CASE
A 60-year-old female was admitted to a hospital in Missouri with back pain and pathological fractures of multiple thoracic vertebrae. Four months prior to presentation, the patient began experiencing low back pain without inciting trauma. Over the next 2 months, the pain continued to worsen and she experienced a 20-lb weight loss and drenching night sweats, prompting her to seek care. During the initial encounter, the patient denied experiencing fevers. She was not immunocompromised and had no risk factors for tuberculosis and no history of recent travel, hospitalization, or invasive procedures. Magnetic resonance imaging (MRI) revealed abnormal enhancement and edema involving the T6, T7, and T8 vertebral bodies with associated prevertebral and postvertebral soft tissue enhancement and edema. A fluoroscopy-guided T7 vertebral body biopsy revealed acute and chronic osteomyelitis with no evidence of malignancy, prompting an infectious disease consult.
Two sets of blood cultures (VersaTREK REDOX 1 and REDOX 2 media per set) were obtained, and they signaled positive on the blood culture instrument (VersaTREK; Thermo Fisher Scientific). The aerobic bottles from both sets yielded Gram-positive cocci in clusters that were identified as Staphylococcus species by the VERIGENE Gram-Positive Blood Culture Test (BC-GP) (Nanosphere/Luminex). Anaerobic medium bottles remained negative. Subculture to sheep blood agar revealed beta-hemolytic colonies that were subsequently identified as Staphylococcus schleiferi by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) with an acceptable species-level score of 2.151 (Bruker Biotyper, Reference Database 5.0; Bruker Daltonics, Billerica, MA).
A specimen from the T7 vertebral bone biopsy was sent for culture. Gram staining of the specimen revealed rare polymorphonuclear leukocytes, and no organisms were seen. Aerobic media (sheep blood agar, chocolate agar, and MacConkey agar) were held for 3 days, and anaerobic media (brucella blood agar, laked blood kanamycin vancomycin agar, and phenyethyl alcohol agar) were held for 5 days and remained negative for growth for the duration of incubation. A transesophageal echocardiogram was negative for vegetations on the heart valves, and the patient was initiated on intravenous (i.v.) vancomycin, which was subsequently narrowed to i.v. ceftriaxone once susceptibility testing was available. Two follow-up blood cultures taken 24 h after the initiation of antimicrobial therapy were positive for the same microorganism. The bacteremia was ultimately cleared after another 2 days, and the patient was discharged home on i.v. ceftriaxone for 6 weeks. Surveillance blood cultures taken 2 weeks after the completion of therapy were negative, and at a subsequent outpatient clinic follow-up at 3 months, she reported complete resolution of her back pain and night sweats. Follow-up imaging of her spine revealed stable vertebral fractures with no disease progression.
A thorough review of the patient's social history revealed that she had a pet Labrador retriever with a chronic ear infection for the past 8 months. A call to the veterinarian confirmed purulent otitis, and pus from the dog's ear was obtained for culture, which grew S. schleiferi. The patient recalled cleaning the dog's ear with her bare hands. She was employed as a housekeeper and had intermittent skin cracks at her fingertips due to exposure to chemicals, which we postulated might have been the portal of entry for the microbe.
DISCUSSION
Several bacteria that cause human infections are associated with dogs, including Pasteurella canis, Capnocytophaga canimorsus, Streptococcus canis, and Staphylococcus intermedius group (SIG). These organisms are often associated with wound infections from dog bites, as they are common inhabitants of the oral mucosa of dogs. S. schleiferi bacteria are thought to be part of the normal skin flora of dogs and are commonly associated with canine otitis externa and pyoderma (1). This species is divided into two subspecies, S. schleiferi subsp. schleiferi and S. schleiferi subsp. coagulans. S. schleiferi subsp. schleiferi is more commonly associated with human infections, including skin and soft tissue infections, device infections, osteomyelitis, and bacteremia (2). Few cases of human infection caused by S. schleiferi subsp. coagulans have been documented, although wound infections, endocarditis, and device infections have been reported. Immunosuppression, malignancies, and recent surgical procedures are important comorbidities reported in prior case series and point toward nosocomial acquisition (3). However, the association of S. schleiferi with canine and human infections supports the possibility that transmission from dogs to humans can occur, as is likely in the case presented here.
S. schleiferi bacteria are facultatively anaerobic, Gram-positive cocci. Colonies are round and nonpigmented and are beta-hemolytic on sheep blood agar (Fig. 1). Several biochemical reactions differentiate S. schleiferi from other Staphylococcus species that cause human infections. S. schleiferi is characteristically pyrrolidonyl arylamidase (PYR) positive, ornithine decarboxylase negative, alkaline phosphatase positive, and susceptible to novobiocin and polymyxin B (4).
FIG 1.
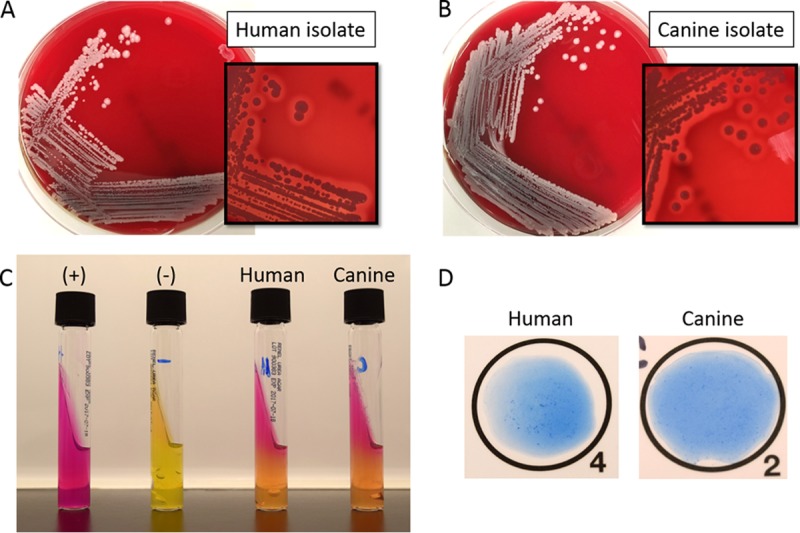
(A and B) S. schleiferi growth on sheep blood agar after 24 h of incubation of the human (A) and canine (B) isolates. Hemolysis is visible in the smaller image. (C) Christensen's medium for the detection of urease production. (+) and (−) indicate positive and negative controls, respectively. (D) Latex agglutination (StaphTEX) of the human and canine isolates.
Identification systems vary in their performance characteristics for S. schleiferi identification. One study found that 22 isolates of S. schleiferi were identified 100% of the time by MALDI-TOF with both the Bruker Biotyper and the Vitek2 system (GP no. 21342; bioMérieux) (5). In our case, the Bruker Biotyper MALDI-TOF identified both the human and canine isolates to the species level with acceptable scores of 2.143 and 2.245, respectively. Both isolates were also assayed on the Vitek MS and identified as S. schleiferi with a score of 99.9%. Neither system identified the isolates to the subspecies level. This species is also included in the database of MicroScan systems and the API Staph biochemical profiling system (bioMérieux); however, little is known about the performance of these systems for the identification of S. schleiferi. Notably, identification systems often recommend additional testing such as sugar fermentation to confirm an identification of S. schleiferi.
Differentiation of the two subspecies is possible through biochemical testing with the tube coagulase and urease reactions. S. schleiferi subsp. schleiferi is slide coagulase (clumping factor) positive, tube coagulase (free coagulase) negative, and urease negative, while S. schleiferi subsp. coagulans is negative by slide coagulase testing and positive by urease and tube coagulase testing. Interestingly, recent genetic analysis of several clinically relevant Staphylococcus species found that, unlike Staphylococcus aureus, S. schleiferi does not possess the coa gene that encodes the staphylocoagulase protein. Thus, the coagulase activity of S. schleiferi is likely mediated by other mechanisms (6). The patient and canine isolates described in the case presentation exhibited the same morphological and biochemical characteristics, further supporting a zoonotic source for the patient's infection (Fig. 1). Both isolates were positive by tube coagulase and for PYR and urease production. Although we did not report the human isolate to the subspecies level, these characteristics supported the idea that both the human and canine isolates were likely S. schleiferi subsp. coagulans because of the positive urease and tube coagulase reactions. Latex agglutination testing (StaphTEX; Hardy Diagnostics) produced a questionable weakly positive result with both the human and canine isolates in our case (Fig. 1). Similar to another clumping factor-positive species, Staphylococcus lugdunensis, S. schleiferi lacks protein A and may produce a rough or “stringy” reaction if tested by latex agglutination.
Because of the microbiological and pathological characteristics of S. schleiferi, the incidence of infections caused by this organism may be underrecognized and underreported. Gram-positive cocci in clusters with a positive tube coagulase (S. schleiferi subsp. coagulans) or slide agglutination (S. schleiferi subsp. schleiferi) reaction may be misidentified as S. aureus. Unlike S. aureus, S. schleiferi does not acidify maltose, mannitol, or sucrose. In addition, a positive reaction for PYR should raise suspicion that an isolate is not S. aureus. Coagulase-positive staphylococcal isolates that are PYR positive fit the description of S. schleiferi subsp. coagulans or SIG. These species can be separated biochemically by fermentation of sucrose and mannose, which is positive for SIG. Importantly, MALDI-TOF MS can accurately separate S. schleiferi from other coagulase-positive staphylococcal organisms.
Beta-lactam resistance mediated by mecA (PBP2a) in staphylococcal isolates is a major concern in human and veterinary infections. In one study, 57% of the clinical isolates of S. schleiferi from dogs were methicillin resistant (1). While cefoxitin is used as the surrogate for methicillin resistance in S. aureus, there is evidence that it is not a sensitive predictor of methicillin resistance in S. schleiferi and SIG. Instead, oxacillin disk diffusion is more effective at predicting methicillin resistance in SIG and S. schleiferi (7). The performance of an alternative method of mecA detection, rapid detection of the mecA gene product PBP2a, has been assessed for S. schleiferi. An evaluation of the Alere PBP2a immunochromatographic assay found that it was 100% sensitive and specific for the differentiation of mecA-positive and -negative clinical isolates of S. schleiferi subsp. coagulans with mecA PCR as a gold standard (8). The human and canine isolates in our case were tested by disk diffusion. Both isolates were pansusceptible to all of the antibiotics tested in our panel, providing supporting evidence that the human and canine isolates were from the same source (Table 1). While there are no interpretive criteria for oxacillin disk diffusion, the oxacillin result would be interpreted as susceptible when using the SIG criterion of ≥18 mm as susceptible (Table 1) (9).
TABLE 1.
Antimicrobial susceptibility results of S. schleiferi isolates
| Antibiotic | Human |
Canine |
||
|---|---|---|---|---|
| Zone size (mm) or MIC | Interpretation | Zone size (mm) or MIC | Interpretation | |
| Cefoxitin | 30 | Susceptible | 32 | Susceptible |
| Oxacillin | 23 | —a | 22 | —a |
| Erythromycin | 27 | Susceptible | 28 | Susceptible |
| Clindamycin | 26 | Susceptible | 27 | Susceptible |
| Trimethoprim-sulfamethoxazole | 20 | Susceptible | 19 | Susceptible |
| Rifampin | 32 | Susceptible | 32 | Susceptible |
| Doxycycline | 29 | Susceptible | 29 | Susceptible |
| Linezolid | 30 | Susceptible | 31 | Susceptible |
| Daptomycin | 0.064b | Susceptible | 0.064b | Susceptible |
No CLSI disk diffusion breakpoints are available for oxacillin.
Etest MIC (μg/ml).
In summary, S. schleiferi is an uncommon but important cause of human infection that may be mistaken for S. aureus or other Staphylococcus species. While MALDI-TOF MS analysis can differentiate most Staphylococcus species, additional biochemical testing may be necessary for clinical laboratories that do not use these systems. Differentiation of the species is important, as the presence of mecA is common in S. schleiferi, but methods for detection of methicillin resistance differ between staphylococcal species. Last, a thorough history of exposures, including animal contact, is important for interpreting microbiological results in a clinical context.
SELF-ASSESSMENT QUESTIONS
- What is detected by a tube coagulase test using rabbit plasma?
- Free coagulase
- Bound coagulase
- Protein A
- Hemolysin
- A Gram-positive coccus in clusters tests positive for catalase and PYR but negative for urease and ornithine decarboxylase. What is the most likely identity of this organism?
- S. aureus
- S. schleiferi subsp. schleiferi
- S. schleiferi subsp. coagulans
- S. lugdunensis
- Which of the following methods is a rapid test that could be used to predict methicillin resistance in S. schleiferi?
- PBP2a testing by immunochromatographic assay
- Cefinase test
- Cefoxitin disk diffusion
- Oxacillin disk diffusion
For answers to the self-assessment questions and take-home points, see page 3309 in this issue (https://doi.org/10.1128/JCM.00512-17).
ACKNOWLEDGMENT
We thank William Fales (Veterinary Medical Diagnostic Laboratory at the College of Veterinary Medicine, University of Missouri, Columbia, MO) for providing the canine S. schleiferi isolate for comparison and additional analysis.
REFERENCES
- 1.Cain CL, Morris DO, Rankin SC. 2011. Clinical characterization of Staphylococcus schleiferi infections and identification of risk factors for acquisition of oxacillin-resistant strains in dogs: 225 cases (2003–2009). J Am Vet Med Assoc 239:1566–1573. doi: 10.2460/javma.239.12.1566. [DOI] [PubMed] [Google Scholar]
- 2.Davis MF, Cain CL, Brazil AM, Rankin SC. 2013. Two coagulase-negative staphylococci emerging as potential zoonotic pathogens: wolves in sheep's clothing? Front Microbiol 4:123. doi: 10.3389/fmicb.2013.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hernández JL, Calvo J, Sota R, Agüero J, García-Palomo JD, Fariñas MC. 2001. Clinical and microbiological characteristics of 28 patients with Staphylococcus schleiferi infection. Eur J Clin Microbiol Infect Dis 20:153–158. doi: 10.1007/s100960100467. [DOI] [PubMed] [Google Scholar]
- 4.Becker K, Skov RL, Von Eiff C. 2015. Staphylococcus, Micrococcus, and other catalase-positive cocci, p 354–382. In Jorgensen JH, Pfaller MA, Carroll KC, Funke G, Landry ML, Richter SS, Warnock DW (ed), Manual of clinical microbiology, 11th ed, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 5.Delport JA, Peters G, Diagre D, Lannigan R, John M. 2015. Identification of coagulase-negative staphylococci by the Bruker MALDI-TOF Biotyper compared to the Vitek 2 and MIS gas liquid chromatography. J Bacteriol Mycol Open Access 1(1):00003. doi: 10.15406/jbmoa.2015.01.00003. [DOI] [Google Scholar]
- 6.Misic AM, Cain CL, Morris DO, Rankin SC, Beiting DP. 2016. Divergent isoprenoid biosynthesis pathways in Staphylococcus species constitute a drug target for treating infections in companion animals. mSphere 1:e00258-16. doi: 10.1128/mSphere.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bemis DA, Jones RD, Hiatt LE, Ofori ED, Rohrbach BW, Frank LA, Kania SA. 2006. Comparison of tests to detect oxacillin resistance in Staphylococcus intermedius, Staphylococcus schleiferi, and Staphylococcus aureus isolates from canine hosts. J Clin Microbiol 44:3374–3376. doi: 10.1128/JCM.01336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnold AR, Burnham CA, Ford BA, Lawhon SD, McAllister SK, Lonsway D, Albrecht V, Jerris RC, Rasheed JK, Limbago B, Burd EM, Westblade LF. 2016. Evaluation of an immunochromatographic assay for rapid detection of penicillin-binding protein 2a in human and animal Staphylococcus intermedius group, Staphylococcus lugdunensis, and Staphylococcus schleiferi clinical isolates. J Clin Microbiol 54:745–748. doi: 10.1128/JCM.02869-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CLSI. 2016. Performance standards for antimicrobial susceptibility testing; twenty-sixth informational supplement. CLSI document M100-S26. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]


