Abstract
Neurexins (NRXNs) are presynaptic terminal proteins and candidate neurodevelopmental disorder susceptibility genes; mutations presumably upset synaptic stabilization and function. However, analysis of human cortical tissue samples by RNAseq and quantitative real-time PCR at 8–12 postconceptional weeks, prior to extensive synapse formation, showed expression of all three NRXNs as well as several potential binding partners. However, the levels of expression were not identical; NRXN1 increased with age and NRXN2 levels were consistently higher than for NRXN3. Immunohistochemistry for each NRXN also revealed different expression patterns at this stage of development. NRXN1 and NRXN3 immunoreactivity was generally strongest in the cortical plate and increased in the ventricular zone with age, but was weak in the synaptogenic presubplate (pSP) and marginal zone. On the other hand, NRXN2 colocalized with synaptophysin in neurites of the pSP, but especially with GAP43 and CASK in growing axons of the intermediate zone. Alternative splicing modifies the role of NRXNs and we found evidence by RNAseq for exon skipping at splice site 4 and concomitant expression of KHDBRS proteins which control this splicing. NRXN2 may play a part in early cortical synaptogenesis, but NRXNs could have diverse roles in development including axon guidance, and intercellular communication between proliferating cells and/or migrating neurons.
Keywords: cortical development, neurexins, neurodevelopmental disorders, subplate
Introduction
Neurodevelopmental conditions such as autism spectrum disorders (ASDs) schizophrenia and intellectual disability are highly heritable and can be regarded as disorders in formation of complex neural circuitry which, although specified by genetic instructions, relies on adaptive mechanisms to fine tune connectivity. A large number of candidate susceptibility genes have been identified (Pinto et al. 2010; Betancur 2011; Cukier et al. 2014; Kavanagh et al. 2015) many coding for proteins that interact in synapse formation, stabilization and function, including neurexins (NRXNs) as well as their postsynaptic binding partners (Ye et al. 2010; Reichelt et al. 2012; Bang and Owczarek 2013) focusing attention on the synapse and aberrant circuitry (Südhof 2008). However, synapses and their proteins are ubiquitous in the brain, whereas circuit alterations identified postmortem, or by fMRI and EEG, suggest localized deficits in the frontal and temporal lobes of the cerebral cortex in particular, as well as in the cerebellum and basal ganglia (Courchesne et al. 2007; Miyata et al. 2009; Kubota et al. 2013; Catani et al. 2016). Perhaps selected circuits are particularly sensitive to synaptic dysfunction. Alternatively, uncovering susceptibility gene expression patterns during early development may suggest additional roles key to understanding these localized deficits.
The NRXN genes are among the largest in the genome, exceeding 1 Mb, and differentially spliced into thousands of isoforms (Missler and Südhof 1998; Rowen et al. 2002; Schreiner et al. 2014). Each NRXN gene codes for a longer α and shorter β transcript transcribed from separate promoters (Ushkaryov et al. 1994). The α transcript contains six laminin/NRXN/sex-hormone binding globulin (LNS) domains and three epidermal growth factor (EGF)-like domains while β transcripts have the same transmembrane and cytoplasmic domain but only the sixth LNS domain (Ullrich et al 1995; Missler and Südhof 1998). Six differential splicing sites have been identified in α NRXNs (splice sites 1–6) two of which, 4 and 5, are also present in β NRXNs (Schreiner et al. 2014; see Supplementary Fig. 1 ).
α NRXNs are involved in Ca2+-dependent neurotransmitter release (Missler et al. 2003) their intracellular PDZ domain binding to proteins such as calcium/calmodulin-dependent serine protein kinase (CASK) which couple NRXN-mediated cell adhesion to synaptic vesicle exocytosis machinery (Hata et al. 1996). However, they are not required for synapse formation (Missler et al. 2003). β NRXNs also recruit other PDZ domain proteins to the presynaptic membrane but chiefly bind to neuroligin (NLGN) proteins across the synaptic cleft via their extracellular domain (Ichtchenko et al. 1995; Koehnke et al. 2010) as do certain α NRXN splice variants (Boucard et al. 2005; Reissner et al. 2013). These events are regulated by alternative splicing of NRXN genes (Ichtchenko et al. 1995; Ichtchenko et al. 1996). Different roles in synaptogenesis and synaptic transmission have been attributed to α and β NRXNs, due to separate ligand interactions (Petrenko et al. 1996; Reissner et al. 2013).
Binding affinities differ between various pairs of NRXNs and NLGNs, controlled by alternative splicing of both binding partners (Comoletti et al. 2006). In vertebrates, NRXNs (and NLGNs) are synthesized throughout the brain in all excitatory and inhibitory neurons (Ichtchenko et al. 1995, 1996; Ullrich et al. 1995). The different α- and β NRXNs are coexpressed in the same class of neuron (Ullrich et al. 1995); however, each type of NRXN (1, 2, or 3; Ullrich et al. 1995) and also different α splice variants, mRNA and protein (Schreiner et al. 2014; Schreiner et al. 2015) are differentially distributed between different neuronal types. In rodents, the STAR (Signal Transduction and Activation of RNA) proteins KHDBRS1, 2, and 3 (also known as SAM68, SLM1, and SLM2/T-STAR, respectively) regulate alternative splicing of Nrxns (Iijima et al. 2011; Klein et al. 2013; Ehrmann et al. 2013) and determine affinity for different binding partners (Vuong et al. 2016).
Numerous studies have found deletions, truncations, and copy number variants in NRXN genes at a higher frequency in neurodevelopmental disorders than in the control groups. The evidence is strongest for NRXN1. Two original studies detected both NRXN1β gene variants (Feng et al. 2006) and α gene variants (Friedman et al. 2006) in ASD patients. Subsequently multiple studies have found evidence for NRXN1 as a candidate susceptibility gene for ASD, schizophrenia and intellectual disability (Szatmari et al. 2007; Kim et al. 2008; Kirov et al. 2008; Glessner et al. 2009; Rujescu et al. 2009; Ching et al. 2010; Gauthier et al. 2011; Viñas Jornet et al. 2014). Mutations in NRXN2 and 3 have also been implicated although the evidence is less compelling; an identified NRXN2 gene mutation was found to result in a truncated protein which did not bind its NLGN partner in vitro (Gauthier et al. 2011) and there is supporting evidence from genome wide association studies (Cukier et al. 2014). NRXN3 mutations have been detected in autistic children from four families (Vaags et al. 2012) and a subsequent association study linked NRXN3 polymorphisms with schizophrenia (Hu et al. 2013). In mice knock-out of Nrxn1α-encoded isoforms targeting the first coding exon resulted in a mild ASD-related phenotype (Etherton et al. 2009; Grayton et al. 2013). Knock-out of Nrxn2 exon 23, shared by α and β isoforms, resulted in deficits in social interaction, social memory and anxiety-like behavior (Dachtler et al. 2014; Born et al. 2015).
Preliminary studies suggest that NRXNs and associated genes are all expressed at the earliest stages of cortical plate (CP) formation (Ip et al. 2010; Kang et al. 2011) when very few synapses are present in the human cerebral cortex (Kostović and Rakic 1990). This suggests additional roles for NRXNs although an alternative (but not mutually exclusive) explanation would be that perhaps correct functioning of precocious synaptic circuitry present in the presubplate (pSP) and marginal zone (MZ) is crucial to cortical development. To explore these possibilities further, we have studied the expression of the three NRXN genes at these early stages and provided additional information on the expression of NRXN binding partners, and regulators of NRXN alternative splicing, to gain a more complete picture of how mutations in NRXNs may perturb human cortical development. Some of these findings have already been published in abstract form (Harkin et al. 2015).
Methods and Materials
Human Tissue
Human fetal brain samples between 8 and 12 postconceptional weeks (PCWs) were obtained from the MRC-Wellcome Trust Human Developmental Biology Resource (HDBR, http://www.hdbr.org; Gerrelli et al 2015). Brains were collected from terminations of pregnancy with maternal written consent and approval from the Newcastle and North Tyneside NHS Health Authority Joint Ethics Committee. Age was determined by the assessment of external morphology (O'Rahilly et al 1987; Bullen and Wilson 1997) and by comparing foot and heel to knee length to a standard growth chart (Hern 1984). Adult brain sections were obtained from the Newcastle Brain Tissue Resource (NBTR). Brains were collected with written consent from donors postmortem with approval from the Newcastle and North Tyneside NHS Health Authority Joint Ethics Committee. Both the HDBR and NBTR are regulated by the UK Human Tissue Authority (HTA; www.hta.gov.uk) and operate in accordance with the relevant HTA Codes of Practice.
Whole fetal brains were isolated from the skull and the meninges were removed. The hemispheres were split apart allowing removal of the choroid plexus and subcortical structures leaving only the cerebral cortex. One or both hemispheres were then divided into six sections. Each hemisphere represented an independent sample. The temporal lobe, including lateral and medial walls was removed and labeled section 6. The remaining cortex was divided into five sections of equal width from the anterior (A) to the posterior (P) pole of the cortex including lateral and medial cortical walls (labeled 1–5). Sections 1, 3, 5, and 6 were used for RNA extraction and corresponded to anterior, central (C), posterior and temporal (T) regions (see Fig. 1). Sections were immediately frozen and stored at –80°C and subsequently used to extract RNA. Other fetal brains were fixed in buffered 4% paraformaldehyde solution (Sigma Aldrich, Dorset, UK) and embedded in paraffin wax before sectioning.
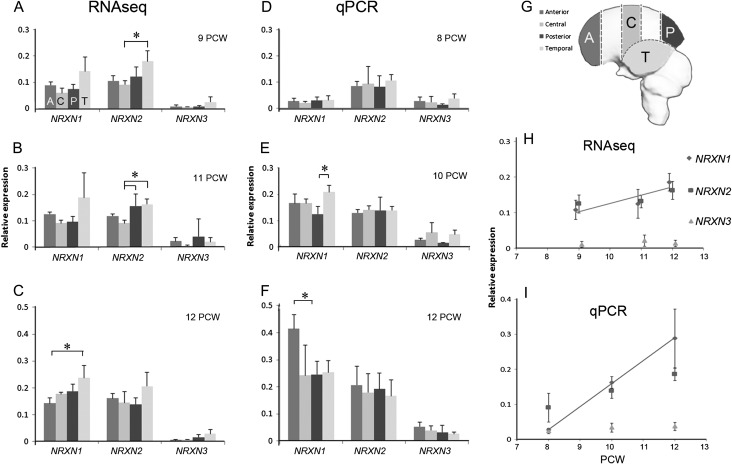
Figure 1.
The differences in NRXN expression with age and cortical region. (A–C) Expression by RNAseq relative to reference genes and (D–F) expression by qPCR (relative to the same reference genes), in four different cortical regions; anterior (A), central (C), posterior (P), and temporal (T) at different ages postconception. (G) The precise locations of the four sampling regions. Error bars represent standard errors of the mean. Statistically significant differences in expression between regions at a given age are marked with an asterisk (one-way ANOVA, P < 0.05). RNAseq and qPCR data revealed broadly the same expression patterns with a slight tendency toward increased expression in the temporal cortex at all ages. (H and I) Changes in expression with age for all pooled cortical samples. Blue lines mark statistically significant positive correlations showing NRXN1 expression increases with age. NRXN2 expression remains relatively high throughout, NRXN3 expression relatively low.
RNA Extraction and Reverse Transcription
RNA was extracted from sections 1 (A), 3 (C), 5 (P), and 6 (T) using the QIAgen RNeasy mini kit (Qiagen, Manchester, UK) according to the manufacturer's protocol. Sixty-three samples were taken in all at 9, 11, and 12 PCW (see Table 1). The concentration and quality of RNA was measured using the Nanodrop 8000 (Thermo Fisher Scientific, Cramlington, UK) to detect spectrophotometric absorbance at 260 nm. To control for RNA purity and degradation, 260/230 and 260/280 spectrophotometric ratios were only accepted between the ranges of 1.8 and 2.1. A selection of the RNA was run on the Bioanalyser 2100 (Agilent, Stockport, UK) and all RIN values measured were above 7.
Table 1.
A summary of the number of tissue samples taken at each age and location for either RNAseq analysis or qPCR
| Age (PCW) | Number of samples | ||||
|---|---|---|---|---|---|
| Anterior | Central | Posterior | Temporal | Total | |
| RNAseq | |||||
| 9 | 4 | 3 | 4 | 8 | 19 |
| 11 | 4 | 3 | 4 | 8 | 19 |
| 12 | 5 | 3 | 6 | 10 | 24 |
| 62 | |||||
| qPCR | |||||
| 8 | 5 | 5 | 5 | 3 | 18 |
| 10 | 5 | 5 | 5 | 3 | 18 |
| 11 | 5 | 5 | 5 | 5 | 20 |
| 56 | |||||
At each age/location samples were taken from a minimum of two brains as samples taken from each hemisphere were counted separately.
To reverse transcribe the RNA to cDNA, 2 µg of random primers (Promega, Southhampton, UK), up to 5 µg of RNA, and 1 µl dNTPs (10 mM) were combined and the volume made up to 13 µl using RNase free water. The mixture was heated at 65°C for 5 min and cooled on ice for a minimum of 1 min. Four microliters 5× first strand buffer, 1 µL (200 U/µL) Superscript Reverse Transcriptase (Invitrogen, Thermo Fisher Scientific), 1 µL of 1 M dithiothreitol and 1 µL RNaseOut Recombinant Ribonuclease Inhibitor (Invitrogen, 40 U/µL) were added to the same tube and incubated at 25°C for 5 min before being heated to 50°C for 60 min. The reaction was inactivated by heating to 70°C for 15 min and the cDNA was stored at –20°C. Total cDNA concentration was measured using the Nanodrop 8000 (Thermo Fisher Scientific) to detect spectrophotometric absorbance at 260 nm and 260/230, 260/280 spectrophotometric ratios were between the accepted range of 1.7–2.1. cDNA samples were then diluted to a concentration of 100 ng/µL for use in PCR.
RNAseq Methodology
Full details of the origins, collection, preparation, sequencing, and analysis of the human fetal RNA samples are provided by Lindsay et al. (2016). The entire RNAseq dataset from which data were extracted for this study has been deposited at www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-4840. Briefly, cDNA was generated from the RNAs using Illumina's Stranded mRNA Sample Prep Kit. Four hundred nanograms of total RNA was used as the input for each sample. The concentration of each library was determined using the KAPA quantitative real-time PCR (qPCR) kit (KK4835) and triplicate reactions using three independent 106-fold dilutions of the libraries. The size profile of approximately 15% of the libraries was evaluated using an Agilent Bioanalyzer DNA 1000 chip. The average final library size was between 272 and 467 bp (includes 120 nucleotides of adapter sequence). The libraries were sequenced on an Illumina HiSeq2000. The high-quality reads were then mapped to the human reference genome hg38 with Tophat2 (Kim et al. 2013). Reads aligned to genes and exons were counted with BEDTools (Quinlan and Hall 2010). Read length was 101 bp prior to trimming and, in the majority of cases, 85 bp after trimming with no reads of <20 bp retained. The minimum number of reads examined per sample was 63 million and the average was 90 million. Differentially expressed genes and exons were then identified with the Bionconductor package DESeq2 (Love et al. 2014).
Quantitative Real-Time PCR
In order to validate the RNAseq findings, the expression levels of the three NRXN genes and three reference genes (β-ACTIN, GAPDH, and SDHA) (Vandesompele et al. 2002) were measured by qPCR from 56 samples taken from the four sampled sections at 8, 10, or 12 PCW (see Table 2). Primers were designed according to the standard criteria for PCR primer design using the Primer3 (v. 0.4.0) designing program (http://frodo.wi.mit.edu/primer3/). Primers synthesis and product sequencing for quality control were performed by Eurofins MWG Operon1 (http://www.eurofinsgenomics.eu). Table 2 presents the sequence and product size of each primer used. A SYBR Green-based rtPCR assay was performed in 7900HT Fast Real-Time PCR system (Applied Biosystems, Warrington, UK). A total volume of 10 μL qPCR reaction was set up in triplicates, containing 5 μL of 2× SYBR Green qPCR Master Mix (Invitrogen), 1 μL of the diluted cDNA template, 0.5 μL of each primer (10 ρmol/μL), and 3 μL of Molecular Biology grade water. A negative control was incorporated by replacing the cDNA template with Molecular Biology grade water (VWR International, Lutterworth, UK). A standard thermal cycle protocol was used as previously described (Ip et al. 2010). The results of each reaction were analyzed by uploading raw data to the Real-Time PCR Miner web site (http://www.miner.ewindup.info). This calculated the reaction efficiency and the fractional cycle number at threshold (CT) for each reaction (Zhao and Fernald 2005).
Table 2.
List of primers for qPCR
| Gene | Forward primer 5′–3′ | Reverse primer 5′–3′ | Amplicon size (bp) |
|---|---|---|---|
| NRXN1 | aggacattgacccctgtgag | ccttcatcccggtttctgta | 205 |
| NRXN2 | catcctcctctacgccatgt | ttgttcttcttggccttgct | 165 |
| NRXN3 | gctgagaacaaccccaata | atgctggctgtagagcgatt | 179 |
| *BACTIN | ctacaatgagctgcgtgtggc | caggtccagacgcaggatggc | 271 |
| *GAPDH | tgcaccaccaactgcttagc | ggcatggactgtggtcatgag | 87 |
| *SDHA | tgggaacaagagggcatctg | ccaccactgcatcaaattcatg | 86 |
*Reference genes BACTIN, β-actin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; SDHA, succinate dehydrogenase complex subunit A.
Quantitative Comparison of RNAseq and qPCR Data, and Individual Exon Expression by RNAseq
To make expression data comparable between different methodologies (Al-Jaberi et al. 2015), we divided the expression level of the gene of interest, Reads Per Kilobase of transcript per Million mapped reads (RPKM) for RNAseq, and CT values for qPCR, by the geometric mean expression of three reference genes (β-ACTIN, GAPDH, and SDHA; Vandesompele et al. 2002) to give relative expression levels in different regions of the cortex at different ages which were then compared by one-way ANOVA followed by Tukey's post hoc test. Expression of the three reference genes was found, on average, to change little across the different regions of the cortex or with age, by either RNAseq or qPCR (Supplementary Fig. 2) validating the choice of these genes.
Expression levels of NRXN-related genes, KHDBRS genes, and individual NRXN exons (6, 17, 19, and 20; see Supplementary Fig. 1) were quantified in terms of normalized RPKM per exon for between 16 and 33 samples for each gene from across the cortex, right, and/or left hemisphere from 3 fetuses at 9 PCW, 5 fetuses at 11 PCW, and 4 fetuses at 12 PCW. The differences in expression between genes or exons were tested by one-way ANOVA followed by Tukey's post hoc test.
Immunohistochemistry
Deparaffinised coronal and sagittal sections collected on slides were immersed in 1.5 % hydrogen peroxide/methanol solution (Merck Millipore, Watford, UK) to block activity of endogenous peroxidases followed by heat-mediated antigen retrieval treatment in citrate buffer (pH 6.0) before incubating sections with 10% of the appropriate normal serum (Vector Laboratories, Peterborough, UK) in Tris-buffered saline (TBS) for 10 min. For immunoperoxidase, staining sections were then incubated with a primary antibody (see Table 3 for sources of antibodies) in TBS solution overnight at 4°C. Sections were then washed and incubated for 30 min with the appropriate biotinylated secondary antibody (Vector Laboratories, Peterborough, UK), washed then incubated for 30 min with Vectastain Elite ABC kit (Vector Labs) and developed using 3,3′-diaminobenzidine (Vector Labs). Sections were dehydrated, cleared, and coverslipped.
Table 3.
Primary antibodies employed
| Antigen | Supplier | Previous use in human | Dilution | Species |
|---|---|---|---|---|
| NRXN1α+β | Santa Cruz. Sc14334 | Western blot: Kim et al. (2008). | 1/300 | Goat polyclonal |
| Immunohistochemistry: Zhang et al. (2013). | ||||
| NRXN2α | AbCam. Ab34245 | Immunohistochemistry: Borsics et al. (2010). | 1/2000 | Rabbit polyclonal |
| NRXN3α+β | Sigma-Alldrich Prestige. HPA 002727 | Tested by human protein atlas including immunohistochemistry. | 1/300 | Rabbit polyclonal |
| CASK | AbCam. ab126609 | Immunohistochemistry: Wei et al. (2014). | 1/350 | Rabbit monoclonal |
| KHDBRS1 | Santa Cruz. Sc333 | 1/1500 | Rabbit polyclonal | |
| KHDBRS2 | Dr Peter Scheiffele | 1/10 000 | Rabbit polyclonal | |
| KHDBRS3 | Dr Peter Scheiffele | 1/5000 | Guinea pig polyclonal | |
| PAX6 | Covance, PRB-278P | Immunohistochemistry: Bayatti et al. (2008). | 1/1500 | Rabbit polyclonal |
| TBR1 | AbCam Ab31940 | Immunohistochemistry: Bayatti et al. (2008). | 1/1500 | Rabbit polyclonal |
| GAP43 | Sigma-Alldrich G9264 | Immunohistochemistry: Bayatti et al. (2008). | 1/10 000 | Mouse monoclonal |
| SYP | Sigma-Alldrich | Immunohistochemistry: Bayatti et al. (2008). | 1/1000 | Mouse monoclonal |
For antigenic sites of anti-NRXN antibodies, see Supplementary Fig. 1. anti-NRXN1 has also been tested against recombinant mouse NRXN1 expressed in a human kidney cell line (Cheng et al. 2009).
For immunofluorescent double labeling, including using two antibodies from the same species, the following method was employed (Goto et al. 2015; Harkin et al. 2016). Sections were incubated with the first primary antibody as before, washed and incubated with the appropriate ImmPRESS HRP IgG (Peroxidase) Polymer Detection Kit (Vector Labs) for 1 h, washed then incubated for 30 min with tyramide signal amplification (TSA) fluorescein plus system reagent (Perkin Elmer, Buckingham, UK). After washing but before application of a second primary antibody, sections were subjected to heat-mediated antigen retrieval in boiling citrate buffer (pH 6.0). This removed the first set of primary and secondary antibodies employed but left fluorescent tyramide covalently bound to the tissue section. The method above was repeated for the detection of this second primary antibody except that TSA rhodamine plus was used for detection. Sections were then washed, counterstained with 4′,6-diamidino-2-phenylindole, dihydrochloride (Thermo Fisher Scientific) and coverslipped with Vectashield (Vector Labs).
Results
Quantitative Analysis of mRNA Reveals Distinct Patterns of NRXN Gene Expression
Sixty-three samples of RNA, taken from four different regions of the cortical surface (see Fig. 1G) at either 9, 11, or 12 PCW were sequenced and mapped. All three NRXNs were expressed in all regions and at all ages, although NRXN3 was consistently expressed at lower levels than NRXN 1 and 2 (Fig. 1A–C). NRXN 1 and 2 were found to be highly expressed (top quartile of all protein coding genes); however, NRXN3 was expressed at levels we would still expect to be detectable by histological methods (Al-Jaberi et al. 2015). For all three NRXNS, at all ages, expression tended to be higher in the temporal cortex, but this only reached statistical significance for NRXN2 at 9 PCW and 11 PCW compared to central regions of the cortex (Fig. 1A,B) and for posterior cortex compared to central cortex at 11 PCW (Fig. 1B). For NRXN1, at 12 PCW, expression was also significantly higher in temporal cortex compared to anterior cortex (Fig. 1C). In order to confirm these results, a separate set of 56 samples of RNA were taken for qPCR analysis, although at slightly different age points (8, 10, and 12 PCW, Fig. 1D–F).
Results from qPCR very largely confirmed the findings by RNAseq except that increased expression in the temporal cortex was not as marked, although for NRXN1 it was significantly higher than expression in the posterior cortex at 10 PCW (Fig. 1E). Somewhat anomalously, expression of NRXN1 in the anterior cortex was markedly high at 12 PCW (Fig. 1F). What was apparent from the qPCR study, with its wider age range, was that whereas expression of NRXN2 remained consistently high over time, NRXN1 expression increased with age. Results from all regional samples were pooled and plotted against age for both RNAseq and qPCR. It was clear that in both cases, NRXN one expression increased significantly with age (Fig. 1H,I). However, NRXN2 expression remained constantly high, and NRXN3 expression consistently lower.
In summary, there were no marked differences in expression between different regions of the cortex detected by either RNAseq or qPCR although expression tended to be higher in temporal cortex, and to a lesser extent frontal cortex, than in central or posterior regions. Although it has been suggested that fronto-temporal circuitry may be primarily “at risk” in neurodevelopmental conditions (Scott-Van Zeeland et al. 2010; Catani et al. 2016), we have found no strong evidence that NRXNs are predominantly expressed there, such that mutations in NRXN genes might have a more a deleterious effect in these cortical regions.
Expression of NRXN-Associated Genes
RNAseq data were then analyzed to determine the levels of expression of proteins associated with NRXNs at synapses, many of which are also candidate susceptibility genes for neurodevelopmental disorders (Ye et al. 2010; Bang and Owczarek 2013). We categorized them according to their location at the synapse, the level of expression and developmental stage (see Table 4). We found that many, but not all, were expressed at similarly high levels as NRXNs 1 and 2, suggesting that they could be potential binding partners for NRXNs in the developing cortex. Expression of postsynaptic binding partner NLGN2 was twice that of NRXNs 1 and 2 (Table 4 and data not shown). NGLN3 and leucine-rich repeat transmembrane 2 (LRRTM2) were highly expressed and NGLN1 increased in expression with age. LRRTMs 3 and 4, NLGN4Y and X were moderately expressed but LRRTM1 showed lower expression. At the presynaptic membrane cerebellin 1 (CBLN1) was highly expressed at the earlier stage. Genes associated with the postsynaptic density, Disks large homolog 4 (DLG4) and SH3 and multiple Ankyrin repeat domains 1 (SHANK1) were highly expressed although SHANK2 and SHANK3 exhibited moderate levels of expression. Genes normally associated with synaptic vesicles, synaptophysin (SYP) and synapse-associated protein 25 kDa (SNAP25) were very highly expressed and CASK, which connects neurexins to the exocytotic machinery, also showed high levels of expression similar to NRXNs 1 and 2 (Table 4).
Table 4.
Summary of expression of mRNA for neurexins and associated proteins
| Protein location | Expression (normalized RPKM) | 9 PCW | 11–12 PCW |
|---|---|---|---|
| Presynaptic/extracellular | High: 5–25% (40–160) | CBLN1 | NRXN1 |
| NRXN1 | NRXN2 | ||
| NRXN2 | |||
| Moderate: 25–50% (10–40) | NXPH1 | NXPH1 | |
| CBLN1 | |||
| NRXN3 | |||
| Low: 50–75% (0.4–10) | CBLN2 | CBLN2 | |
| NRXN3 | |||
| Presynaptic/intraterminal | Very high: top 5% (>160) | SNAP25 | SNAP25 |
| SYP | SYP | ||
| High: 5–25% (40–160) | CASK | CASK | |
| Postsynaptic/extracellular | Very high: top 5% (>160) | NLGN2 | NLGN2 |
| High: 5–25% (40–160) | LRRTM2 | LRRTM2 | |
| NLGN3 | NLGN1 | ||
| NLGN3 | |||
| Moderate: 25–50% (10–40) | LRRTM3 | LRRTM3 | |
| LRRTM4 | LRTMM4 | ||
| NLGN1 | NLGN4X | ||
| NLGN4X | NLGN4Y* | ||
| NLGN4Y* | |||
| Low: 50–75% (0.4–10) | LRRTM1 | LRRTM1 | |
| Postsynaptic/intracellular | Very high: top 5% (>160) | DLG4 | DLG4 |
| High: 5–25% (40–160) | SHANK1 | SHANK1 | |
| Moderate: 25–50% (10–40) | SHANK2 | SHANK2 | |
| SHANK3 | SHANK3 |
Protein location describes its location in the mature central nervous system. Extracellular means at least part of the molecule is an extracellular domain. Expression is the mean across all cortical regions. Percentage values indicate into which quartile range for all protein coding expression data the expression values fall. RPKM values are indicative and not exact. *NLGNY means from male samples only, female samples gave mean expression 0.11 range 0.07–0.45. NXPH, neurexophilin.
Localization of Expression of NRXN Proteins by Immunohistochemistry
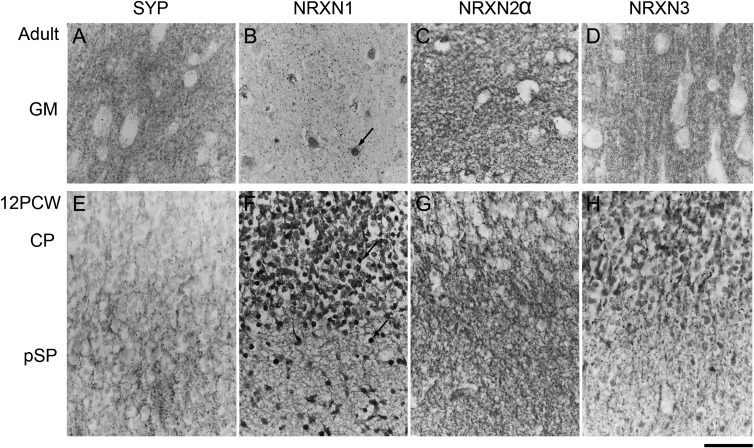
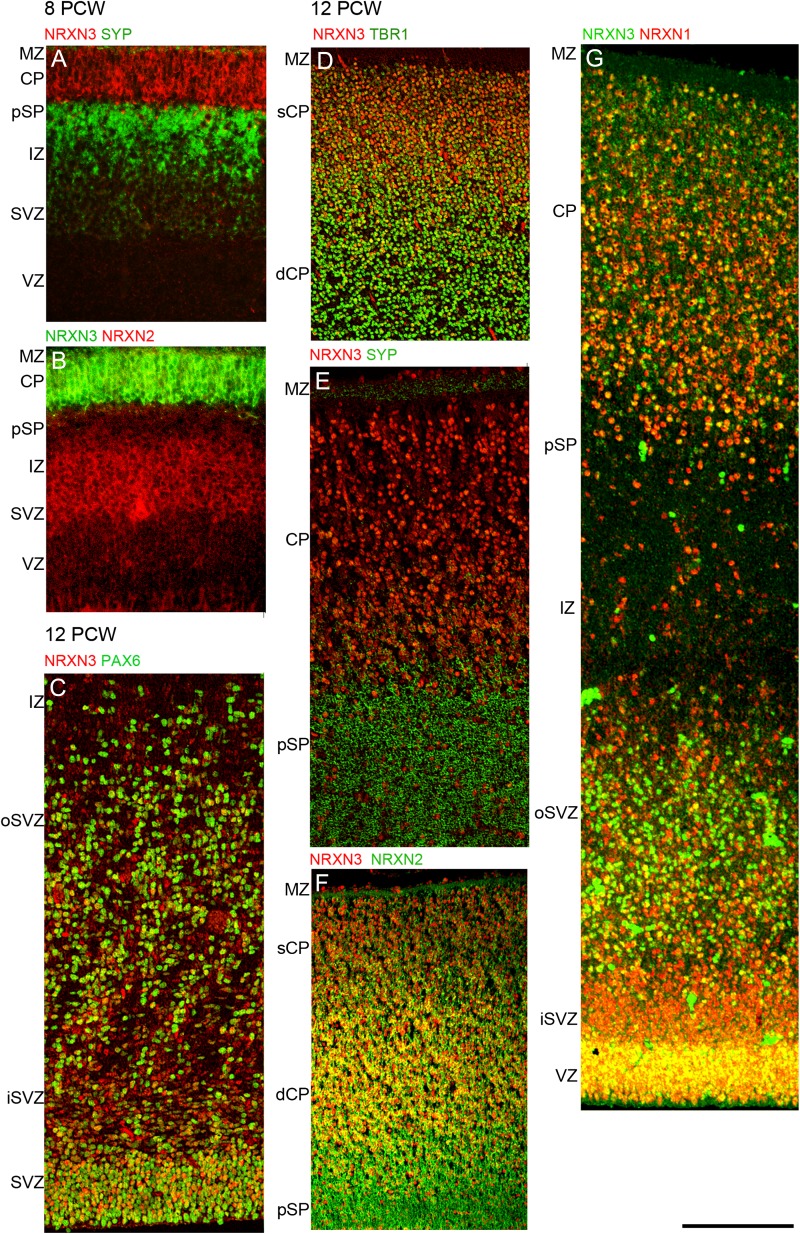
The specificity of our antibodies was first tested by immunostaining sections of adult human cerebral cortex in comparison with SYP immunoreactivity. Our antibodies to NRXN1 and three were generated against peptide sequences common to both α and β isoforms, but the antibody to NRXN2 was raised against the α form only (Supplementary Fig. 1). All anti-NRXN antibodies and anti-SYP produced immunoreactivity in gray matter neuropil which was generally punctate in appearance, suggestive of presynaptic terminals and/or synaptic contacts (Fig. 2A–D). However, the density of punctate staining was lower for NRXN1 and there was additional immunoreactivity in many cell nuclei. NRXN2α immunoreactivity also appeared in the cytoplasm of some cells including oligodendrocyte-like cells in the white matter (not shown). The density of immunoreactivity was greatly reduced in the white compared to gray matter for all four proteins (not shown). In sections of 12 PCW fetal cortex, SYP immunoreactivity revealed a small amount of neuropil in the pSP likely to contain the earliest synapses (Fig. 2E; Bayatti et al. 2008). Only NRXN2α immunoreactivity was strongly expressed in the neuropil of this region (Fig. 2G). Both NRXN1 and NRXN3 were expressed in the cell bodies, their immediate processes and, particularly for NRXN1, some nuclei of many immunoreactive cells in the pSP and the overlying CP. Neuropil staining was very weak for NRXN1 but some punctate staining in the pSP and CP was detectable for NRXN3. Because of the unexpected nuclear labeling observed with anti-NRXN1 antibody, we tested immunostaining with the antibody in the presence of blocking peptide according to the manufacturer's instructions (Santa Cruz). All immunostaining was abolished suggesting that the NRXN1 antigenic site was present in the nucleus (Supplementary Fig. 3).
Figure 2.
Immunohistochemistry for SYP and the NRXNs in paraffin sections from adult (A–D) and fetal (E–H) cerebral cortex. (A) Punctate SYP immunoreactivity confined to presumptive synaptic terminals in the gray matter neuropil. In the fetal cortex (E) such punctate staining is largely confined to the pSP. All three NRXNs also exhibited punctate staining (B–D) in adult cortex, although at a lower density for NRXN1 with evidence of nuclear staining (arrow, B). In the fetal cortex, NRXN1 was expressed in cell bodies, processes, and some nuclei (arrows) of many immunoreactive cells in the pSP and CP but without punctate staining (F). NRXN2, however, exhibited punctate staining in the pSP in particular (G). NRXN3 was expressed in cell bodies and processes mostly in the CP, with some punctate immunoreactivity, largely in the pSP (H). Scale bar = 50 µm.
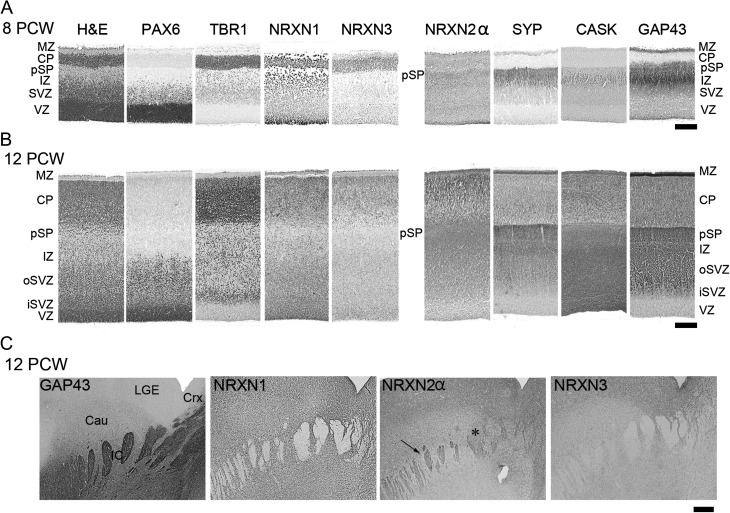
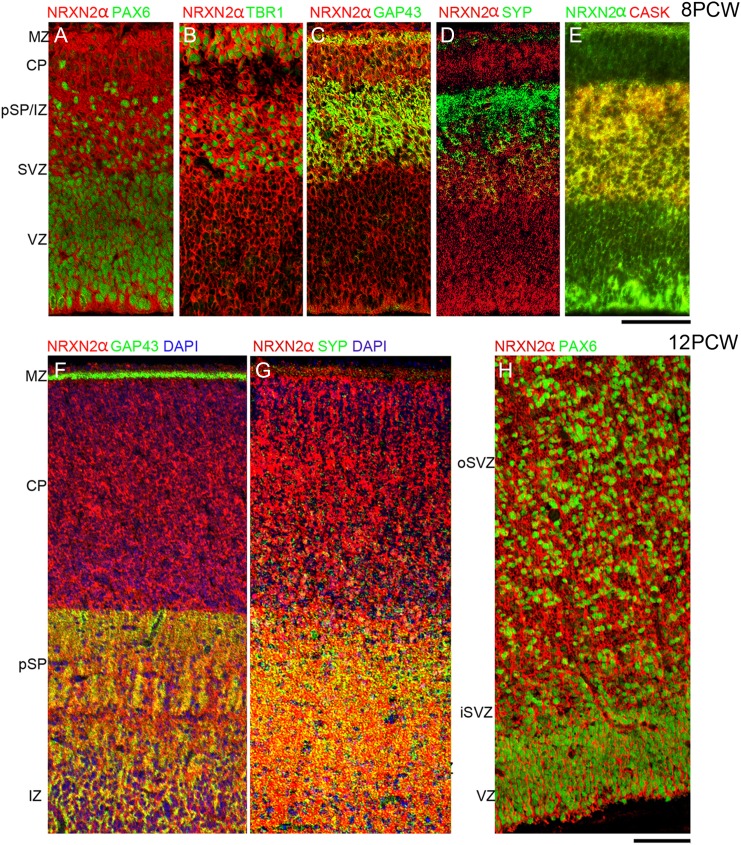
We proceeded to investigate NRXN immunoreactivity over the age range 8–12 PCW in comparison to the expression of other markers for the different cortical layers (Fig. 3). As has been previously demonstrated (Bayatti et al. 2008), PAX6 identifies radial glial cells in the proliferative ventricular (VZ) and subventricular (SVZ) zones, TBR1 immunoreactivity identifies postmitotic neurons in the SVZ, migrating through the intermediate zone (IZ) and in the pSP CP and outer MZ. GAP43 and SYP immunoreactivity are localized to growing axons, growth cones and newly forming synaptic terminals and delineate the MZ and pSP (SYP more than GAP43) and IZ (GAP43 more than SYP).
Figure 3.
A comparison of NRXN immunoreactivity with other cell markers across the cortical wall at 8 PCW (A) and 12 PCW (B). H&E, hemotoxylin and eosin stained; o, outer; i, inner. Note that PAX6 revealed radial glial progenitor cells; TBR1, postmitotic neurons; SYP, CASK and GAP43 neurites of the pSP and IZ. NRXN1 and 3 predominantly localized to layers with a high-cellular density, whereas only NRXN2α predominantly colocalized with synaptogenic zones and growing axons. (C) Internal capsule in the ventral forebrain in which bundles of growing axons were GAP43 positive, NRXN1 and 3 negative, and partially positive for NRXN2α (arrow) although some axon bundles appeared negative (*). Scale bars: A and B, 200 µm; C, 500 µm.
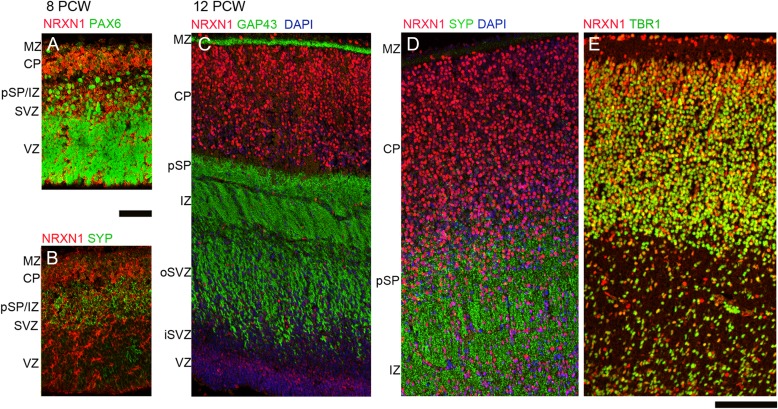
Each NRXN showed a distinct pattern of immunoreactivity. At 8 PCW, NRXN1 expression was strongest in cells lining the apical surface of the VZ, in the pSP, and in the CP, particularly at the border with the MZ, and in the MZ itself and was predominantly cytoplasmic or membranous, rather than nuclear (Figs 3A and 4A). Immunofluorescent double labeling revealed NRXN1 expression around the margins of PAX6-positive progenitor cells in the VZ (Fig. 4A) but no strong colocalization with SYP in the pSP (Fig. 4B). Similarly, by 12 PCW, there was no expression in the network of neurites in the MZ, pSP, and IZ that was strongly immunopositive for GAP43 and SYP (Fig. 4C,D). However, NRXN1 was expressed in many but not all cell bodies, in all layers of cortical wall and, expression was strongest in the VZ (Figs 3B and 4C) and in the immature neurons of the MZ and the superficial CP and appeared to be expressed in cell nuclei (Figs 3B and 4D,E). Many cells coexpressed TBR1 and NRXN1 but NRXN1 expression predominated in the superficial CP, TBR1 in the lower CP, with cells expressing one, the other or both in the pSP (Fig. 4E). There was no evidence for more intense NRXN1 immunoreactivity in the frontal or temporal lobes (not shown).
Figure 4.
Colocalization of NRXN1 immunoreactivity (red) with phenotypic markers (green) by immunofluorescence. At 8 PCW NRXN1 immunoreactivity was present in cytoplasm/membranes around PAX6-positive radial glia, particularly near the apical ventricular surface (A) but showed little colocalization with SYP in the pSP at either 8 PCW (B) or 12 PCW (D) nor with GAP43 in growing axons of the IZ (C). However, there was strong colocalization (orange–yellow) with TBR1 in the postmitotic neurons of the CP. Scale bars = 100 µm.
NRXN2α immunoreactivity was expressed strongly throughout the cortical wall at 8 PCW (Fig. 3A) both in and around cells, but not in their nuclei, and in networks of processes away from the cell bodies. It was colocalized both with PAX6-positive progenitors and TBR1-positive postmitotic neurons (Fig. 5A,B). Although present in the pSP it showed comparatively weak colocalization with SYP but exhibited stronger expression and colocalization with GAP43, particularly in the IZ (Figs 3A and 5C,D). It was also strongly coexpressed with CASK, particularly in the IZ (Fig. 5E). This pattern of expression was little changed by 12 PCW (Fig. 5F–H) although colocalization with SYP was possibly more evident by this stage. Expression of CASK became much stronger in the proliferative SVZ and VZ at 12 PCW where it was colocalized with all three NRXNs (Fig. 3B).
Figure 5.
Colocalization of NRXN2 immunoreactivity (red) with phenotypic markers (green) by immunofluorescence. At 8 PCW NRXN2 was ubiquitously expressed and surrounded both PAX6-positive nuclei of progenitor cells (A) and TBR1-positive nuclei of postmitotic neurons (B) (green). In the pSP and IZ, NRXN2 showed stronger colocalization (yellow) with GAP43 (C) and particularly CASK (E) than with SYP (D). At 12 PCW NRXN2 and GAP43 are strongly colocalized (yellow) in the pSP and IZ but not the MZ (F). NRXN2 also colocalized with SYP in pSP and IZ (G). As at 8 PCW, many cells coexpressed NRXN2 in the cytoplasm and processes surrounding PAX6-positive nuclei, especially in the oSVZ and VZ (H). iSVZ and oSVZ, inner and outer subventricular zone. Scale bars = 100 µm in A–E and F–H.
At 8 PCW, NRXN3 immunoreactivity was almost entirely confined to the CP and MZ (Figs 3A and 6A,B). By 12 PCW, there was an increase in nuclear staining as was observed for NRXN1. NRXN3 immunoreactivity was also detected in the proliferative zones where it colocalized with PAX6-positive progenitors in the upper part of the VZ in particular (Figs 3B and 6C) and with NRXN2α in the VZ, and to a lesser extent, in the SVZ (Fig. 6F). NRXN2α expression was present at the apical surface of the VZ where NRXN3 was absent. In the CP, NRXN3 expression was colocalized with TBR1, NRXN1, and NRXN 2α (Fig. 6D,F,G) but in all cases NRXN3 expression was relatively stronger in the superficial part of the CP near the border with the MZ, where TBR1 in particular was more weakly expressed (Fig. 6D). There was very little colocalization between NRXN3 and SYP in the pSP and MZ, although NRXN3 positive cells were present in these layers (Figs 3B and 6E).
Figure 6.
Colocalization of NRXN3 immunoreactivity (red) with phenotypic markers (green) by immunofluorescence. At 8 PCW NRXN3 expression was almost completely confined to the CP and separated from SYP immunoreactivity (A) showing only weak colocalization with NRXN2 in the CP (pale green, B). At 12 PCW NRXN3 was expressed in the proliferative zones and colocalized (yellow) with PAX6 (C) and in the CP was colocalized with TBR1, especially in the deeper layer (dCP) (D). NRXN3 did not colocalize with SYP (E) but was coexpressed with NRXN2 in the dCP (F). NRXN1 and 3 were colocalized throughout the cortical wall at 12 PCW but especially strongly in the CP and VZ (G). Scale bar = 200 µm.
Expression of KHDBRS mRNA and Proteins and Alternative Splicing of NRXNs
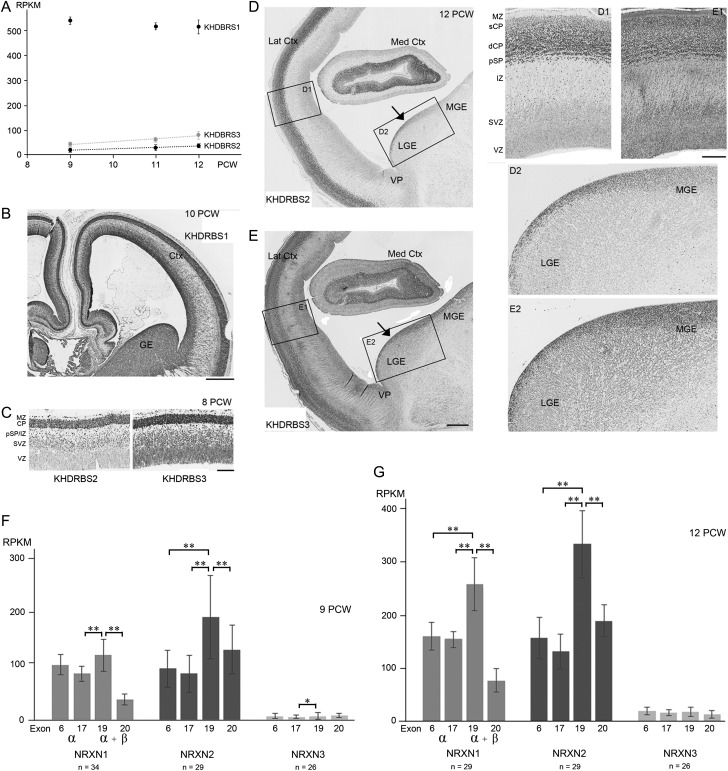
From the RNAseq data, it can be seen that all three KHDBRS genes were expressed in the developing cortex (Fig. 7A). KHDBRS1 showed very high levels of expression, four to five times greater than the highest expression levels measured for NRXN1 and 2. KHDBRS2 and 3 exhibited expression levels comparable to NRXNs, although KHDBRS3 showed slightly higher expression than KHDBRS2. There was no significant change in expression levels with age or with cortical location (Fig. 7A).
Figure 7.
KHDRBS 1–3 expression in human early fetal forebrain and suppression of NRXN exon 20. (A) RNAseq analysis found very high expression of KHDBRS1 maintained with age, and moderate expression of KHDRBS2 and 3 which increased with age (dotted trend lines indicate a significant correlation, P < 0.001). (B) Patterns of KHDRBS immunoreactivity reflected mRNA expression; KHDBRS1 immunoreactivity was intense throughout the telencephalon at 10 PCW. (C) At 8 PCW, many KHDBRS2 positive cells were observed in the MZ, CP, and pSP, isolated cells were also seen in the IZ and SVZ; there was stronger immunostaining for KHDBRS3 in the CP, MZ, and pSP/IZ, with widespread moderate immunoreactivity in the SVZ and VZ. At 12 PCW, KHDBRS2 immunoreactive cells were predominant in the deeper part of the CP (dCP), pSP and MZ, but rare in the superficial CP (sCP). Scattered positive cells were observed in the IZ and SVZ along with a small number in the VZ (D and D1). For KHDBRS3, a complex pattern of immunostaining was observed in the cortex with more immunopositive cells in the sCP and close to the boundary with the pSP, but with a band of weaker immunoreactivity in the middle of the CP. Weak to moderately immunoreactive cells were present throughout the IZ and proliferative layers (E and E1). KHDBRS2 expression was greater in the proliferative zone of the LGE than the MGE with a few scattered positive cells present in the postmitotic mantle zone (D and D2). KHDBRS3 immunopositive cells were present throughout the proliferative zones of the LGE and MGE, with scattered cells observed in the postmitotic mantle zone (E and E2). Med and Lat Crtx, medial and lateral cortex; VP, ventral pallium, border between cortex and ganglionic eminences. (F and G) Expression of four exons, present in all three NRXNs, were chosen for analysis; 6 and 17 that are in the alpha (α) isoforms only of NRXN proteins, and 19 and 20 translated in both alpha and beta (β) isoforms. KHDBRS proteins splice NRXN mRNA to exclude exon 20. Statistically significant reductions in exons 6 or 17 compared to 19 indicate less expression of α compared to the β isoforms, and significant reductions in 20 compared to 19 indicates suppression of exon 20 expression by KHDBRS proteins (*P < 0.05, **P < 0.01 by Tukey's post hoc test). Error bars indicate standard errors of the mean. Scale bars = 750 µm for B; 200 µm for C; 1 mm for D and E; 250 µm for D1, D2, E1, and E2.
In general terms, protein expression reflected mRNA expression. Immunoreactivity for KHDBRS1 (SAM68) was intense and appeared to be present in every cell, proliferative and postmitotic, and throughout all compartments of the cerebral cortex and ventral telencephalon, between 8 and 12 PCW (Fig. 7B). KHDBRS2 (SLM1) showed the lowest levels of immunoreactivity, in concordance with the findings from RNAseq. At 8 PCW most cells of the MZ, CP, and pSP were immunopositive for KHDBRS2. Isolated immunostained cells were also seen in the IZ and SVZ but not the VZ. By 12 PCW, KHDBRS2 immunoreactive cells were predominant in the deeper part of the CP, pSP, and MZ, but rare in the superficial CP. Scattered positive cells were observed in the IZ and SVZ along with a small number in the VZ. In the ventral telencephalon, KHDBRS2 expression was greater in the proliferative zone of the lateral ganglionic eminence (LGE) than the medial ganglionic eminence (MGE) a few scattered positive cells were also seen in the postmitotic mantle zone (Fig. 7D).
KHDBRS3 (SLM2/T-STAR) showed higher levels of immunoreactivity than KHDRBS2. At 8 PCW, in addition to strong immunostaining in the CP, MZ, and pSP/IZ, there was widespread moderate immunoreactivity in the proliferative layers (Fig. 7C). By 12 PCW, a complex pattern of immunostaining had emerged in the CP, with more immunopositive cells in the superficial layer and also close to the boundary with the pSP, but with a band of weaker immunoreactivity in the middle of the CP. Weak to moderately immunoreactive cells were present throughout the IZ and proliferative layers (Fig. 7E). In the ganglionic eminences, immunopositive cells were present throughout the proliferative zones of the LGE and MGE, with scattered cells present in the postmitotic mantle zone (Fig. 7E). In the mouse, KHDBRS2 and 3 are expressed in distinct populations of cortical interneurons with KHDBRS2 restricted to a small subpopulation of calbindin positive interneurons (Iijima et al. 2014). The reduced expression of KHDBRS2 compared to KHDBRS3, particularly in the MGE suggests that in human also that KHDBRS proteins may segregate to different interneuron precursors, but this was not explored further. However, the extensive expression of KHDBRS2 protein in the CP differs from the lack of expression of this protein by the glutamatergic neurons of the adult mouse neocortex (Iijima et al. 2014).
Having established expression of KHDBRS proteins we reexamined the RNAseq data at the level of expression of individual exons of the NRXN genes in cortical samples at 9 and 12 PCW. Exons 6 and 17, which belong to α NRXNs only, and exons 19 and 20, which are found in both α and β isoforms, were chosen. Alternative splicing by KHDBRS proteins occurs at alternative splice site (AS) 4 and results in suppression of expression of exon 20 compared to exon 19 (Rowen et al. 2002). One-way ANOVA established that there were statistically significant differences in expression between exons for all three NRXNs (at 9 PCW for NRXN1, P < 0.0001; NRXN2, P < 0.001; NRXN3, P < 0.03; at 12 PCW for NRXN1, P < 0.0001; NRXN2, P < 0.0001; NRXN3, P < 0.04). Post hoc comparisons revealed that at 9 PCW expression of NRXN1 exon 19 was significantly higher than expression of exon 17 or exon 20 (P < 0.01) suggesting that a modest amount of NRXN1β was transcribed and there was a significant two-thirds reduction in expression of exon 20 (Fig. 7F). By 12 PCW, the differences in expression between exon 19 and exons 6 and 17 were significant (P < 0.01) and larger than at 9 PCW, showing that NRXN1β expression had increased. Exon 20 continued to show a high degree of suppression of expression (Fig. 7F,G).
For NRXN2, at both ages studied, expression of exon 19 was significantly higher than all the other exons studied (P < 0.01). Relative production of NRXN2β compared to α isoforms seems to be greater than for NRXN1 isoforms especially at 9 PCW, but the degree of suppression of exon 20 is not as marked as for NRXN1 (Fig. 7F,G). For NRXN3, the expression levels of each exon were low and not markedly different, except that we observed significantly greater expression of exon 19 compared to 17 at 9 PCW; evidence for some production of the NRXN3β isoform, at least at this stage of development (Fig. 7F,G).
Discussion
This study has confirmed that all three NRXN genes show significant levels of expression in the early developing human cerebral cortex prior to extensive synapse formation, as suggested by preliminary gene expression studies (Ip et al. 2010; Kang et al. 2011), and as was previously shown in the mouse by in situ hybridization (Puschel and Betz 1995). Each NRXN had a unique expression pattern, which suggests that they each may have different functions. NRXN1 and 3 protein showed broadly similar patterns of expression, strongest in the CP, although only NRXN1 expression increased with age whereas NRXN3 expression was relatively low. This is in partial disagreement with Konopka et al. (2012) who found that NRXN1 was predominantly expressed in postmitotic cortical cells whereas NRXN3 was predominantly expressed in proliferating cells in both mid-gestation tissue sections and normal human neural progenitor cells differentiating in vitro. More recently, Jenkins et al. (2015) have demonstrated by qPCR that NRXN1 expression, in both α and β isoforms, increases in expression from 14 PCW until birth in the prefrontal cortex.
NRXN2 was more highly expressed at earlier stages than NRXN1 or NRXN3 throughout, and although present throughout the cortical wall NRXN2α was the only NRXN to strongly colocalized with neurites in the IZ and synaptogenic pSP. This diversity in expression patterns is unexpected as it is generally believed that there is no functional differentiation between α NRXNs based on “rescue” experiments in transgenic mice (Zhang et al. 2005) although this only probed their function in neurotransmitter release at the synapse. However, there are other respects in which NRXNs 1 and three diverge from NRXN2; a phylogenetic tree for the NRXN protein family demonstrates that NRXNs 1 and 3 are more closely related to each other than either is to NRXN2 (Reissner et al. 2013). Whereas NRXNs 1 and 3 are unusually long genes, greater than one Mega base pairs (Mbp) in length, the NRXN2 gene, despite having a very similar number of exons and amino acids in the protein, is only 0.117 Mbp in length (Rowen et al. 2002; Tabuchi and Südhof 2002). The smaller length of NRXN2 may permit transcription to be completed within a cell cycle in dividing cells (Rowen et al. 2002: Reissner et al. 2013) explaining its increased expression compared to NRXNs 1 and 3 at earlier stages of development and in proliferative layers. The largest gene NRXN3, on the other hand, showed the greatest degree of restriction to postmitotic layers.
Regulation of Alternative Splicing at AS4 and Synapse Formation
It is well established that presynaptic NRXNs on growing axons can interact with NLGNs expressed on growing dendrites to stabilize the postsynaptic site and establish vesicle accumulation at the presynaptic site (Graf et al. 2004; Chen et al. 2010). Although synapses are rare in the early developing cortex, they can be detected by electron microscopy in the MZ and pSP (but not the CP) and increase substantially over the period of the present study (Kostović and Rakic 1990). Proteins associated with synapses such as SYP, vesicular GABA transporter, and CASK have also been detected in the pSP at these ages by immunohistochemistry (Bayatti et al. 2008; Figs 3–6, present study). Therefore, it is reasonable to suppose that NRXN expression could reflect the nascent synaptogenesis occurring at this stage.
Regulation of Nrxn exon 20 expression at AS4 generates + or – isoforms that show differential binding to cell-type-specific postsynaptic partners (Baudouin and Scheiffele 2010; Vuong et al. 2016). KHDBRS proteins bring about splicing at AS4 and there is evidence of Khdbrs3 regulated exon skipping during mouse forebrain development (Ehrmann et al. 2013). NRXNβ AS4– isoforms preferentially bind NLGN1 concentrated at glutamatergic synapses, whereas NRXNα AS4– and NRXNβ AS4+ isoforms preferentially bind NLGN2 at GABAergic and glycinergic synapses (Chih et al. 2006). Thus, exclusion of exon 20 in different α or β isoforms might be required to induce formation of both major classes of synapse in the cortex. KHDBRS1 was found to be abundantly expressed in the developing cortex, but in mouse, KHDBRS1 only affects Nrxn splicing in the presence of depolarizing neuronal activity (Iijima et al. 2011). As in the 16–20 PCW human cortex, there is only significant neuronal activity in the subplate and the CP is largely quiescent (Moore et al. 2009), it is not expected that KHDBRS1 makes a significant contribution to NRXN splicing in our study.
However, both KHDBRS2 and 3 constitutively affect Nrxn splicing: ablation of either leads to a severe reduction in the Nrxn AS4– isoform in neurons or brain areas where they are typically expressed ( Ehrmann et al. 2013; Iijima et al. 2014; Traunmuller et al. 2014). We observed expression of both these proteins, particularly in the CP and pSP, along with significant reduction in exon 20 expression, which we predict could occur in NRXN1α and NRXN2α at all ages studied, NRXN3α at 12 PCW and also in β isoforms of NRXN1 and 2 (Fig. 7F,G). GABAergic synapses are likely to be present in the human pSP at this stage of development (Bayatti et al. 2008) and our data suggest that both NRXNα AS4– and NRXNβ AS4+ isoforms and NLGN2, which interact in their formation (Chih et al. 2006: Vuong et al. 2016) could be expressed in the pSP at this stage of development. NRXN2α immunoreactivity was observed within synaptogenic zones. NRXNα appears important for GABAergic synapse assembly as triple Nrxnα knock-out mice show a 50% reduction in cortical GABAergic synapse density (Missler et al. 2003). Early cortical network activity also relies upon glutamate receptor mediated activity (Kilb et al. 2011) and our mRNA expression data suggest the proteins required for glutamatergic synapse formation, NLGN1, NRXN1β AS4–, and NRXN2β AS4– (Chih et al. 2006) could also be present, although their protein expression was not examined in this study.
NRXNs, Vesicles and Axon Growth
Alpha NRXNs interact with components of the exocytosis machinery indirectly via binding to CASK and synaptotagmin (O'Connor et al. 1993; Hata et al. 1996; Reissner et al. 2013). They are one of the receptors for α-latrotoxin which causes uncontrolled exocytosis (Südhof 2001) and knock-out of Nrxnα greatly reduces spontaneous and evoked neurotransmitter release at all synapses in the mouse brain (Missler et al. 2003). Although NRXN2α colocalized with SYP, our marker for presynaptic terminals, in the pSP and MZ, there appeared to be stronger colocalization with GAP43 in the IZ of the cortex dorsally and the internal capsule ventrally (Fig. 3) where growing axons mostly emanating from CP neurons are located at this stage (Ip et al. 2011). There was also strong colocalization of NRXN2α and CASK in growing axons. The fusion of intracellular vesicles with the external membrane at the growth cone provides a mechanism for addition of membrane and guidance receptors to the leading filopodia (Tsaneva-Atanasova et al. 2009) and there is increasing evidence for the role of synapse-related SNARE (soluble N-ethylmaleimide-sensitive fusion attachment protein receptor) proteins in the growth of neurites (Kunwar et al. 2010; Zylbersztejn and Galli 2011). NRXN1β has also been shown to promote neuritic outgrowth in concert with NGLN1 and activation of fibroblast growth factor receptor-1 (Gjørlund et al. 2012).
NRXNs and Other Forms of Intercellular Connections
NRXNs appear not to be just confined to synapses of the nervous system. NRXNs can form a stoichiometric complex with dystroglycan, a ubiquitously expressed transmembrane protein linking cytoskeletal actin to the extracellular matrix. Dystroglycan extracted from brain, heart, and skeletal muscle can interact with NRXN1 (Sugita et al. 2001) and it was found that cardiac isoforms of NRXN3 participate in a complex involving dystroglycan and proteins of the extracellular matrix, presumably involved in intercellular connections (Occhi et al. 2002). Alternative splicing at the α isoform-specific AS2 regulates binding of NRXNαs to dystroglycan (Sugita et al 2001). Dystroglycan expression in the developing mouse cerebral cortex at embryonic day 15, equivalent to the stage of development studied here, is confined to both the CP and the apical surface of the VZ (Lathia et al. 2007) reminiscent of the pattern of NRXN1 immunoreactivity we have observed (Figs 3 and 4). The competitive interaction of dystroglycan and neurexophilin1 with NRXN1α controls the rate of proliferation of human hematopoietic stem cells (Kinzfogel et al. 2011). As radial glial cells predominantly undergo cell division at the apical surface of the VZ (Taverna and Huttner 2010; Harkin et al. 2016) and neurexophilin1 is expressed in the cortex at this stage of development (Table 2), we hypothesize that NRXN1α in particular may interact with the extracellular matrix at tight junctions at the apical surface of the VZ, participating in the control of progenitor cell proliferation, and may also interact with dystroglycan in the CP.
NRXNs and Cell Migration
Expression of NRXN protein in cells of the VZ, SVZ, and IZ may be an indication of its requirement in cell migration away from the proliferative zones. Indeed, NRXN expression also tended to be higher in the outer part of the CP, where new neurons are arriving, compared to the inner CP where older cells have settled having finished migration (Bystron et al. 2008). Cell adhesion molecules are important for neuronal migration (Maness and Schachner 2007) and molecules such as cadherins, for instance, also found at the synapse in the mature nervous system, are regarded as integral to the migration process (Redies et al. 2012). Certainly, the knock down of Cntnap2, which is similar in structure to NRXNs and classed as an autism susceptibility gene, causes abnormal neuronal migration, affecting both cortical projection neurons and interneurons, in transgenic mice (Peñagarikano et al. 2011) resulting in epilepsy and autism-like behavioral deficits. In cases of human CNTNAP2 gene mutation, cell migration defects, epilepsy, ASD, and language deficits have all been reported (Strauss et al. 2006; Alarcón et al. 2008; Scott-Van Zeeland et al. 2010). The large extracellular portion of CNTNAP2, like the NRXNs (Supplementary Fig. 1), is composed of protein–protein interaction domains including laminin G and EGF repeats ( Poliak et al. 1999). CNTNAP2 interacts extracellularly with CNTN2 (TAG-1) which is expressed early in mouse development and blocking its function results in migration abnormalities of cortical pioneer neurons and GABAergic interneurons (Denaxa et al. 2005; Morante-Oria et al. 2003). Potentially, NRXNs may have as yet unappreciated roles in cell migration, and by extension, neurite outgrowth, processes which largely share the same molecular machinery (Maness and Schachner 2007).
Nuclear Localization of NRXN 1 and 3
The observation of NRXN-like immunoreactivity in the nucleus was unexpected. This is unlikely to be artefactual as, for NRXN1 at least, it was abolished with blocking peptide. Also the detection of nuclear NRXN expression increased with age which would not be expected for nonspecific staining. Finally, both antibodies employed recognize antigenic sties in the highly conserved c-terminal regions of NRXN1 and NRXN3, rather than sites in the ubiquitous protein domains of the extracellular parts of these molecules. Synaptic proteins are often colocalized to the synapse and the nucleus (Jordan and Kreutz 2009). For instance, CASK is known to translocate to the nucleus where CASK interacts with TBR1 and regulates the TBR1-dependent transcription (Hseuh et al. 2000; Wang et al. 2004). CASK immunoreactivity increased in the CP and proliferative regions at 12 PCW, the same age at which anti-NRXN immunoreactivity increased in these zones. CASK binds indirectly to the c-terminal regions of the NRXNs (Hata et al. 1996). Whether the C-terminal portion of NRXNs translocate to the nucleus, with or without CASK, remains a possibility to be further investigated.
NRXN Mutations and Neurodevelopmental Disorders
To what extent has uncovering expression patterns during early development suggested roles for NRXNs key to understanding how mutations may cause deficits in brain development? Genetic studies of neurodevelopmental conditions have shown that the great majority of mutations affect NRXN1α expression, leaving β expression intact (Reichelt et al. 2012). We have found that NRXN1 was expressed in substantial amounts in the developing cortex, with a higher proportion likely to be expressed in the α form, especially at 9 PCW where there is protein expression in the VZ. Therefore, mutations in NRXN1α in early cortical development could cause subtle alterations in rates and quantity of neuroblast production from radial glia.
A NRXN2 gene mutation in an ASD patient caused the production of a truncated protein which, in vitro, did not bind its NLGN partner or induce synaptic differentiation (Gauthier et al. 2011). The present study has shown that NRXN2α is the isoform most likely to be involved in induction of synapse differentiation in the human pSP and MZ; therefore, it is plausible that NRXN2 mutations may cause alterations in subplate development. Even at the early stages studied here, synaptic connectivity in the pSP may be generating synchronous oscillatory network activity in a first step toward the subplate developing into a hub for network activity to guide cortical development (Kanold and Luhmann 2010). Disruptions to the subplate have been implicated in epilepsy, ASD, and schizophrenia (Bunney et al. 1997; Kostović et al. 2015; Hutsler and Casanova 2016). However, we have provided equally strong evidence for involvement of NRXN2α in axon outgrowth and pathfinding (see above). It has also been argued recently that dysregulation of axonal growth and guidance should be given greater prominence when considering the etiology of ASD, as many candidate ASD-susceptibility genes impinge upon these processes (McFadden and Minshew 2013). Finally, it is worth noting that NRXN3 shows the lowest levels and the most restricted pattern of expression, and also the weakest association of gene polymorphisms with neurodevelopmental disorders (see Introduction section).
Conclusion
Without discounting the importance of the contribution of NRXN mutations to synaptic dysfunction in neurodevelopmental conditions, the present study suggests that there may be potential roles for NRXNs in many fundamental developmental processes in the cerebral cortex. All three NRXNs are expressed at the earliest stages of human cortical development and show distinct expression patterns. Functional experiments should be designed to probe potential roles in cell migration and axon guidance, axon outgrowth, regulation of neuronal proliferation, and early development of subplate circuitry.
Supplementary Material
Supplementary material are available at Cerebral Cortex online.
Funding
The human fetal material was provided by the Joint UK MRC/Wellcome Trust (grant # 099175/Z/12/Z) Human Developmental Biology Resource (www.hdbr.org). The RNAseq study was funded by a grant from UK MRC (grant # MC/PC/13047).
Supplementary Material
Notes
Lauren Harkin was in receipt of a studentship from the Anatomical Society. Ayman Alzu'Bi is in receipt of a studentship from Yarmouk University, Jordan. Alexandra Ferrara was supported by a bursary from the British Division of the International Academy of Pathology. We are grateful to Dr Peter Scheiffele for the generous gift of antibodies to KHDBRS2 and 3 and to the staff of the HDBR for their skilled assistance. Conflict of Interest: None declared.
References
- Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, et al. 2008. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 82:150–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Jaberi N, Lindsay S, Sarma S, Bayatti N, Clowry GJ. 2015. The early fetal development of human neocortical gabaergic interneurons. Cereb Cortex. 25:635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang ML, Owczarek S. 2013. A matter of balance: role of neurexin and neuroligin at the synapse. Neurochem Res. 38:1174–1189. [DOI] [PubMed] [Google Scholar]
- Baudouin S, Scheiffele P. 2010. SnapShot: neuroligin–neurexin complexes. Cell. 141:908. [DOI] [PubMed] [Google Scholar]
- Bayatti N, Moss JA, Sun L, Ambrose P, Ward JFH, Lindsay S, Clowry GJ. 2008. A molecular neuroanatomical study of the developing human neocortex from 8 to 17 post conceptional weeks revealing the early differentiation of the subplate and subventricular zone. Cereb Cortex. 18:1536–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur C. 2011. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 1380:42–77. [DOI] [PubMed] [Google Scholar]
- Born G, Grayton HM, Langhorst H, Dudanova I, Rohlmann A, Woodward BW, Collier DA, Fernandes C, Missler M. 2015. Genetic targeting of NRXN2 in mice unveils role in excitatory cortical synapse function and social behaviors. Front Synaptic Neurosci. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsics T, Lundberg E, Geerts D, Koomoa DL, Koster J, Wester K, Bachmann AS. 2010. Subcellular distribution and expression of prenylated Rab acceptor 1 domain family, member 2 (PRAF2) in malignant glioma: Influence on cell survival and migration. Cancer Sci. 101:1624–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Südhof TC. 2005. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 48:229–236. [DOI] [PubMed] [Google Scholar]
- Bullen P, Wilson D. 1997. The Carnegie staging of human embryos: a practical guide In: Strachan T, Lindsay S, Wilson DI, editors. Molecular genetics of early human development. Oxford: Bios Scientific Publishers. [Google Scholar]
- Bunney BG, Potkin SG, Bunney WE. 1997. Neuropathological studies of brain tissue in schizophrenia. J Psychiatr Res. 31:159–173. [DOI] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, Rakic P. 2008. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosi. 9:110–122. [DOI] [PubMed] [Google Scholar]
- Catani M, Dell'Acqua F Budisavlijevic S, Howell H, Thiebaut de Schotten M, Froudist-Walsh S, D'Anna L, Thompson A, Sandrone S, Bullmore ET et al. 2016. Frontal networks in adults with autism spectrum disorder Brain. 139:616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SX, Tari PK, She K, Haas K. 2010. Neurexin-neuroligin cell adhesion complexes contribute to synaptotropic dendritogenesis via growth stabilization mechanisms in vivo. Neuron. 67:967–983. [DOI] [PubMed] [Google Scholar]
- Cheng S-B, Amici SA, Ren X-Q, McKay SB, Treuil MW, Lindstrom JM, Rao J, Anand R. 2009. Presynaptic targetting of α4β2 nicotinic acetylcholine receptors Is regulated by neurexin-1β. J Biol Chem. 28:23251–23259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Gollan L, Scheiffele P. 2006. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 51:171–178. [DOI] [PubMed] [Google Scholar]
- Ching MS, Shen Y, Tan WH, Jeste SS, Morrow EM, Chen X, Mukaddes NM, Yoo SY, Hanson E, Hundley R, et al. 2010. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am J Med Genet B Neuropsychiatr Genet. 153B:937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoletti D, Flynn RE, Boucard AA, Demeler B, Schirf V, Shi J, Jennings LL, Newlin HR, Südhof TC, Taylor P. 2006. Gene selection, alternative splicing, and post-translational processing regulate neuroligin selectivity for beta-neurexins. Biochemistry. 24:12816–12827. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Pierce K, Schumann CM, Redcay E, Buckwalter JA, Kennedy DP, Morgan J. 2007. Mapping early brain development in autism. Neuron. 56:319–413. [DOI] [PubMed] [Google Scholar]
- Cukier HN, Dueker ND, Slifer SH, Lee JM, Whitehead PL, Lalanne E, Leyva N, Konidari I, Gentry RC, Hulme WF, et al. 2014. Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Mol Autism. 10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dachtler J, Glasper J, Cohen RN, Ivorra JL, Swiffen DJ, Jackson AJ, Harte MK, Rodgers RJ, Clapcote SJ. 2014. Deletion of α-neurexin II results in autism-related behaviors in mice. Transl Psychiatry. 4:e484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denaxa M, Kyriakopoulou K, Theodorakis K, Trichas G, Vidaki M, Takeda Y, Watanabe K, Karagogeos D. 2005. The adhesion molecule TAG-1 is required for proper migration of the superficial migratory stream in the medulla but not of cortical interneurons. Dev Biol. 288:87–99. [DOI] [PubMed] [Google Scholar]
- Ehrmann I, Dalgliesh C, Liu Y, Danilenko M, Crosier M, Overman L, Arthur HM, Lindsay S, Clowry GJ, Venables J, et al. 2013. The tissue specific RNA binding protein T-STAR controls regional splicing patterns of neurexin pre mRNAs in the brain. PLoS Genet. 9:e1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton MR, Blaiss CA, Powell CM, Südhof TC. 2009. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci USA. 106:17998–18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Schroer R, Yan J, Song W, Yang C, Bockholt A, Cook EH Jr, Skinner C, Schwartz CE, Sommer SS. 2006. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 409:10–13. [DOI] [PubMed] [Google Scholar]
- Friedman JM, Baross A, Delaney AD, Arbour L, Armstrong L, Asano J, Bailey DK, Barber S, Birch P, Brown-John M, et al. 2006. Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am J Hum Genet. 79:500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Siddiqui TJ, Huashan P, Yokomaku D, Hamdan FF, Champagne N, Lapointe M, Spiegelman D, Noreau A, Lafrenière RG, et al. 2011. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum Genet. 130:563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrelli D, Lisgo S, Copp AJ, Lindsay S. 2015. Enabling research with human embryonic and fetal tissue resources. Development. 142:3073–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjørlund MD, Nielsen J, Pankratova S, Li S, Korshunova I, Bock E, Berezin V. 2012. Neuroligin-1 induces neurite outgrowth through interaction with neurexin-1beta and activation of fibroblast growth factor receptor-1. Faseb J. 26:4174–4186. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes E, Brune CW, Bradfield JP, et al. 2009. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 459:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S, Morigaki R, Okita S, Nagahiro S, Kaji R. 2015. Development of a highly sensitive immuno-histochemical method to detect neurochemical molecules in formalin-fixed and paraffin embedded tissues from autopsied human brains. Front Neuroanat. 9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. 2004. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 119:1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayton HM, Missler M, Collier DA, Fernandes C. 2013. Altered social behaviours in neurexin 1α knockout mice resemble core symptoms in neurodevelopmental disorders. PLoS One. 8:e67114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata Y, Butz S, Südhof TC. 1996. CASK: a novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J Neurosci. 14:2488–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkin LF, Gerrelli D, Diaz DCG, Santos C, Alzu'bi A, Austin CA, Clowry GJ. 2016. Distinct expression patterns for type II topoisomerases IIA and IIB in the early foetal human telencephalon. J Anat. 228:452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkin LF, Gullon EA, Lindsay S, Clowry GJ. 2015. The expression of autism susceptibility genes in the earliest stages of human cerebral cortex development. Int J Dev Neurosci. 47:75.29887103 [Google Scholar]
- Hern WM. 1984. Correlation of fetal age and measurements between 10 and 26 weeks of gestation. Obstet Gynecol. 63:26–32. [PubMed] [Google Scholar]
- Hu X, Zhang J, Jin C, Mi W, Wang F, Ma W, Ma C, Yang Y, Li W, Zhang H, et al. 2013. Association study of NRXN3 polymorphisms with schizophrenia and risperidone-induced bodyweight gain in Chinese Han population. Prog Neuropsychopharmacol Biol Psychiatry. 43:197–20. [DOI] [PubMed] [Google Scholar]
- Hsueh YP, Wang TF, Yang FC, Sheng M. 2000. Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature. 404:298–302. [DOI] [PubMed] [Google Scholar]
- Hutsler JJ, Casanova MF. 2016. Cortical construction in autism spectrum disorder: columns, connectivity and the subplate. Neuropathol Appl Neurobiol. 42:115–134. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K, Hata Y, Nguyen T, Ullrich B, Missler M, Moomaw C, Südhof TC. 1995. Neuroligin 1: a splice site-specific ligand for β-neurexins. Cell. 81:435–443. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K, Nguyen T, Südhof TC. 1996. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J Biol Chem. 271:2676–2682. [DOI] [PubMed] [Google Scholar]
- Iijima T, Iijima Y, Witte H, Scheiffele P. 2014. Neuronal cell type-specific alternative splicing is regulated by the KH domain protein SLM1. J Cell Biol. 204:331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima T, Wu K, Witte H, Hanno-Iijima Y, Glatter T, Richard S, Scheiffele P. 2011. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 147:1601–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip BK, Bayatti N, Howard NJ, Lindsay S, Clowry GJ. 2011. The corticofugal neuron-associated genes ROBO1, SRGAP1 and CTIP2 exhibit an anterior to posterior gradient of expression in early foetal human neocortex development. Cereb Cortex. 21:1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip BK, Wappler I, Peters H, Lindsay S, Clowry GJ, Bayatti N. 2010. Investigating gradients of gene expression involved in early human cortical development. J Anat. 217:300–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins AK, Paterson C, Wang Y, Hyde TM, Kleinman JE, Law AJ. 2015. Neurexin 1 (NRXN1) splice isoform expression during human neocortical development and aging. Mol Psychiatry. 21:701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan BE, Kreutz MR. 2009. Nucleocytoplasmic protein shuttling: the direct route in synapse-to-nucleus signaling. Trends Neurosci. 32:392–401. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AMM, Pletikos M, Meyer KA, Sedmak G, et al. 2011. Spatio-temporal transcriptome of the human brain. Nature. 478:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanold PO, Luhmann HJ. 2010. The subplate and early cortical circuits. Annu Rev Neurosci. 33:23–48. [DOI] [PubMed] [Google Scholar]
- Kavanagh DH, Tansey KE, O'Donovan MC, Owen MJ. 2015. Schizophrenia genetics: emerging themes for a complex disorder. Mol Psychiatry. 20:72–76. [DOI] [PubMed] [Google Scholar]
- Kilb W, Kirischuk S, Luhmann HJ. 2011. Electrical activity patterns and the functional maturation of the neocortex. Eur J Neurosci. 34:1677–1686. [DOI] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, Lally E, Weiss LA, Najm J, Kutsche K, et al. 2008. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 82:199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzfogel J, Hangoc G, Broxmeyer HE. 2011. Neurexophilin 1 supresses the proliferation of hematopoietic progenitor cells. Blood. 118:565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, O'Donovan MC, Erdogan F, Owen MJ, Ropers HH, et al. 2008. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 17:458–465. [DOI] [PubMed] [Google Scholar]
- Klein ME, Younts TJ, Castillo PE, Jordan BA. 2013. RNA-binding protein Sam68 controls synapse number and local β-actin mRNA metabolism in dendrites. Proc Natl Acad Sci USA. 110:3125–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehnke J, Katsamba PS, Ahlsen G, Bahna F, Vendome J, Honig B, Shapiro L, Jin X. 2010. Splice form dependence of beta-neurexin/neuroligin binding interactions. Neuron. 67:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Wexler E, Rosen E, Mukamel Z, Osborn GE, Chen L, Lu D, Gao F, Gao K, Lowe JK, et al. 2012. Modeling the functional genomics of autism using human neurons. Mol Psychiatr. 17:202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostović I, Rakic P. 1990. Developmental history of the transient subplate zone in the visual and somatosensory cortex of the macaque monkey and human brain. J Comp Neurol. 297:441–470. [DOI] [PubMed] [Google Scholar]
- Kostović I, Sedmak G, Vukšić M, Judaš M. 2015. The relevance of human fetal subplate zone for developmental neuropathology of neuronal migration disorders and cortical dysplasia. CNS Neurosci Therapeut. 21:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota M, Miyata J, Sasamoto A, Sugihara G, Yoshida H, Kawada R, Fujimoto S, Tanaka Y, Sawamoto N, Fukuyama H, et al. 2013. Thalamocortical disconnection in the orbitofrontal region associated with cortical thinning in schizophrenia. JAMA Psychiatry. 70:12–21. [DOI] [PubMed] [Google Scholar]
- Kunwar AJ, Rickmann M, Backofen B, Browski SM, Rosenbusch J, Schoning S, Fleischmann T, Krieglstein K, von Mollard GF. 2010. Proc Nat Acad Sci USA. 108:2575–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Patton B, Eckley DM, Magnus T, Mughal MR, Sasaki T, Caldwell MA, Rao MS, Mattson MP, Ffrench-Constant C. 2007. Patterns of laminins and integrins in the embryonic ventricular zone of the CNS. J Comp Neurol. 505:630–643. [DOI] [PubMed] [Google Scholar]
- Lindsay S, Xu Y, Lisgo S, Harkin LF, Copp A, Gerrelli D, Clowry GJ, Talbot A, Wilson I, Coxhead J, et al. 2016. A unique resource for global and individual gene expression studies during early human brain development. Front Neuroanat. 10:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M, Anders S, Huber W. 2014. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maness PF, Schachner M. 2007. Neural recognition molecules of the immunoglobulin superfamily: signalling transducers of axon guidance and neuronal migration. Nature Neurosci. 10:19–26. [DOI] [PubMed] [Google Scholar]
- McFadden K, Minshew NJ. 2013. Evidence for dysregulation of axonal growth and guidance in the etiology of ASD. Front Human Neurosci. 7:671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Südhof TC. 1998. Neurexins: three genes and 1001 products. Trends Genet. 14:20–26. [DOI] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, Südhof TC. 2003. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 214:939–948. [DOI] [PubMed] [Google Scholar]
- Moore AR, Filipovic R, Mo Z, Rasband MN, Zecevic N, Antic SD. 2009. Electrical excitability of early neurons in the human cerebral cortex during the second trimester of gestation. Cereb Cortex. 19:1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante-Oria J, Carleton A, Ortino B, Kremer EJ, Fairén A, Lledo PM. 2003. Subpallial origin of a population of projecting pioneer neurons during corticogenesis. Proc Natl Acad Sci USA. 100:12468–12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata J, Hirao K, Namiki C, Fujiwara H, Shimizu M, Fukuyama H, Sawamoto N, Hayashi T, Murai T. 2009. Reduced white matter integrity correlated with cortico-subcortical gray matter deficits in schizophrenia. Schizophr Res. 111:78–85. [DOI] [PubMed] [Google Scholar]
- Occhi G, Rampazzo A, Beffagna G, Danieli GA. 2002. Identification and characterization of heart-specific splicing of human neurexin 3 mRNA (NRXN3). Biochem Biophys Res Commun. 298:151–155. [DOI] [PubMed] [Google Scholar]
- O'Connor VM, Shamotienko O, Grishin E, Betz H. 1993. On the structure of the ‘synaptosecretosome’. Evidence for a neurexin/synaptotagmin/syntaxin/Ca2+ channel complex. FEBS Lett. 326:255–260. [DOI] [PubMed] [Google Scholar]
- O'Rahilly R, Müller F, Hutchins GM, Moore GW. 1987. Computer ranking of the sequence of appearance of 73 features of the brain and related structures in staged human embryos during the sixth week of development. Am J Anat. 180:69–86. [DOI] [PubMed] [Google Scholar]
- Peñagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, et al. 2011. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 147:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko AG, Ullrich B, Missler M, Krasnoperov V, Rosahl TW, Südhof TC. 1996. Structure and evolution of neurexophilin. J Neurosci. 16:4360–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. 2010. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 466:368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliak S, Gollan L, Martinez R, Custer A, Einheber S, Salzer JL, Trimmer JS, Shrager P, Peles E. 1999. Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron. 24:1037–1047. [DOI] [PubMed] [Google Scholar]
- Puschel AW, Betz H. 1995. Neurexins are differentially expressed in the embryonic nervous system of mice. J Neurosci. 15:2849–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redies C, Hertel N, Hubner CA. 2012. Cadherins and neuropsychiatric disorders. Brain Res. 1470:130–144. [DOI] [PubMed] [Google Scholar]
- Reichelt AC, Rodgers RJ, Clapcote SJ. 2012. The role of neurexins in schizophrenia and autistic spectrum disorder. Neuropharmacol. 62:1519–1526. [DOI] [PubMed] [Google Scholar]
- Reissner C, Runkel F, Missler M. 2013. Neurexins. Genome Biol. 13:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowen L, Young J, Birditt B, Kaur A, Madan A, Philipps DL, Qin S, Minx P, Wilson RK, Hood L, Graveley BR. 2002. Analysis of the human neurexin genes: alternative splicing and the generation of protein diversity. Genomics. 14:587–597. [DOI] [PubMed] [Google Scholar]
- Rujescu D, Ingason A, Cichon S, Pietilainen OP, Barnes MR, Toulopoulou, Picchioni M, Vassos E, Ettinger U, Bramon E, et al. 2009. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 18:988–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner D, Nguyen T-M, Russo G, Heber S, Patrignani A, Ahrné E, Scheiffele P. 2014. Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron. 84:386–298. [DOI] [PubMed] [Google Scholar]
- Schreiner D, Simicevic J, Ahrné E, Schmidt A, Scheiffele P. 2015. Quantitative isoform-profiling of highly diversified recognition molecules. Elife. 18:e07794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Van Zeeland AA, Abrahams BS, Alvarez-Retuerto AI, Sonnenblick LI, Rudie JD, Ghahremani D, Mumford JA, Poldrack RA, Dapretto M, Geschwind DH, et al. 2010. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci Transl Med. 2:56RA80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. 2006. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 354:1370–1377. [DOI] [PubMed] [Google Scholar]
- Südhof TC. 2001. alpha-latrotoxin and its receptors: neurexins and CIRL/latrophilins. Ann Rev Neurosci. 24:933–962. [DOI] [PubMed] [Google Scholar]
- Südhof TC. 2008. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 455:903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S, Saito F, Tang J, Satz J, Campbell K, Südhof TC. 2001. A stoichiometric complex of neurexins and dystroglycan in brain. J Cell Biol. 154:435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. 2007. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 39:319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Südhof TC. 2002. Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics. 79(8):49–59. [DOI] [PubMed] [Google Scholar]
- Taverna E, Huttner WB. 2010. Neural progenitor nuclei IN motion. Neuron. 67:906–914. [DOI] [PubMed] [Google Scholar]
- Traunmuller L, Bornmann C, Scheiffele P. 2014. Alternative splicing coupled nonsense mediated decay generates neuronal cell type specific expression of SLM proteins. J Neurosci . 34:16755–16761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaneva-Atanasova K, Burgo A, Galli T, Holcman D. 2009. Quantifying neurite growth mediated by interactions among secretory vesicles, microtubules, and actin networks. Biophys J. 96:840–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich B, Ushkaryov YA, Südhof TC. 1995. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 14:497–507. [DOI] [PubMed] [Google Scholar]
- Ushkaryov YA, Hata Y, Ichtchenko K, Moomaw C, Afendis S, Slaughter CA, Südhof TC. 1994. Conserved domain structure of beta-neurexins. Unusual cleaved signal sequences in receptor-like neuronal cell-surface proteins. J Biol Chem. 269:11987–11992. [PubMed] [Google Scholar]
- Vaags AK, Lionel AC, Sato D, Goodenberger M, Stein QP, Curran S, Ogilvie C, Ahn JW, Drmic I, Senman L, et al. 2012. Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am J Hum Genet. 90:133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viñas-Jornet M, Esteba-Castillo S, Gabau E, Ribas-Vidal N, Baena N, San J, Ruiz A, Coll MD, Novell R, Guitart M. 2014. A common cognitive, psychiatric, and dysmorphic phenotype in carriers of NRXN1 deletion. Mol Genet Genomic Med. 2:512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong CK, Black DL, Zheng S. 2016. The neurogenetics of alternative splicing. Nature Neurosci Rev. 17:265–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TF, Ding CN, Wang GS, Luo SC, Lin YL, Ruan Y, Hevner R, Rubenstein JL, Hsueh YP. 2004. Identification of Tbr-1/CASK complex target genes in neurons. J Neurochem. 91:1483–1492. [DOI] [PubMed] [Google Scholar]
- Wei JL, Fu ZX, Fang M, Zhou QY, Zhao QN, Guo JB, Lu WD, Wang H. 2014. High expression of CASK correlates with progression and poor prognosis of colorectal cancer. Tumour Biol. 35:9185–9194. [DOI] [PubMed] [Google Scholar]
- Ye H, Liu J, Wu JY. 2010. Cell adhesion molecules and their involvement in autism spectrum disorder. Neurosignals. 18:62–71. [DOI] [PubMed] [Google Scholar]
- Zhang W, Rohlmann A, Sargsyan V, Aramuni G, Hammer RE, Südhof TC, Missler M. 2005. Extracellular domains of alpha-neurexins participate in regulating synaptic transmission by selectively affecting N- and P/Q-type Ca2+ channels. J Neurosci. 25:4330–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang J, Li A, Liu H, Zhang W, Cui X, Wang K. 2013. Expression of neurexin and neuroligin in the enteric nervous system and their down-regulated expression levels in Hirschsprung disease. Mol Bio Rep. 40:2969–2975. [DOI] [PubMed] [Google Scholar]
- Zhao S, Fernald RD. 2005. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 12:1047–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylbersztejn K, Galli T. 2011. Vesicular traffic in cell navigation. FEBS J. 278:4497–4505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.