Abstract
Objectives
In this preliminary study, we examined whether imaging-based phenotypes are associated with reported predictive gene signatures in hepatocellular carcinoma (HCC).
Methods
38 patients (M/F 30/8, mean age 61 y) who underwent pre-operative CT or MR imaging before surgery as well as transcriptome profiling were included in this IRB approved single centre retrospective study. Eleven qualitative and 4 quantitative imaging traits (size, enhancement ratios, wash-out ratio, tumour-to-liver contrast ratios) were assessed by 3 observers and were correlated with 13 previously reported HCC gene signatures by using logistic regression analysis.
Results
39 HCC tumours (mean size 5.7 ± 3.2 cm) were assessed. Significant positive associations were observed between certain imaging traits and gene signatures of aggressive HCC phenotype (G3-Boyault, Proliferation-Chiang profiles, CK19-Villanueva, S1/S2-Hoshida) with odds ratios ranging from 4.44–12.73 (P <0.045). Infiltrative pattern at imaging was significantly associated with signatures of microvascular invasion and aggressive phenotype. Significant but weak associations were also observed between each of enhancement ratios and tumour-to-liver contrast ratios and certain gene expression profiles.
Conclusions
This preliminary study demonstrates a correlation between phenotypic imaging traits with gene signatures of aggressive HCC, which warrants further prospective validation to establish imaging-based surrogate markers of molecular phenotypes in HCC.
Keywords: Hepatocellular carcinoma, Genomics, Magnetic resonance imaging, Computed tomography, Biomarkers
Introduction
Genomics technologies enable survey of the expression of thousands of genes, and have shown great potential for establishing tumour diagnosis, prognosis and response to therapy [1–3]. Hepatocellular carcinoma (HCC) tumours have been intensively profiled for genome-wide gene expression, deoxyribonucleic acid (DNA) copy number alterations, DNA methylation, and gene mutations [4–16] which may have a role for the prediction of prognosis, microvascular invasion and treatment response. These advanced genomics methods require tissue sampling, which is invasive, and rarely clinically indicated in HCC. Moreover, they require specialized equipment, which limit their widespread use. A surrogate marker of gene expression using imaging would be of major interest, as imaging is noninvasive, repeatable, and can sample the whole lesion, as well as multiple lesions in the same patient.
Imaging plays an essential role in HCC screening, diagnosis and staging [17, 18]. There is limited data correlating imaging with genomic analysis in HCC [19–21]. Two of the prior studies (from the same group) have shown that dynamic imaging traits using computed tomography (CT) correlate with global gene expression programs of HCC. Segal et al described that the combination of 28 imaging traits (assessed in 28 HCCs with CT) can reconstruct 78% of the global gene expression profiles [19]. In the other study, CT-based phenotypes were found to correlate with a doxorubicin drug response gene expression program, with vascular invasion and tumor stage in 30 HCCs [20]. Using magnetic resonance (MR) imaging, a recent study showed that signal intensity on hepatobiliary phase after injection of Gd-EOB-DTPA correlates with gene expression [21]. A refinement in the understanding of gene expression prognostic signatures in HCC occurred during recent years [4–16], allowing to explore a more comprehensive radiogenomic and biologically/clinically relevant correlation.
The objective of this preliminary study was to correlate phenotypic traits assessed with CT or MR imaging with gene expression profiles predictive of aggressive phenotypic features (vascular invasion) or poor outcome obtained in patients with HCC prior to resection or liver transplantation.
PATIENTS AND METHODS
Patients
This retrospective single centre study was HIPAA (health insurance portability and accountability) compliant and approved by our institutional review board. Informed consent was waived due to the retrospective nature of the study. Consecutive patients from our centre, included in a clinical study assessing the role of genomics for prediction of HCC recurrence after surgical resection or transplantation, and with an available pre-surgical CT or MR imaging were selected. Thirty-eight patients (M/F 30/8, mean age 61 y, range 45–82 y) with 39 HCCs were included in the study, as part of a cohort previously reported [22]. Table 1 summarizes the clinical characteristics of this cohort, including Barcelona Clinic Liver Cancer (BCLC) and American Joint Committee on Cancer (AJCC) stages. All patients underwent contrast-enhanced imaging, including CT (n=26) or MR imaging (n=12) before liver resection (n=36) or liver transplantation (n=2). The mean interval time between imaging and surgery was 19 days (range: 4–106 days). All HCC lesions were treatment-naive.
Table 1.
Clinical characteristics, imaging traits and pathologic traits assessed in 39 HCCs diagnosed in 38 patients.
| Parameter | Classification | Results |
|---|---|---|
| Age (y) | Mean ± SD (range) | 61 ± 9 (45-82) |
| Sex | M/F | 30/8 |
| Aetiology | HCV/HCV+NASH | 36/2 |
| Advanced fibrosis or cirrhosis | Y/N | 30/8 |
| Child-Pugh scores | A/B/C | 30/6/2 |
| BCLC stage | A/B/C | 26/2/10 |
| AJCC stage* | T1/T2/T3/T4 | 5/19/12/1 |
| Imaging traits (CT/MR imaging: 26/12 patients) | ||
| □ T2 hyperintensity** | Yes/No | 11/1 |
| □ Fat content** | Yes/No | 1/11 |
| □ Single vs. multiple HCCs | Single/multiple | 37/2 |
| □ Tumour necrosis | Yes/No | 15/24 |
| □ Enhancement characteristics | Wash-in / wash-out | 29 |
| Wash-in / no wash-out | 7 | |
| Hypovascular | 3 | |
| □ Tumour morphology | Nodular/Infiltrative pattern | 30/9 |
| □ Macrovascular invasion | Yes/No | 8/31 |
| □ Mosaic appearance | Yes/No | 30/9 |
| □ Internal arteries | Yes/No | 31/8 |
| □ Capsule/pseudocapsule | Yes/No | 18/21 |
| □ Extra-nodular growth*** | Yes/No | 9/9 |
| □ Tumour size (cm) | Mean ± SD (range) | 5.7 ± 3.2 (1.5-14.0) |
| □ Enhancement ratio AP (%) | Mean ± SD | 83.88 ± 49.52 |
| □ Enhancement ratio PVP (%) | Mean ± SD | 115.25 ± 47.75 |
| □ Wash-out ratio (%) | Mean ± SD | −26.43 ± 32.96 |
| □ Tumour-to-liver contrast ratio AP (%) | Mean ± SD | 27.04 ± 35.46 |
| □ Tumour-to-liver contrast ratio PVP (%) | Mean ± SD | −10.42 ± 18.76 |
| Pathologic traits* | ||
| □ Tumour size (cm) | Mean ± SD (range) | 5.7 ± 3.4 (1.2-15.0) |
| □ Grade | WD/MD/PD | 5/17/16 |
| □ Microvascular invasion | Yes/No | 30/8 |
Pathologic information was not available in one patient
Assessed in 12 patients with MR imaging
Assessed in 18 tumours with capsule/pseudocapsule,
WD = well differentiated, MD= moderately-differentiated, PD= poorly-differentiated, AP = arterial phase, PVP = portal venous phase
Imaging
CT technique
26 patients underwent multi-detector CT scans of the abdomen, using different systems [HighSpeed CT/i GE (n=5), LightSpeed Ultra GE (n=5), Somatom Emotion 6 Siemens (n=5), Somatom Sensation 16 Siemens (n=6), Somatom Sensation Cardiac 64 Siemens (n=5)]. CT examinations included non-contrast and contrast-enhanced imaging through the liver, including arterial phase (using bolus tracking), portal venous phase (70 seconds) and late venous phase (at 180 sec, obtained in 5 patients) after initiation of the bolus of intravenous contrast material (100 mL of Isovue 370, Bracco Diagnostics, Princeton, NJ). CT imaging parameters were kVp 110–130, mAs 140–280, collimation and reconstruction interval 5/1.5 mm.
MR imaging technique
12 patients underwent MR imaging of the abdomen, using various 1.5T clinical systems [GE Healthcare (n=6), Siemens Healthcare (n=6)]. The following sequences were obtained: coronal T2 HASTE, axial fat suppressed FSE T2, T1 GRE in- and opposed-phase, and dynamic contrast-enhanced axial 3D GRE T1-weighted images before and after administration of an extracellular gadolinium contrast agent (0.1 mmol/kg of Gadopentetate Dimeglumine, Magnevist). Arterial phase images were obtained using timing run or bolus tracking methods. Portal venous and equilibrium phase images were obtained at 60 and 180 sec post-contrast injection.
Image analysis
Eleven qualitative imaging traits were assessed in consensus by two experienced abdominal radiologists (observer 1 and observer 2, BT and SK, with 13 and 10 years experience in abdominal imaging, respectively), using a PACS (Centricity v3.0, GE). The observers assessed the enhancement characteristics: wash-in (defined as an enhancement of the lesion of the arterial phase higher than the one of the liver parenchyma), wash-out (defined as a lesion hypodense/hypointense compared to the liver parenchyma on the portal venous and/or delayed phases) and hypovascular (absence of wash-in). Based on enhancement, lesions were classified as typical (wash-in and wash-out) or atypical (wash-in without wash-out or hypovascular). Lesion morphology was described as nodular or infiltrative (Fig. 1). On MR imaging, the presence of intra-lesional fat (defined as drop of the signal in the lesion on opposed-phase compared to in-phase images) and a T2 hyperintensity were evaluated. For MR imaging and CT, macrovascular invasion, mosaic pattern, internal arteries, capsule or pseudocapsule and extra nodular growth (defined as incomplete capsule) were also evaluated. The maximum tumour diameter was measured on post-contrast images obtained during the arterial phase.
Fig. 1.
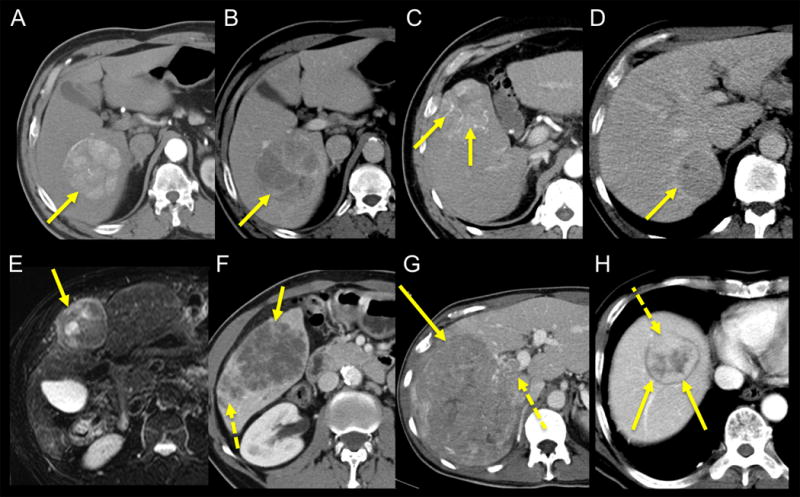
Phenotypic imaging traits in HCC illustrated in different patients with HCC. A, B: 58 year-old male. Contrast-enhanced CT obtained during arterial (A) and portal venous (B) phases demonstrates HCC in right hepatic lobe (arrows) with wash-in/wash-out pattern and mosaic appearance, without capsule/pseudocapsule. C: 68 year-old male. Contrast-enhanced CT obtained during arterial phase demonstrates HCC in segment 5 with internal arteries (arrows) (D). D: 51 year-old male. Contrast-enhanced CT obtained during portal venous phase demonstrates HCC in segment 6 with pseudo-capsule (arrow). E: 69 year-old male. Fat suppressed T2-weighted MR image demonstrates mildly T2 hyperintense encapsulated HCC in segment 4, with mosaic appearance (arrow). F: 62 year-old male. Contrast-enhanced CT obtained during portal venous phase demonstrates infiltrative HCC in right posterior lobe (arrow) with internal necrosis and satellite lesions posteriorly (dashed arrow). G: 57 year-old male. CT obtained during portal venous phase demonstrates large infiltrative HCC in right hepatic lobe (arrow) with right portal vein invasion (dashed arrow). H: 69 year-old male. CT obtained during portal venous phase demonstrates encapsulated HCC in hepatic dome (arrows) with extra-nodular growth anteriorly (dashed arrow).
Additionally, regions of interest (ROIs) were placed by a 3rd observer (MW) on the whole index lesions as large as possible to fit lesion size, avoiding vessels and artefacts using Osirix Software (Osirix ver. 4.1.2, pixmeo). A second ROI was drawn in the right hepatic lobe away from tumour. The tumour and liver signal intensities (on MR imaging, arbitrary units) or attenuations (on CT, Hounsfield units) were measured on post-contrast arterial and portal venous phases in order to compute tumour-to-liver contrast ratios (CR); and the tumour signal intensity (SI, arbitrary units) on MR imaging, or attenuation (att, Hounsfield units) on CT were measured on pre- and post-contrast images in order to calculate arterial and portal venous tumour enhancement ratios [ER AP (arterial phase)/ ER PVP (portal venous phase)] and the washout ratio (WOR), as below:
Tumour-to-liver CR (%)= (Lesion MRSI or CTatt – Liver MRSI or CTatt)/ Liver MRSI or CTatt
ER (%)= (MRSI or CTatt post-contrast - MRSI or CTatt pre-contrast)/ MRSI or CTatt pre-contrast
WOR (%)= (MRSI or CTatt AP - MRSI or CTatt PVP)/ MRSI or CTatt AP
Two MR imaging examinations were obtained at outside institutions, and were read off digitized film copies, therefore quantitative evaluation for these two cases was not possible.
Pathologic analysis
All of the surgical specimens were routinely processed, stained by hematoxylin and eosin, and were retrospectively reviewed by a liver pathologist (MIF) blinded to the imaging data. The maximum tumour size, tumour grade (well, moderately or poorly differentiated), and presence of microvascular invasion were assessed.
Analysis of gene expression profiles
Genome-wide gene-expression profiles were generated by using HG-U133 plus 2 DNA microarray (Affymetrix) and Human HT-12 beadarray (Illumina) as previously described (dataset is available at NCBI Gene Expression Omnibus database, accession number: GSE20140) [6]. A comprehensive collection of 13 HCC gene signatures in the literature capturing a variety of clinical, biological, and histological phenotypes was analyzed (Supplementary content) [23]. To circumvent the issue of DNA microarray platform difference that hampers direct merging of the gene expression matrices due to experimental batch difference, presence of each gene signature was separately determined in each dataset by using nearest template prediction algorithm [24] implemented in GenePattern genomic analysis toolkit (www.broadinstitute.org/genepattern) based on prediction confidence p-value cut-off of 0.05. Subsequently, the prediction results from the two datasets were merged for assessment of correlation with the imaging and other clinical features.
Statistical analysis
Quantitative data are presented as mean ± standard deviation (range). Qualitative data are presented as number of cases (percentage of cases). Correlation of the presence of the gene signatures with the imaging patterns, imaging quantitative parameters, pathology, and clinical features were quantitatively determined as odds ratios and p-values computed by logistic regression analysis, and visualized by Gene-E module of GenePattern. No sub-analysis of CT and MR imaging dataset was performed because of the small sample size of the study. All data analyses were performed by using GenePattern and R statistical language (www.r-project.org).
RESULTS
Patients’ characteristics
Clinico-pathological characteristics of our patient population are presented in Table 1. Most patients were males, all of them had chronic hepatitis C virus infection, and 30/38 (78.9%) had advanced fibrosis or cirrhosis. Most were classified as BCLC A (25/38=65.7%) or AJCC T2 (19/37=51.3%) (Pathologic information was not available in one patient).
Imaging and pathology characteristics
Imaging and pathology characteristics are presented in Table 1. Thirty-nine HCCs were assessed (mean size 5.7 ± 3.2 cm). In one patient with two HCCs (post-transplantation), both lesions were analysed for genomics signatures.
Typical enhancement profile (wash-in/wash-out) was observed in the majority of lesions (29/39=74.3%). The majority of lesions were nodular in morphology (30/39=76.9%), while 9 were infiltrative (Fig. 1). A capsule/pseudocapsule was observed in 18/39 (46.1%) lesions. On pathology, most of the lesions were poorly and moderately differentiated and 30/39 (76.9%) of lesions showed microvascular invasion. ER at the arterial and portal venous phases, WOR and tumour-to-liver contrast ratios are listed in Table 1.
Imaging/pathology and genomics correlations (Fig. 2, Table 2, Table 3)
Fig. 2.
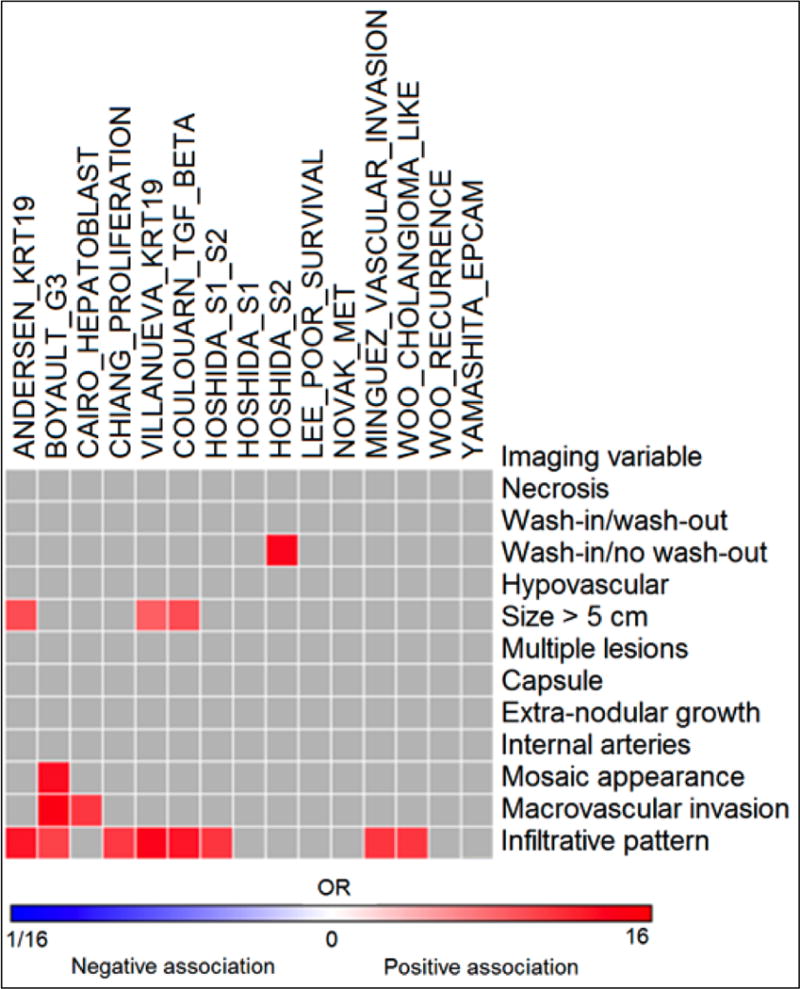
Heatmap of imaging-genomics correlations in 39 HCCs (see also Table 2). High odds ratios were observed for certain phenotypic traits. For example, infiltrative pattern was associated with several gene signatures, such as Minguez_Vascular invasion [9], Boyault_G3 [25], Cairo_Hepatoblastoma [47], Coulouarn_TGF beta [28], Chiang_Proliferation [22] and Hoshida S1/S2 expression [12]. Odds ratios with p <0.05 are shown in the Heatmap.
Table 2.
Association between imaging traits and gene expression profiles (from logistic regression analysis). Data are listed in ascending odds ratio (OR) values for each variable. Only associations with confidence intervals (CI) not overlapping with 1 and P <0.05 are shown.
| Imaging trait | Genomic variable | OR | 95% CI | P |
|---|---|---|---|---|
| Infiltrative pattern | Boyault_G3 [25] | 6.05 | 1.06-34.38 | 0.042 |
| Chiang_Proliferation [22] | 6.57 | 1.30–33.33 | 0.023 | |
| Hoshida_S1/S2 [12] | 7.00 | 1.22–40.09 | 0.029 | |
| Minguez_Vascular invasion [9] | 7.00 | 1.22–40.09 | 0.029 | |
| Woo_Cholangiocarcinoma_like [26] | 7.00 | 1.22–40.09 | 0.029 | |
| Andersen_KRT19 [27] | 9.63 | 1.64–56.37 | 0.012 | |
| Coulouarn_TGF-beta [28] | 9.63 | 1.64–56.37 | 0.012 | |
| Villanueva_KRT19 [6] | 11.50 | 1.93–68.52 | 0.007 | |
| Macrovascular invasion | Cairo_Hepatoblastoma [47] | 6.75 | 1.19–38.41 | 0.031 |
| Boyault_G3 [25] | 12.73 | 1.38–117.27 | 0.025 | |
| Size > 5 cm | Villanueva_KRT19 [6] | 4.44 | 1.08–18.36 | 0.039 |
| Andersen_KRT19 [27] | 5.50 | 1.32–22.86 | 0.019 | |
| Coulouarn_TGF-beta [28] | 5.50 | 1.32–22.86 | 0.019 | |
| Mosaic appearance | Boyault_G3 [25] | 10.46 | 1.16–94.48 | 0.037 |
| Wash-in / no wash-out | Hoshida_S2 [12] | 11.25 | 1.42–89.26 | 0.022 |
| Enhancement ratio (arterial phase) | Yamashita_EpCAM [48] | 0.96 | 0.93–0.99 | 0.033 |
| Cairo_Hepatoblastoma [47] | 0.96 | 0.93–0.99 | 0.044 | |
| Tumour-to-liver contrast ratio (arterial phase) | Yamashita_EpCAM [48] | 0.96 | 0.94–0.99 | 0.046 |
| Cairo_Hepatoblastoma [37] | 0.95 | 0.91–0.99 | 0.023 | |
| Woo_Cholangiocarcinoma_like [26] | 0.97 | 0.95–0.99 | 0.037 | |
| Tumour-to-liver contrast ratio (portal venous phase) | Andersen_KRT19 [27] | 0.91 | 0.85–0.98 | 0.017 |
| Boyault_G3 [25] | 0.92 | 0.86–0.98 | 0.022 | |
| Cairo_Hepatoblastoma [47] | 0.92 | 0.86–0.99 | 0.031 | |
| Villanueva_KRT19 [6] | 0.91 | 0.84–0.98 | 0.017 | |
| Coulouarn_TGF-beta [28] | 0.90 | 0.83–0.98 | 0.014 | |
| Hoshida_S1 [12] | 0.93 | 0.87–0.99 | 0.03 | |
| Lee_Poor survival [49] | 0.93 | 0.87–0.99 | 0.03 | |
| Novak_Met [50] | 0.93 | 0.88–0.99 | 0.043 | |
| Villanueva_KRT19 [6] | 0.94 | 0.89–0.99 | 0.045 | |
| Woo_Cholangiocarcinoma_like [26] | 0.91 | 0.85–0.99 | 0.019 | |
| Woo_Recurrence [51] | 0.89 | 0.81–0.97 | 0.012 |
Table 3.
Association between clinicopathologic stage and gene expression profiles (from logistic regression analysis). Data are listed in ascending odds ratio (OR) values for each clinicopathologic stage. Only associations with confidence intervals (CI) not overlapping with 1 and P <0.05 are shown.
| Tumour stage | Genomic variable | OR | 95% CI | P |
|---|---|---|---|---|
| AJCC | Minguez_Vascular invasion [9] | 4.50 | 1.05-19.22 | 0.042 |
| Villanueva_KRT19 [6] | 4.80 | 1.13-20.46 | 0.034 | |
| Novak_Met [50] | 6.00 | 1.18-30.58 | 0.031 | |
| Andersen_KRT19 [27] | 6.75 | 1.51-30.16 | 0.012 | |
| Coulouarn_TGF-beta [28] | 6.75 | 1.51-30.16 | 0.012 | |
| Boyault_G3 [25] | 13.36 | 2.33-76.48 | 0.004 | |
| BCLC | Villanueva_KRT19 [6] | 5.07 | 1.19-21.51 | 0.028 |
| Novak_Met [50] | 6.29 | 1.24-31.96 | 0.027 | |
| Boyault_G3 [25] | 7.08 | 1.52-33.03 | 0.013 |
AJCC: American joint committee on cancer
BCLC: Barcelona Clinic Liver Cancer
AJCC: T1/T2 vs. T3/T4
BCLC: A vs. B/C
Significant positive associations were observed between imaging traits such as infiltrative pattern, mosaic appearance, presence of macrovascular invasion, size >5 cm and gene signatures (odds ratios range 4.44–12.73, P ≤0.042 from univariate analysis). The infiltrative pattern showed the highest number of positive associations with several gene signatures (Boyault_G3 [25], Chiang_Proliferation [22], Hoshida_S1/S2 [12], Minguez_Vascular invasion [9], Woo_Cholangiocarcinoma_like [26], Andersen_KRT19 [27], Coulouarn_TGF-beta [28], Villanueva_KRT19 [6]) (Table 2, Fig. 3, Supplementary Table 1). The highest correlation was observed between macrovascular invasion on imaging and Boyault_G3 expression [25] (odds ratio 12.73, P=0.025). We found also a positive correlation between the Hoshida_S2 [12] and atypical enhancement pattern (wash-in without wash-out). Negative weak associations were found between ER at the AP and certain gene signatures; and between tumour-to-liver contrast ratios and several gene signatures (Table 2). No significant association was found between WOR and ER at the PVP and gene signatures. We also observed positive associations between each of clinicopathologic stages/pathologic grade/size with multiple gene signatures (Table 3, Table 4, Supplementary Table 2, Supplementary Table 3).
Fig. 3.
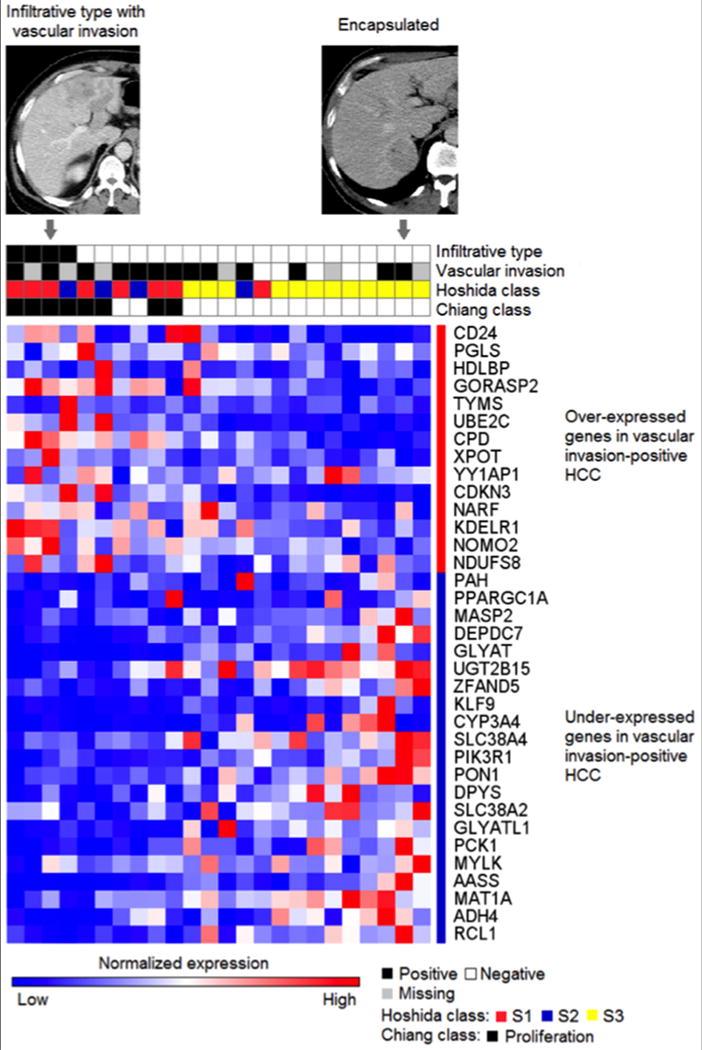
Representative expression pattern of vascular invasion signature genes shown with imaging and histology features for the samples profiled with the Affymetrix DNA microarray platform (n=24). Transcriptome subclasses of more aggressive HCC, Hoshida S1/S2 classes [12] and Chiang_Proliferation class [22], show overlap with the Minguez_Vascular invasion signature [9]. Representative contrast-enhanced CT images of infiltrative and encapsulated types are shown.
Table 4.
Association between histopathologic findings and gene expression profiles (from logistic regression analysis). Data are listed in ascending odds ratio (OR) values for each histopathologic finding. Only associations with CI not overlapping with 1 and P < 0.05 are shown.
| Pathologic traits | Gene signature | OR | 95% CI | P |
|---|---|---|---|---|
| Size >5 cm | Boyault_G3 [25] | 3.93 | 1.03–15.00 | 0.045 |
| Lee_Poor survival [49] | 4.86 | 1.21–19.47 | 0.026 | |
| Chiang_Proliferation [22] | 5.06 | 1.20–21.42 | 0.028 | |
| Villanueva_KRT19 [6] | 6.43 | 1.51–27.45 | 0.012 | |
| Andersen_KRT19 [27] | 8.25 | 1.90–35.91 | 0.005 | |
| Coulouarn_TGF beta [28] | 8.25 | 1.90–35.91 | 0.005 | |
| Tumour grade (poorly differentiated) | Hoshida_S1 [12] | 4.50 | 1.04–19.39 | 0.044 |
| Yamashita_EpCAM [48] | 4.50 | 1.04–19.39 | 0.044 | |
| Boyault_G3 [25] | 4.71 | 1.18–18.86 | 0.028 | |
| Lee_Poor survival [49] | 5.67 | 1.37–23.46 | 0.017 | |
| Minguez_Vascular invasion [9] | 5.87 | 1.43–24.11 | 0.014 | |
| Woo_Cholangiocarcinoma_like [26] | 5.87 | 1.43–24.11 | 0.014 | |
| Andersen_KRT19 [27] | 9.90 | 2.18–44.98 | 0.003 | |
| Coulouarn_TGF beta [28] | 9.90 | 2.18–44.98 | 0.003 | |
| Hoshida_S1/S2 [12] | 10.20 | 2.26–46.09 | 0.003 | |
| Chiang_Proliferation [22] | 10.56 | 2.17–51.42 | 0.004 | |
| Villanueva_KRT19 [6] | 13.93 | 2.78–69.88 | 0.001 | |
| Cairo_Hepatoblastoma [47] | 16.33 | 1.75–152.80 | 0.014 | |
| Novak_Met [50] | 21.00 | 2.25–195.79 | 0.008 |
DISCUSSION
In this initial study, we have correlated imaging phenotypes with gene expression profiles in 39 HCCs. We have observed positive associations between imaging phenotypes of aggressive disease and certain gene expression signatures.
Genomic and molecular features of HCC tumours and their biological, histological, and prognostic implication has been extensively studied by analysing transcriptome deregulations, structural aberrations of genomic DNA, and epigenetic deregulations [29]. These studies revealed molecular subclasses and signatures that depict heterogeneity of HCC tumours across patients, and highlighted the necessity to determine molecular characteristics of each tumour for potential personalized/stratified management of patients. For example, molecular signatures of pathway deregulations may inform indication of molecular targeted agents as anti-HCC drugs as companion biomarkers or identify patients to be enrolled in clinical trials of experimental therapies [16]. However, genomic profiling analysis requires tissue sampling, which is invasive, and is not mandatory for HCC diagnosis in case of typical HCC in a cirrhotic liver [30]. Therefore, less invasive surrogates of the molecular signatures will have a great potential in translating the genomic discoveries to refine clinical patient care. In the current study, we aimed at exploring practical clinical utility of imaging, using basic imaging patterns, easily identified, as surrogate markers of clinically relevant genomic/molecular signatures. We restricted our analysis to clinically well-established imaging features and molecular signatures in literature representing well-characterized biological, histological, and prognostic phenotypes in HCC.
First, we found significant positive associations between imaging traits of aggressive disease such as infiltrative pattern, mosaic appearance, presence of macrovascular invasion, large size and signatures with aggressive genotype. The infiltrative type is one of the major imaging features strongly associated with signatures with increased cellular proliferation (Boyault_G3, Coulouarn_TGF-beta, Chiang_proliferation, Hoshida_S1/S2), expression of biliary lineage markers such as cytokeratine 19 (Andersen_KRT19, Villanueva_KRT19, Woo-cholangiocarcinoma_like), and vascular invasion (Minguez_vascular invasion) [9]. Although it should be confirmed in larger studies, the high association between the infiltrative type and the vascular invasion gene expression suggests that the imaging-based determination of infiltrative type serves as a surrogate marker indicating high likelihood of microvascular invasion. Indeed, it is well known that infiltrative HCC has worst outcome than nodular HCC and is associated with vascular invasion [31, 32]. Nowadays, the prediction of microvascular invasion can be determined only based on post-surgical histological assessment, even if some recent publications showed that imaging patterns may be able to predict it [33, 34]. As the Metroticket Investigator Study Group proposed to use two distinct charts in predicting prognosis after liver transplantation for HCC according to the presence or absence of micro-vascular invasion [35], non-invasive imaging-based surrogate marker of microvascular invasion may potentially have a significant impact by enabling pre-surgical estimation of microvascular invasion to make more precise prognostic prediction and more rational donor organ allocation.
The presence of macrovascular invasion was associated with the molecular signatures of aggressive HCC tumours, including Boyault_G3 signature [36], in which the signature was tightly associated with the vascular invasion, distant metastases and staging of the tumour. Moreover, when this signature was present, the prognosis was impaired, as it is in case of macrovascular invasion on imaging [37]. A size larger than 5 cm is also an independent factor of poor prognosis [38]. As expected, in our cohort, the presence of a HCC larger than 5 cm was associated with 3 signatures of aggressive disease. Finally, the mosaic pattern was associated with a signature of proliferative lesion. The mosaic pattern reflects heterogeneity of the lesion and is mostly identified in large lesions [39], which are more often poorly differentiated and highly proliferative.
Surprisingly, an atypical pattern of HCC enhancement (presence of wash-in without wash-out) was associated with the Hoshida_S2 signature [30, 40]. Hoshida_S2 is a molecular subclass of HCC tumours defined by meta-analysis of transcriptome datasets representing worldwide HCC populations, which is characterized by increased cellular proliferation and over-expression of a cellular stemness marker gene (EpCAM) [12]. The atypical HCC without wash-out are in most of the cases, early HCC or well-differentiated HCC, with a good prognosis [41, 42] while the Hoshida-S2 signature is characterized by poor proliferation and is associated with a poor prognosis [12]. Thus, these counterintuitive results require further confirmation in a larger cohort of patients.
The correlations between enhancement ratios, tumour-to-liver contrast ratios, even if significant, were weak, and should be validated prospectively using quantitative DCE-MR imaging [43].
Our study has several limitations. First, in addition to its retrospective design, the sample size was small, mixing CT and MR imaging. This reflects our preliminary experience, and the observed associations between imaging and genomics should be verified in a larger study. Second, additional quantitative and qualitative imaging features such as diffusion [44], perfusion [43] and hypoxia [45] may need to be evaluated for their association with molecular and biological phenotypes of HCC tumours [46]. Third, we included only patients who had chronic HCV, which mean that our population may not represent a classic HCC population. Fourth, the image analysis was done in consensus which did not allow assessing for interobserver reproducibility.
In conclusion, this preliminary radiogenomics correlation study demonstrates potential clinical value of phenotypic imaging traits as surrogate markers of molecular phenotypes in HCC. These preliminary findings need to be verified in a larger study.
Supplementary Material
Key points.
There are associations between imaging and gene signatures of aggressive hepatocellular carcinoma.
Infiltrative type is associated with gene signatures of microvascular invasion and aggressiveness.
Infiltrative type may be a surrogate marker of microvascular invasion gene signature.
Acknowledgments
Grant support:
BT: NIH Grants U01 CA172320, 1R01 DK087877
YH: NIH Grant R01 DK099558, EU Grant ERC-2014-AdG-671231 HEPCIR, Irma T. Hirschl Trust, and U.S. Department of Defense W81XWH-16-1-0363
MW: Fondation ARC; contract grant number: SAE20140601302
JML: U.S. Department of Defense Grant (CA150272P3), European Commission Framework Program 7 (HEPTROMIC, proposal number 259744) and Horizon 2020 Program (HEPCAR, proposal number 667273-2), Asociación Española Contra el Cáncer (AECC), Samuel Waxman Cancer Research Foundation, Spanish National Health Institute (SAF2013-41027) and Grup de Recerca Consolidat –- Recerca Translacional en Oncologia Hepàtica. AGAUR (Generalitat de Catalunya), SGR 1162.
Abbreviations and acronyms
- AJCC
American joint committee on cancer
- AP
arterial phase
- BCLC
Barcelona clinic liver cancer
- CT
computed tomography
- ER
enhancement ratio
- HCC
hepatocellular carcinoma
- HIPAA
health insurance portability and accountability
- MR
magnetic resonance
- PVP
portal venous phase
- SI
signal intensity
- WOR
washout ratio;
References
- 1.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 3.van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 4.Hoshida Y, Toffanin S, Lachenmayer A, Villanueva A, Minguez B, Llovet JM. Molecular Classification and Novel Targets in Hepatocellular Carcinoma: Recent Advancements. Semin Liver Dis. 2010;30:35–51. doi: 10.1055/s-0030-1247131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villanueva A, Hoshida Y, Llovet JM. Hepatocellular carcinoma enters the sequencing era. Gastroenterology. 2011;141:1943–1945. doi: 10.1053/j.gastro.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villanueva A, Hoshida Y, Battiston C, et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501–1512. doi: 10.1053/j.gastro.2011.02.006. e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villanueva A, Hoshida Y. Depicting the role of TP53 in hepatocellular carcinoma progression. J Hepatol. 2011;55:724–725. doi: 10.1016/j.jhep.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 8.Toffanin S, Hoshida Y, Lachenmayer A, et al. MicroRNA-based classification of hepatocellular carcinoma and oncogenic role of miR-517a. Gastroenterology. 2011;140:1618–1628. doi: 10.1053/j.gastro.2011.02.009. e1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minguez B, Hoshida Y, Villanueva A, et al. Gene-expression signature of vascular invasion in hepatocellular carcinoma. J Hepatol. 2011;55:1325–1331. doi: 10.1016/j.jhep.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villanueva A, Hoshida Y, Toffanin S, et al. New strategies in hepatocellular carcinoma: genomic prognostic markers. Clin Cancer Res. 2010;16:4688–4694. doi: 10.1158/1078-0432.CCR-09-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoshida Y, Villanueva A, Llovet JM. Molecular profiling to predict hepatocellular carcinoma outcome. Expert Rev Gastroenterol Hepatol. 2009;3:101–103. doi: 10.1586/egh.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoshida Y, Nijman SM, Kobayashi M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villanueva A, Chiang DY, Newell P, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–1983. doi: 10.1053/j.gastro.2008.08.008. 1983 e1971-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007;27:55–76. doi: 10.1055/s-2006-960171. [DOI] [PubMed] [Google Scholar]
- 16.Nault JC, De Reynies A, Villanueva A, et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176–187. doi: 10.1053/j.gastro.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 17.Choi JY, Lee JM, Sirlin CB. CT and MR Imaging Diagnosis and Staging of Hepatocellular Carcinoma: Part II. Extracellular Agents, Hepatobiliary Agents, and Ancillary Imaging Features. Radiology. 2014;273:30–50. doi: 10.1148/radiol.14132362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi JY, Lee JM, Sirlin CB. CT and MR imaging diagnosis and staging of hepatocellular carcinoma: part I. Development, growth, and spread: key pathologic and imaging aspects. Radiology. 2014;272:635–654. doi: 10.1148/radiol.14132361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 20.Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol. 2007;18:821–831. doi: 10.1016/j.jvir.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 21.Miura T, Ban D, Tanaka S, et al. Distinct clinicopathological phenotype of hepatocellular carcinoma with ethoxybenzyl-magnetic resonance imaging hyperintensity: association with gene expression signature. Am J Surg. 2015;210:561–569. doi: 10.1016/j.amjsurg.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 22.Chiang DY, Villanueva A, Hoshida Y, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshida Y, Moeini A, Alsinet C, Kojima K, Villanueva A. Gene signatures in the management of hepatocellular carcinoma. Semin Oncol. 2012;39:473–485. doi: 10.1053/j.seminoncol.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Hoshida Y. Nearest template prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS One. 2010;5:e15543. doi: 10.1371/journal.pone.0015543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyault S, Rickman DS, de Reynies A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 26.Woo HG, Lee JH, Yoon JH, et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 2010;70:3034–3041. doi: 10.1158/0008-5472.CAN-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen JB, Loi R, Perra A, et al. Progenitor-derived hepatocellular carcinoma model in the rat. Hepatology. 2010;51:1401–1409. doi: 10.1002/hep.23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47:2059–2067. doi: 10.1002/hep.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoshida Y, Moeini A, Alsinet C, Kojima K, Villanueva A. Gene signatures in the management of hepatocellular carcinoma. Semin Oncol. 2012;39:473–485. doi: 10.1053/j.seminoncol.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Bruix J, Sherman M. American Association for the Study of Liver D. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yopp AC, Mokdad A, Zhu H, et al. Infiltrative Hepatocellular Carcinoma: Natural History and Comparison with Multifocal, Nodular Hepatocellular Carcinoma. Ann Surg Oncol. 2015;(22 Suppl 3):1075–1082. doi: 10.1245/s10434-015-4786-7. [DOI] [PubMed] [Google Scholar]
- 32.Mehta N, Fidelman N, Sarkar M, Yao FY. Factors associated with outcomes and response to therapy in patients with infiltrative hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2013;11:572–578. doi: 10.1016/j.cgh.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renzulli M, Brocchi S, Cucchetti A, et al. Can Current Preoperative Imaging Be Used to Detect Microvascular Invasion of Hepatocellular Carcinoma? Radiology. 2015:150998. doi: 10.1148/radiol.2015150998. [DOI] [PubMed] [Google Scholar]
- 34.Banerjee S, Wang DS, Kim HJ, et al. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology. 2015;62:792–800. doi: 10.1002/hep.27877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazzaferro V, Llovet JM, Miceli R, et al. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol. 2009;10:35–43. doi: 10.1016/S1470-2045(08)70284-5. [DOI] [PubMed] [Google Scholar]
- 36.Goossens N, Sun X, Hoshida Y. Molecular classification of hepatocellular carcinoma: potential therapeutic implications. Hepat Oncol. 2015;2:371–379. doi: 10.2217/hep.15.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franssen B, Alshebeeb K, Tabrizian P, et al. Differences in surgical outcomes between hepatitis B- and hepatitis C-related hepatocellular carcinoma: a retrospective analysis of a single North American center. Ann Surg. 2014;260:650–656. doi: 10.1097/SLA.0000000000000917. discussion 656-658. [DOI] [PubMed] [Google Scholar]
- 38.Liu PH, Hsu CY, Hsia CY, et al. Prognosis of hepatocellular carcinoma: Assessment of eleven staging systems. J Hepatol. 2016;64:601–608. doi: 10.1016/j.jhep.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 39.van den Bos IC, Hussain SM, Dwarkasing RS, et al. MR imaging of hepatocellular carcinoma: relationship between lesion size and imaging findings, including signal intensity and dynamic enhancement patterns. J Magn Reson Imaging. 2007;26:1548–1555. doi: 10.1002/jmri.21046. [DOI] [PubMed] [Google Scholar]
- 40.Ronzoni A, Artioli D, Scardina R, et al. Role of MDCT in the diagnosis of hepatocellular carcinoma in patients with cirrhosis undergoing orthotopic liver transplantation. AJR Am J Roentgenol. 2007;189:792–798. doi: 10.2214/AJR.07.2040. [DOI] [PubMed] [Google Scholar]
- 41.Witjes CD, Willemssen FE, Verheij J, et al. Histological differentiation grade and microvascular invasion of hepatocellular carcinoma predicted by dynamic contrast-enhanced MRI. J Magn Reson Imaging. 2012;36:641–647. doi: 10.1002/jmri.23681. [DOI] [PubMed] [Google Scholar]
- 42.Enomoto S, Tamai H, Shingaki N, et al. Assessment of hepatocellular carcinomas using conventional magnetic resonance imaging correlated with histological differentiation and a serum marker of poor prognosis. Hepatol Int. 2011;5:730–737. doi: 10.1007/s12072-010-9245-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taouli B, Johnson RS, Hajdu CH, et al. Hepatocellular carcinoma: perfusion quantification with dynamic contrast-enhanced MRI. AJR Am J Roentgenol. 2013;201:795–800. doi: 10.2214/AJR.12.9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taouli B, Koh DM. Diffusion-weighted MR imaging of the liver. Radiology. 2010;254:47–66. doi: 10.1148/radiol.09090021. [DOI] [PubMed] [Google Scholar]
- 45.Bane O, Besa C, Wagner M, et al. Feasibility and reproducibility of BOLD and TOLD measurements in the liver with oxygen and carbogen gas challenge in healthy volunteers and patients with hepatocellular carcinoma. J Magn Reson Imaging. 2016;43:866–876. doi: 10.1002/jmri.25051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hectors SJ, Wagner M, Bane O, et al. Quantification of hepatocellular carcinoma heterogeneity with multiparametric magnetic resonance imaging. Scientific Reports. doi: 10.1038/s41598-017-02706-z. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cairo S, Armengol C, De Reynies A, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 48.Yamashita T, Forgues M, Wang W, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 49.Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 50.Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woo HG, Park ES, Cheon JH, et al. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin Cancer Res. 2008;14:2056–2064. doi: 10.1158/1078-0432.CCR-07-1473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


