Abstract
An estimated 71 million people worldwide are infected with hepatitis C virus (HCV). The lack of small animal models has impeded studies of antiviral immune mechanisms. Here we show that an HCV-related hepacivirus discovered in Norway rats can establish high titer hepatotropic infections in laboratory mice with immunological features resembling those seen in human viral hepatitis. While immune-compromised mice developed persistent infection, immune-competent mice cleared the virus within 3–5 weeks. Acute clearance was T cell dependent and associated with liver injury. Transient depletion of CD4+ T cells prior to infection resulted in chronic infection, characterized by high levels of intrahepatic regulatory T cells and expression of inhibitory molecules on intrahepatic CD8+ T cells. Natural killer cells controlled early infection but were not essential for viral clearance. This model may provide mechanistic insights into hepatic antiviral immunity, a prerequisite for the development of HCV vaccines.
Main text
Hepatitis C virus (HCV), a major cause of human liver cirrhosis and cancer, is narrowly restricted to the human liver (1). Currently, there are no immune-competent small animal models for HCV and this limits the study of host-virus interactions and the development of vaccine strategies (2). A prophylactic and protective vaccine against HCV, which will likely be needed for global HCV eradication, does not exist (3).
Several HCV-related hepaciviruses have been discovered in horses, bats, and wild rodents (4). In 2014, a hepacivirus was identified in Norway rats from New York City (5) and named Norway rat hepacivirus (NrHV) or rodent hepacivirus-nr-1 (RHV-nr-1). Similar to HCV in humans, NrHV can establish a hepatotropic infection in rats (5). Rats represent a natural context in which to study NrHV. However, given numerous genetic variants and tools available for mice that permit deep mechanistic studies we aimed to develop a mouse model of NrHV infection speculating that NrHV might infect laboratory mice, given their close phylogenetic relationship to rats.
We first explored whether NrHV could establish infection in the immune-compromised mouse strains NRG (NOD-Rag1−/− IL2Rγ−/−), A129 (IFNRαβ−/−) and AG129 (IFNRαβ−/− IFNRγ−/−) that lack adaptive immunity, type I, and type I/II interferon signaling, respectively. We infected 4-week-old mice intravenously with 104 genome equivalents (GE) of NrHV derived from the serum of an infected laboratory rat. NrHV established a high titer (106–108 GE/ml serum) chronic infection in these mice (Fig. 1A). Mice lacking MAVS (mitochondrial antiviral signaling protein) cleared the virus within 3 weeks post infection (p.i.) (Fig. 1A).
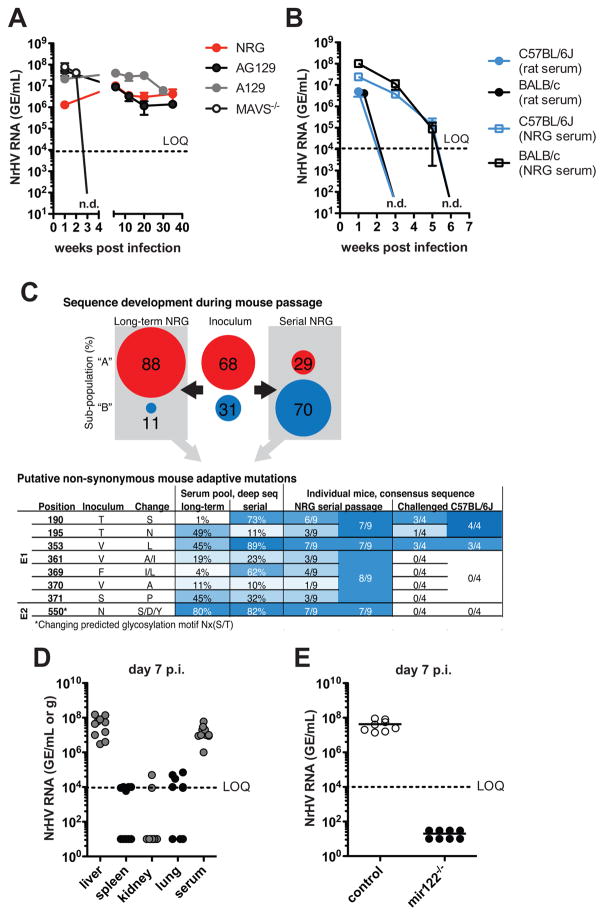
Fig. 1. NrHV establishes hepatotropic infection in common laboratory mice.
4-week-old mice were infected i.v. with 104 GE of NrHV. Viremia (NrHV RNA GE/ml) was analyzed by RT-qPCR. LOQ: limit of quantification: n.d.: not detectable. (A) Viremia in NRG, A129, AG129 and MAVS−/− mice infected with rat-serum-derived NrHV. (B) Viremia in C57BL/6J and BALB/c mice infected with either rat- or NRG-serum-derived virus. (C) NrHV sequence development during mouse passage (see fig. S1). Percentages of two identified sub-populations in the rat inoculum and long-term and serial pools are shown. Below, putative mouse adaptive positions (all variants <1% in the inoculum). Fraction of variants, determined by deep sequencing of the pools or consensus ORF sequences from the 9 mice of the last serial passage and 4 C57BL/6J mice challenged with the serial pool (week 1 p.i.) are shown. (D) Viral loads in tissue and serum of C57BL/6J at day 7 p.i. (E) Viremia in miR-122−/− mice and controls at day 7 p.i.. Panels (A–B) and (D–E) show representative data from 2–5 independent experiments with 4–5 mice per group (mean ±SEM).
Intravenous infection of the immune-competent mouse strains C57BL/6J and BALB/c with 104 GE resulted in a high titer (106–108 GE/ml serum) acute resolving infection (Fig. 1B). NrHV derived from rat serum was cleared significantly faster than NrHV passaged one time through NRG mice (Fig. 1B), indicating that NrHV can adapt to the mouse host. To test the extent of NrHV adaptation in NRG mice we performed either 12-week long-term adaptation in 1 mouse or serial passage adaptation through 5 mice (4-week infection of each mouse) (fig. S1A). We then challenged naïve NRG and C57BL/6J mice with 9x104 GE of either the pooled adapted (long-term pool or serial pool) or the parental virus. Adapted viruses showed 0.5–1 log higher viral titers at week 1 p.i. and persisted longer in C57BL/6J mice than did the parental virus (fig. S1B), suggesting increased viral fitness in the mouse host.
Comparing the consensus NrHV genome open reading frame (ORF) sequences of the rat inoculum with the adapted viruses revealed changes in 2 and 59 nucleotide positions in the long-term and serial passage pool, respectively (fig. S1C). Phylogenetic analysis of full-ORF clones revealed the presence of two sub-populations in the inoculum; the minor one was selected during the serial passage (Fig. 1C and fig. S1D). Single coding mutations in viral envelope proteins E1 and E2 occurred in both pools and in individual NRG mice from passage 5 of the serial adaptation (Fig. 1C and fig. S1E and tables S1–2). The E1 mutation V353L, combined with either T190S (serial pool) or T195N (long term pool) represent putative mouse adaptive mutations, as they were maintained in challenged C57BL/6J mice. Mutations at amino acid position 550, combined with at least one mutation in the cluster 361/369/370/371, were selected in NRG mice, but were immediately lost in C57BL/6J mice. Changing position 550 would disrupt a predicted Nx(S/T) glycosylation site possibly de-shielding neutralization epitopes as observed for HCV (6). For subsequent experiments we used serial pool virus as our source of NrHV.
Our results indicate that NrHV is both highly infectious and hepatotropic in mice. Even a low dose infection with 10 GE resulted in high titer viremia. The dose did not influence the outcome of infection, as mice infected with 104,103,102 and 10 GE cleared the virus with similar kinetics. In contrast, age influenced clearance: 4-week-old mice typically cleared the virus by week 5 p.i., whereas 2–6 month-old mice cleared the virus by week 3 p.i. (fig. S2, A to C). We consistently detected high viral titers in liver tissue (106–108 NrHV GE/gram of tissue) but not in spleen, kidney and lung (Fig. 1D). NrHV replication was dependent on miR-122, a liver-specific microRNA required for HCV replication (7, 8) (Fig. 1E).
Like HCV in humans, NrHV infection was not associated with signs of acute disease or mortality. Chronically infected NRG and AG129 mice showed minimal to mild liver inflammation at week 35 p.i. (fig. S3).
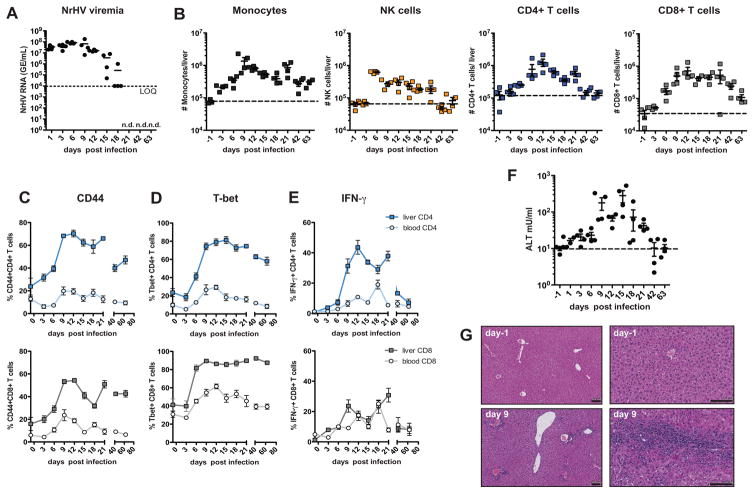
To identify the cellular mediators of NrHV clearance, we characterized the immune response during acute resolving NrHV infection in 8-week-old C57BL/6J mice. High titer viremia was detectable as early as 24 h p.i.; titers started to decline at day 15 p.i. and were undetectable at day 21 p.i. (Fig. 2A). Early acute infection (day 3–9 p.i.) was associated with an expansion of intrahepatic Ly6C+monocytes and NKp46+NK cells (Fig. 2B). Starting at day 9 p.i. we observed a substantial increase in proliferating (Ki67+) intrahepatic CD4+ and CD8+ T cells. These cells were characterized by a CD44+ effector phenotype with an antiviral type 1 differentiation signature (9) as indicated by high T-bet expression and IFN-γ production (Fig. 2, B to E and fig. S4, A and B). CD8+ T cells also showed a significant up-regulation of granzyme B (fig. S4C). The T cell response was predominant in the liver and less pronounced in peripheral blood and spleen (Fig. 2 and fig. S4). High levels of intrahepatic effector T cells coincided with a decline in viremia starting at day 15 p.i. and were associated with elevated alanine transaminase (ALT) levels (Fig. 2F), indicating T cell mediated liver injury. Hepatic leukocyte infiltration was confirmed by histology (Fig. 2G). We detected NrHV-specific IFN-γ production by CD4+ and CD8+ T cells in response to peptide pools covering the NrHV proteins NS3 and NS4 at multiple time points p.i. with a peak response by intrahepatic CD8+ T cells against the NS4 pool at day 21 pi. (fig. S5). These results are consistent with a strong and broadly directed virus-specific T cell response during acute resolving NrHV infection.
Fig. 2. NrHV clearance in immune-competent mice is associated with a strong intrahepatic antiviral immune response.
8-week-old C57BL/6J mice were infected i.v. with 104 GE of NrHV. (A) Viremia during acute resolving infection. (B) Flow cytometric analysis of total numbers of hepatic Ly6C+monocytes, NKp46+NK cells, CD3+CD4+ T cells and CD3+CD8+ T cells during acute infection. Dotted lines indicate baseline levels. (C–E) Flow cytometric analysis of peripheral and hepatic T cells during acute NrHV infection. Frequencies of CD44+ effector cells (C), T-bet+ cells (D) and IFN-γ producing cells (PMA/Ionomycin stimulation) (E) within the CD4+ (upper panels) and CD8+ (lower panels) T cell subsets are shown. (F) ALT levels in serum of mice. Dotted line indicates baseline level. (G) Representative H&E histology at day −1 and day 9 p.i. Scale bars, 100μm. Representative data from 2 independent experiments with 4 mice per group (mean ±SEM) are shown.
T cells play an important role in HCV and hepatitis B virus (HBV) infection in humans and in the LCMV (lymphocytic choriomeningitis virus) mouse model (10, 11). T cell depletion studies in HCV infected chimpanzees showed that these cells are critical for viral clearance during primary and secondary infection (12, 13).
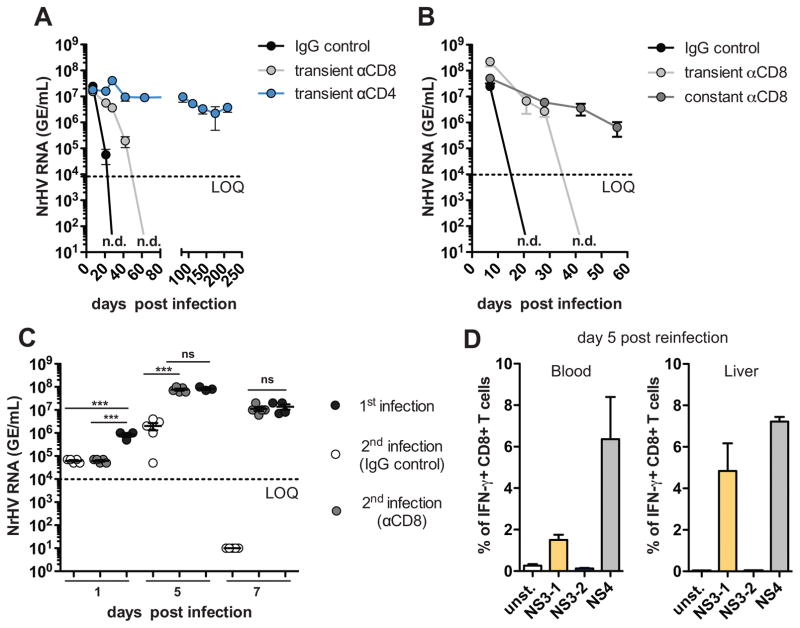
To analyze if T cells play an essential role in NrHV clearance we depleted them in mice (fig. S6, A and B). Transient CD4+ T cell depletion initiated prior to infection resulted in chronic infection (analyzed until day 210 p.i.) in C57BL/6J and BALB/c mice even after the CD4+ cell population recovered. Transient CD8+ T cell depletion led to delayed viral clearance as compared to controls (Fig. 3A, fig. S7, A and B). Mice that were constantly (every 10 days p.i.) depleted of CD8+ T cells failed to clear the virus (Fig. 3B).
Fig. 3. Clearance of NrHV infection is T cell dependent.
(A) 8-week-old C57BL/6J mice were transiently depleted of CD4+ or CD8+ T cells using antibodies (day 4 prior; day 7 and 28 p.i.) and infected with 104 GE of NrHV. Viremia was analyzed until day 210 p.i. (B) Mice were either transiently or constantly (day 4 prior; every 10 days p.i.) depleted of CD8+ T cells and infected with 104 GE NrHV. Viremia was analyzed until day 56 p.i. (C) Mice that cleared NrHV infection more than 4 months prior were reinfected with 104 GE of the same inoculum. One group of mice was depleted of CD8+ T cells 4 days prior to reinfection. Primary infection in age-matched mice served as a control. Viremia at days 1, 5 and 7 p.i. is shown. (D) NrHV-specific IFN-γ production of CD8+ T cells at day 5 post reinfection. Cells were stimulated ex vivo with NrHV NS3 or NS4 peptide pools. Representative data from 2–5 independent experiments with 4–5 mice per group (mean ±SEM) are shown. Statistics: unpaired students t-test: *** ≤ 0.0001; ns, not significant.
We next analyzed NrHV infection during recovery of CD4+ or CD8+ T cell after transient depletion (4 days prior to infection) in C57BL/6J mice (fig. S6C). In CD8+ T cell depleted mice, intrahepatic T cell recovery (fig. S8A) was associated with the emergence of CD44+IFN-γ+ effector CD8+ T cells, elevated ALT levels, and viral clearance. This was preceded by the induction of intrahepatic CD44+IFN-γ+ CD4+ T cells (fig. S8, C and D). In contrast, in CD4+ T cell depleted mice, intrahepatic cell recovery (fig. S8B) yielded minimal induction of CD44+IFN-γ+ CD4+ T cells. The presence of CD8+ effector cells induced in the absence of CD4 help was not associated with elevated ALT levels or viral clearance but resulted in chronicity (fig. S8, E and F).
These results indicate that CD4+ T cells are crucial for the initiation of a successful antiviral adaptive immune response, and that CD8+ T cells are main effectors in NrHV clearance.
In NrHV infection, memory CD8+ T cells also limited secondary infection (Fig. 3C). Mice that had cleared NrHV infection 4–7 months earlier became re-infected upon secondary challenge with the same inoculum but rapidly cleared the virus within 7 days (Fig. 3C). This was accompanied by a strong NrHV-specific CD8+ T cell recall response at day 5 p.i. (Fig. 3D). When we depleted CD8+ T cells 4 days prior to secondary infection, mice showed higher viral titer at day 5 p.i. compared to controls and could not clear the secondary infection by day 7 p.i. (Fig. 3C).
Neutralizing antibodies (ntAbs) might also play a role in limiting secondary infection. We observed significantly lower viral titers 24h post reinfection as compared to primary infection (Fig. 3C). During primary infection there was an increase of hepatic B cells and elevated frequencies of splenic and hepatic follicular T helper cells (fig. S9, A and B). Anti-NrHV NS3 antibodies were detected starting at day 21 p.i. (fig. S9C). Pre-incubation of 104, 103 or 102 GE NrHV with serum from mice that previously cleared infection mostly prevented infection of naïve mice with 102 GE but not with 103 or 104 GE. (fig. S9D). This suggests that ntAbs are produced but at too low frequency to completely prevent NrHV reinfection. This is similar to HCV infection, where previously infected humans and chimpanzees can be reinfected with the same HCV strain (14).
We also investigated the potential role of NK cells in viral clearance. During NrHV infection we observed increased numbers, activation (CD69 up-regulation) and IFN-γ production of hepatic NK cells starting at day 3 p.i. (Fig. 2B, fig. S10, A to C). This was attributed to an expansion of conventional CD49b+ NK cells while the number of liver-resident CD49a+ NK cells (15) remained stable leading to a substantial change of the hepatic CD49b+ to CD49a+ NK cell ratio over the course of infection (fig. S10, D and E). NK cell depletion resulted in significantly elevated viremia at day 3 p.i.. However the kinetics of viral clearance and the extent of liver injury were similar in NK cell depleted mice and controls (fig. S10, F and G). Thus, NK cells may contribute to the control of NrHV early in infection but they are not required for NrHV clearance.
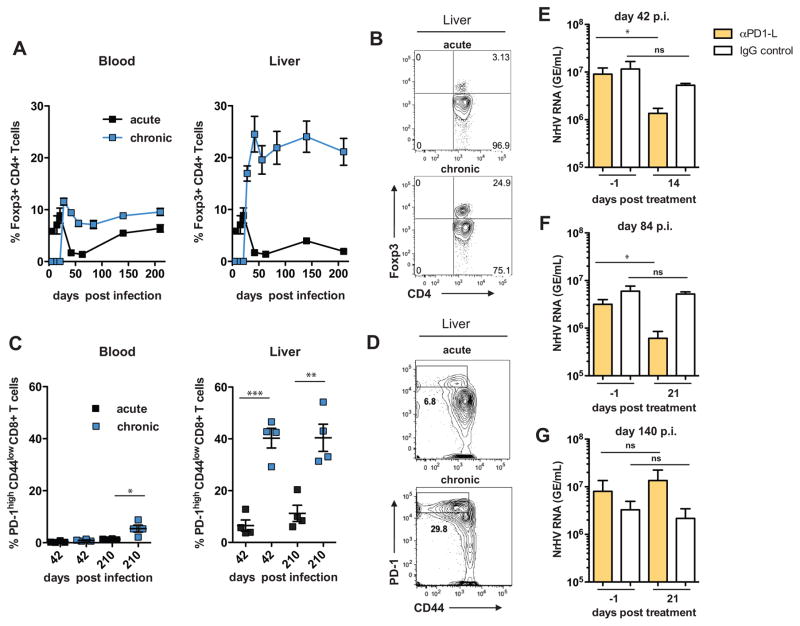
Chronic viral hepatitis in humans and chronic LCMV infection in mice are characterized by antigen-specific T cell dysfunction and exhaustion (10, 11). Mechanisms contributing to this phenomenon include the up-regulation of checkpoint inhibitors (e.g. PD-1 (programmed cell death 1)) or suppression by regulatory Foxp3+CD4+ T cells (Tregs) (10, 11). After transient CD4+ T cell depletion, NrHV established a long-term chronic infection in immune-competent mice (Fig. 3A) that was associated with mild liver inflammation (fig. S11). We found that chronic infection in contrast to acute clearance coincided with the emergence of intrahepatic Tregs that remained at high levels throughout infection (Fig. 4, A and B). Thus, suppression of antiviral immune responses by Tregs might play a role in the establishment of chronic NrHV infection.
Fig. 4. Chronic NrHV infection is associated with hepatic CD8+ T cell exhaustion.
Flow cytometric analysis of C57BL/6J mice that developed chronic NrHV infection after transient CD4+ T cell depletion. (A) Foxp3+CD4+ Treg frequencies within the CD4+ T cell subset in blood (left) and liver (right) during acute resolving and chronic NrHV infection. (B) Representative flow cytometry plots showing hepatic Tregs at day 210 p.i. (C) Frequencies of PD-1highCD44low CD8+ T cells at day 42 and 210 p.i. in blood (left) and liver (right). (D) Representative flow cytometry plots showing hepatic PD-1highCD44low CD8+ T cells at day 210 p.i. (E–G) Chronically NrHV infected C57BL/6J mice were treated with a PD1-L blocking antibody or appropriate IgG control starting at day 42 (E), day 84 (F) or day 140 (G) p.i.. Viremia was analyzed at day 1 prior to the start of treatment and compared to viremia at day 14 (E) or day 21 (F–G) post treatment. Representative or combined data from 2–5 independent experiments with 4–5 mice per group (mean ±SEM) are shown. Statistics: unpaired and paired students t-test: * ≤ 0.01, ** ≤ 0.001, *** ≤ 0.0001.
Chronic NrHV infection also displayed elevated frequencies of intrahepatic CD8+ T cells with an exhausted phenotype characterized by PD-1highCD44low surface expression (Fig. 4, C and D), co-expression of the inhibitory receptors 2B4 and Tim-3, and high expression levels of the transcription factor eomesodermin (fig. S12A) (16, 17). The frequencies of these cells were lower in acute resolving mice, suggesting that chronic NrHV infection may lead to T cell exhaustion (Fig. 4C).
Checkpoint inhibitor blockade (e.g., the inhibition of PD-1:PD-1 ligand (PD-1L) interactions), is a promising immunotherapy that can invigorate exhausted T cells (18). PD-1/PD-1L blockade showed mixed results when tested in HCV-infected chimpanzees and patients so its efficacy in the setting of chronic viral hepatitis is still unclear (19, 20). We thus tested whether PD-1L blockade could reduce viremia during chronic NrHV infection in mice (fig. S6D). Blockade at day 42 p.i. significantly reduced viremia (0.5–1 log) at day 14 post start of treatment while blockade at day 84 p.i. reduced viremia only at day 21 post start of treatment. At an even later time point (day 140 p.i.) no decrease in viremia was observed (Fig. 4, E to G, fig. S12, B and C). These results suggest that blockade of the PD-1:PD1-L pathway can reduce NrHV viral loads only during early chronic infection.
In summary, we have developed an immune-competent inbred mouse model of an HCV-related hepacivirus. Because NrHV can adapt to infect mice with diverse genetic backgrounds, this model can potentially help unravel mechanisms of hepacivirus host adaptation, immune evasion, and the development of liver disease. It can also be used to select for viral variants that can establish chronic infection in immune-competent mice. Given the similarities between NrHV infection in mice and HCV infection in humans, this model might prove valuable in the future for the development and testing of HCV vaccines.
Supplementary Material
Acknowledgments
We thank M. MacDonald, M. Saeed and W. Schneider for manuscript editing. This work was supported by NIH grants R01AI072613, R01CA057973, and R01AI131688-01, The Starr Foundation, the Greenberg Medical Research Institute and several generous donors (C.M.R.); NIH grant AI107631 and the Nationwide Children’s Hospital Research Institute (A.K); the Danish Council for Independent Research (6110-00595 and 6111-00314 (T.K.H.S); 4004-00598 (J.B.)); The Novo Nordisk Foundation (NNF15OC0017404 (T.K.H.S.) and NNF14OC001253 (J.B.)); The Lundbeck Foundation (R192-2015-1154 (T.K.H.S), R221-2016-1455 (J.B.)); and NIH grant R01A193244 (K.G). The HCV-related hepacivirus NrHV is available from the authors under a material transfer agreement. The ORF consensus sequence of the NrHV rat inoculum was deposited at GenBank (accession no: MF113386). The authors declare no conflict of interest.
Footnotes
References
- 1.Lindenbach BD, Rice CM. The ins and outs of hepatitis C virus entry and assembly. Nature reviews Microbiology. 2013;11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billerbeck E, de Jong Y, Dorner M, de la Fuente C, Ploss A. Animal models for hepatitis C. Curr Top Microbiol Immunol. 2013;369:49–86. doi: 10.1007/978-3-642-27340-7_3. [DOI] [PubMed] [Google Scholar]
- 3.Walker CM, Grakoui A. Hepatitis C virus: why do we need a vaccine to prevent a curable persistent infection? Curr Opin Immunol. 2015;35:137–143. doi: 10.1016/j.coi.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheel TK, Simmonds P, Kapoor A. Surveying the global virome: identification and characterization of HCV-related animal hepaciviruses. Antiviral research. 2015;115:83–93. doi: 10.1016/j.antiviral.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Firth C, et al. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. mBio. 2014;5:e01933–01914. doi: 10.1128/mBio.01933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cashman SB, Marsden BD, Dustin LB. The Humoral Immune Response to HCV: Understanding is Key to Vaccine Development. Front Immunol. 2014;5:550. doi: 10.3389/fimmu.2014.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 8.Hsu SH, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehermann B. Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nat Med. 2013;19:859–868. doi: 10.1038/nm.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zehn D, Wherry EJ. Immune Memory and Exhaustion: Clinically Relevant Lessons from the LCMV Model. Advances in experimental medicine and biology. 2015;850:137–152. doi: 10.1007/978-3-319-15774-0_10. [DOI] [PubMed] [Google Scholar]
- 12.Shoukry NH, et al. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. The Journal of experimental medicine. 2003;197:1645–1655. doi: 10.1084/jem.20030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grakoui A, et al. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302:659–662. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 14.Bukh J, et al. Previously infected chimpanzees are not consistently protected against reinfection or persistent infection after reexposure to the identical hepatitis C virus strain. J Virol. 2008;82:8183–8195. doi: 10.1128/JVI.00142-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng H, et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest. 2013;123:1444–1456. doi: 10.1172/JCI66381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackburn SD, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nature immunology. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paley MA, et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 2012;338:1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pauken KE, Wherry EJ. Overcoming T cell exhaustion in infection and cancer. Trends in immunology. 2015;36:265–276. doi: 10.1016/j.it.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardiner D, et al. A randomized, double-blind, placebo-controlled assessment of BMS-936558, a fully human monoclonal antibody to programmed death-1 (PD-1), in patients with chronic hepatitis C virus infection. PLoS One. 2013;8:e63818. doi: 10.1371/journal.pone.0063818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller MJ, et al. Immunotherapy of chronic hepatitis C virus infection with antibodies against programmed cell death-1 (PD-1) Proc Natl Acad Sci U S A. 2013;110:15001–15006. doi: 10.1073/pnas.1312772110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fahnoe U, et al. Creation of Functional Viruses from Non-Functional cDNA Clones Obtained from an RNA Virus Population by the Use of Ancestral Reconstruction. PLoS One. 2015;10:e0140912. doi: 10.1371/journal.pone.0140912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verbist B, et al. ViVaMBC: estimating viral sequence variation in complex populations from illumina deep-sequencing data using model-based clustering. BMC bioinformatics. 2015;16:59. doi: 10.1186/s12859-015-0458-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.