Abstract
Collagen IV scaffolds assemble through an intricate pathway that begins intracellularly and is completed extracellularly. Multiple intracellular enzymes act in concert to assemble collagen IV protomers, the building blocks of collagen IV scaffolds. After being secreted from cells, protomers are activated to initiate oligomerization, forming insoluble networks that are structurally reinforced with covalent crosslinks. Within these networks, embedded binding sites along the length of the protomer lead to the “decoration” of collagen IV triple helix with numerous functional molecules. We refer to these networks as “smart” scaffolds, which as a component of the basement membrane enable the development and function of multicellular tissues in all animal phyla. In this review, we present key molecular mechanisms that drive the assembly of collagen IV smart scaffolds.
Keywords: collagen IV, extracellular matrix, chloride, scaffold, basement membrane
Abbreviations
- BM
basement membrane
- ECM
extracellular matrix
- PBS
phosphate‐buffer saline
- SDS‐PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TBS
tris‐buffer saline
- PTM
post translational modification
Introduction
Type IV collagen is a unique component of the basement membrane (BM) present in all animal phyla.1, 2, 3, 4 It is a member of the collagen superfamily that comprises 28 different types in vertebrates.5, 6, 7, 8 Unlike other vertebrate collagens, collagen IV occurs only in the BM and contains up to six genetically distinct α‐chains designated α1(IV) to α6(IV). Three helical polypeptide α‐chains combine to form a collagen IV protomer. For example, two α1 chains and one α2 chain combine to form the α112 collagen IV protomer. Out of the many potential combinations, the α‐chains interact and assemble with a remarkable specificity to form only three distinct protomers, that is α112, α345, and α556.7 The α1(IV) and α2(IV) chains were first to be described and thus called “classical” chains. They are present in the BM of all tissues, whereas the other four chains have restricted tissue distribution during development.7 For example, the α3(IV), α4(IV), and α5(IV) chains are present in the glomerular basement membrane of the kidney and in the BM of lung, testis, and eye, whereas the α5(IV) and α6(IV) chains are found in the BM of skin, smooth muscle, and the kidney.9 Most mutations in COL4A1 and COL4A2, the genes encoding α1(IV) or α2(IV), respectively, cause multisystem disorders with heterogeneous pathogenic mechanisms and often lead to embryonic lethality.10, 11, 12, 13, 14 Mutations in COL4A3, COL4A4, and COL4A5, the genes encoding α3(IV), α4(IV), or α5(IV) chains, respectively, lead to renal failure and deafness in adult patients with Alport's syndrome.15, 16
Once secreted into the extracellular space, the triple‐helical protomers self‐associate to form distinct networks providing a molecular scaffold for interactions between other BM components such as laminin networks, perlecans, and proteoglycans to form a mature BM. At the cellular level, collagen IV scaffolds are found underlying epithelial cells and surrounding Schwann cells, myocytes, and adipocytes to name a few examples (Fig. 1). The scaffolds provide a structural support for nearby cells, and they tether other extracellular molecules, including growth factors, laminins, proteoglycans, and nidogens. This complex, embedded in the BM, possesses a diverse set of biologic functions, including cell adhesion, migration, development, tissue regeneration and wound healing, immobilization of growth factors and enzymes, and in molecular sieving.17, 18, 19 Such biologic activity places great importance on understanding how collagen IV smart scaffolds are assembled.
Figure 1.
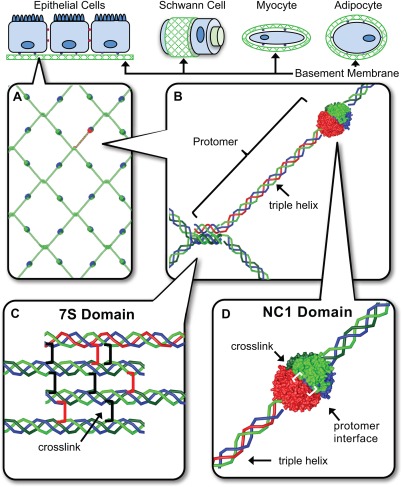
The NC1 and 7S domains are the primary junction points in collagen IV network assembly in BMs. BMs interact directly with most eukaryotic cell types enabling tissue functions. (A) Heterotrimeric collagen IV protomers are composed of three α chain monomers. (B) In network assembly, four collagen IV protomers associate at their 7S domains (C) where as two protomers self‐associate at their NC1 domains. (D).
Structurally, collagen IV scaffolds are composed of networked heterotrimeric collagen IV protomers [Fig. 1(A)], containing a C‐terminal trimer of NC1 domains, triple helix, and N‐terminal 7S domain. Key protomer junctions include head‐to‐head and tail‐to‐tail interactions between collagen IV NC1 domains and 7S domains, respectively [Fig. 1(B–D)]. Recent discoveries have painted an elaborate picture of key molecular mechanisms driving intracellular protomer assembly; these include intracellular protomer assembly, protomer secretion from cells, extracellular network assembly, and novel covalent crosslinking. We herein review major steps in building functional collagen IV smart scaffolds.
Overview of Scaffold Assembly
Collagen IV scaffolds are synthesized through a pathway of intracellular and extracellular mechanisms where the intracellular steps include assembly of heterotrimeric protomers while further protomer assembly into a three‐dimensional (3D) scaffold occurs extracellularly. Scaffold assembly involves multiple enzymes collectively underscoring the resources invested by the cell in the formation of collagen IV scaffolds. Protomers form through the self‐assembly of three collagen IV α‐chains. NC1 domains nucleate protomer assembly9 through a mechanism that regulates chain selection and triggers winding of the triple helix toward the N‐termini20 (Fig. 2). Protomer assembly is also assisted by post‐translational modifications and chaperone functions through the activities of prolyl 3‐hydroxylase 2 (P3H2) and heat shock protein 47 (HSP47), respectively.21, 22, 23 Upon secretion assembled protomers adjoin through their NC1 domains and 7S domains, providing key junctions at the protomer termini, while triple helices interact through lateral interactions that form supercoils.24 Oligomerization at the NC1 domain is driven through ionic Cl− driven activation of a molecular switch within individual protomers enabling binding of a neighboring protomer (Fig. 2).20 The 7S domains assemble into dodecameric structures (e.g., heterotrimeric 7S domains from a complex of four independent protomers). As a result of this NC1‐ and 7S‐directed oligomerization, newly secreted protomers are incorporated into nascent BMs.
Figure 2.
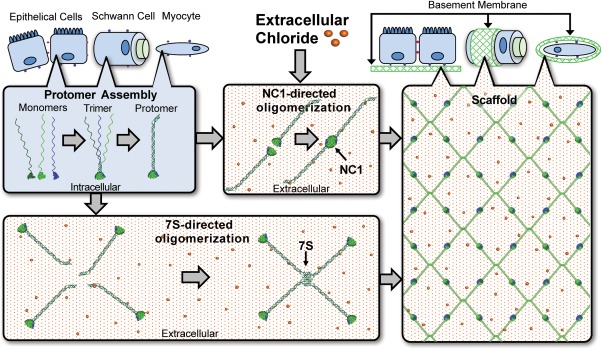
The NC1‐ and 7S‐mediated assembly drive collagen IV scaffold assembly. Collagen IV monomers are transcribed and assemble into protomers intracellularly. The NC1 and 7S domains facility assembly of protomers into higher order scaffold structures extracellularly. The higher Cl− concentration of the ECM is required for NC1‐directed oligomerization.
Networks of collagen IV protomers are reinforced with covalent crosslinks in the NC1 and 7S regions in order to function as a scaffold. NC1 hexamers are reinforced with sulfilimine crosslinks (‐S=N‐) between Met93 and Hyl211 on opposing NC1 domains [Fig. 1(D)],25 being formed by a heme peroxidase embedded within BMs called peroxidasin (PXDN).26, 27 In parallel, 7S dodecamers are crosslinked by lysyl oxidase‐like 2 (LOXL2) within BMs.28 Loss of either sulfilimine crosslinks or the LOX2 crosslinks can disrupt collagen IV scaffolds, the encompassing BMs, and nearby tissues.26, 28, 29 In summary, scaffold assembly is a highly regulated process involving specific molecular mechanisms. Key stages of this assembly at the atomic and molecular levels are presented below.
Stages of Scaffold Assembly
NC1 trimerization
NC1 trimerization is a seminal event in protomer assembly that governs chain selection, registration, and stoichiometry.9, 20 Although there are six collagen IV chains (α1–6) allowing for numerous potential trimeric protomers, the chain composition of assembled protomers is restricted to only three combinations: α112, α345, and α556. NC1 trimers nucleate assembly of N‐terminal triple helices (Fig. 3), which involves aligning the three α‐chains to establish the correct downstream binding sites in the protomer.30, 31 This is achieved through specific recognition motifs within NC1 domains that enable the self‐selection of the appropriate binding partner.
Figure 3.
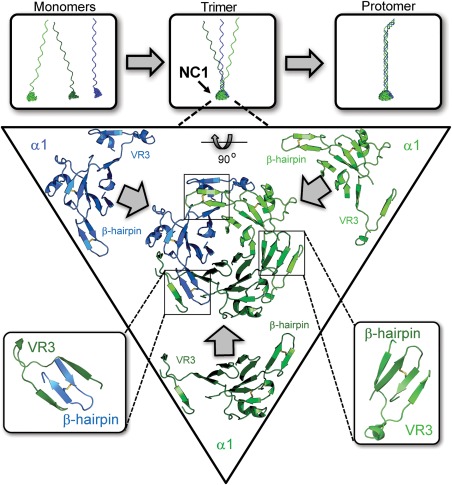
NC1 domains direct protomer assembly. Each collagen IV protomer is formed from three individual monomers. The NC1 domain controls protomer stoichiometry, initiates triple helix formation, and mediates triple helical chain register. Within each NC1 trimer, the monomers recognize one another through a domain‐swapping mechanism in which β‐hairpin motif of one monomer is strand‐swapped into a docking site formed by VR3 of its binding partner. Cartoon representation of the trimerization domain viewed down the threefold rotation axis and rotated by 90° about the horizontal axis.
The mechanism of NC1 self‐assembly involves domain‐swapping interactions where each domain extends a β‐hairpin motif into a complementary docking site located within a groove on the adjoining domain (Fig. 3).32, 33 Among the six α‐chains, three variable regions are found within the NC1 domain. Of these, variable region 3 (VR3) is located near the β‐hairpin motif of the adjoining NC1 domain and appears to strongly influence side‐to‐side NC1 interactions and chain selection.33 Extensive noncovalent interactions are found among the VR3, β‐hairpin motif, and the rest of the NC1–NC1 interface. Specifically, the initial monomer–monomer association of the protomer assembly is governed predominantly by nonpolar interactions; chain specificity is controlled by the β‐hairpin motif and its docking partner, the VR3 region (Fig. 3).32, 33, 34 For the α112 NC1 trimer, kinetic studies indicate that trimerization is initiated by side‐to‐side binding between the α2 and α1 NC1 domains. The α2 NC1 domain β‐hairpin loop binds to the α1 VR3 region forming a stronger interaction compared to analogous interactions in the trimer, that is α1 β‐hairpin to α2 VR3 or α1 β‐hairpin to α1 VR3. Thus, the α2 VR3 appears to initiate formation of the α112 NC1 trimer.33 Sequence comparisons suggest the α4 and α6 chains may nucleate trimerization of the α345 and α556 NC1 trimers, respectively.32
NC1‐directed oligomerization
Collagen IV NC1 domains not only govern protomer assembly within the cell, they also prevent aberrant intracellular scaffold assembly, and direct oligomerization in the extracellular space. Upon secretion from cells, protomers are exposed to higher Cl− concentrations (ca. 12 mM inside muscle cells vs. 100 mM in serum)35 that trigger NC1‐directed oligomerization as an initial step of network assembly. Cl− is required for NC1 oligomerization in vitro as well as the production of collagen IV networks in cell culture. The precise molecular “switch” controlling NC1 oligomerization has been recently described with atomic detail.20
Within the cell where free Cl− levels are relatively low, the side‐chain charge of residue Arg76 is balanced by forming an intramolecular salt‐bridge with residue Asp78 and to a lesser extent Glu40 [Fig. 4(A)]. The strong bias of sequentially proximal side chains to form salt‐bridges is well known36, 37 in turn decreasing conformational entropy by loop closure.38 The Arg76‐Asp78 salt‐bridge functionally blocks protomer oligomerization. Therefore, disruption of the Arg76–Asp78 association is a requisite step preceding protomer oligomerization. The stability of the intramolecular Arg76–Asp78 is modulated by the nonspecific electrostatic screening properties of Cl−. Once a protomer is excreted from the cell and exposed to the extracellular Cl− levels, the occupancy of the Arg76–Asp78 salt‐bridge is greatly reduced [Fig. 4(B)]. The molecular switch allowing protomer oligomerization at the NC1 domain is effectively turned on when a chloride ion binds a nest motif (residues 75–79) within each monomer NC1.34, 39 Bound Cl− functions in part as a structural wedge blocking Arg76–Asp78 reassociation by coordinating their amide backbones, limiting side chain conformations [Fig. 4(C)].
Figure 4.
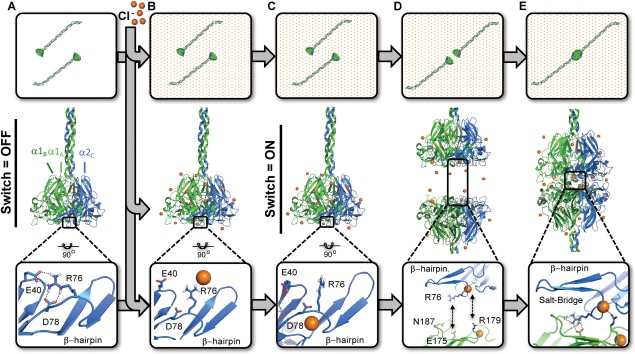
Cl− triggers a molecular switch enabling NC1‐directed oligomerization. In the absence of Cl−, Arg76 forms intramolecular salt‐bridge with Asp78 and/or Glu40 (A). Extracellular Cl− disrupts Arg76‐Asp78 salt bridge by electrostatic screening (B). Cl− binds each monomer by coordination with the Arg76 backbone amide, thus orienting the side toward an opposing NC1 timer (C). Reorientation of the Arg76 side chain allows for two protomers to bind one another through a Arg76‐Glu175:Asn187 salt‐bridge (D). Each bound Cl− ion can also coordinate Arg179 of the opposing protomer for a total up to 12 Cl− mediated interactions per hexamer (E).
With the Arg76–Asp78 salt bridge broken, the electrostatically rich protomer interface is free to bind another protomer where residue Arg76 forms the crux of six essential intermolecular salt‐bridges with Glu175 and Asn187. Residue Arg76 bridges the protomer interface forming a bidentate side‐on interprotomer interaction with Glu175. In addition Arg76 “networks” with Asn187 by hydrogen bonding in an “end‐on” configuration [Fig. 4(D)] forming what is termed a “bridging‐networked” salt‐bridge.40 These salt‐bridges are rare in composition while novel in application within NC1 domains. To date, this salt‐bridge motif is found only in three other structures: Acyl‐CoA oxidase (1IS2),41 α‐l‐arabinofuranosidase (1WD3),42 and malate dehydrogenase43 (1BMD). In contrast with in all three structures, the Glu‐Arg salt‐bridge was “networked” by hydrogen‐bonding an intradomain Asn side‐chain, whereas the “bridging” functionality is exclusive to the collagen IV NC1 domain. In addition to the salt‐bridge, the Cl− ion can form 6 additional intermolecular electrostatic interactions directly with Arg179 to further stabilize the protomer–protomer interface [Fig. 4(E)]. In total six salt‐bridges and six electrostatic interactions are dependent on Cl− binding.
Sulfilimine crosslinking by PXDN
NC1 hexamers are critical junctures within collagen IV scaffolds and sulfilimine crosslinks are key to their structural integrity. Following NC1‐directed oligomerization, NC1 domains are covalently bound across the protomer interface at Met93 and Hyl211 with novel sulfilimine crosslinks (‐S=N‐) (Fig. 5).25 To date, this is the only known occurrence of sulfilimine bonds within native biomolecules. Sulfilimine crosslinks add critical structural rigidity to the collagen IV scaffold, enabling the network to withstand physiologic tensile forces within BMs. In contrast, loss of sulfilimine crosslinks display altered BM and tissue morphology, aberrant embryogenesis, and lethality in Drosophila.26, 29
Figure 5.
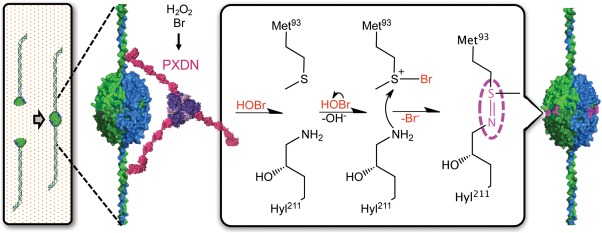
Proposed chemical mechanism of sulfilimine formation by HOBr. BM‐embedded PXDN catalyzes the conversion of Br− and oxidant into HOBr within the local environment of collagen IV NC1 hexamers. Formation of sulfilimine crosslinks is proposed to proceed through the initial formation of a bromosulfonium ion (Br‐S+) at Met93, which reacts with the ɛ‐NH2 on Hyl211. This triggers debromination of Met93 and deprotonation of Hyl211 to yield the sulfilimine crosslink.
Sulfilimine formation requires the concerted activity of collagen IV, Br−, PXDN, and oxidant making each component necessary for BM assembly and tissue development. PXDN is an animal heme peroxidase embedded within BMs near its collagen IV substrate, which locally generates hypohalous acids (Fig. 5) similar to other heme peroxidases, for example eosinophil peroxidase. PXDN uses hydrogen peroxidase to catalyze the conversion of Br− into hypobromous acid (HOBr), in turn serving as the oxidative intermediate of sulfilimine crosslinking.26, 29 The requirement for Br− during sulfilimine formation derives from the selectivity of the bromosulfonium reaction intermediate. The chemical character of bromine uniquely creates an energetically favorable reaction between the S‐Br intermediate and Hyl211. The S‐Br molecular orbital structure facilitates selective reactivity with an amine nucleophile to form the crosslink. PXDN harnesses this HOBr‐based selectivity during crosslinking while apparently avoiding oxidative damage to the BM.29
7S crosslinking by LOX‐L2
Elegant rotary shadowing‐electron microscopy studies on the supramolecular organization of collagen IV established that four adjoining triple‐helical protomers associate in a parallel and anti‐parallel fashion through their amino‐termini forming a 110 nm overlap known as the 7S dodecamer (Fig. 6).44 The assembly process is described as a spontaneous process in which the 7S domains self‐associate noncovalently into dodecamers. This event is followed by the formation of disulfide and nondisulfide crosslinks that stabilize the interaction between the four molecules conferring the 7S dodecamer its unusual resistance to proteolytic enzymes such as collagenase, which greatly facilitates its purification from different tissues and biochemical characterization.45
Figure 6.
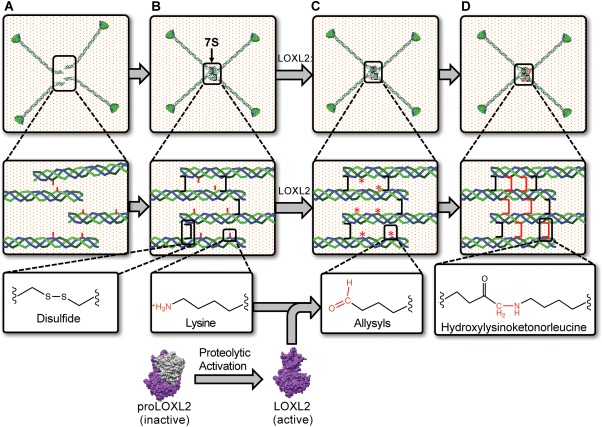
LOXL2 mediated crosslinking of 7S dodecamer. The assembly of four triple helical protomers occurs in the extracellular space. 7S domains contain intrinsic sequences that drive their assembly into a tetramer of trimers that is dodecameric. (A) The 7S dodecamer is stabilized by intramolecular (not shown) and intermolecular (black bracket) disulfide crosslinks. (B) As collagen IV networks are deposited into the BM, proteolytically activated LOXL268 promotes the formation of lysyl‐derived crosslinks in the 7S dodecamer (C) by catalysis of the oxidative deamination of the ɛ‐amino group of lysyl residues (red sticks), generating allysine (*). Allysines spontaneously form crosslinks (red brackets) with neighboring lysyl or hydroxylysyl residues. (D) As hydroxylysine is abundant in collagen IV, hydroxylysyl‐derived crosslinks such as hydroxylysinoketonorleucine is likely to be formed in 7S dodecamer. The occurrence of intra‐ and intermolecular crosslinks in the 7S dodecamer, including disulfides and LOXL2‐mediated hydroxylysinoketonorleucine, provides collagen IV scaffolds with the required strength to stabilize BMs.
As each protomer is a heterotrimeric molecule composed of two α1‐like chains and one α2 chain, the 7S dodecamer, a tetramer of trimers, is composed of eight α1‐like and 4 α2‐like protomers. Protein sequencing studies revealed that human 7S domains of α1(IV) and α2(IV) are comprised of 145 and 158 amino acids, respectively.46, 47, 48 The sequence is predominantly collagenous in nature and shares many features of the triple helical domain including a large number of post‐translationally modified amino acid residues such as hydroxylated proline and lysine residues as well as O‐glycosylated hydroxylysine residues.49 The sequence of the 7S domain begins with a noncollagenous domain comprised of about 15 residues containing four conserved cysteine residues, which are thought to spontaneously form intra‐ and interprotomer disulfide crosslinks upon 7S dodecamer assembly.47, 48 Furthermore, antiparallel protomers may form interprotomer disulfides between one of the four N‐terminal cysteine residues (likely Cys14) with an unusual cysteine residue located at position 98 (Cys98) in an interruption of the triple helical sequence on the opposite interacting protomer.47 Cys98 is present in all six α‐chains of collagen IV and neighbors a conserved asparagine residue (Asn99) that serves as an attachment site for a N‐linked carbohydrate moiety that appears to play a role in 7S alignment.50
Within the 7S dodecamer nondisulfide crosslinks are formed by condensation of lysyl residues oxidized by LOXL2.28 LOXL2 is a copper‐dependent amine oxidase enzyme that catalyzes the oxidative deamination of lysine and/or hydroxylysine residues in collagens and elastin. Although it belongs to a family of lysyl oxidases composed by five members (LOX, LOXL1–4), LOXL2 is the only one associated with collagen IV crosslinking and BM structures in different tissues.51 Emphasizing the importance of LOXL2‐catalyzed crosslinking in the function and maintenance of collagen IV scaffolds, knockdown of LOXL2 perturbed collagen IV network assembly and decreased endothelial cell tube formation in an in vitro model system of tubulogenesis.52 Notably, LOXL2 crosslinking activity is restricted to the antiparallel molecules in the N‐terminus, whereas the NC1 domains sulfilimine bonds are catalyzed by PXDN.26 Thus, LOXL2 is now considered a member of the molecular machinery required for collagen IV scaffold biosynthesis.
LOXL2‐catalyzed crosslinking of the 7S dodecamer likely occurs on the outside of cells once protomers are assembled and deposited as supramolecular scaffolds. LOXL2 catalyzes the formation of reactive aldehyde lysine (allysine), which forms crosslinks by condensing with other allysine (aldol condensation) or with the ɛ‐amino group of lysine/hydroxylysine residues through the formation of a Schiff base (Fig. 6).53 Experiments using 2,4‐dinitrophenylhydrazine showed the presence of carbonyl‐containing hydroxylysine‐derived divalent crosslinks such as dihydroxylysinonorleucine (DHLNL) in 7S dodecamers incubated with recombinant LOXL2.28 In fibrillar collagens these divalent crosslinks are the precursors to more mature complex crosslinks such as pyrroles and pyridinolines.54 However, these structures have not been identified in hydrolysates of BMs using standard crosslink analysis.55 Experiments using tritiated sodium borohydride to monitor levels of reducible crosslinks in BMs established that DHLNL crosslinks reach a high level in tissues of young animals, but as tissues age they decline progressively to undetectable levels.55 Furthermore, as the amount of divalent crosslinks alone is too low to explain the number of crosslinked 7S subunits, it is believed that multivalent lysyl‐derived crosslinks of unknown chemical structures, different to pyrroles and pyridinolines found in fibrillar collagens, are formed in BM collagen IV. Thus, even though LOXL2 mediates the initial formation of divalent hydroxylysine‐derived crosslinks in the 7S dodecamer in BMs, these seem to follow a different maturation pathway.
The supramolecular organization of protomers and registration of 7S sequences likely determines the location of lysyl‐derived crosslinks. The available 3D models of the 7S dodecamer based on rotary shadowing‐electron microscopy and alignment of amino acidic sequences suggested the most likely location of disulfide and lysyl derived crosslinks. Protomers interacting in a parallel manner have multiple possibilities to form lysyl‐lysine crosslinks. For an anti‐parallel interaction between the 7S domains of two α1(IV) chains, Glanville proposed the formation of intermolecular lysyl‐lysine crosslink between Lys11 and Lys102 pairs.47
Identification of the location of disulfide and lysyl‐derived crosslink sites may be instrumental for the construction of an accurate 3D model of the 7S dodecamer. X‐ray crystallography and mass spectrometry played an important role in the identification of sulfilimine crosslinks reinforcing the trimer–trimer interface in NC1 hexamers. Unlike NC1 domain sequences, however, 7S polypeptides are highly post‐translationally modified and heterogeneous, making determination of the 3D crystal structure and mass spectrometry analyses challenging. As amino acid sequence information alone is not enough for the identification of lysyl‐derived crosslinks by mass spectrometry, extensive mass spectrometry analyses, including different types of gas‐phase fragmentation to overcome the anomalous behavior of hydroxylated and glycosylated collagenous peptides, were required to generate post translational modification (PTM) maps of the 7S domains. Completion of these PTM maps will facilitate mass spectrometry identification and localization of lysyl‐derived crosslinks, which will allow the generation of a more accurate 3D molecular model to use as a tool for the study of the mechanism of assembly of the 7S dodecamer.
Collagen IV Scaffolds
As a core structural backbone of BMs, collagen IV scaffolds provide structural support and molecular organization to the matrix. Collagen IV protomers possess multiple binding sites, including sites for laminin networks, bone morphogenic protein (BMP), nidogen, heparin sulfate, and laminins.31 Within BMs, this results in the “decoration” of collagen IV scaffolds with a diverse spectrum of functional molecules (Fig. 7). As shown with BMP signaling in Drosophila, collagen IV scaffolds are key for immobilizing growth factors and maintaining growth factor gradients.56, 57, 58
Figure 7.
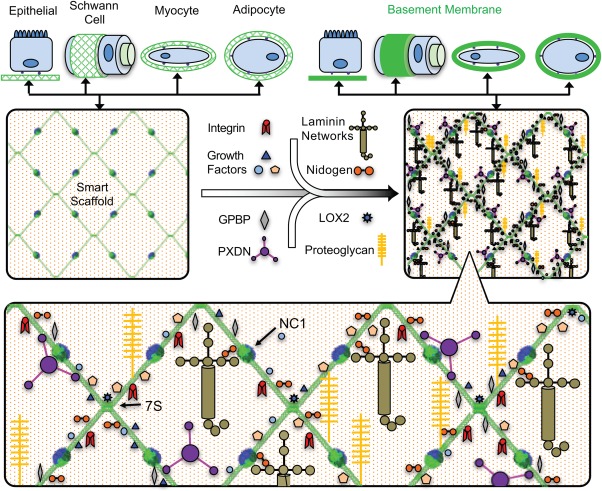
Collagen IV functions as a smart scaffold. Binding motifs are embedded along the length of the triple helix for binding integrins, and various macromolecules. This protein complex influences cell adhesion, migration, development, tissue regeneration, wound healing, and plays a role in molecular sieving.
Scaffold assembly is critical to the functionality of collagen IV. Disruption of the assembly process, either intracellularly or extracellularly, can alter the function of these scaffolds with potentially severe biologic effects. For example, embryonic lethality is reported following genetic loss of collagen IV in multiple species59, 60, 61 or even inhibition of sulfilimine crosslinking through PXDN mutation or Br‐deficiency in Drosophila.1, 26, 29 Further, collagen IV scaffolds act as ligands for cellular receptors, by binding integrins (α1β1 and α2β1) and discoidin domain receptor 1 (DDR1).7, 30, 62, 63 Interestingly, both types of receptors interact with triple‐helical collagen IV protomers but not with the isolated α‐chains, demonstrating the importance of the triple helix as a ligand.30, 64
At the tissue level, collagen IV scaffolds are key players in tissue sculpting such as its role as a “molecular corset” during Drosophila egg chamber elongation.65, 66 Moreover, removal of collagen IV networks through enzymatic digestion results in severe loss of tissue architecture in Drosophila wing.67 Thus, maintaining the structural integrity of smart scaffolds is critical tissue functionality.
Conclusion
In summary, the assembly of collagen IV scaffolds has emerged as an intricately controlled pathway occurring in both the intracellular and the extracellular environments. Investigations into the scaffold assembly process have unveiled remarkable molecular and enzymatic mechanisms. The intracellular assembly of collagen IV protomers, their secretion into the extracellular space, and subsequent assembly into networks are dependent on chloride cofactors, bromide, LOXL2, and PXDN enzymes. The assembled networks harbor multiple binding sites that spatially and temporally organization extracellular molecules. We refer to these networks as “smart” scaffolds, which as a component of the BM enable the development and function of multicellular tissues in all animal phyla.
Acknowledgments
I am honored and humbled to receive the 2017 Carl Branden award from the Protein Society for our discoveries on collagen and our development of the Aspirnaut educational pipeline for students from disadvantaged background. I accept the award on behalf of my research team—students, fellows, collaborators and Aspirnauts. This review on collagen IV smart scaffolds highlights the numerous discoveries by these talented scientists over a period of five decades. A key turning point was the crystal structure of the noncollagenous (NC1) domain hexamer that bridges triple‐helical protomers within the scaffolds. This advance led to our discovery of the novel sulfilimine bond, a function for the enzyme PXDN, and a function for chloride and bromide in scaffold assembly. The pivotal importance of X‐ray crystallography in our work is reminiscent of the pioneering crystallography work of Carl Branden.
Over the past decade, our research team has leveraged research progress to address the national need for increased diversity in the science, technology, engineering, and mathematics (STEM) workforce. We developed a model science education pipeline, called the Aspirnaut K‐20 STEM Pipeline for Diversity (www.aspirnaut.org). The pipeline is illuminated by educational enhancements, research experiences, and continual mentoring. More than 3000 elementary students in rural schools participated in the beaming of science kits through Skype, and with undergraduates and research fellows partnering with classroom teachers. One hundred and ten high school students (the Aspirnauts) from disadvantaged backgrounds in 13 states across America have participated in a 6‐week discovery science experience as summer research interns at Vanderbilt University Medical Center. Ninety undergraduate students, from disadvantaged backgrounds and from 19 colleges and universities across America, have participated in an 8‐week summer discovery science experience. These students contributed to the advancement of collagen IV biology and chemistry, in parallel their research experience has provided guidance for their career development.
Finally, we express our deep appreciation to Linda Langley and Doug Strickland for their contributions and inspiration in our pursuit to understand the molecular basis of Goodpasture's syndrome and Alport syndrome.
Billy Hudson is the winner of the 2017 Carl Brändén Award.
References
- 1. Fidler AL, Vanacore RM, Chetyrkin SV, Pedchenko VK, Bhave G, Yin VP, Stothers CL, Rose KL, McDonald WH, Clark TA, BorzaD B, Steele RE, Ivy MT, Aspirnauts T, Hudson JK, Hudson BG (2014) A unique covalent bond in basement membrane is a primordial innovation for tissue evolution. Proc Natl Acad Sci USA 111:331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fidler AL, Darris CE, Chetyrkin SV, Pedchenko VK, Boudko SP, Brown KL, Jerome WG, Hudson JK, Rokas A, Hudson BG (2017) Collagen IV and basement membrane at the evolutionary dawn of metazoan tissues. Elife 6:e24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kefalides NA (1966) A collagen of unusual composition and a glycoprotein isolated from canine glomerular basement membrane. Biochem Biophys Res Commun 22:26–32. [DOI] [PubMed] [Google Scholar]
- 4. Kefalides NA (1973) Structure and biosynthesis of basement membranes. Int Rev Connect Tissue Res 6:63–104. [DOI] [PubMed] [Google Scholar]
- 5. Myllyharju J, Kivirikko KI (2004) Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet 20:33–43. [DOI] [PubMed] [Google Scholar]
- 6. Veit G, Kobbe B, Keene DR, Paulsson M, Koch M, Wagener R (2006) Collagen XXVIII, a novel von Willebrand factor A domain‐containing protein with many imperfections in the collagenous domain. J Biol Chem 281:3494–3504. [DOI] [PubMed] [Google Scholar]
- 7. Khoshnoodi J, Pedchenko V, Hudson BG (2008) Mammalian collagen IV. Microsc Res Tech 71:357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ricard‐Blum S, Ruggiero F (2005) The collagen superfamily: from the extracellular matrix to the cell membrane. Pathol Biol 53:430–442. [DOI] [PubMed] [Google Scholar]
- 9. Boutaud A, Borza DB, Bondar O, Gunwar S, Netzer KO, Singh N, Ninomiya Y, Sado Y, Noelken ME, Hudson BG (2000) Type IV collagen of the glomerular basement membrane. Evidence that the chain specificity of network assembly is encoded by the noncollagenous NC1 domains. J Biol Chem 275:30716–30724. [DOI] [PubMed] [Google Scholar]
- 10. Mao M, Alavi MV, Labelle‐Dumais C, Gould DB (2015) Type IV collagens and basement membrane diseases: Cell biology and pathogenic mechanisms. Curr Top Membr 76:61–116. [DOI] [PubMed] [Google Scholar]
- 11. Kuo DS, Labelle‐Dumais C, Gould DB (2012) COL4A1 and COL4A2 mutations and disease: insights into pathogenic mechanisms and potential therapeutic targets. Hum Mol Genet 21:R97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jeanne M, Gould DB (2017) Genotype‐phenotype correlations in pathology caused by collagen type IV alpha 1 and 2 mutations. Matrix Biol 57:29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guiraud S, Migeon T, Ferry A, Chen Z, Ouchelouche S, Verpont MC, Sado V, Allamand V, Ronco P, Plaisier E (2017) HANAC Col4a1 mutation in mice leads to skeletal muscle alterations due to a primary vascular defect. Am J Pathol 187:505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Z, Migeon T, Verpont MC, Zaidan M, Sado Y, Kerjaschki D, Ronco P, Plaisier E (2016) HANAC syndrome Col4a1 mutation causes neonate glomerular hyperpermeability and adult glomerulocystic kidney disease. J Am Soc Nephrol 27:1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hudson BG, Tryggvason K, Sundaramoorthy M, Neilson EG (2003) Alport's syndrome, Goodpasture's syndrome, and type IV collagen. N Engl J Med 348:2543–2556. [DOI] [PubMed] [Google Scholar]
- 16. Hudson BG (2004) The molecular basis of Goodpasture and Alport syndromes: beacons for the discovery of the collagen IV family. J Am Soc Nephrol 15:2514–2527. [DOI] [PubMed] [Google Scholar]
- 17. Good MC, Zalatan JG, Lim WA (2011) Scaffold proteins: hubs for controlling the flow of cellular information. Science 332:680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trappmann B, Gautrot JE, Connelly JT, Strange DG, Li Y, Oyen ML, Cohen Stuart MA, Boehm H, Li B, Vogel V, Spatz JP, Watt FM, Huck WTS (2012) Extracellular‐matrix tethering regulates stem‐cell fate. Nat Mater 11:642–649. [DOI] [PubMed] [Google Scholar]
- 19. Greenwald EC, Redden JM, Dodge‐Kafka KL, Saucerman JJ (2014) Scaffold state switching amplifies, accelerates, and insulates protein kinase C signaling. J Biol Chem 289:2353–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cummings CF, Pedchenko V, Brown KL, Colon S, Rafi M, Jones‐Paris C, Pokydeshava E, Liu M, Pastor‐Pareja JC, Stothers C, Ero‐Tolliver IA, McCall AS, Vanacore R, Bhave G, Santoro S, Blackwell TS, Zent R, Pozzi A, Hudson BG (2016) Extracellular chloride signals collagen IV network assembly during basement membrane formation. J Cell Biol 213:479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pokidysheva E, Boudko S, Vranka J, Zientek K, Maddox K, Moser M, Fassler R, Ware J, Bachinger HP (2014) Biological role of prolyl 3‐hydroxylation in type IV collagen. Proc Natl Acad Sci USA 111:161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nagai N, Hosokawa M, Itohara S, Adachi E, Matsushita T, Hosokawa N, Nagata K (2000) Embryonic lethality of molecular chaperone hsp47 knockout mice is associated with defects in collagen biosynthesis. J Cell Biol 150:1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsuoka Y, Kubota H, Adachi E, Nagai N, Marutani T, Hosokawa N, Nagata K (2004) Insufficient folding of type IV collagen and formation of abnormal basement membrane‐like structure in embryoid bodies derived from Hsp47‐null embryonic stem cells. Mol Biol Cell 15:4467–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yurchenco PD, Ruben GC (1987) Basement membrane structure in situ: evidence for lateral associations in the type IV collagen network. J Cell Biol 105:2559–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vanacore R, Ham AJ, Voehler M, Sanders CR, Conrads TP, Veenstra TD, Sharpless KB, Dawson PE, Hudson BG (2009) A sulfilimine bond identified in collagen IV. Science 325:1230–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bhave G, Cummings CF, Vanacore RM, Kumagai‐Cresse C, Ero‐Tolliver IA, Rafi M, Kang J‐S, Pedchenko V, Fessler LI, Fessler JH, Hudson BG (2012) Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. Nat Chem Biol 8:784–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nelson RE, Fessler LI, Takagi Y, Blumberg B, Keene DR, Olson PF, Parker CG, Fessler JH (1994) Peroxidasin: a novel enzyme‐matrix protein of Drosophila development. EMBO J 13:3438–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anazco C, Lopez‐Jimenez AJ, Rafi M, Vega‐Montoto L, Zhang MZ, Hudson BG, Vanacore RM (2016) Lysyl oxidase‐like‐2 cross‐links collagen IV of glomerular basement membrane. J Biol Chem 291:25999–26012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCall AS, Cummings CF, Bhave G, Vanacore R, Page‐McCaw A, Hudson BG (2014) Bromine is an essential trace element for assembly of collagen IV scaffolds in tissue development and architecture. Cell 157:1380–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eble JA, Golbik R, Mann K, Kuhn K (1993) The alpha 1 beta 1 integrin recognition site of the basement membrane collagen molecule [alpha 1(IV)]2 alpha 2(IV). EMBO J 12:4795–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parkin JD, San Antonio JD, Pedchenko V, Hudson B, Jensen ST, Savige J (2011) Mapping structural landmarks, ligand binding sites, and missense mutations to the collagen IV heterotrimers predicts major functional domains, novel interactions, and variation in phenotypes in inherited diseases affecting basement membranes. Hum Mutat 32:127–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khoshnoodi J, Cartailler JP, Alvares K, Veis A, Hudson BG (2006) Molecular recognition in the assembly of collagens: terminal noncollagenous domains are key recognition modules in the formation of triple helical protomers. J Biol Chem 281:38117–38121. [DOI] [PubMed] [Google Scholar]
- 33. Khoshnoodi J, Sigmundsson K, Cartailler JP, Bondar O, Sundaramoorthy M, Hudson BG (2006) Mechanism of chain selection in the assembly of collagen IV: a prominent role for the alpha2 chain. J Biol Chem 281:6058–6069. [DOI] [PubMed] [Google Scholar]
- 34. Sundaramoorthy M, Meiyappan M, Todd P, Hudson BG (2002) Crystal structure of NC1 domains. Structural basis for type IV collagen assembly in basement membranes. J Biol Chem 277:31142–31153. [DOI] [PubMed] [Google Scholar]
- 35. Ziomber A, Machnik A, Dahlmann A, Dietsch P, Beck FX, Wagner H, Hilgers KF, Luft FC, Eckardt K‐U, Titze J (2008) Sodium‐, potassium‐, chloride‐, and bicarbonate‐related effects on blood pressure and electrolyte homeostasis in deoxycorticosterone acetate‐treated rats. Am J Physiol Renal Physiol 295:F1752–F1763. [DOI] [PubMed] [Google Scholar]
- 36. Kumar S, Nussinov R (1999) Salt bridge stability in monomeric proteins. J Mol Biol 293:1241–1255. [DOI] [PubMed] [Google Scholar]
- 37. Sarakatsannis JN, Duan Y (2005) Statistical characterization of salt bridges in proteins. Proteins 60:732–739. [DOI] [PubMed] [Google Scholar]
- 38. Bastolla U, Demetrius L (2005) Stability constraints and protein evolution: the role of chain length, composition and disulfide bonds. Protein Eng Des Sel 18:405–415. [DOI] [PubMed] [Google Scholar]
- 39. Vanacore RM, Shanmugasundararaj S, Friedman DB, Bondar O, Hudson BG, Sundaramoorthy M (2004) The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain‐1 by noncovalent forces and the absence of Met‐Lys cross‐links. J Biol Chem 279:44723–44730. [DOI] [PubMed] [Google Scholar]
- 40. Donald JE, Kulp DW, DeGrado WF (2011) Salt bridges: Geometrically specific, designable interactions. Proteins 79:898–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakajima Y, Miyahara I, Hirotsu K, Nishina Y, Shiga K, Setoyama C, Tamaoki H, Miura R (2002) Three‐dimensional structure of the flavoenzyme acyl‐CoA oxidase‐II from rat liver, the peroxisomal counterpart of mitochondrial acyl‐CoA dehydrogenase. J Biochem 131:365–374. [DOI] [PubMed] [Google Scholar]
- 42. Miyanaga A, Koseki T, Matsuzawa H, Wakagi T, Shoun H, Fushinobu S (2004) Crystal structure of a family 54 alpha‐L‐arabinofuranosidase reveals a novel carbohydrate‐binding module that can bind arabinose. J Biol Chem 279:44907–44914. [DOI] [PubMed] [Google Scholar]
- 43. Kelly CA, Nishiyama M, Ohnishi Y, Beppu T, Birktoft JJ (1993) Determinants of protein thermostability observed in the 1.9‐A crystal structure of malate dehydrogenase from the thermophilic bacterium Thermus flavus . Biochemistry 32:3913–3922. [DOI] [PubMed] [Google Scholar]
- 44. Kuhn K, Wiedemann H, Timpl R, Risteli J, Dieringer H, Voss T, Glanville RW (1981) Macromolecular structure of basement membrane collagens. FEBS Lett 125:123–128. [DOI] [PubMed] [Google Scholar]
- 45. Duncan KG, Fessler LI, Bachinger HP, Fessler JH (1983) Procollagen IV. Association to tetramers. J Biol Chem 258:5869–5877. [PubMed] [Google Scholar]
- 46. Risteli J, Bachinger HP, Engel J, Furthmayr H, Timpl R (1980) 7‐S collagen: characterization of an unusual basement membrane structure. Eur J Biochem 108:239–250. [DOI] [PubMed] [Google Scholar]
- 47. Glanville RW, Qian RQ, Siebold B, Risteli J, Kuhn K (1985) Amino acid sequence of the N‐terminal aggregation and cross‐linking region (7S domain) of the alpha 1 (IV) chain of human basement membrane collagen. Eur J Biochem 152:213–219. [DOI] [PubMed] [Google Scholar]
- 48. Qian RG, Glanville RW (1984) Separation and characterization of two polypeptide chains from the 7S cross‐linking domain of basement‐membrane (type IV) collagen. Biochem J 222:447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Basak T, Vega‐Montoto L, Zimmerman LJ, Tabb DL, Hudson BG, Vanacore RM (2016) Comprehensive characterization of glycosylation and hydroxylation of basement membrane collagen IV by high‐resolution mass spectrometry. J Proteome Res 15:245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Langeveld JP, Noelken ME, Hard K, Todd P, Vliegenthart JF, Rouse J, Hudson BG (1991) Bovine glomerular basement membrane. Location and structure of the asparagine‐linked oligosaccharide units and their potential role in the assembly of the 7 S collagen IV tetramer. J Biol Chem 266:2622–2631. [PubMed] [Google Scholar]
- 51. Csiszar K (2001) Lysyl oxidases: a novel multifunctional amine oxidase family. Prog Nucleic Acid Res Mol Biol 70:1–32. [DOI] [PubMed] [Google Scholar]
- 52. Bignon M, Pichol‐Thievend C, Hardouin J, Malbouyres M, Brechot N, Nasciutti L, Barret A, Teillon J, Guillon E, Etienne E, Caron M, Joubert‐Caron R, Monnot C, Ruggiero F, Muller L, Germain S (2011) Lysyl oxidase‐like protein‐2 regulates sprouting angiogenesis and type IV collagen assembly in the endothelial basement membrane. Blood 118:3979–3989. [DOI] [PubMed] [Google Scholar]
- 53. Kagan HM, Ryvkin F Lysyl oxidase and lysyl oxidase‐like enzymes In: Mecham RP, Ed. (2011) The Extracellular Matrix: an Overview. New York: Springer; pp 303–335. [Google Scholar]
- 54. Eyre DR, Paz MA, Gallop PM (1984) Cross‐linking in collagen and elastin. Annu Rev Biochem 53:717–748. [DOI] [PubMed] [Google Scholar]
- 55. Bailey AJ, Sims TJ, Light N (1984) Cross‐linking in type IV collagen. Biochem J 218:713–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang X, Harris RE, Bayston LJ, Ashe HL (2008) Type IV collagens regulate BMP signalling in Drosophila. Nature 455:72–77. [DOI] [PubMed] [Google Scholar]
- 57. Bunt S, Hooley C, Hu N, Scahill C, Weavers H, Skaer H (2010) Hemocyte‐secreted type IV collagen enhances BMP signaling to guide renal tubule morphogenesis in Drosophila. Dev Cell 19:296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sawala A, Sutcliffe C, Ashe HL (2012) Multistep molecular mechanism for bone morphogenetic protein extracellular transport in the Drosophila embryo. Proc Natl Acad Sci USA 109:11222–11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Poschl E, Schlotzer‐Schrehardt U, Brachvogel B, Saito K, Ninomiya Y, Mayer U (2004) Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development 131:1619–1628. [DOI] [PubMed] [Google Scholar]
- 60. Gupta MC, Graham PL, Kramer JM (1997) Characterization of alpha1(IV) collagen mutations in Caenorhabditis elegans and the effects of alpha1 and alpha2(IV) mutations on type IV collagen distribution. J Cell Biol 137:1185–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Borchiellini C, Coulon J, Le Parco Y (1996) The function of type IV collagen during Drosophila muscle development. Mech Dev 58:179–191. [DOI] [PubMed] [Google Scholar]
- 62. Fu HL, Valiathan RR, Arkwright R, Sohail A, Mihai C, Kumarasiri M, Mahasenan KV, Mobashery S, Huang P, Agarwal G, Fridman R (2013) Discoidin domain receptors: unique receptor tyrosine kinases in collagen‐mediated signaling. J Biol Chem 288:7430–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Valiathan RR, Marco M, Leitinger B, Kleer CG, Fridman R (2012) Discoidin domain receptor tyrosine kinases: new players in cancer progression. Cancer Metastasis Rev 31:295–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC (2000) Structural basis of collagen recognition by integrin alpha2beta1. Cell 101:47–56. [DOI] [PubMed] [Google Scholar]
- 65. Haigo SL, Bilder D (2011) Global tissue revolutions in a morphogenetic movement controlling elongation. Science 331:1071–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bilder D, Haigo SL (2012) Expanding the morphogenetic repertoire: perspectives from the Drosophila egg. Dev Cell 22:12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pastor‐Pareja JC, Xu T (2011) Shaping cells and organs in Drosophila by opposing roles of fat body‐secreted collagen IV and perlecan. Dev Cell 21:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. López‐Jiménez A, Basak T, Vanacore RM (in press) Proteolytic Processing of Lysyl Oxidase like‐2 in the Extracellular Matrix is Required for Crosslinking of Basement Membrane Collagen IV. J Biol Chem doi:10.1074/jbc.M117.798603 [DOI] [PMC free article] [PubMed] [Google Scholar]


