Abstract
DNA polymerases synthesize new DNA during DNA replication and repair, and their ability to do so faithfully is essential to maintaining genomic integrity. DNA polymerase beta (Pol β) functions in Base Excision Repair (BER) to fill in single-nucleotide gaps, and variants of Pol β have been associated with cancer. Specifically, the E288K Pol β variant has been found in colon tumors and has been shown to display sequence-specific mutator activity. In order to probe the mechanism that may underlie E288K’s loss of fidelity, a fluorescence resonance energy transfer (FRET) system was employed which utilizes a fluorophore on the fingers domain of Pol β and a quencher on the DNA substrate. Our results show that E288K utilizes an overall mechanism similar to wild type (WT) Pol β when incorporating correct dNTP. However, when inserting the correct dNTP, E288K exhibits a faster rate of fingers closing combined with a slower rate of nucleotide release compared to WT Pol β. We also detect enzyme closure upon mixing with the incorrect dNTP for E288K but not WT Pol β. Taken together, our results suggest that E288K Pol β incorporates all dNTPs more readily than WT due to an inherent defect that results in rapid isomerization of dNTPs within its active site. Structural modeling implies that this inherent defect is due to interaction of E288K with DNA, resulting in a stable closed enzyme structure.
Graphical abstract
INTRODUCTION
DNA is under constant assault from both endogenous and exogenous sources of damage. Cells handle a diverse array of lesions by maintaining many DNA repair pathways, each targeting specific types of damage. One such pathway is BER, which addresses approximately 20,000 lesions per cell per day and is a pathway that is conserved from bacteria to humans1, 2. In short patch BER, the lesion is first recognized by a DNA glycosylase, which removes the damaged base leaving an abasic site3. If the DNA glycosylase is monofunctional, AP endonuclease 1 (APE 1) cleaves the backbone of the helix on the 5′ side of the abasic site, generating a single-nucleotide gap that has a 3′OH and a 5′ deoxyribose phosphate (dRP)3. Pol β removes the dRP group and then fills in the single nucleotide gap3. If a bifunctional glycosylase removes the damaged base, end remodeling takes place that is catalyzed by enzymes including APE 1 and polynucleotide kinase (PNK) to generate a 3′OH and 5′ phosphate. After Pol β fills the gap, the nick is sealed by Ligase III/XRCC13. In the minor long patch BER pathway, Pol β synthesizes DNA beyond the single base pair gap, displacing the downstream strand and creating a flap that is then cleaved by FEN14.
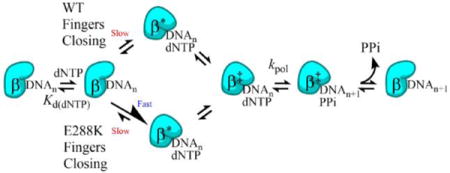
Pol β is a 39 kDa protein that contains 4 domains—a thumb domain with a helix-hairpin-helix motif that binds DNA; a fingers domain which binds incoming dNTP; a palm domain containing the active site; and a 8 kDa domain that has lyase activity (Figure 1)5. Given Pol β’s role as a repair polymerase, its mechanism and fidelity are of particular interest and importance because if Pol β cannot correctly fill in the DNA gap, genomic integrity can be compromised5. The first step in Pol β’s mechanism of nucleotide incorporation (Scheme 1) is binding DNA to create the binary complex6. Then the binary complex binds to the incoming dNTP, forming the ternary complex6. Upon correct dNTP binding, the fingers move from an open to a closed conformation7. This conformational change consists of the fingers rotating 30° and moving approximately 12Å5. While it is widely accepted that the WT Pol β-DNA-dNTP ternary complex closes upon binding correct dNTP, the nature of the ternary complex with incorrect dNTP remains somewhat controversial8–10. Observation in “real time” of crystals of Pol β bound to the incorrect dNTP suggest that catalysis occurs from a closed conformation in which the O3′ of the primer is poorly positioned as shown by time-resolved crystallography. The high B factors associated with these structures indicate elevated dynamics that lead to incorporation of the incorrect dNTP8. Crystal structures of Pol β with preformed mismatches also show evidence for strain in the primer terminus11. Combined structural and modeling studies by a different group suggest that incorporation of specific mismatches may occur from an open ternary complex12. NMR characterization of matched and mismatched complexes demonstrates significantly increased structural flexibility for a mismatched primer terminus, suggesting that misincorporation by Pol β may occur from an open ternary complex9.
Figure 1. The human Pol β variant E288K and the AEDANS-Dabcyl FRET system (Protein Data Bank codes 4KLE and 3ISB).
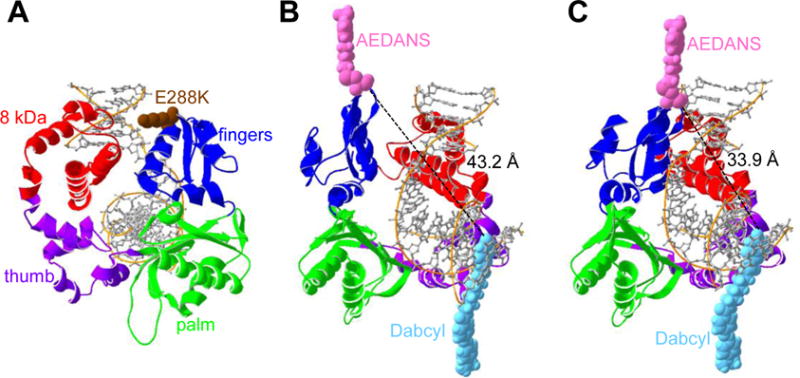
(A) The four domains of Pol β (4KLE): fingers domain (blue), palm domain (green), the thumb domain (purple), and the 8kDa domain (red). Residue 288 (brown), located at the tip of the fingers domain at the end of helix N, has been mutated to lysine to show the E288K variant. (B and C) Pol β used in this FRET system includes an AEDANS label on residue V303C (pink) and a Dabcyl quencher (light blue) on the DNA substrate. (B) Pol β in the open conformation (3ISB), in which the AEDANS and Dabcyl have a calculated distance of 43.2 Å, which is greater than the Förster radius of 40 Å. (C) In the closed conformation (4KLE), the AEDANS moves closer (33.9 Å) to the Dabcyl and results in a lower signal. Distances are determined using Swiss-Pdb Viewer, Swiss Institute of Bioinformatics.
Scheme 1. Pol β’s mechanism of nucleotide incorporation.
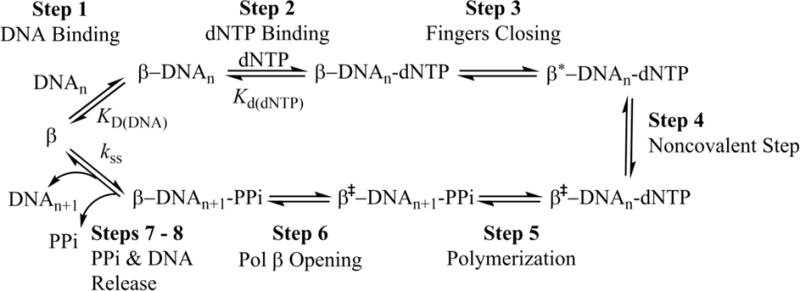
Step 1: Pol β first binds single-nucleotide gapped DNA (DNAn) to form the binary complex. Step 2: The binary complex binds dNTP. Step 3: The fingers domain moves to a closed conformation (β′) Step 4: A non-covalent step occurs. Step 5: The nucleotidyl transfer reaction is carried out to add the dNTP to the single-nucleotide gapped DNA, forming nicked DNA (DNAn+1) and pyrophosphate (PPi). Step 6: Pol β opens with PPi still bound. Steps 7 – 8: PPi is released followed by DNA product release (Scheme adapted from Towle-Weicksel et. al.10).
After binding to the correct dNTP and fingers closure, a non-covalent step occurs, followed by chemistry10. Next the fingers domain reopens and either during or after this movement, pyrophosphate (PPi) is released, followed by DNA product dissociation8, 13. Importantly, recent postcatalytic structures of Pol β show that PPi is present within the active site in an open complex, suggesting that opening occurs prior to PPi release14.
Pol β mutations have been linked to cancer and it has been found that about 30% of human tumors express variants of Pol β15. It is thought that Pol β variants contribute to the etiology of cancer by carrying out aberrant BER, which induces mutations and genomic instability that can in turn result in cancer15. One of the Pol β variants that was found in human colon tumors16 and more recently in a subtype of invasive breast cancers17, but was not seen as a polymorphism in healthy individuals18, was E288K (Figure 1A). Further experimentation with E288K Pol β revealed that this variant had a mutator phenotype when inserting dCTP opposite template A19.
To investigate the mechanism underlying this loss of fidelity of Pol β variant E288K, a FRET system developed by Towle-Weicksel et al. was employed10. The Pol β protein was labeled on the fingers domain at position V303C with a donor fluorophore and the acceptor used in this FRET system was a Dabcyl quencher located 8 nucleotides upstream of the templating base on the DNA substrate10 (Figure 1). This setup allowed for monitoring of the movement of the fingers domain as the enzyme closes relative to the DNA substrate (Figures 1B and 1C). We found that E288K shares a mechanism of correct nucleotide insertion with WT, having neither additional nor missing steps. However, E288K has a faster rate of fingers closing and slower nucleotide release than WT Pol β. Our results indicate that E288K’s closed ternary complex is more stable compared to that of WT Pol β, and suggest that the reverse rate of fingers closing, which is likely facilitating nucleotide release, plays an important role in governing fidelity.
EXPERIMENTAL SECTION
Generation of E288K Pol β Variant
Site-directed mutagenesis (Strategene) was used to introduce the E288K mutation into a human tagless Pol β gene (in a modified pET28a vector), which already contained V303C, C267S, and C239S mutations. The plasmid was transformed into Escherichia coli Rosetta 2 DE3 cells, which were then stored in 15% glycerol at −80°C for future use.
Purification of Pol β and Labeling with IAEDANS
Purification of E288K and WT Pol β was conducted as previously described20. Subsequent labeling with IAEDANS was performed according to the protocol used by Towle-Weicksel et al.10.
Generation of DNA Oligonucleotides
Deoxyoligonucleotides were ordered from the Keck Oligo Synthesis Resource (Yale University) and purified using polyacrylamide gel electrophoresis. The desired DNA substrates (Table 1) were created by annealing as previously described21, but with the primer:template:downstream ratio changed to 1:1.2:1.5.
Table 1.
DNA substrates used in this study.
| Name | Sequence | ||
|---|---|---|---|
| extA | 5′ | GCCTCGCAGCCGGCAGATGCGCOHGTCGGTCGATCCAATGCCGTCC CGGAGCGTCGGCCGXCTACGCGACAGCCAGCTAGGTTACGGCAGG |
5′ |
| ddA | 5′ | GCCTCGCAGCCGGCAGATGCGCH GTCGGTCGATCCAATGCCGTCC CGGAGCGTCGGCCGXCTACGCGACAGCCAGCTAGGTTACGGCAGG |
5′ |
The bold, underlined bases are the templating bases.
X represents the Dabcyl residue.
Circular Dichroism
The secondary structures of AEDANS labeled WT and E288K Pol β were compared using a circular dichroism spectrophotometer (Applied Photophysics Chirascan) to measure the ellipticity of a 3 μM protein solution in 10 mM dibasic sodium phosphate buffer from 195–260 nm.
Pre-steady-state Bursts
A mixture containing 200 nM Pol β and 600 nM radiolabeled extA DNA was combined with an equal volume of a solution with 200 μM dTTP, 10 mM MgCl2 using a KinTek Chemical Quench Flow Apparatus. The reaction was allowed to proceed at 37°C in buffer C (50 mM Tris pH 8, 100 mM NaCl, 10% glycerol) for a given amount of time before it was quenched with 0.5 M EDTA and combined with 90% formamide sequencing dye. A timecourse from 0.02 seconds to 3 seconds was obtained, and the samples were resolved on a 20% polyacrylamide sequencing gel. The gel was visualized using autoradiography, the n (substrate) and n+1 (product—substrate with one nucleotide added) bands were quantified using ImageQuant software, and the amount of product was plotted versus time. The data were fitted to the biphasic burst equation
| (1) |
where t is time, kobs is the observed pre-steady state burst rate, kss is the steady state rate.
Polymerization Single Turnover
Polymerization reactions with 750 nM Pol β, 50 nM radiolabeled extA DNA, 10 mM MgCl2, and a given amount of dNTP (concentration series ranging from 0.5–100 μM for correct dNTP and 1–1500 μM for incorrect dNTP) were carried out in buffer C at 37°C. Reactions were quenched with 0.5 M EDTA after incubation for a given duration (ranging from 0.08 seconds to 60 minutes). Experiments were carried out by hand to obtain time points from 40 seconds to 60 minutes, and on the KinTek Chemical Quench Flow Apparatus to obtain time points from 0.08 seconds to 60 seconds. Quenched reactions were combined with 90% formamide dye, run on a 20% polyacrylamide sequencing gel, visualized and quantified as described above, and the data from each dNTP concentration (for a given dNTP) were fitted to the single exponential equation
| (2) |
The kobs from these fits were plotted versus [dNTP] and fitted to the hyperbolic equation
| (3) |
where kpol is the polymerization rate of the enzyme and Kd(dNTP) is the equilibrium dissociation constant of the incoming dNTP from the Pol β-DNA binary complex.
Stopped-flow FRET
All experiments were conducted on the stopped-flow SX-20 (Applied Photophysics) with samples excited at 336 nm and emission filtered with a 400 nm longpass filter. The temperature was 37°C and the voltage was set between 400 V and 500 V such that the emission recorded with buffer was roughly 1.5–2.0 V. Data were collected using the pre-trigger setting and continued for 10 s. Artifacts from the initial flow and mixing of the solutions and instrument deadtime were accounted for. Each trace reported is an average of multiple traces. Forward reaction experiments were set up such that a solution of Pol β, DNA (ddA or extA), and MgCl2 in buffer C was mixed with an equal volume of a solution containing dNTP and MgCl2 in buffer C. The final mixture contained 500 nM Pol β, 200 nM ddA or 100 nM extA DNA, 10 mM MgCl2, and various concentrations of dNTP.
For reverse-opening experiments, a solution containing a ternary complex of 500 nM AEDANS labeled Pol β, 200 nM ddA, 1 μM dTTP and 10 mM MgCl2 in buffer C was mixed with a solution containing 10-fold excess of an unlabeled Pol β-extA binary complex.
KinTek Explorer Modeling
Fluorescence traces were analyzed using KinTek Explorer global fitting software in order to determine mechanisms and obtain rate constants22. The experiment was set up in two mixing steps, the first with enzyme and DNA in respective concentrations pre-incubated together for 100 seconds, followed by mixing with dNTP for 10 seconds. Data for each experiment was imported into the dNTP mixing step as a concentration series and sigma was estimated by fitting to a double exponential (for ddA experiments) or quadruple exponential (for extA experiments). A concentration Series Scaling Factor (multiplier) was used and observables were entered as a*(E+b*ED+c*EDN+d*END…). Various models were used to try to fit the data, with either 3, 4, or 5 steps for ddA experiments or 5, 6, or 7 steps for extA experiments. The Chi Square/Degrees of Freedom (DoF) values were used to evaluate which model best fit the data. The DNA binding step was constrained so that k−/k+ reflected the KD (DNA) determined by electrophoretic mobility shift assays (approximate WT KD (DNA) = 5 nM; E288K KD (DNA) = 19 nM), the dNTP binding step was constrained such that k−/k+ reflected the Kd(dNTP) determined in single turnover experiments, the rate of chemistry was set to the kpol obtained via single turnover, and the reverse polymerization rate was fixed at 0. The value of kss from pre-steady-state bursts were used as an approximate target for the step after chemistry, while the fingers closing was to be fast, (i.e. starting value before fitting ~100 s−1).
RESULTS
AEDANS-Labeled WT and E288K Display Pre-steady-state Burst Activity
In order to observe movements of Pol β during catalysis, the protein and DNA (Table 1) were labeled with a fluorophore and quencher, respectively. The donor fluorophore ((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid (IAEDANS) was attached to the fingers domain of the protein using a sulfide bond10. In order to allow for this attachment, residue 303 was changed from valine to cysteine10. Furthermore, to prevent unwanted labeling at Pol β’s endogenous cysteines, C267 and C239 were both converted to serine10. These three mutations did not impact the activity of Pol β10, and in the current study V303C/C267/C239S Pol β will henceforth be referred to as wild type (WT).
The E288K Pol β variant was generated from the POL B construct with the V303C/C267S/C239S alterations using site-directed mutagenesis and will simply be referred to as E288K. Purification of both WT and the E288K variant Pol β was successfully carried out, as verified by SDS-PAGE analysis, and labeling with AEDANS was conducted. Circular dichroism analysis (Figure 2) implied that the secondary structure of the labeled protein is retained in the E288K variant. All labeled protein preparations were tested for pre-steady-state burst activity before use in additional experiments. Both WT and E288K displayed a rapid burst of product formation and a linear phase (Figure 3). For both E288K and WT, the values obtained for kobs were usually ~15 s−1, while kss was typically ~1 s−1.
Figure 2. WT and E288K have similar secondary structures.
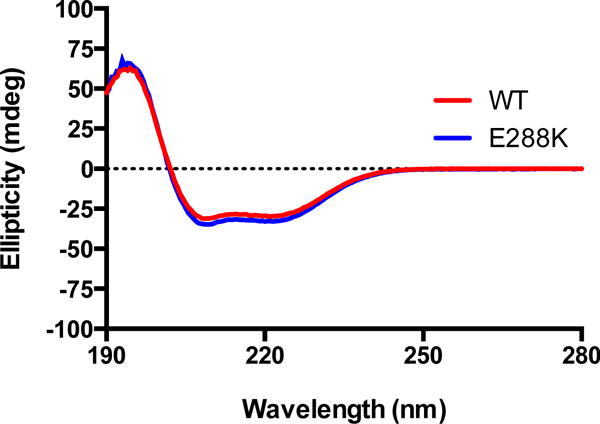
The ellipticity of a 3 μM protein solution in 10 mM dibasic sodium phosphate buffer was measured from 195–260 nm for AEDANS-labeled WT and E288K Pol β.
Figure 3. WT and E288K display pre-steady-state burst activity.
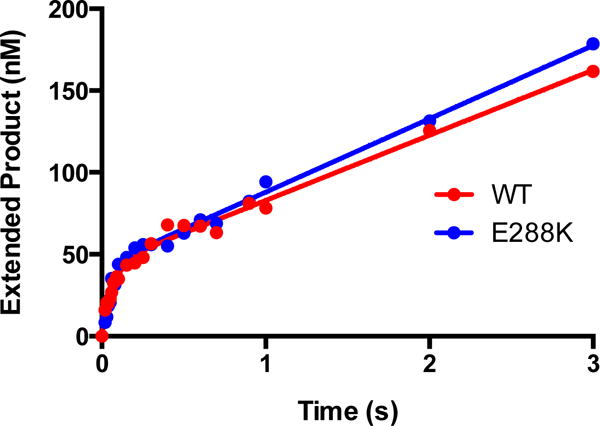
Representative plots are displayed. The biphasic nature of the pre-steady state activity of WT and E288K on extA DNA are evident in plotted data (dots). Curves show the best fit to Equation 1 for each data set. The two bursts shown here are representative of at least ten, but the parameters of the fits shown are WT: kobs=14±2 s−1, kss=0.9±0.1s−1; E288K: kobs=15±2 s−1, kss=1.0±0.1s−1.
AEDANS Labeled E288K Retains Sequence-specific Mutator Activity
In order to determine if the AEDANS-labeled E288K protein exhibits mutator activity opposite template A, single turnover experiments were conducted with AEDANS-labeled WT and E288K proteins with the correct dNTP (dTTP) and all three incorrect dNTPs for both WT and E288K. Values of kpol and Kd(dNTP) were derived from the data (Figures S1 and S2), and these values as well as the parameters calculated from them are reported in Table 2. Most importantly, the single turnover results confirm the mutator activity observed by Murphy et al19. Here, E288K is a sequence specific mutator inserting dCTP opposite template dA with a fidelity that is 5.2 fold lower than WT, which is consistent with the 6-fold difference observed previously19. The results also show that E288K catalyzes nucleotidyl transfer faster than WT, as the kpol for E288K was greater than that for WT for any given dNTP (Table 2). Furthermore, in nearly all conditions, discrimination of correct versus incorrect dNTP takes place at the level of kpol as opposed to Kd(dNTP). Finally, in any given dNTP condition, E288K’s catalytic efficiency is higher than that of WT.
Table 2.
Single turnover kinetic data for WT and E288K Pol β on single nucleotide-gapped DNA.
| Sequencea | Protein |
kpol (s−1) |
Kd(dNTP) (μM) |
D kpolb | D Kd(dNTP)c | Efficiencyd (M−1s−1) |
Fidelitye | X-foldf |
|---|---|---|---|---|---|---|---|---|
| A:dT | WT | 10.9±0.3 | 3.6±0.5 | 3.01×106 | ||||
| E288K | 22.6±1.1 | 5.8±1.2 | 3.87×106 | |||||
|
| ||||||||
| A:dG | WT | 0.11±0.003 | 492±30 | 102 | 137 | 215 | 1.40×104 | |
| E288K | 0.14±0.008 | 129±26 | 162 | 22 | 1.08×103 | 3.60×103 | 3.9 | |
|
| ||||||||
| A:dA | WT | 0.03±0.001 | 182±31 | 384 | 50 | 156 | 1.93×104 | |
| E288K | 0.08±0.004 | 181±32 | 289 | 31 | 432 | 8.98×103 | 2.2 | |
|
| ||||||||
| A:dC | WT | 0.07±0.002 | 89±10 | 149 | 25 | 818 | 3.69×103 | |
| E288K | 0.15±0.004 | 27±4 | 154 | 5 | 5.46×103 | 710 | 5.2 | |
The primer-template is extA, with the template base A; templating base:incoming dNTP is shown in the Table.
Discrimination of kpol = kpol(correct)/kpol(incorrect)
Discrimination of Kd(dNTP) = Kd(dNTP)(incorrect)/Kd(dNTP)(correct)
Efficiency = kpol/Kd(dNTP)
Fidelity = (correct efficiency + incorrect efficiency)/incorrect efficiency
X-fold = WT fidelity/E288K fidelity
FRET Data Indicate the Presence of Non-covalent Step for both WT and E288K
In order to investigate any pre-chemistry steps that occur along the catalytic pathway of Pol β, FRET experiments were conducted using non-extendable (ddA) DNA. In these experiments, a solution of Pol β, ddA DNA, and Mg2+ was rapidly mixed with a solution of correct dNTP (dTTP) and Mg2+, yielding traces for both WT and E288K that show a quench in fluorescence without recovery (Figure 4A). This quench likely results from the binary complex binding dNTP and subsequent fingers closing. Because the primer DNA does not have a free 3′OH, nucleotidyl transfer does not occur and the enzyme remains in a closed state, resulting in maintenance of the lower fluorescence level.
Figure 4. FRET demonstrates non-covalent step in E288K correct incorporation mechanism.
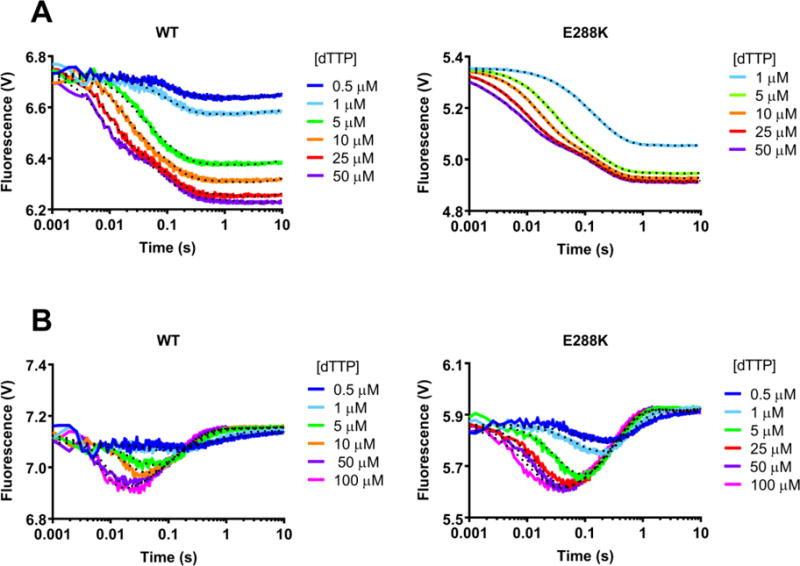
Using the stopped flow apparatus, a solution of AEDANS-labeled WT or E288K Pol β and ddA (A) or extA DNA (B) DNA was mixed with the given dNTP in the presence of Mg2+, excited at 336 nm, and then fluorescence was measured for 10 seconds. Each trace shown is an average of 4 recordings. Data shown (solid curves) is modeled using KinTek Global Explorer. Fits obtained by KinTek Global Explorer are indicated by the dotted curves. Figure S3 shows the Chi Squared analysis of these FRET experiments with ddA and extA DNA and indicates that WT and E288K are best described by a mechanism that includes a non-covalent step before chemistry.
These traces were modeled using KinTek Global Explorer software, to determine how many steps were present in the kinetic pathway represented by the data and the corresponding rates of those steps. The data for both WT and E288K were best fit by a 4-step mechanism (Figure S3). Using Chi Square/(DoF) as a measure of goodness-of-fit (Chi Square/DoF = 1 indicates a better fit), the presence of a fingers closing step followed by one non-covalent step in the mechanism for WT confirmed what was previously found by Towle-Weicksel et al., and we found E288K is similar to WT in this regard (Figure S3)10. The rate constants from modeling the 4-step mechanism are reported in Table 3. The rates of each step are similar for WT and E288K with the important exception of reverse of fingers closing (k−3) which is much faster for WT (84 s−1) than for E288K (9 s−1).
Table 3.
Rates obtained from modeling the FRET data using KinTek Global Explorer
| Step | Reactiona | Constantb | Non-extendable DNA | Extendable DNA | ||
|---|---|---|---|---|---|---|
| WT | E288K | WT | E288K | |||
| DNA Binding | E + D → ED | KD (DNA)# | 5 | 19 | 5 | 19 |
| ED → E + D | ||||||
|
| ||||||
| dNTP Binding | ED → N + EDN | Kd(dNTP)* | 3.6±0.5 | 5.8±1.2 | 3.6±0.5 | 5.8±1.2 |
| EDN → ED + N | ||||||
|
| ||||||
| Fingers Closing | EDN → END | k+3 | 129±18 | 114±8 | 84±44 | 90±21 |
| END → EDN | k−3 | 84±9 | 9±2 | 148±27 | 9±3 | |
|
| ||||||
| Non-covalent Step | END → NDE | k+4 | 18±1 | 6.5±0.3 | 17±13 | 10±2 |
| NDE → END | k−4 | 0.59±0.05 | 0.26±0.03 | 1.4±2.0 | 41±13 | |
|
| ||||||
| Polymer-ization | NDE → EP | k+5 | 10.9±0.3 | 22.6±1.1 | ||
| EP → NDE | k−5 | 0 | 0 | |||
|
| ||||||
| Post Chemistry | EP → E + P | k+6 | 0.45±0.11 | 0.3±0.2 | ||
| E + P → EP | k−6⁋ | 0.02±0.07 | 0.03±0.2 | |||
Reaction as modeled in KinTek Explorer. E represents enzyme, D represents DNA substrate, N represents nucleotide, and P represents product.
Rate constants all have units of s−1 exceptK has units of μM−1s−1. # has units of nM and * has units of μM.
The Reverse Rate of Fingers Closing is Slow for E288K
Next, the full mechanism of nucleotide incorporation using extendable DNA (extA) was probed with FRET. In these experiments, a solution of Pol β, extA DNA, and Mg2+ was mixed with a solution of correct dTTP and Mg2+. Both WT and E288K showed an initial quench in fluorescence followed by a recovery and maintenance of fluorescence (Figure 4B). The initial quench occurs as the enzyme binds dNTP and the fingers close. Fluorescence recovery occurs after polymerization is executed and the fingers open.
Using KinTek Global Explorer and the Chi Square/DoF evaluation (Figure S3) of various mechanisms, it was determined that the traces for WT and E288K both fit best to a 6-step mechanism as shown in Scheme 1, with the steps modeled as 1) DNA binding (this is modeled as a pre-mix step) 2) dNTP binding 3) fingers closing 4) non-covalent step 5) polymerization and with post-chemistry steps combined as 6) fingers opening. Again, the 6-step mechanism in this condition confirms the presence of the non-covalent step which was observed for WT by Towle-Weicksel et al. and establishes that E288K follows a pathway similar to WT when inserting correct dNTP10.
The rates obtained from modeling the 6-step mechanism in KinTek Global Explorer can be found in Table 3. The reverse rate of fingers closing, previously suggested to be nucleotide release23, 24 was much faster for WT (148 s−1) than for E288K (9 s−1). In contrast, the reverse rate of the noncovalent step subsequent to fingers closing was higher for the E288K mutant than WT Pol β.
Nucleotide Release is Slower for E288K than WT Pol β
Discrimination against incorrect dNTPs was shown to occur by rapid release of mismatched dNTPs by the highly accurate and rapid T7 DNA polymerase24. To determine if nucleotide release functions in substrate discrimination by Pol β, we attempted to directly measure k−4 and k−3, given that our model suggested that they were slower for E288K than WT Pol β. We performed these experiments for WT and E288K by mixing a solution containing a Pol β-ddA-correct dTTP ternary complex with a 10-fold excess of an unlabeled Pol β-extA binary complex as described23. This method enabled us to measure the rate of release of the nucleotide from the tight ternary complex in a competition experiment. The unlabeled secondary complex rapidly traps the free and released dTTP. The fluorescence traces of the reverse closing for both WT and E288K are shown in Figure 5.
Figure 5. Reverse closing FRET using a trap experiment.
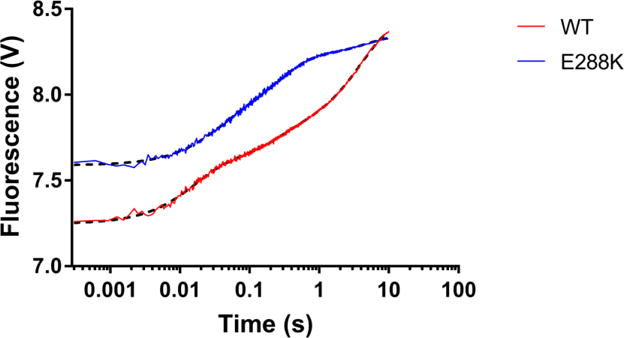
A solution containing AEDANS labeled Pol β-ddA-correct dTTP and 10 mM Mg2+ in ternary complex was mixed with a 10-fold excess of an unlabeled Pol β-extA binary complex, excited at 336 nM and fluorescence was recorded for 10 seconds. As the unlabeled secondary complex rapidly trapped and sequestered the free and released dTTP, an increase in fluorescence can be observed. Each trace (red for WT and blue for E288K) is an average of at least 4 recordings. Fits obtained by GraphPad Prism are indicated by the black dotted curves. The reverse rates measured using KinTek Global Explorer for WT were k−4=0.25s−1, k−3=65 s−1, k−2=26s−1 and for E288K were k−4=0.21s−1, k−3=13.3 s−1, k−2=64s−1, where k−4 corresponds to the reverse non-covalent step, k−3 is the reverse of fingers closing and k−2 is dNTP dissociation.
The traces for both WT and E288K fit best to a triple-exponential function that we modeled as follows
where step (1) is defined as the reverse non-covalent step, k−4, step (2) is the reverse of fingers closing, i.e. fingers opening, k−3, and step (3) is the release of the dNTP, k−2. We used Kintek Global Explorer to model our reverse reaction to determine rate orders. We constrained the dNTP binding ratio K2 to the Kd(dNTP) obtained from single turnover experiments. The best-fit rates we obtained are shown in Table S1 and Scheme 2. Clearly, k−3 is indeed much slower for E288K (13.3 s−1) than for WT Pol β (65 s−1), whereas k−4 for WT and E288K is similar. The percent contribution calculated for each step are shown in Table S1.
Scheme 2. Rates obtained from modeling the FRET data with fixed reverse rates using KinTek Global Explorer.
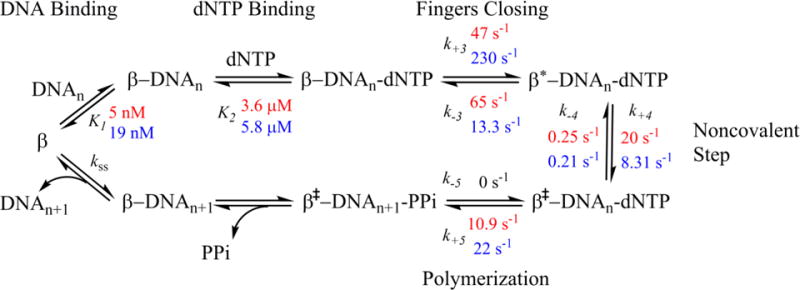
Using non-extendable ddA, WT rates are in and E288K rates are in .
We re-modeled our WT and E288K data using KinTek Global Explorer and fixed our measured reverse rates from the competition experiments. The forward rates for fingers closing, k+3, for both ddA E288K (230 s−1) and extA E288K (119 s−1) were much faster than WT (47 s−1 and 60 s−1 respectively) (Scheme 2). When taken together with the reverse rate (k−3), our results indicate that perhaps E288K is adopting a more stable closed conformational state upon binding dNTP and moving in a forward direction towards the chemistry step, k+5. In contrast, k−3 for WT in the extendable reaction is similar to k+3 for incorporation of correct dNTP, suggesting that much of the bound nucleotide may not get incorporated.
A Decreased Fluorescence Signal is Observed for E288K in the Presence of Incorrect dNTP
In order to investigate the mechanism of misincorporation, FRET experiments were conducted with individual incorrect dNTPs, first with ddA DNA (Figure 6). For WT (Figure 6A), the traces for all incorrect dNTPs show no quenching, as originally observed in our previous study10. In contrast, slight quenching is observed with E288K (Figure 6B) for all incorrect dNTPs in the same shape as correct dNTP, but with a substantially smaller magnitude. We were unable to model these reactions because the changes in fluorescence were too shallow. Qualitatively, these results indicate that WT does not form a closed ternary complex with incorrect dNTP (within the first 10 seconds), while E288K does, at least partially. Alternatively, a larger percentage of E288K versus WT molecules close such that a quench is observed for E288K in our ensemble reactions.
Figure 6. FRET with incorrect dNTP and non-extendable DNA shows quenching for E288K but not WT.
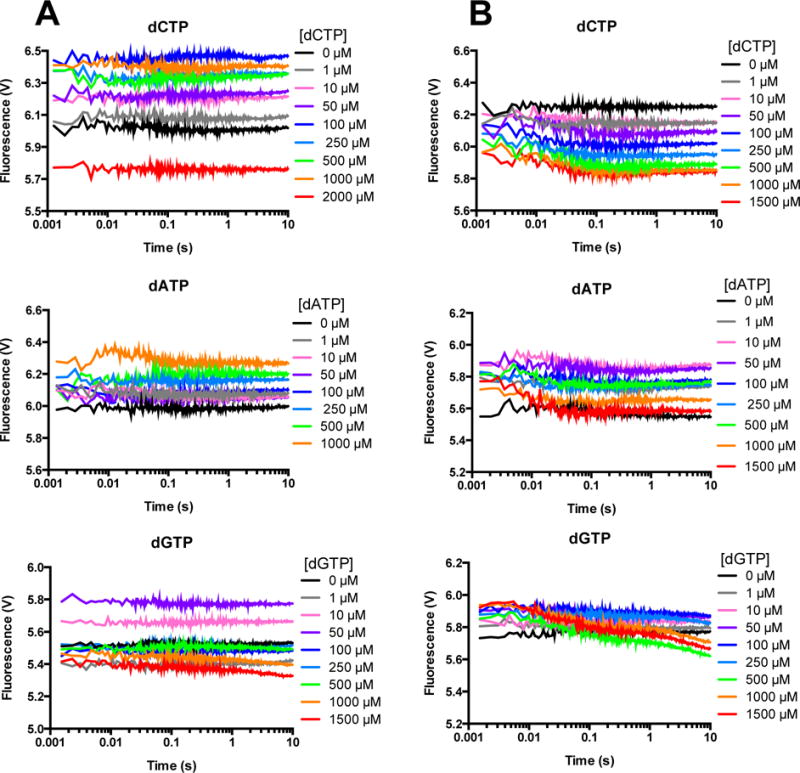
A solution of AEDANS-labeled WT (A) or E288K (B) Pol β and ddA DNA was mixed with the indicated incorrect dNTP in the presence of Mg2+, excited at 336 nm, and fluorescence was measured for 10 seconds. Each trace shown is an average of 4 recordings.
Finally, FRET experiments with individual incorrect dNTPs were conducted with extA DNA (Figure 7), yielding flat traces for all dNTPs for both WT (Figure 7A) and E288K (Figure 7B), with the exception of dCTP, in which slight curvature is observed for the highest 3 concentrations. In previous work with WT Pol β and this FRET system, but with a different primer-termplate sequence, we did not observe quenching with incorrect dNTP and extendable DNA10.
Figure 7. FRET with incorrect dNTP and extendable DNA shows no quenching.
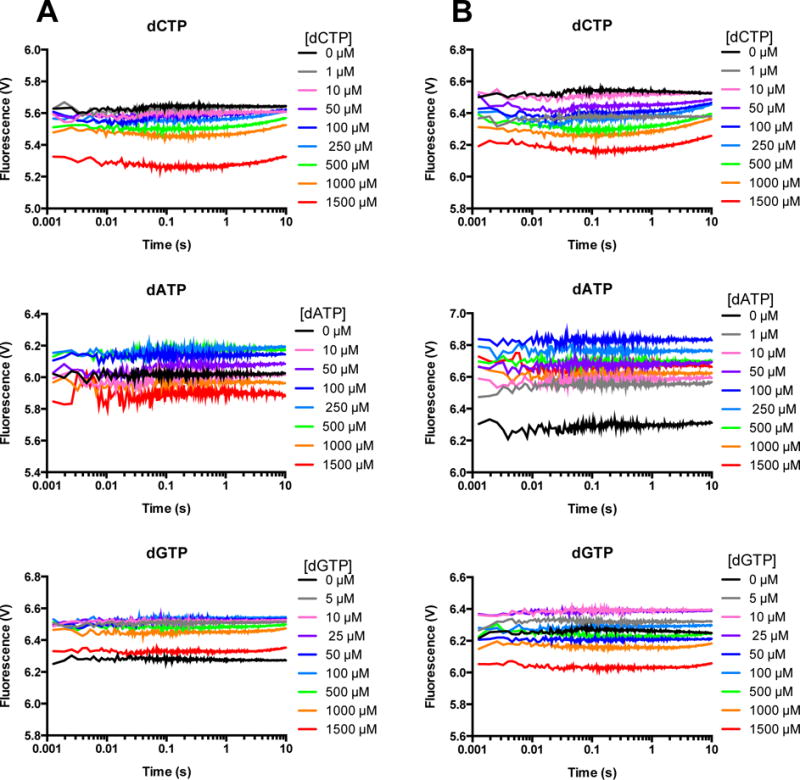
A solution of AEDANS-labeled WT (A) or E288K (B) Pol β and extA DNA was mixed with the indicated incorrect dNTP in the presence of Mg2+, excited at 336 nm, and fluorescence was measured for 10 seconds. Each trace shown is an average of 4 recordings.
DISCUSSION
FRET was used to compare the mechanism of nucleotide incorporation of WT to that of E288K in search of differences that might underlie E288K’s decreased fidelity. Based on FRET data of the forward reaction and the subsequent KinTek Global Explorer analysis, it appears E288K follows the same overall reaction mechanism as WT Pol β when inserting correct dNTP. E288K data are best fitted by a 6-step mechanism, just like WT, which includes pre-chemical fingers closing and a subsequent non-covalent step. However, the rates obtained for various steps along the reaction pathway reveal important disparities between E288K and WT Pol β that may shed light on the ability of E288K to act as a mutator polymerase. Specifically, we show that the forward rate of fingers closing, k+3, for E288K is significantly faster than that of WT Pol β. We also show that k−3 is slower for E288K than for WT Pol β. Thus, binding of the correct dNTP by E288K results in the majority of substrate being incorporated by this enzyme. However, this is not the case for WT Pol β. In addition, E288K has a faster rate of polymerization than WT when inserting either correct or incorrect dNTP. Taken together, our results are consistent with the interpretation that E288K is a mutator variant as a result of defective nucleotide release, permitting it to incorporate both correct and incorrect dNTPs more readily than WT Pol β.
E288K Closed Conformation is Stable
The slower k−3 rate observed for E288K in comparison with WT implies that the closed E288K ternary complex is less flexible than that of WT Pol β. The most likely explanation for this decreased flexibility is simulated in Figure 8 using PyMol25 which shows the potential positions of the newly introduced basic K288 residue with respect to the template DNA strand upstream from the nucleotide insertion site. It is possible that in the E288K ternary complex, there is an attractive interaction between the basic K288 and the negatively charged DNA backbone. The left panel of Figure 8 shows the position of E288 in the WT ternary state of Pol β. The right panel of Figure 8 shows the potential position of K288 simulated via mutagenesis in Coot26, displaying three standard lysine rotamers from the geometry library. Two of the rotamers would be in position to hydrogen bond to the non-bridging oxygens of the +5 position on the DNA while contact to the +4 position would likely require a conformational shift or be solvent mediated. Any of these potential interactions would contribute to stabilizing and perhaps limiting the flexibility of a closed ternary complex, and would not be present in WT, as shown in the left panel of Figure 8 (PDB ID 2FMS)27, due to the acidic Glu residue at this position.
Figure 8. The position of E288 in the ternary state for Pol β.
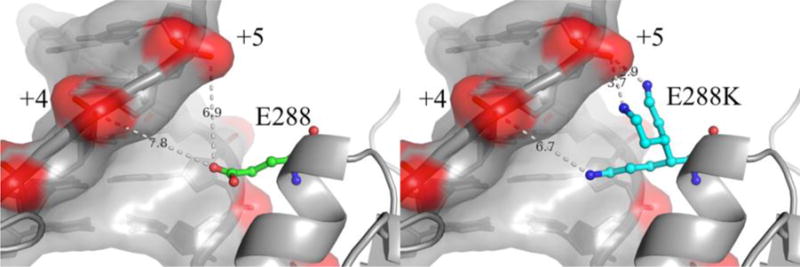
Left shows the position of E288 in the ternary complex (PDB ID 2FMS) in relation to the DNA template strand upstream from the nucleotide insertion site. Right shows the potential position of K288 simulated via mutagenesis in Coot and displaying three standard lysine rotamers from the geometry library. Two of the rotamers would be in position to hydrogen bond to the non-bridging oxygens of the +5 position while contact to the +4 position would likely require a conformational shift or be solvent mediated. Both figures were made using PyMOL25.
Nucleotide Release is Important for Accurate DNA Synthesis
Previous work from other laboratories (see for example23, 28 and reviewed in29) has shown that the rate of nucleotide release or reverse conformational change (k−3 in our case) impacts polymerase specificity. Rapid isomerization of the correct nucleotide within the polymerase active site favors rapid incorporation of the dNTP substrate. In the case of Pol β, results from this study and a previous one10 suggest that k−3 for the WT enzyme is fast and probably equivalent to the forward rate, k+3 of fingers closing. Chemistry (k+5) also appears to be rate limiting in the case of Pol β. This might be a basis for the relatively low specificity of Pol β compared to other enzymes, especially replicative polymerases. Like Klenow fragment, the Pol β reaction mechanism also appears to involve a second noncovalent step subsequent to fingers closing, the nature of which is not known at this time30–32. In contrast to WT Pol β, the forward rate of fingers closing, k+3 is fast and the k−3 is slow for E288K, suggesting that this polymerase is inherently built for rapid incorporation of the dNTP substrate, at least in part due to the interaction of E288K with DNA. Thus, dNTP isomerization, regardless of the nature of the dNTP, may be more rapid within the active site of E288K, ultimately favoring higher levels of misincorporation compared to WT Pol β.
In previous studies we have been unable to detect a stable closed form of Pol β in the presence of incorrect dNTP10. Our results are consistent with misincorporation by Pol β taking place in an open or semi-open form of the ternary complex, as suggested by others9, 12, 33, 34. However, we show that the E288K mutator variant exhibits some closing with incorrect dNTP and ddA DNA substrate. This is consistent with the idea that K288 is facilitating a more stable closed complex overall even in the presence of incorrect dNTP (Figure 8).
To the best of our knowledge only three other mutator variants of DNA polymerases have been characterized for their abilities to undergo precatalytic conformational changes32, 34. The E710A and E710Q Klenow fragment mutator enzymes appear to be defective in a step preceeding fingers closing, whereas the Y766A variant results in an enzyme that appears to be unable to undergo fingers closing in the presence of correctly paired A:dTTP and T:dATP, implicating fingers closing as an important parameter of specificity that is governed by enzyme structure35. Our results with E288K are consistent with the idea that a type of monitoring of the dNTP fit during fingers closing is important for specificity. However, in contrast to the findings with the mutators of Klenow fragment, E288K Pol β appears to be defective in its ability to reject incorrect dNTPs during a precatalytic conformational change.
Summary
An in-depth investigation of the mechanism of nucleotide incorporation of human E288K Pol β has been presented here. We showed that E288K incorporates the correct dNTP using a mechanism similar to WT. However, E288K incorporates dNTPs more readily than WT, as E288K has an inherently slower rate of dNTP release, ultimately leading to lower fidelity. We also extend our previous results by showing that the ternary complex of WT Pol β does not close in a stable manner in the presence of an incorrect dNTP substrate. Our results provide evidence that at least one precatalytic conformational change in the Pol β reaction pathway is important for substrate discrimination. Our results are also consistent with the suggestion that incorrect incorporation by Pol β is catalyzed by an open or semi-open form of the enzyme.
Supplementary Material
Acknowledgments
This research was supported by R01 CA080830 from the National Cancer Institute and T32 CA193200, also from the National Cancer Institute. We would also like to thank Kenneth A. Johnson from University of Texas at Austin for his help and advice on modeling some of our data using Kintek Global Explorer.
Footnotes
Supporting Information. One table and three supporting figures are supplied as Supporting Information in PDF format and are available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interest.
References
- 1.Wallace SS. Base excision repair: a critical player in many games. DNA Repair (Amst) 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 3.Krokan HE, Bjoras M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace SS, Murphy DL, Sweasy JB. Base excision repair and cancer. Cancer Lett. 2012;327:73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamtich J, Sweasy JB. DNA polymerase family X: function, structure, and cellular roles. Biochim Biophys Acta. 2010;1804:1136–1150. doi: 10.1016/j.bbapap.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanabe K, Bohn EW, Wilson SH. Steady-state kinetics of mouse DNA polymerase beta. Biochemistry. 1979;18:3401–3406. doi: 10.1021/bi00582a029. [DOI] [PubMed] [Google Scholar]
- 7.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science (New York, NY) 1994;264:1891–1903. [PubMed] [Google Scholar]
- 8.Freudenthal BD, Beard WA, Shock DD, Wilson SH. Observing a DNA polymerase choose right from wrong. Cell. 2013;154:157–168. doi: 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moscato B, Swain M, Loria JP. Induced Fit in the Selection of Correct versus Incorrect Nucleotides by DNA Polymerase beta. Biochemistry. 2016;55:382–395. doi: 10.1021/acs.biochem.5b01213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Towle-Weicksel JB, Dalal S, Sohl CD, Doublie S, Anderson KS, Sweasy JB. Fluorescence resonance energy transfer studies of DNA polymerase beta: the critical role of fingers domain movements and a novel non-covalent step during nucleotide selection. The Journal of biological chemistry. 2014;289:16541–16550. doi: 10.1074/jbc.M114.561878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batra VK, Beard WA, Shock DD, Pedersen LC, Wilson SH. Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation. Mol Cell. 2008;30:315–324. doi: 10.1016/j.molcel.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koag MC, Lee S. Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta. J Am Chem Soc. 2014;136:5709–5721. doi: 10.1021/ja500172d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arndt JW, Gong W, Zhong X, Showalter AK, Liu J, Dunlap CA, Lin Z, Paxson C, Tsai MD, Chan MK. Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes. Biochemistry. 2001;40:5368–5375. doi: 10.1021/bi002176j. [DOI] [PubMed] [Google Scholar]
- 14.Reed AJ, Vyas R, Raper AT, Suo Z. Structural Insights into the Post-Chemistry Steps of Nucleotide Incorporation Catalyzed by a DNA Polymerase. J Am Chem Soc. 2017;139:465–471. doi: 10.1021/jacs.6b11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase beta and cancer? Cell Cycle. 2004;3:998–1001. [PubMed] [Google Scholar]
- 16.Donigan KA, Sun KW, Nemec AA, Murphy DL, Cong X, Northrup V, Zelterman D, Sweasy JB. Human POLB gene is mutated in high percentage of colorectal tumors. The Journal of biological chemistry. 2012;287:23830–23839. doi: 10.1074/jbc.M111.324947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, Bowlby R, Shen H, Hayat S, Fieldhouse R, Lester SC, Tse GM, Factor RE, Collins LC, Allison KH, Chen YY, Jensen K, Johnson NB, Oesterreich S, Mills GB, Cherniack AD, Robertson G, Benz C, Sander C, Laird PW, Hoadley KA, King TA, T. R. Network. Perou CM. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohrenweiser HW, Xi T, Vazquez-Matias J, Jones IM. Identification of 127 amino acid substitution variants in screening 37 DNA repair genes in humans. Cancer Epidemiol Biomarkers Prev. 2002;11:1054–1064. [PubMed] [Google Scholar]
- 19.Murphy DL, Donigan KA, Jaeger J, Sweasy JB. The E288K colon tumor variant of DNA polymerase beta is a sequence specific mutator. Biochemistry. 2012;51:5269–5275. doi: 10.1021/bi3003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckenroth BE, Towle-Weicksel JB, Sweasy JB, Doublie S. The E295K cancer variant of human polymerase beta favors the mismatch conformational pathway during nucleotide selection. The Journal of biological chemistry. 2013;288:34850–34860. doi: 10.1074/jbc.M113.510891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy DL, Kosa J, Jaeger J, Sweasy JB. The Asp285 variant of DNA polymerase beta extends mispaired primer termini via increased nucleotide binding. Biochemistry. 2008;47:8048–8057. doi: 10.1021/bi702104y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson KA, Simpson ZB, Blom T. Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 23.Kellinger MW, Johnson KA. Nucleotide-dependent conformational change governs specificity and analog discrimination by HIV reverse transcriptase. Proc Natl Acad Sci U S A. 2010;107:7734–7739. doi: 10.1073/pnas.0913946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothwell PJ, Mitaksov V, Waksman G. Motions of the fingers subdomain of klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Molecular cell. 2005;19:345–355. doi: 10.1016/j.molcel.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC, Wilson SH. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure (London, England: 1993) 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bakhtina M, Roettger MP, Tsai MD. Contribution of the reverse rate of the conformational step to polymerase beta fidelity. Biochemistry. 2009;48:3197–3208. doi: 10.1021/bi802119f. [DOI] [PubMed] [Google Scholar]
- 29.Johnson KA. Role of induced fit in enzyme specificity: a molecular forward/reverse switch. J Biol Chem. 2008;283:26297–26301. doi: 10.1074/jbc.R800034200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purohit V, Grindley ND, Joyce CM. Use of 2-aminopurine fluorescence to examine conformational changes during nucleotide incorporation by DNA polymerase I (Klenow fragment) Biochemistry. 2003;42:10200–10211. doi: 10.1021/bi0341206. [DOI] [PubMed] [Google Scholar]
- 31.Bermek O, Grindley ND, Joyce CM. Distinct roles of the active-site Mg2+ ligands, Asp882 and Asp705, of DNA polymerase I (Klenow fragment) during the prechemistry conformational transitions. J Biol Chem. 2011;286:3755–3766. doi: 10.1074/jbc.M110.167593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joyce CM, Potapova O, Delucia AM, Huang X, Basu VP, Grindley ND. Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry. 2008;47:6103–6116. doi: 10.1021/bi7021848. [DOI] [PubMed] [Google Scholar]
- 33.Santoso Y, Joyce CM, Potapova O, Le Reste L, Hohlbein J, Torella JP, Grindley ND, Kapanidis AN. Conformational transitions in DNA polymerase I revealed by single-molecule FRET. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:715–720. doi: 10.1073/pnas.0910909107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minnick DT, Liu L, Grindley ND, Kunkel TA, Joyce CM. Discrimination against purine-pyrimidine mispairs in the polymerase active site of DNA polymerase I: a structural explanation. Proc Natl Acad Sci U S A. 2002;99:1194–1199. doi: 10.1073/pnas.032457899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bermek O, Grindley ND, Joyce CM. Prechemistry nucleotide selection checkpoints in the reaction pathway of DNA polymerase I and roles of glu710 and tyr766. Biochemistry. 2013;52:6258–6274. doi: 10.1021/bi400837k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


