Abstract
Lead exposure remains a global public health issue, and the recent Flint water crisis has renewed public concern about lead toxicity. The toxicity of lead has been well established in a variety of systems and organs. The gut microbiome has been shown to be highly involved in many critical physiological processes, including food digestion, immune system development and metabolic homeostasis. However, despite the key role of the gut microbiome in human health, the functional impact of lead exposure on the gut microbiome has not been studied. The aim of this study is to define gut microbiome toxicity induced by lead exposure in C57BL/6 mice using multi-omics approaches, including 16S rRNA sequencing, whole genome metagenomics sequencing and gas chromatography-mass spectrometry (GC-MS) metabolomics. 16S rRNA sequencing revealed that lead exposure altered the gut microbiome trajectory and phylogenetic diversity. Metagenomics sequencing and metabolomics profiling showed that numerous metabolic pathways, including vitamin E, bile acids, nitrogen metabolism, energy metabolism, oxidative stress and the defense/detoxification mechanism, were significantly disturbed by lead exposure. These perturbed molecules and pathways may have important implications for lead toxicity in the host. Taken together, these results demonstrated that lead exposure not only altered gut microbiome community structures/diversity but also greatly affected metabolic functions, leading to gut microbiome toxicity.
Keywords: gut microbiome toxicity, omics, lead, metabolic functions
Graphical Abstract
Introduction
As one of the oldest known and most widely studied metals, lead has been recognized as a toxin since antiquity. The primary sources of lead exposure are water, food, soil, paint, leaded gasoline, food preparation utensils and electronic waste.1–3 The phasing out of lead in gasoline and the restriction of the amount of lead permitted in paint substantially lowered the blood lead levels in the U.S. population.4 However, the Flint water crisis has renewed public awareness of lead toxicity. After the city’s water source was changed from Lake Huron to the Flint River, increases were observed in the lead concentration in Flint’s water supply and the blood lead levels in children.5 Children are more susceptible to lead toxicity than adults, and cognitive development problems make childhood lead exposure a special public health concern.6 In addition to its neurobehavioral toxicity, lead exposure has destructive effects on the reproductive, hematopoietic, cardiovascular, gastrointestinal, circulatory, immunological and renal systems and deleterious effects on the urinary tract, liver and kidney.1, 7–10 Lead causes oxidative stress and impairs the antioxidant defense systems.11 Antioxidants, such as herbs, bioactive peptides, vitamin E and N-acetylcysteine, have been demonstrated to protect against lead toxicity.12, 13
The gut microbiome, which is also known as our “second genome”, has received much attention recently due to its involvement in human health and disease.14 The gut microbiome plays a key role in gut permeability and inflammation, energy harvest and lipid metabolism.15, 16 Disruption or dysbiosis of gut bacteria is associated with diverse diseases, such as allergies, gastric cancer, autism, obesity, anorexia, Crohn’s disease, inflammatory bowel disease and diabetes.17 Host-microbiome metabolic interactions can trigger biological effects both locally and systemically.18 For example, the gut microbiome produces short-chain fatty acids (acetate, propionate and butyrate), vitamins and other cofactors through the digestion of polysaccharides.19 The gut microbiome also produces metabolites through amino acid fermentation, such as cresol and indole, which can be toxic to the host.20 Likewise, the gut microbiome biotransforms primary hydrophilic bile acids to secondary hydrophobic bile acids in the large intestine via deconjugation, oxidation, epimerization and dihydroxylation.21 Bile acids can facilitate fat and fat-soluble vitamin absorption and maintain cholesterol homeostasis.22 Furthermore, bile acids are viewed as signaling molecules that bind to nuclear receptor FXR and the G-protein-coupled receptor TGR5.21, 23
The gut microbiome can be affected by many factors, including genetics, gender, age and antibiotics. Recent studies showed that the gut microbiome was disturbed by heavy metals, such as arsenic, cadmium and lead.24–27 A previous study investigated the impact of eight weeks of oral lead ingestion at a 100 or 500 ppm concentration on the gut bacterial compositions of mice via pyrosequencing of the 16S rRNA gene.28 However, the functional effects of lead on the gut microbiome remain unknown. Functional characterization of gut microbiome changes is clearly more important and informative than bacterial composition profiling by itself. Therefore, in the present study, we applied 16S rRNA sequencing, whole genome metagenomics sequencing and gas chromatography-mass spectrometry (GC-MS) metabolomics profiling to explore the effects of lead exposure on the gut microbiome and its functions and to define gut microbiome toxicity. The 16S rRNA sequencing provided the phylogenetic and taxonomic information needed to infer the microbial community structures. Whole genome metagenomics sequencing not only identifies the abundance and diversity of the microbial community but also reveals the gene contents and functional potential of genes encoded in the whole genomes of the microbial community.29 This approach can be coupled with metabolomic profiling to better understand the impact of lead exposure on the metabolic functions of the gut microbiome. Taken together, these system-level approaches reveal that lead exposure has a profound effect on gut microbiome development and numerous key metabolites and metabolic/cellular pathways of gut bacteria, leading to gut microbiome toxicity.
Materials and Methods
Animals and exposure
Specific pathogen-free C57BL/6 female mice (~7 weeks old) were purchased from Jackson Laboratory and housed at the University of Georgia animal facility for one week before the experiment. The mice were allowed to consume tap water ad libitum. Before and throughout the experimental period, the mice were housed under environmental conditions of 22°C, 40–70% humidity and a 12:12 hr light:dark cycle and were provided a standard rodent pellet diet. At the start of experimentation, the mice were randomly assigned to either a control or a lead-treated group (n=5). Lead chloride (Sigma-Aldrich, catalog number 449865) was administered to the mice in their drinking water at a concentration of 10 ppm (based on the lead content) for a study period of 13 weeks. This concentration was equivalent to ~2 mg/kg body weight/day. The control mice received water alone. The animal protocol was approved by the University of Georgia Institutional Animal Care and Use Committee. The animals were treated humanely with attention to the alleviation of their suffering.
16S rRNA gene sequencing
16S rRNA gene sequencing was performed as previously described using fecal pellets collected before exposure (day 0) and 4 weeks (week 4) and 13 weeks post-exposure (week 13).27 Briefly, total DNA was extracted from the fecal pellets collected during necropsy using the PowerSoil® DNA Isolation Kit according to the manufacturer’s instructions. For 16S rRNA gene sequencing, DNA was amplified using the 515F and 806R primers,30 which target the V4 region of the 16S rRNA gene, followed by normalization and barcoding. The resulting DNA was pooled, quantified using the Qubit 2.0 Fluorometer and sequenced at the Georgia Genomics Facility using an Illumina MiSeq (500 cycles v2 kit). Operational taxonomic unit (OTU) picking and the diversity analysis were performed with the Quantitative Insights into Microbial Ecology (QIIME) software package.
Metagenomics sequencing
DNA (10 ng/μL) was fragmented using the Bioruptor UCD-300 sonication device, followed by sequencing library construction using the KAPA Hyper Prep Kit according to the manufacturer’s instructions. The resulting DNA was pooled, quantified and sequenced at the Georgia Genomics Facility using an Illumina NextSeq High Output Flow Cell. Raw FASTQ files were imported into the MG-RAST metagenomics analysis server (version 3.5). The sequences were assigned to the M5NR Subsystems database for functional analysis with a maximum e-value cutoff of 10−5, a 75% minimum identity cutoff and a minimum alignment length cutoff of 35. Notably, metagenomics reveals changes only at the gene level and not at the protein level. The relative abundance of specific genes was determined by the MG-RAST pipeline with multiple steps to address inter-sample variability, including normalization using DEseq, log 2 transformation and scaling (0–1).
Metabolomics analysis
Metabolites were isolated using the previously described method with modifications27. Briefly, two fecal pellets (~20 mg) were vortexed with 1 mL of methanol/chloroform/water solution (2:2:1) for 20 minutes, followed by centrifugation at 1,000 x g for 15 minutes. The resulting upper and lower phases were transferred to a flat-bottomed high-performance liquid chromatography (HPLC) vials and dried for 4 hours in a SpeedVac, followed by derivatization with methoxyamine-HCl and N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA). Then, the samples were injected into an Agilent GC-MS that was running in full scan mode. XCMS was used for peak-picking, alignment and extraction of the peak intensities. Molecular features with significant changes (p<0.05 and fold change >1.5) were identified using the National Institute of Standards and Technology (NIST) Standard Reference Database and searched against the Human Metabolome Database to obtain function and pathway information.
Statistical analysis
Differences in the gut microbiome composition were analyzed using a nonparametric test in the Metastats software as previously described.31 Principle coordinate analysis (PCoA) was used to compare gut microbiome profiles between the control and treated samples and examine differences in beta diversity based on the UniFrac distance metric.32 Alpha diversity was also analyzed to examine the species richness in given samples. A two-tailed Welch’s t-test was used to initially profile individual metabolite differences between the control and treatment groups (p<0.05), as described elsewhere.27, 33 Partial least squares discriminant analysis (PLS-DA) was used to compare the metabolomics profiles between the control and lead-treated groups.
Results
Lead exposure disrupted gut microbiome trajectories
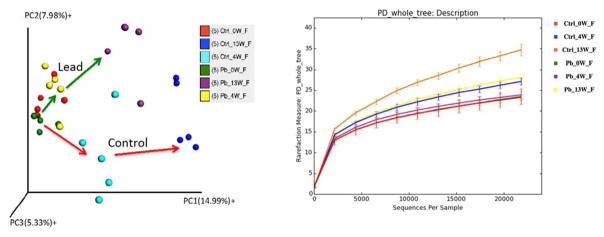
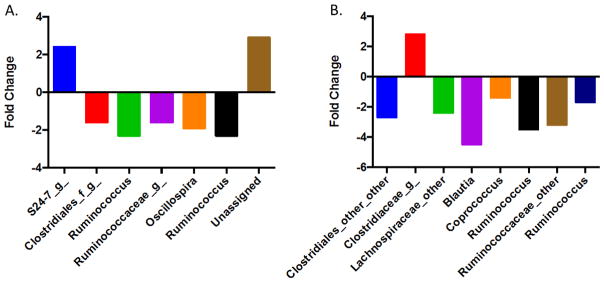
First, we used 16S rRNA gene sequencing to examine changes in the gut bacterial compositions over time using beta and alpha diversity metrics. Beta diversity evaluates the diversity in a microbial community between samples, whereas alpha diversity reflects the species richness in given samples. One of the key features of the gut microbiome is that it is a highly dynamic system, with the gut microbiome composition or abundance changing over time (Figure S1). A 3D PCoA plot (Figure 1A) showed that the trajectories of the gut microbiome community structures at the genus level were readily differentiated between the control and lead-treated mice. At day 0, the gut microbiome community structures were similar for all animals at baseline. Conversely, the microbial community structures were significantly different between the control and lead-treated animals at weeks 4 and 13. The different trajectories between the control and treated animals indicated a strong effect of lead on disrupting gut microbiome development. Consistently, the phylogenetic diversity or species richness of the control animals increased over time based on the phylogenetic diversity (PD) whole tree, which is an alpha diversity metric (Figure 1B). However, the alpha diversity increased to a much lesser extent in the lead-treated mice than in the control animals. The alpha diversity of the lead-treated animals at week 13 was equivalent to that of the control mice at week 4. These dynamic changes in alpha diversity demonstrated that disruption of the gut microbiome had already occurred at week 4 post-exposure, which was the earliest time point assessed. Notably, the alpha diversity was not significantly increased at week 4 compared to that at week 0 for the lead-treated animals, indicating that the development of phylogenetic diversity was severely inhibited by lead exposure. Figure 2 illustrated the fold changes of significantly perturbed gut bacterial genera that were induced by lead exposure at weeks 4 and 13 compared to the controls (see Tables S1 and S2 for individual variations in all bacterial genera). The majority of the altered bacterial genera were reduced due to lead exposure.
Figure 1.
Lead exposure disturbed the trajectories of gut microbiome development as assessed by beta diversity metrics (A). Lead exposure also reduced/inhibited the phylogenetic diversity of the gut bacteria as examined using alpha diversity metrics (B). Here, 16S rRNA gene sequencing was performed, followed by bacterial taxonomic assignment via QIIME. Beta diversity evaluated the diversity in the microbial community between samples, whereas alpha diversity reflected the species richness.
Figure 2.
The fold changes of selected significantly changed gut bacterial genera between the controls and lead-treated mice (A. 4 weeks post-lead exposure; B. 13 weeks post-lead exposure). The fold changes were calculated using the group means for each bacterial genus.
Lead exposure reduced vitamin E and bile acids in the gut microbiome
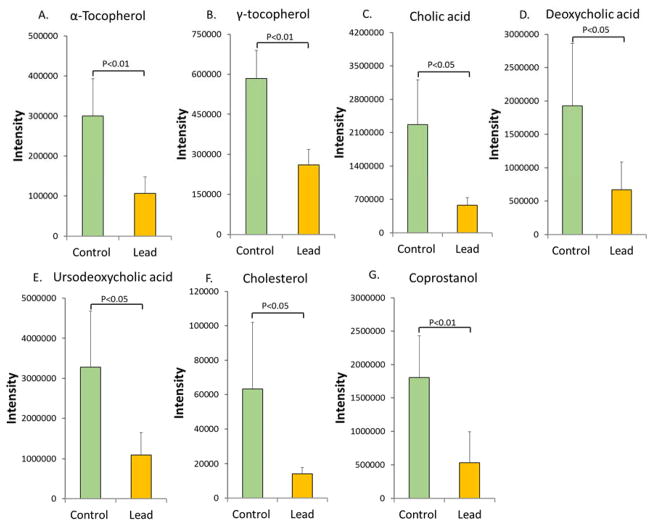
Since the gut microbiome plays a key role in metabolic processing, next we used metabolomics to examine metabolic changes associated with gut microbiome perturbations. Lead exposure perturbed the metabolic profiles of the gut microbiome, leading to the identification of 1314 molecular features with p<0.05 and fold changes >1.5. The control and lead-treated groups could be readily differentiated using metabolite features (Figure S2), and excellent separation of the control and lead-treated animals was achieved using the first two components of the PLS-DA. Metabolites with diverse structures were identified (Table S3). For example, two forms of vitamin E (α-tocopherol and γ-tocopherol), the primary bile acids cholic acid (CA) and ursodeoxycholic acid (UDCA), the secondary bile acid deoxycholic acid (DCA) and cholesterol and its derivative coprostanol were significantly down-regulated, as shown in Figure 3.
Figure 3.
The abundance of key metabolites was measured by GC-MS to examine the impact of lead exposure on the metabolic functions of the gut microbiome. Vitamin E, bile acids and cholesterol and its derivative were significantly reduced in mice after exposure to lead for 13 weeks (A. α-tocopherol, B. γ-tocopherol, C. cholic acid, D. deoxycholic acid, E. ursodeoxycholic acid; F. cholesterol; and G. coprostanol).
Lead exposure altered the nitrogen metabolism of gut bacteria
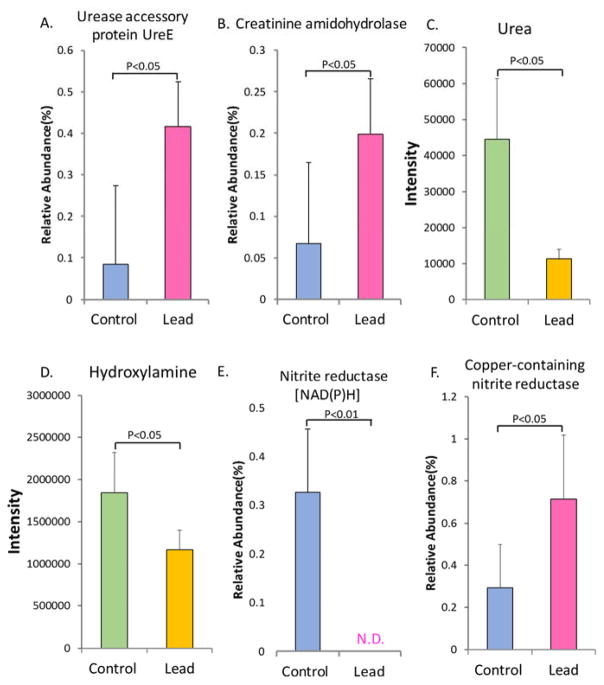
Nitrogen metabolism is a key component of the metabolic functions in the gut microbiome. We used metagenomics and metabolomics to examine whether lead exposure altered pathways and metabolites related to nitrogen metabolism. As shown in Figure 4A, the lead-treated group had a higher level of UreE gene than the control group. UreE is an accessory protein of urease and plays a key role in urease activation.34, 35 The abundance of gene encoding creatinine amidohydrolase was also increased (Figure 4B); this gene is involved in nitrogen metabolism by catalyzing creatinine to creatine.36, 37 Urea was significantly reduced in the gut bacteria after lead exposure (Figure 4C). Lead exposure also led to a significant decrease in hydroxylamine (Figure 4D), which is a precursor of nitrite in bacteria. Simultaneously, the gene encoding nitrite reductase [NAD(P)H] (Figure 4E), which reduces nitrite to ammonium, decreased, whereas the gene encoding copper-containing nitrite reductase, which catalyzes nitrite to nitric oxide, increased (Figure 4F).
Figure 4.
Metagenomics and metabolomics analyses showing that lead exposure significantly alters the nitrogen metabolism of the gut bacteria, as evidenced by the perturbed key genes and metabolites (A. urease accessory protein UreE; B. creatinine amidohydrolase; C. urea; D. hydroxylamine; E. nitrite reductase [NAD(P)H]; and F. copper-containing nitrite reductase). N.D.: non-detectable.
Lead exposure altered energy metabolism
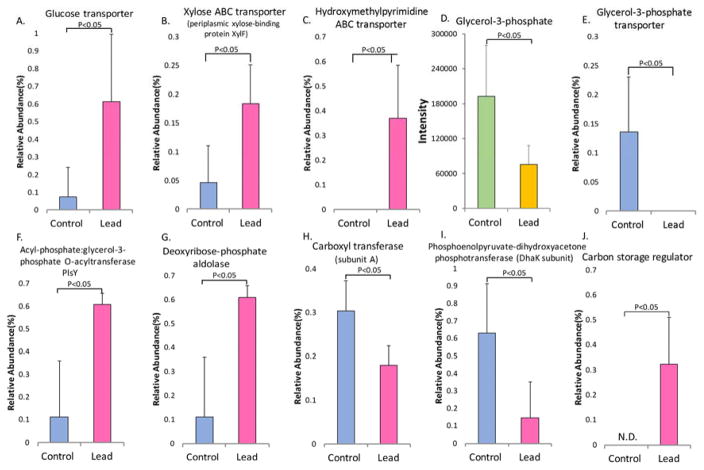
The gut microbiome plays a key role in regulating energy metabolism and harvest for the host. Therefore, examining the effects of lead on energy metabolism by the gut microbiome would be highly informative. The impact of lead exposure on energy metabolism is shown in Figure 5. Carbon metabolism was significantly perturbed by lead exposure, with saccharide transport being highly activated. The abundance of related genes, including the glucose transporter, xylose ABC transporter and hydroxymethyl pyrimidine ABC transporter, was increased in the gut bacteria of the lead-treated mice (Figures 5A–C). A key metabolite of cellular energy metabolism (glycerol-3-phosphate) was reduced (−2.5-fold, Figure 5D). The gene encoding the glycerol-3-phosphate transporter decreased (Figure 5E), whereas the gene encoding the acyl-phosphate:glycerol-3-phosphate O-acyltransferase PlsY increased following lead exposure; this latter enzyme uses glycerol-3-phosphate as a substrate (Figure 5F).38 Conversely, the gene encoding deoxyribose-phosphate aldolase increased, as shown in Figure 5G; this enzyme can catalyze the lysis of 2-deoxyribose-phosphate to eventually produce glycerol-3-phosphate. Two genes encoding pyruvate carboxyl transferase subunit A and phosphoenolpyruvate-dihydroxyacetone phosphotransferase, which are involved in gluconeogenesis, were down-regulated (Figure 5H and 5I).39, 40 Correspondingly, the carbon storage regulator, which negatively regulates gluconeogenesis, was significantly increased (Figure 5J). This finding indicates that gluconeogenesis may be inhibited after lead exposure.
Figure 5.
Energy metabolism, which is a key metabolic function of the gut microbiome, was disturbed by lead exposure, as demonstrated by the altered abundance of a number of bacterial genes and the key metabolite glycerol-3-phosphate. N.D.: non-detectable. Note: the relative abundance reflects only the gene levels and not the protein levels.
Lead induced oxidative stress and activated the defense/detoxification mechanism in gut bacteria
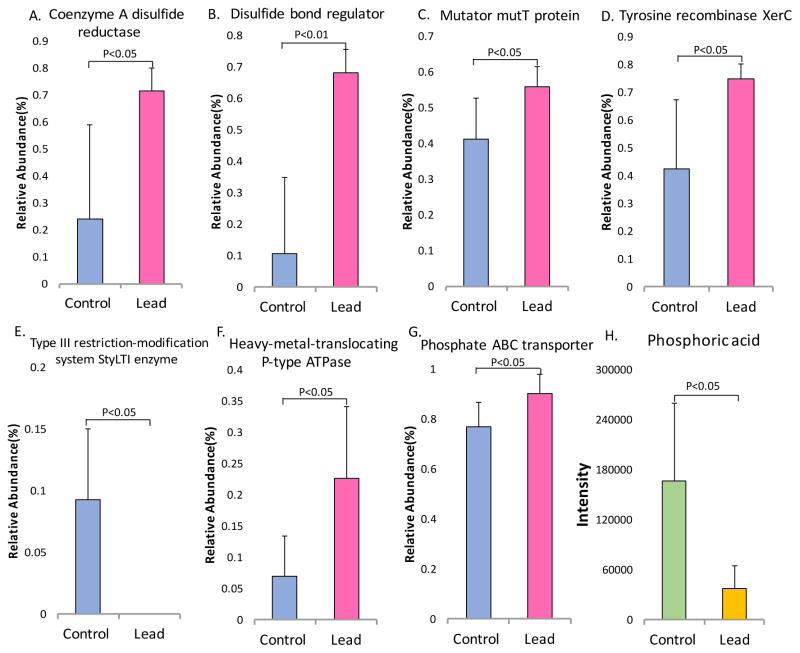
Lead is known to induce oxidative stress, but whether lead can also cause oxidative stress responses in the gut microbiome is unknown. Therefore, we combined metagenomics and metabolomics to define the role of lead exposure in oxidative stress and the associated detoxification mechanisms in the gut microbiome. Figure 6 shows that lead exposure induced oxidative stress and activated the defense mechanism in the gut microbiota. The primary toxic effects of lead are due to oxidative stress.41, 42 As shown in Figure 6, the abundance levels of several genes involved in resisting oxidative stress were perturbed after lead exposure. For instance, the gene encoding coenzyme A disulfide reductase (CoADR) was induced by lead exposure (Figure 6A). CoADR is a key gene in redox systems in some bacteria that plays a key role in maintaining the reduced state of cells and is involved in the robust oxygen defense systems.43, 44 Another related gene (the disulfide-bond regulator) was also increased in the lead-treated animals (Figure 6B). Additionally, MutT was increased in the lead-treated mice (Figure 6C); this gene encodes a hydrolase that converts 8-oxodGTP to 8-oxodGMP and thus is involved in preventing the incorporation of the 8-oxo-Gua lesion into the DNA.45, 46 In addition to the induction of DNA damage by oxidative stress, the abundance of some genes involved in DNA modification changed after lead exposure. For example, the tyrosine recombinase gene XerC was significantly increased in the gut bacteria of the lead-treated mice, whereas the gene encoding the type III restriction-modification system StyLTI enzyme was reduced (Figures 6D and E).
Figure 6.
Alterations in key genes and metabolites involved in oxidative stress and the defense response indicate that lead exposure activates cellular defense genes (A and B), DNA repair systems (C, D and E), and detoxification pathways (F, G and H).
Defense mechanisms were activated in gut bacteria in response to lead exposure. For example, the gene encoding the heavy metal-translocating P-type ATPase, which is involved in lead efflux in bacteria, was up-regulated (Figure 6F).47, 48 Likewise, the gene encoding the phosphate ABC transporter was significantly increased (Figure 6G), which was consistent with the highly reduced phosphoric acid level (Figure 6H). These findings suggest that gut bacteria may increase lead precipitation by increasing the extracellular phosphoric acid concentration.49, 50
Discussion
We combined 16S rRNA gene sequencing, whole genome metagenomics sequencing and metabolomics to study the effects of lead exposure on the gut microbiome and its metabolic functions. We demonstrated that lead exposure changed the gut microbiome taxonomic composition and the functional metagenome and metabolic profiles in C57BL/6 mice. These results highlighted the functional impact of lead exposure on the gut microbiome. Moreover, lead perturbed the trajectories of the gut microbiome over time and significantly reduced or inhibited the normal development of gut bacterial phylogenetic diversity. Additionally, we identified a number of important perturbed metabolites and metabolic pathways. These metabolic and cellular signaling pathways include bile acids, vitamins, nitrogen metabolism, oxidative stress and defense mechanisms and energy metabolism, which may have important implications for lead-induced diseases.
We clearly demonstrated that lead exposure altered gut microbiome community structures. In particular, lead interfered with the normal development of gut bacteria over time. We showed that lead exposure decreased or inhibited the establishment of normal phylogenetic diversity in the gut microbiome, as illustrated in Figure 2, which was an extremely important observation. The establishment and development of the gut microbiome is a temporal and sensitive process in children. Having a normal gut microbiome is critical for many physiological processes, including immune system development, metabolic processing, energy production, food digestion and epithelial homeostasis. Lead exposure in children is common, and children were found to have increased blood lead levels in the recent Flint water crisis. Thus, lead could significantly impact gut microbiome development in children, which may contribute to many lead-induced diseases in later life. Clearly, further study is warranted to shed light on this possibility.
We found that the abundance of both primary and secondary bile acids was significantly reduced by lead exposure (Figure 3). The gut microbiome can transform primary hydrophilic bile acids into secondary hydrophilic bile acids in the large intestine through deconjugation, dehydroxylation and dehydrogenation.51 Here, we observed that the secondary bile acid DCA was decreased by 2.9-fold in the lead-treated mice relative to that in the control mice. DCA is biochemically synthesized from the primary bile acid CA, which was reduced 3.9-fold in the lead-treated mice compared to that in the controls (Figure 3C). Clearly, bile acid homeostasis is significantly altered by lead exposure, which may affect many other biological processes due to the key roles of bile acids. As signaling molecules, bile acids bind or activate host nuclear receptors to regulate a number of downstream signaling pathways.20 Bile acids exert both hepatic and extra-hepatic effects ranging from regulating their own biosynthesis to lipid, cholesterol, glucose, lipoprotein, and energy metabolism, local gut mucosal defenses and inflammatory responses.23, 52 Primary bile acids are formed from cholesterol by multiple enzymes through the modification of the sterol ring and oxidation and shortening of the side chains.53 In turn, the formation of bile acids maintains cholesterol homeostasis.54 Here, we observed that cholesterol was decreased by 4.5-fold in the lead-treated mice (Figure 3F). In addition to cholesterol reduction, we also observed that the cholesterol derivative coprostanol was decreased in a corresponding manner (Figure 3E). A previous study identified specific gut bacteria associated with fecal cholesterol and coprostanol using integrated metabolomics and microbiome analyses.55 Specifically, a total of 63 gut microbes were identified, including a number of Ruminococcus and Lachnospiraceae species.55 In the present study, we found that Ruminococcus and several Lachnospiraceae genera were significantly reduced (Figure 2). Thus, lead exposure greatly affected the gut bacterial genera that are highly involved in regulating the homeostasis of cholesterol and its derivatives.
Vitamin E is part of a family of eight essential fat-soluble nutrients. Four of the compounds carry a tocol structure, and the other four carry a tocotrienol structure.56 A reduction in two forms of vitamin E was observed in the lead-treated mice compared with the levels in the control mice. The α-tocopherol and γ-tocopherol levels were decreased by 2.8 and 2.2-fold, respectively. Vitamin E exhibits both antioxidant and non-antioxidant activities, such as gene expression modulation, cell proliferation, platelet aggregation and bone mass regulation.57 Vitamin E absorption can be affected by many factors, including gut permeability, dietary fat availability and a bile salt-dependent carboxyl ester hydrolase.56, 58, 59 Since the gut microbiome affects gut permeability,60 lipid metabolism61 and the bile acid pool,52 the disturbance of the gut microbiome by lead may affect vitamin E homeostasis. Perturbed bile acid pools in lead-treated mice may be attributed to alterations in bile salt-dependent enzymatic hydrolysis for vitamin E. Consequently, the absorption of vitamin E could be disrupted. Consistent with our results, a previous study showed that the monocolonization of germ-free mice affected vitamin E metabolism.62 Likewise, previous studies showed that vitamin E could protect cells against lead-induced oxidative stress;2, 13 thus, the reduction in vitamin E may also reflect oxidative stress in the gut bacteria arising from lead exposure. Interestingly, a few previous human studies found an inverse relationship between the vitamin E level and the lead concentration in the blood.63–65 Here, we found reduced vitamin E levels in the gut microbiome in the lead-exposed animals, suggesting a potential link between vitamin E in the gut microbiome and host tissues. Future studies are needed to elucidate the role of the gut microbiome in the lead-perturbed homeostasis of vitamin E in host tissues.
In mammals, nitric oxide (NO) is involved in various physiological and pathophysiological events in multiple organs, including the gastrointestinal tract.66, 67 Endogenous NO in the gut regulates the mucosal blood flow, mucus generation, gut motility and host defense responses.66 Recent studies have revealed that gut bacteria can also generate NO and may play an important role in gut physiology.68 As shown in Figure 4, we found that the gene encoding copper-containing nitrite reductase, which catalyzes the conversion of nitrite to NO, increased after lead exposure. Conversely, we observed decreased abundance of the nitrite reductase [NAD(P)H] gene, which is involved in an alternative nitrite metabolism pathway. These results may indicate that lead exposure increases NO generation in the gut bacteria.
The energy metabolism of the gut bacteria was also dramatically perturbed by lead exposure. Our data revealed that genes related to saccharide absorption were highly activated, which should increase the energy levels in the gut bacteria. However, other genes, such as laminarinase, cyclomaltodextrin glucanotransferase, 5-deoxy-glucuronate isomerase, D-malate dehydrogenase and glycerol-3-phosphate transporter, were decreased by lead treatment. These genes generally promote the degradation of saccharides. In addition, the glycerol-3-phosphate level was reduced. Two gluconeogenesis-related genes (pyruvate carboxyl transferase subunit A and phosphoenolpyruvate-dihydroxyacetone phosphotransferase) were decreased after lead exposure, which was consistent with the up-regulation of the carbon storage regulator gene that suppressed gluconeogenesis, glycogen biosynthesis and catabolism and activated glycolysis and acetate metabolism.69, 70 Taken together, these data show that lead exposure disturbs energy production and causes energy starvation in gut bacteria, although the specific perturbed steps of glycolysis or respiration are unknown. The significant decrease in fatty acids, hexadecanoic acid and linoleic acid in lead-treated gut bacteria (Table S3) also partially supports the finding of an energy-deprived condition in the gut bacteria.
Lead is known to induce oxidative stress in different systems.41, 42 Lead can generate reactive oxygen species to cause damage in cells.41 Two primary redox systems are widely adopted by bacteria for protection from oxidative stress: the glutathione thiol/disulfide redox system and the reduced CoA thiol/disulfide redox system.42, 43 Glutathione reductase (GSR) and coenzyme A disulfide reductase (CoADR) are the key enzymes of each redox system, respectively. Coenzyme A disulfide reductase (CoADR) is a key enzyme in an alternative redox system in some bacteria that do not contain the glutathione thiol/disulfide redox system; this system plays an important role in maintaining the reduced state of cells and is involved in robust oxygen defense systems.43, 44 Although we did not observe changes in GSR gene abundance, the up-regulation of the CoADR and disulfide-bond regulator genes supports the hypothesis that lead induces oxidative stress in gut bacteria. Likewise, the increased gene abundance of MutT, which is an oxidative DNA damage repair gene, suggests lead-induced oxidative stress. The consequences of lead-induced oxidative stress in gut bacteria are not well understood yet.
As a ubiquitous environmental toxicant, lead can cause severe cellular dysfunction in animals, plants and bacteria.71 Bacteria have evolved several mechanisms to resist lead, including adsorption by extracellular polysaccharides, precipitation as insoluble phosphates, and efflux to the cell exterior via metal-resistant ion channels.50 The gene encoding the heavy metal-translocating P-type ATPase significantly increased in the gut bacteria under lead exposure. The increased gene abundance of the phosphate ABC transporter gene and the greatly reduced phosphoric acid level indicated that gut bacteria might also resist lead by sequestering it in the form of phosphate salts, as shown in Figure 6. The activation of lead resistance mechanisms suggests an essential role for the gut microbiota in protecting the host from lead exposure.
An increase in multiple amino acids was evident in the lead-treated gut bacteria. Amino acids that increased after lead exposure included glycine, threonine, serine, glutamate, isoleucine, valine and proline. Only alanine was decreased. The metagenomics analysis revealed that a series of genes related to amino acid degradation and synthesis were altered by lead exposure. The abundance of genes encoding asparagine synthetase, glycine/sarcosine/betaine reductase protein A, pyruvate carboxyl transferase subunit A, putrescine carbamoyltransferase and ornithine aminotransferase was decreased, whereas the abundance of the methionine gamma-lyase and aspartate 1-decarboxylase genes was increased in the lead-treated group (Figure S3). No simple conclusion could be provided to explain how lead influenced the amino acid levels due to the complex regulation of amino acid homeostasis. One potential explanation is that lead induces oxidative stress in gut bacteria and increases the levels of oxidized proteins, which will largely be degraded by cells and thus increase the free amino acid levels.42, 72
In summary, we demonstrated that lead exposure significantly altered the gut microbiome trajectories over time, phylogenetic diversity, key metabolites and metabolic and cellular signaling pathways. These perturbed molecules and pathways may have important implications for lead toxicity in the host and lead-induced diseases. Many issues require further study. For example, what is the dose-dependent response to lead exposure, particularly at environmental and human-relevant concentrations? What are the health effects of lead-induced gut microbiome perturbations in the host? Likewise, what are the roles of host responses in mediating the gut microbiome and its functions? Future studies are warranted to address these intriguing questions to better understand the complex functional interactions among lead, the gut microbiome and the host.
Supplementary Material
Acknowledgments
Funding Information
The University of Georgia, University of North Carolina at Chapel Hill and the NIH/NIEHS provided partial financial support (R01ES024950) for this work.
Abbreviations
- GC-MS
Gas chromatography-mass spectrometry
- PCoA
Principle coordinate analysis
- PLS-DA
Partial least squares discriminant analysis
- CA
Cholic acid
- UDCA
Ursodeoxycholic acid
- DCA
Deoxycholic acid
- CoADR
Coenzyme A disulfide reductase
- GSR
Glutathione reductase
Footnotes
Relative abundance of typical gut bacterial genera in the measurements from the control and lead-exposed mice at weeks 13 and 4. The fecal metabolites were significantly changed based on GC-MS metabolomics. Gut microbiome community structures changed at multiple time points (baseline and weeks 4 and 13). Lead exposure perturbed the metabolic profiles of fecal samples from mice. Gut microbial amino acid homeostasis was disturbed by lead exposure. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Lockitch G. Perspectives on lead toxicity. Clinical Biochemistry. 1993;26:371–381. doi: 10.1016/0009-9120(93)90113-k. [DOI] [PubMed] [Google Scholar]
- 2.Patrick L. Lead toxicity, a review of the literature. Part 1: Exposure, evaluation, and treatment. ALTERNATIVE MEDICINE REVIEW. 2006;11:2–22. [PubMed] [Google Scholar]
- 3.Chen M, Ogunseitan OA, Wang J, Chen H, Wang B, Chen S. Evolution of electronic waste toxicity: Trends in innovation and regulation. Environment international. 2016;89–90:147–154. doi: 10.1016/j.envint.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Bellinger DC. Lead Contamination in Flint — An Abject Failure to Protect Public Health. New England Journal of Medicine. 2016 doi: 10.1056/NEJMp1601013. [DOI] [PubMed] [Google Scholar]
- 5.Hanna-Attisha M, LaChance J, Sadler RC, Champney Schnepp A. Elevated Blood Lead Levels in Children Associated With the Flint Drinking Water Crisis: A Spatial Analysis of Risk and Public Health Response. American journal of public health. 2016;106:283–290. doi: 10.2105/AJPH.2015.303003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evens A, Hryhorczuk D, Lanphear BP, Rankin KM, Lewis DA, Forst L, Rosenberg D. The impact of low-level lead toxicity on school performance among children in the Chicago Public Schools: a population-based retrospective cohort study. Environmental health : a global access science source. 2015;14:21. doi: 10.1186/s12940-015-0008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Navas-Acien A, Guallar E, Silbergeld EK, Rothenberg SJ. Lead exposure and cardiovascular disease--a systematic review. Environmental health perspectives. 2007;115:472–482. doi: 10.1289/ehp.9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahamed M, Siddiqui MK. Low level lead exposure and oxidative stress: current opinions. Clinica chimica acta; international journal of clinical chemistry. 2007;383:57–64. doi: 10.1016/j.cca.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 9.Riaz M, Mahmood Z, Shahid M, Saeed MU, Tahir IM, Shah SA, Munir N, El-Ghorab A. Impact of reactive oxygen species on antioxidant capacity of male reproductive system. International journal of immunopathology and pharmacology. 2015 doi: 10.1177/0394632015608994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papanikolaou NC, Hatzidaki EG, Belivanis S, Tzanakakis GN, Tsatsakis AM. Lead toxicity update. A brief review. MEDICAL SCIENCE MONITOR. 2005;11:RA329–RA336. [PubMed] [Google Scholar]
- 11.Stohs SJ, Bagchi D. Oxidative mechanisms in the toxicity of metal ions. Free Radical Biology and Medicine. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- 12.Dua TK, Dewanjee S, Khanra R, Joardar S, Barma S, Das S, Zia-Ul-Haq M, De Feo V. Cytoprotective and Antioxidant Effects of an Edible Herb, Enhydra fluctuans Lour. (Asteraceae), against Experimentally Induced Lead Acetate Intoxication. PLoS One. 2016;11:e0148757. doi: 10.1371/journal.pone.0148757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu PC, Guo YL. Antioxidant nutrients and lead toxicity. Toxicology. 2002;180:33–44. doi: 10.1016/s0300-483x(02)00380-3. [DOI] [PubMed] [Google Scholar]
- 14.Zmora N, Zeevi D, Korem T, Segal E, Elinav E. Taking it Personally: Personalized Utilization of the Human Microbiome in Health and Disease. Cell host & microbe. 2016;19:12–20. doi: 10.1016/j.chom.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 15.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 16.Devaraj S, Hemarajata P, Versalovic J. The human gut microbiome and body metabolism: implications for obesity and diabetes. Clinical chemistry. 2013;59:617–628. doi: 10.1373/clinchem.2012.187617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends in microbiology. 2011;19:349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Karlsson FH, Nookaew I, Petranovic D, Nielsen J. Prospects for systems biology and modeling of the gut microbiome. Trends in biotechnology. 2011;29:251–258. doi: 10.1016/j.tibtech.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Current opinion in gastroenterology. 2014;30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sagar NM, Cree IA, Covington JA, Arasaradnam RP. The interplay of the gut microbiome, bile acids, and volatile organic compounds. Gastroenterology research and practice. 2015;2015:398585. doi: 10.1155/2015/398585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ridlon JM, Bajaj JS. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta pharmaceutica Sinica B. 2015;5:99–105. doi: 10.1016/j.apsb.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breton J, Le Clère K, Daniel C, Sauty M, Nakab L, Chassat T, Dewulf J, Penet S, Carnoy C, Thomas P. Chronic ingestion of cadmium and lead alters the bioavailability of essential and heavy metals, gene expression pathways and genotoxicity in mouse intestine. Arch Toxicol. 2013;87 doi: 10.1007/s00204-013-1032-6. [DOI] [PubMed] [Google Scholar]
- 25.Fazeli M, Hassanzadeh P, Alaei S. Cadmium chloride exhibits a profound toxic effect on bacterial microflora of the mice gastrointestinal tract. Human & experimental toxicology. 2011:30. doi: 10.1177/0960327110369821. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Li Y, Liu K, Shen J. Exposing to cadmium stress cause profound toxic effect on microbiota of the mice intestinal tract. PLoS One. 2014;9:e85323. doi: 10.1371/journal.pone.0085323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu K, Abo RP, Schlieper KA, Graffam ME, Levine S, Wishnok JS, Swenberg JA, Tannenbaum SR, Fox JG. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environmental health perspectives. 2014;122:284–291. doi: 10.1289/ehp.1307429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breton J, Massart S, Vandamme P, De Brandt E, Pot B, Foligné B. Ecotoxicology inside the gut: impact of heavy metals on the mouse microbiome. BMC Pharmacology and Toxicology. 2013;14:1–11. doi: 10.1186/2050-6511-14-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bikel S, Valdez-Lara A, Cornejo-Granados F, Rico K, Canizales-Quinteros S, Soberon X, Del Pozo-Yauner L, Ochoa-Leyva A. Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome. Computational and structural biotechnology journal. 2015;13:390–401. doi: 10.1016/j.csbj.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White JR, Nagarajan N, Pop M. Statistical Methods for Detecting Differentially Abundant Features in Clinical Metagenomic Samples. PLoS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozupone C, Knight R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Applied and Environmental Microbiology. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G, Kay SA. Metabolomics Analysis Reveals Large Effects of Gut Microflora on Mammalian Blood Metabolites. National Academy of Sciences. 2009:3698. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee MH, Pankratz HS, Wang S, Scott RA, Finnegan MG, Johnson MK, Ippolito JA, Christianson DW, Hausinger RP. Purification and characterization of Klebsiella aerogenes UreE protein: A nickel-binding protein that functions in urease metallocenter assembly. Protein Science. 1993;2:1042–1052. doi: 10.1002/pro.5560020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellucci M, Zambelli B, Musiani F, Turano P, Ciurli S. Helicobacter pylori UreE, a urease accessory protein: specific Ni2+-and Zn2+-binding properties and interaction with its cognate UreG. Biochemical Journal. 2009;422:91–100. doi: 10.1042/BJ20090434. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu S, Kim JM, Shinmen Y, Yamada H. Evaluation of two alternative metabolic pathways for creatinine degradation in microorganisms. Archives of microbiology. 1986;145:322–328. [Google Scholar]
- 37.Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiological reviews. 2000;80:1107–1213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
- 38.Geiger O, González-Silva N, López-Lara IM, Sohlenkamp C. Amino acid-containing membrane lipids in bacteria. Progress in lipid research. 2010;49:46–60. doi: 10.1016/j.plipres.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Attwood PV. The structure and the mechanism of action of pyruvate carboxylase. The international journal of biochemistry & cell biology. 1995;27:231–249. doi: 10.1016/1357-2725(94)00087-r. [DOI] [PubMed] [Google Scholar]
- 40.Monniot C, Zébré AC, Aké FMD, Deutscher J, Milohanic E. Novel listerial glycerol dehydrogenase-and phosphoenolpyruvate-dependent dihydroxyacetone kinase system connected to the pentose phosphate pathway. Journal of bacteriology. 2012;194:4972–4982. doi: 10.1128/JB.00801-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verma S, Dubey R. Lead toxicity induces lipid peroxidation and alters the activities of antioxidant enzymes in growing rice plants. Plant Science. 2003;164:645–655. [Google Scholar]
- 42.Flora G, Gupta D, Tiwari A. Toxicity of lead: a review with recent updates. Interdisciplinary toxicology. 2012;5:47–58. doi: 10.2478/v10102-012-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris DR, Ward DE, Feasel JM, Lancaster KM, Murphy RD, Mallet TC, Crane EJ. Discovery and characterization of a Coenzyme A disulfide reductase from Pyrococcus horikoshii. FEBS Journal. 2005;272:1189–1200. doi: 10.1111/j.1742-4658.2005.04555.x. [DOI] [PubMed] [Google Scholar]
- 44.Stock KP, Newton GL, Fahey RC, Davies JE. Coenzyme A Disulfide Reductase, the Primary Low Molecular Weight Disulfide Reductase from Staphylococcus aureus PURIFICATION AND CHARACTERIZATION OF THE NATIVE ENZYME. Journal of Biological Chemistry. 1998;273:5744–5751. doi: 10.1074/jbc.273.10.5744. [DOI] [PubMed] [Google Scholar]
- 45.Miller JH. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annual Reviews in Microbiology. 1996;50:625–643. doi: 10.1146/annurev.micro.50.1.625. [DOI] [PubMed] [Google Scholar]
- 46.Bessman MJ, Frick DN, O’Handley SF. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. Journal of Biological Chemistry. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 47.Sharma R, Rensing C, Rosen BP, Mitra B. The ATP hydrolytic activity of purified ZntA, a Pb (II)/Cd (II)/Zn (II)-translocating ATPase from Escherichia coli. Journal of Biological Chemistry. 2000;275:3873–3878. doi: 10.1074/jbc.275.6.3873. [DOI] [PubMed] [Google Scholar]
- 48.Rensing C, Sun Y, Mitra B, Rosen BP. Pb (II)-translocating P-type ATPases. Journal of Biological Chemistry. 1998;273:32614–32617. doi: 10.1074/jbc.273.49.32614. [DOI] [PubMed] [Google Scholar]
- 49.Cao RX, Ma LQ, Chen M, Singh SP, Harris WG. Phosphate-induced metal immobilization in a contaminated site. Environmental Pollution. 2003;122:19–28. doi: 10.1016/s0269-7491(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 50.Jarosławiecka A, Piotrowska-Seget Z. Lead resistance in micro-organisms. Microbiology. 2014;160:12–25. doi: 10.1099/mic.0.070284-0. [DOI] [PubMed] [Google Scholar]
- 51.Pean N, Doignon I, Tordjmann T. Gut microbiota and bile acids: an old story revisited (again) Clinics and research in hepatology and gastroenterology. 2014;38:129–131. doi: 10.1016/j.clinre.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 52.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nature reviews Drug discovery. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 54.Griffiths WJ, Sjovall J. Bile acids: analysis in biological fluids and tissues. Journal of lipid research. 2010;51:23–41. doi: 10.1194/jlr.R001941-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Antharam VC, McEwen DC, Garrett TJ, Dossey AT, Li EC, Kozlov AN, Mesbah Z, Wang GP. An Integrated Metabolomic and Microbiome Analysis Identified Specific Gut Microbiota Associated with Fecal Cholesterol and Coprostanol in Clostridium difficile Infection. PLoS One. 2016;11:e0148824. doi: 10.1371/journal.pone.0148824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abuasal BS, Qosa H, Sylvester PW, Kaddoumi A. Comparison of the intestinal absorption and bioavailability of gamma-tocotrienol and alpha-tocopherol: in vitro, in situ and in vivo studies. Biopharmaceutics & drug disposition. 2012;33:246–256. doi: 10.1002/bdd.1790. [DOI] [PubMed] [Google Scholar]
- 57.Borel P, Preveraud D, Desmarchelier C. Bioavailability of vitamin E in humans: an update. Nutrition reviews. 2013;71:319–331. doi: 10.1111/nure.12026. [DOI] [PubMed] [Google Scholar]
- 58.Bruno RS, Leonard SW, Park S-i, Zhao Y, Traber MG. Human vitamin E requirements assessed with the use of apples fortified with deuterium-labeled α-tocopheryl acetate. The American Journal of Clinical Nutrition. 2006;83:299–304. doi: 10.1093/ajcn/83.2.299. [DOI] [PubMed] [Google Scholar]
- 59.Lombardo D, Guy O. Studies on the substrate specificity of a carboxyl ester hydrolase from human pancreatic juice. II. Action on cholesterol esters and lipid-soluble vitamin esters. Biochimica et biophysica acta. 1980;611:147–155. doi: 10.1016/0005-2744(80)90050-9. [DOI] [PubMed] [Google Scholar]
- 60.Kelly JR, Kennedy PJ, Cryan JF, Dinan TG, Clarke G, Hyland NP. Breaking down the barriers: the gut microbiome, intestinal permeability and stress-related psychiatric disorders. Frontiers in cellular neuroscience. 2015;9:392. doi: 10.3389/fncel.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loscalzo J. Lipid metabolism by gut microbes and atherosclerosis. Circulation research. 2011;109:127–129. doi: 10.1161/RES.0b013e3182290620. [DOI] [PubMed] [Google Scholar]
- 62.Roager HM, Sulek K, Skov K, Frandsen HL, Smedsgaard J, Wilcks A, Skov TH, Villas-Boas SG, Licht TR. Lactobacillus acidophilus NCFM affects vitamin E acetate metabolism and intestinal bile acid signature in monocolonized mice. Gut Microbes. 2014;5:296–303. doi: 10.4161/gmic.28806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ergurhan-Ilhan I, Cadir B, Koyuncu-Arslan M, Arslan C, Gultepe FM, Ozkan G. Level of oxidative stress and damage in erythrocytes in apprentices indirectly exposed to lead. Pediatr Int. 2008;50:45–50. doi: 10.1111/j.1442-200X.2007.02442.x. [DOI] [PubMed] [Google Scholar]
- 64.Possamai FP, Junior SA, Parisotto EB, Moratelli AM, Inacio DB, Garlet TR, Dal-Pizzol F, Filho DW. Antioxidant intervention compensates oxidative stress in blood of subjects exposed to emissions from a coal electric-power plant in South Brazil. Environ Toxicol Pharmacol. 2010;30:175–180. doi: 10.1016/j.etap.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 65.Lee DH, Lim JS, Song K, Boo Y, Jacobs DR., Jr Graded associations of blood lead and urinary cadmium concentrations with oxidative-stress-related markers in the U.S. population: results from the third National Health and Nutrition Examination Survey. Environ Health Perspect. 2006;114:350–354. doi: 10.1289/ehp.8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lamattina L, García-Mata C, Graziano M, Pagnussat G. Nitric oxide: the versatility of an extensive signal molecule. Annual Review of Plant Biology. 2003;54:109–136. doi: 10.1146/annurev.arplant.54.031902.134752. [DOI] [PubMed] [Google Scholar]
- 67.Cho C. Current roles of nitric oxide in gastrointestinal disorders. Journal of Physiology-Paris. 2001;95:253–256. doi: 10.1016/s0928-4257(01)00034-1. [DOI] [PubMed] [Google Scholar]
- 68.Sobko T, Reinders C, Jansson E, Norin E, Midtvedt T, Lundberg J. Gastrointestinal bacteria generate nitric oxide from nitrate and nitrite. Nitric Oxide. 2005;13:272–278. doi: 10.1016/j.niox.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 69.Revelles O, Millard P, Nougayrède JP, Dobrindt U, Oswald E, Létisse F, Portais JC. The carbon storage regulator (Csr) system exerts a nutrient-specific control over central metabolism in Escherichia coli strain Nissle 1917. PloS one. 2013;8:e66386. doi: 10.1371/journal.pone.0066386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gutiérrez P, Li Y, Osborne MJ, Pomerantseva E, Liu Q, Gehring K. Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. Journal of bacteriology. 2005;187:3496–3501. doi: 10.1128/JB.187.10.3496-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stohs S, Bagchi D. Oxidative mechanisms in the toxicity of metal ions. Free radical biology and medicine. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- 72.Starke-Reed PE, Oliver CN. Protein oxidation and proteolysis during aging and oxidative stress. Archives of Biochemistry and Biophysics. 1989;275:559–567. doi: 10.1016/0003-9861(89)90402-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.