dbGaP) yielding paired tumor/normal samples for 52 patient samples. Data were analyzed as described in the Supplementary Methods.
SU2C RNA-seq analysis
RNA-seq data from the AACR-PCF Stand-Up-To-Cancer study of metastatic CRPC biopsies was obtained from the NCBI dbGaP. RNA-seq Illumina reads were aligned to hg19. Data were analyzed as described in the Supplementary Methods.
Results and Discussion
AR gene correction in 22Rv1 cells
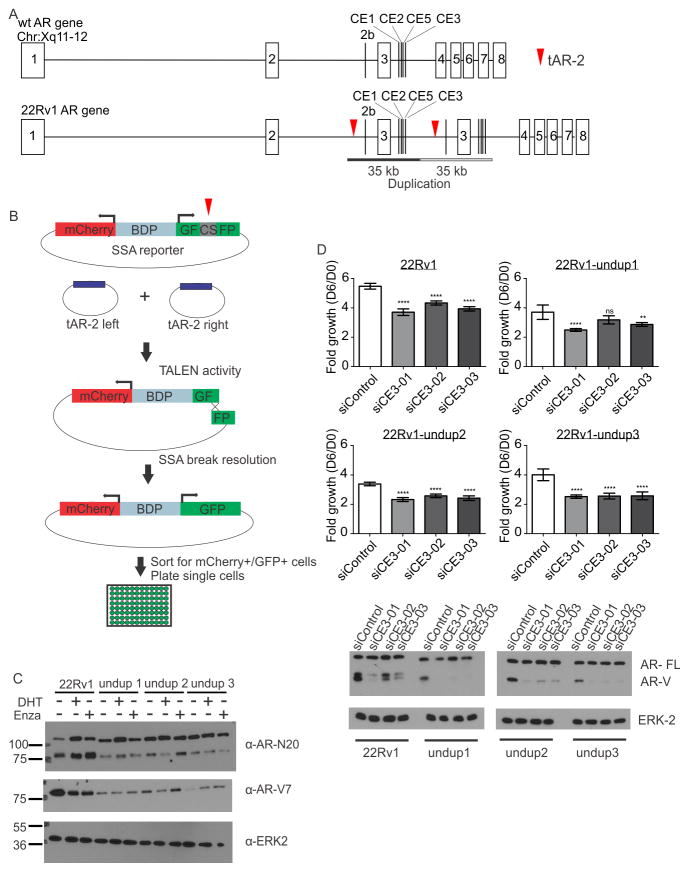
The CRPC 22Rv1 cell line harbors a 35 kb tandem duplication encompassing AR exon 3 and cryptic exons (CEs) that are spliced as the alternative 3′ terminal exons of AR-Vs including AR-V7 (8). Given the link between AR gene rearrangements and AR-V overexpression, we hypothesized that removal of this duplication would eliminate or dramatically reduce AR-V7 expression. To test this, we used transcription activator-like effector nucleases (TALENs), termed tAR-2, to cut DNA in two regions flanking the duplication (Fig. 1A, Supplementary Fig. S1A).
Figure 1.
A. TALEN engineering strategy. tAR-2 TALENs were designed to cut AR (red arrows) and delete the 22Rv1 duplication. B. An mCherry/split GFP reporter harboring a tAR-2 cut site (CS) was used to identify, sort, and plate transfected cells with tAR-2 activity C. Cells were cultured in steroid-deplete medium and treated with 1 nM dihydrotestosterone (DHT), 10 μM enzalutamide (Enza), or vehicle control as indicated. Blots were probed with antibodies specific for AR (N20), AR-V7, or ERK2 (loading control). D. Growth of cells transfected with indicated siRNAs was assessed by crystal violet staining. Bars represent mean fold growth on Day 6 relative to Day 0 (n = 12, error bars = 95% confidence intervals). **** p≤0.0001, ** p ≤ 0.01, ns p>0.05, Mann Whitney U tests. Bottom panels illustrate western blots of lysates from transfected cells, probed with antibodies as in C.
We used a bidirectional mCherry/GFP reporter plasmid to enrich cells displaying tAR-2 activity (Fig. 1B). mCherry+/GFP+ cells were screened by PCR (Supplementary Fig. S1A), and assessed for AR gene copy number using multiplex ligation-dependent probe amplification (Supplementary Fig. S1B). This yielded three 22Rv1 sub-lines, termed 22Rv1-undup1, -undup2, and -undup 3. PCR isolation of break fusion junctions confirmed that 22Rv1-undup1-3 arose from TALEN engineering, as they contained DNA inserted from the bidirectional reporter or tAR-2 plasmids (Supplementary Fig. S2). Paired-end sequencing of AR in 22Rv1-undup1 and 22Rv1-undup3 cells (Supplementary Table S2, Supplementary Data 1) ruled out alternative rearrangements while confirming the parental 22Rv1 H874Y mutation (Supplementary Table S3). We concluded that 22Rv1-undup1-3 were bona fide AR gene-corrected sub-lines of 22Rv1.
Functional AR-V7 expression in AR gene-corrected 22Rv1 cells
Parental 22Rv1 and 22Rv1-undup1-3 sub-lines expressed full length AR (AR-FL) at similar levels under castrate or DHT-stimulated conditions, and after treatment with the AR antagonist enzalutamide (Fig. 1C). Conversely, 22Rv1-undup1-3 sub-lines displayed reduced AR-V7 expression (Fig. 1C). AR-FL electrophoretic mobility in 22Rv1-undup1-3 lysates was faster, consistent with AR-FL no longer harboring an extra zinc-finger in the DNA binding domain due to the 22Rv1 tandem duplication (8). Additionally, AR-FL in 22Rv1-undup3 cells displayed strong nuclear localization upon stimulation with 1 nM of the synthetic androgen mibolerone, indicating normal nuclear translocation (Supplementary Fig. S3). These data confirmed that AR gene rearrangements were driving AR-V overexpression in 22Rv1 cells. However, AR-V7 expression was not abolished, suggesting a rearrangement-independent mechanism of AR-V7 synthesis.
Knockdown of AR-V7 in 22Rv1-undup1-3 sublines using siRNAs targeted to CE3 reduced expression of AR transcriptional targets FKBP5, PSA, TMPRSS2, and hK2 under castrate conditions (Supplementary Figure S4A–S4E). Additionally, knockdown of AR-V7 inhibited androgen-independent growth (Fig. 1D). Collectively, these findings indicated that AR-V7 in 22Rv1-undup1-3 sub-lines was expressed at much lower levels than parental 22Rv1 cells, but this expression remained sufficient for AR-V7 to transcriptionally activate AR target genes and support growth in an the absence of androgens.
Regulation of AR splicing by alternative polyadenylation
To understand the mechanisms regulating AR-V7 expression in 22Rv1-undup-1-3 sub-lines, we first assessed CE3 splice acceptor site recognition. A previous study found that AR exon 3/CE3 splicing in an AR minigene reporter was driven by splicing factors U2AF2, SRSF1, and PTBP1 (14). We knocked down these factors and found that they did not specifically inhibit expression of endogenous AR-V7 in 22Rv1 or 22Rv1-undup1-3 sub-lines (Supplementary Fig. S5A–S5D).
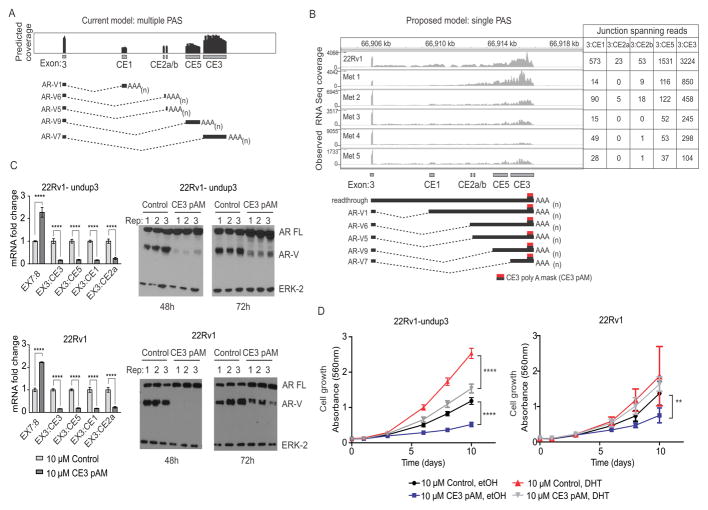
Having ruled out splice acceptor site recognition as a main mechanism, we next addressed alternative polyadenylation. The current model for splicing of AR-V7 and other AR-Vs is that each cryptic exon in AR intron 3 harbors discrete splice acceptor and polyadenylation sites (PAS) (15). This model would predict RNA-seq coverage beginning at each splice acceptor, and terminating at each PAS (Fig. 2A). However, when we analyzed RNA-seq read coverage from 22Rv1 cells and CRPC metastases, we observed patterns that were inconsistent with this model (Fig 2B). For example, we noted signal attenuation at the 3′ end of CE3 consistent with mRNA cleavage and polyadenylation, but lack of signal initiation at the CE3 splice acceptor site, despite abundant exon 3/CE3 splice junctions (Fig. 2B). Similarly, CE1, CE2, and CE5 also lacked distinct 5′ and 3′ termini, despite abundant exon 3/CE1, 3/CE2, and 3/CE5 splice junctions (Fig. 2B).
Figure 2.
Blockade of the AR CE3 PAS inhibits AR-V expression. A. Schematic of predicted AR RNA-seq coverage based on use of annotated splice junctions and PASs in AR cryptic exons (CEs). B. Observed AR RNA-seq coverage from 22Rv1 and metastatic biopsies (top) consistent with a new model wherein multiple AR-Vs arise from use of multiple splice acceptor sites that terminate at a single PAS (bottom). The location at which CE3 pAM morpholino binds to induce steric blockade is shown. C. Left panels: 22Rv1-undup3 (top) and 22Rv1 (bottom) cells were transfected with 10 μM control or CE3-pAM. Mean mRNA fold change of AR and AR-Vs relative to actin control is shown (n=6, error bars=95% confidence intervals). **** p≤0.0001, unpaired t-tests. Right panels: Western blot of lysates from transfected cells, probed with antibodies to AR NTD or ERK-2 (loading control). D. Crystal violet staining to assess growth of 22Rv1-undup3 (left) and 22Rv1 (right) cells transfected as in panel C maintained in androgen-deplete medium (CSS) in the presence of 1 nM DHT or vehicle control (ethanol). Data points represent means (n=6, 22Rv1-undup1, n=4, 22Rv1; error bars = 95% confidence intervals). **** p≤ 0.0001, ** p ≤ 0.01, ns p>0.05 p-values in a two-tailed t-test.
We developed an alternative model, wherein AR-V mRNAs would arise from the use of annotated splice acceptor sites within AR intron 3, but would all terminate at a single PAS that had been annotated previously for CE3 (Fig. 2B) (15). To test this model, we designed a morpholino termed CE3 poly A mask (pAM) to impose a steric blockade by hybridization to the CE3 PAS (Fig. 2B). Consistent with a single PAS model, 22Rv1 and 22Rv1-undup3 cells transfected with CE3 pAM displayed a reduction in mRNA and protein expression of AR-V7 as well as AR-V9 (EX3:CE5), AR-V1 (EX3:CE1), and AR-V6 (EX3:CE2a), accompanied by an increase in expression of AR-FL (Fig. 2C). We also observed a specific reduction of AR-V mRNAs in VCaP and LN95 cells treated with a vivo-morpholino targeting the same sequence as CE3 pAM, which indicates this effect was not restricted to the 22Rv1 genetic background (Supplementary Fig. S6A–S6B). Another prediction arising from this model is that this entire cohort of AR-V mRNAs would be susceptible to knockdown with siRNA targeting AR exon CE3, which has been assumed to target AR-V7 specifically (7,10,15,16). Indeed, transfection of 22Rv1 and VCaP cells with siRNAs targeted to AR exon CE3 resulted in knockdown of AR-V1, AR-V6, AR-V9, and AR-V7 mRNA expression (Supplementary Fig. S7A–S7B).
Since CE3-pAM reduced expression of multiple AR-Vs, we hypothesized that CE3-pAM would inhibit growth of AR-V-positive prostate cancer cells under castrate conditions. Indeed, we observed a reduction in growth of 22Rv1-undup3 and parental 22Rv1 cells transfected with CE3-pAM under androgen-deplete conditions (Fig. 2D). CE3-pAM also inhibited growth of 22Rv1-undup3 cells under androgen-replete conditions (Fig. 2D).
mRNA 3′ end processing factors are enriched in prostate cancer
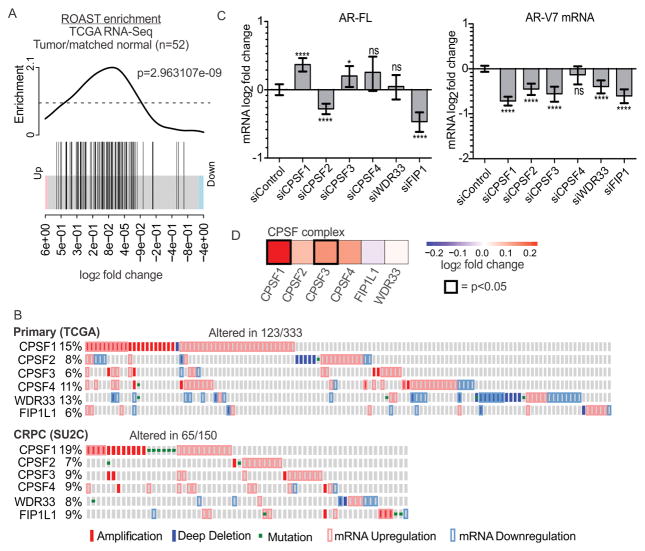
Because these data pointed to regulation of AR-V expression by alternative polyadenylation, we interrogated 52 pairs of normal and matched prostate cancer tissues for altered expression of a set of 91 genes that encode factors involved in mRNA 3′ end processing (12,17,18). Using rotation gene set tests (ROAST), we found that these factors were upregulated in tumor relative to matched normal tissues (Fig. 3A) (18). We queried public datasets for alterations in the 6 core genes encoding the cleavage and polyadenylation specificity factor (CPSF) complex: CPSF1, CPSF2, CPSF3, CPSF4, WDR33, and FIP1. Upon recruitment of this complex to pre-mRNAs, CPSF4 and WDR33 contact the PAS (19,20) and CPSF3 cleaves mRNA downstream of the PAS, triggering polyadenylation (21). CPSF complex factors displayed alterations at the level of genomic copy number, gene mutation, and mRNA abundance, with CPSF1 displaying frequent alterations (11–13) (Fig. 3B). Additionally, we noted trends of co-occurring alterations in CPSF complex factors in localized prostate tumors (Figs 3B and Supplementary Fig. S8).
Figure 3.
CPSF complex alterations in prostate cancer. A. Enrichment analysis of a set of 91 genes involved in mRNA 3′ end processing in 52 tumor/matched normal TCGA RNA-seq samples. B. Oncoprints depicting CPSF core complex gene alterations in primary and metastatic prostate cancer (12,13). C. AR and AR-V mRNA levels in 22Rv1-undup3 cells transfected with siRNAs targeting CPSF core complex genes, relative to 18S internal control. Bars represent means (n=6, error bars = 95% confidence intervals. **** p≤0.0001, ** p ≤ 0.01, ns p>0.05, unpaired t-test. D. Heatmap of log2 fold changes in mRNA expression of CPSF core complex factors in primary prostate cancer compared to normal tissue (52 matched samples).
To test the role of the CPSF complex in regulating AR splicing, we performed a siRNA-based screen, prioritizing factors that would cause an AR splicing switch when knocked-down, as opposed to general reductions in expression of both AR-V7 and AR-FL due to possible non-specific effects. Interestingly, depletion of CPSF1 and CPSF3 led to reduced expression of AR-V7 and increased expression of AR-FL mRNA in 22Rv1-undup3 cells (Fig. 3C and Supplementary Fig. S9). These effects were similar to the splice switch observed in cells treated with CE3-pAM (Fig. 2C).
Next, we analyzed publicly available RNA-seq data for evidence of alterations in CPSF1 or CPSF3 expression (12). We observed that CPSF1 and CPSF3 mRNA levels were upregulated in primary prostate cancer vs. matched normal prostate tissue (Fig. 3D). Since CPSF1 was frequently altered in prostate cancer tissues (Fig. 3B), and had the most robust effect on AR splice switching (Fig. 3C), we nominated CPSF1 for further analysis.
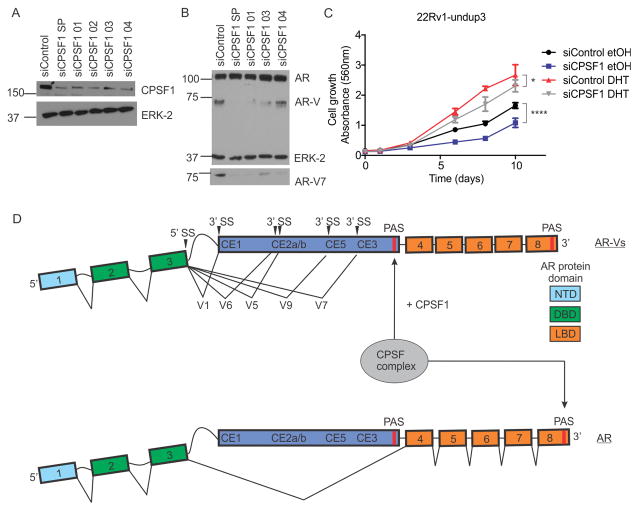
We knocked down CPSF1 in LNCaP-95 cells, which confirmed the increase in AR-FL and decrease in AR-V7 mRNA expression was not restricted to the 22Rv1 genetic background (Supplementary Fig. S10A). Next, we knocked down CPSF1 in 22Rv1-undup3 cells using four CPSF1 siRNAs delivered individually or as a pool (Fig. 4A). One of these reagents (siCPSF 02) was omitted from further study due to toxicity. Knock-down of CPSF1 reduced expression of AR-V7 protein and increased expression of AR-FL protein (Fig. 4B). Accordingly, knock-down of CPSF1 with three independent siRNAs resulted in reduced growth of 22Rv1-undup3 cells under androgen deplete conditions (Fig. 4C, blue). Under androgen-replete conditions, CPSF1 knockdown had a modest effect on 22Rv1-undup3 cell growth, which could be attributed to general effects on mRNA processing by CPSF1 knockdown (Fig. 4C, gray).
Figure 4.
CPSF1 regulates AR-V expression. A. Western blot with lysates from 22Rv1-undup3 transfected with four independent and pooled siRNAs. Blots were probed with antibodies to CPSF1 or ERK-2 (loading control). B. Western blot with lysates from 22Rv1-undup3 cells transfected with CPSFS1 siRNA as in panel A, probed with antibodies to AR, ERK-2 (loading control), or AR-V7. C. Growth of 22Rv1-undup3 cells transfected as with CPSF1 siRNA was assessed under androgen replete (charcoal stripped serum+1 nM DHT) or androgen deplete (charcoal-stripped serum+ethanol, vehicle control) conditions by crystal violet staining. Data points represent means (n = 6, error bars = 95% confidence intervals). **** p ≤ 0.0001,* p≤ 0.05, ns p>0.05, unpaired t-test. D. Model for AR splicing decisions regulated by CPSF1-dependent selection of a single PAS in AR intron 3.
Collectively, the data presented in this study re-annotates AR CE1, CE2, CE5, and CE3 as harboring independent splice acceptor sites but sharing a single PAS that was previously thought to be used exclusively for synthesis of AR-V7 (15). This PAS appears to be the main regulatory point for all AR-Vs utilizing cryptic exons in AR intron 3, as alternative PASs did not appear to become activated following morpholino blockade of the CE3 PAS.
The mechanism by which selection of this PAS is transmitted to upstream splice acceptors remains unknown, but our data nominates CPSF1 in this process (Fig. 4D). CPSF1 has no known RNA target sequence or RNA-binding domain (19,20). Previous studies have shown that CPSF1 interacts with pre-mRNAs over a broad region upstream of PAS, which may include a U-rich motif (22). Polyadenylation is controlled via multiple cis elements including the PAS as well as upstream UGUA and U-rich motifs, and downstream U- and GU-rich motifs (23). Sequence elements governing PAS selection in CE3 remain to be characterized. Our data argue against an absolute requirement of CPSF1 for global cleavage and polyadenylation, as knockdown of CPSF1 led to increased expression of full length AR mRNA and protein, and had modest effects on growth of CRPC cells under androgen-replete conditions. Rather, our data support a model whereby alterations in CPSF1 expression and/or activity promote selection of the intronic AR CE3 PAS, leading to recruitment of the CPSF complex to drive synthesis of multiple AR-V mRNAs, including AR-V7 (Fig. 4D).
This study provides proof-of-principle for targeting selection of the AR CE3 PAS by CPSF1 to prevent coordinate expression of AR-Vs including AR-V7 and overcome therapeutic resistance in prostate cancer. Furthermore, our data reveal frequent deregulation of the CPSF complex in prostate cancer, highlighting the potential for modulating aberrant polyadenylation to achieve therapeutic benefits beyond blockade of AR-Vs.
Supplementary Material
Acknowledgments
Financial Support: This study was funded by a grant from the NIH (R01CA174777) to S.M. Dehm, J.L. Van Etten was supported by T32 CA009138. This work was also supported by a Prostate Cancer Foundation (PCF) Young Investigator Award (to R.Y.) and a PCF Challenge Grant (to S.M.D.).
Footnotes
Disclosure of Potential Conflicts of Interest: S.M.D. has served as a paid consultant/advisor for Astellas/Medivation and Janssen Research & Development, LLC.
References
- 1.Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer. 2011;18(5):R183–96. doi: 10.1530/ERC-11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Nakazawa M, et al. Androgen Receptor Splice Variant 7 and Efficacy of Taxane Chemotherapy in Patients With Metastatic Castration-Resistant Prostate Cancer. JAMA Oncol. 2015;1(5):582–91. doi: 10.1001/jamaoncol.2015.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028–38. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher HI, Lu D, Schreiber NA, Louw J, Graf RP, Vargas HA, et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2016.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henzler C, Li Y, Yang R, McBride T, Ho Y, Sprenger C, et al. Truncation and constitutive activation of the androgen receptor by diverse genomic rearrangements in prostate cancer. Nature Communications. 2016;7:13668. doi: 10.1038/ncomms13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KA, et al. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci U S A. 2013;110(43):17492–7. doi: 10.1073/pnas.1308587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Hwang TH, Oseth LA, Hauge A, Vessella RL, Schmechel SC, et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene. 2012;31(45):4759–67. doi: 10.1038/onc.2011.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Alsagabi M, Fan D, Bova GS, Tewfik AH, Dehm SM. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res. 2011;71(6):2108–17. doi: 10.1158/0008-5472.CAN-10-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uphoff CC, Denkmann SA, Drexler HG. Treatment of mycoplasma contamination in cell cultures with Plasmocin. J Biomed Biotechnol. 2012;2012:267678. doi: 10.1155/2012/267678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73(2):483–9. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry Christopher D, et al. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163(4):1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33(24):3140–50. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69(6):2305–13. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Z, Chen S, Sowalsky AG, Voznesensky OS, Mostaghel EA, Nelson PS, et al. Rapid induction of androgen receptor splice variants by androgen deprivation in prostate cancer. Clin Cancer Res. 2014;20(6):1590–600. doi: 10.1158/1078-0432.CCR-13-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33(3):365–76. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu D, Lim E, Vaillant F, Asselin-Labat ML, Visvader JE, Smyth GK. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26(17):2176–82. doi: 10.1093/bioinformatics/btq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR, 3rd, et al. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev. 2014;28(21):2370–80. doi: 10.1101/gad.250993.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schonemann L, Kuhn U, Martin G, Schafer P, Gruber AR, Keller W, et al. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev. 2014;28(21):2381–93. doi: 10.1101/gad.250985.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10(4):565–73. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murthy KG, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 1995;9(21):2672–83. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 23.Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.