ABSTRACT
The implication of coagulase-negative staphylococci in human diseases is a major issue, particularly in hospital settings wherein these species often act as opportunistic pathogens. In addition, some coagulase-negative staphylococci such as S. lugdunensis have emerged as pathogenic bacteria, implicated in severe infections, particularly, osteoarticular infections, foreign-body-associated infections, bacteremia, and endocarditis. In vitro studies have shown the presence of several putative virulence factors such as adhesion factors, biofilm production, and proteolytic factors that might explain clinical manifestations. Taken together, the clinical and microbiological data might change the way clinicians and microbiologists look at S. lugdunensis in clinical samples.
KEYWORDS: virulence, osteoarticular infections, protease, biofilm, endocarditis, Staphylococcus lugdunensis
INTRODUCTION
Staphylococcus lugdunensis has emerged since the 1990s as a distinctive coagulase-negative staphylococcus (CoNS), implicated in a wide range of severe infections. This bacterium produces a large variety of putative virulence factors. Until recently, the tools used to identify staphylococci at the species level relied on phenotypic methods such as coagulase identification, which helped distinguish S. aureus from other staphylococci. Consequently, the epidemiology of CoNS, which is generally considered less pathogenic than other staphylococci, remained unclear. The identification of coagulase activity refers to two distinctive molecular activities that aim to convert soluble fibrinogen into insoluble fibrin. The first activity involves a free coagulase that leads to prothrombin activation and, ultimately, the conversion of fibrinogen into fibrin. The second activity involves a bound coagulase or clumping factor that matches with two distinctive proteins in S. aureus, clumping factor A and B, that directly convert fibrinogen into fibrin. However, some CoNSs, for example S. lugdunensis, S. schleiferi, and S. sciuri, may produce a bound coagulase that is distinctive from the S. aureus bound coagulase but with similar activity (1). Other CoNSs such as S. pseudointermedius, S. intermedius, S. hyicus, S. delphini, and S. lutrae, produce a free coagulase (1). Phenotypic identification of CoNS is thus challenging, but the implementation of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has provided laboratories with a fast and cost-effective identification tool (2). The production of coagulases and various phenotypic properties of CoNS are no longer obstacles for performing a more systematic identification of CoNS at the species level. This is opportune because CoNSs are emerging as major causes of opportunistic and nosocomial infections. In this context, some CoNSs have emerged as putative virulent species, mainly in retrospective and epidemiological studies, in addition to case reports, with limitations because of the methodology of the analyses (3). The concept of virulence involves two distinctive factors: the clinical severity of infections and in vitro production of virulence factors. The causative link between these two factors remains unknown for CoNS, but it has been extensively explored for S. aureus. Most CoNSs such as S. epidermidis produce various molecular factors, including cytotoxins and adhesion factors that are involved in pathogenicity and help these commensal bacteria become pathogenic (4). Staphylococcus lugdunensis is a specific CoNS with an unusual pathogenicity that recent clinical and in vitro studies have partially explained. These new results will change how microbiologists and clinicians interpret the positivity of clinical samples for this bacterium. Similar to other known CoNSs, S. lugdunensis is a commensal bacterium that colonizes the skin. This review examines the current clinical and microbiological research to understand why S. lugdunensis appears different from other CoNSs. We also attempt to derive conclusions regarding how microbiologists and clinicians should consider this bacterium when a positive sample is obtained in the laboratory.
S. LUGDUNENSIS IDENTIFICATION AND MICROBIOLOGICAL ISSUES HAVE BEEN SOLVED
Staphylococcus lugdunensis grows under aerobic conditions on common media such as blood agar and causes intense beta-hemolysis. The traditionally used tube test and slide agglutination test are now obsolete due to their low sensitivity and specificity. Rapid latex and hemagglutination assays have been developed more recently based on the detection of clumping factor, protein A, and capsule types 5 and 8. Nevertheless, these innovative tools fail to distinguish S. aureus from S. lugdunensis efficiently, because S. lugdunensis might produce a bound coagulase (clumping factor) and yield positive results in up to 65% of cases (5). At the biochemical level, S. lugdunensis can be differentiated from other staphylococci based on showing positivity in pyrrolidonyl arylamidase and ornithine decarboxylase reactions in more than 90% of cases (6). Various manual and automated biochemical test systems are still used in laboratories for the identification of bacteria, including staphylococci. Regarding S. lugdunensis identification, despite its biochemical peculiarities, the reliability of these systems is too low and the accuracy of identification ranges from 70% to 90% depending on the system used and the performance of additional tests (7, 8). In an effort to improve the identification rate, various molecular methods, particularly real-time PCR assays targeting some conserved genes such as gyrA, gap, sodA, and rpoB or 16S and 23S ribosomal DNA, have been successfully proposed (6, 9, 10). Identification by nucleic acid-based approaches appears to be a gold standard for S. lugdunensis, but the implementation of proteomic methods, particularly MALDI-TOF MS, in routine clinical laboratories has allowed a rapid, cost-effective, and reliable identification of bacteria, including staphylococci (2). The growing number of MALDI-TOF MS spectra included in manufacturers' databases has allowed high levels of sensitivity and specificity, approximately 100%, to be achieved and markedly changed the way we look at CoNS, particularly S. lugdunensis (3). More recently, it appears that MALDI-TOF MS is a reliable tool to identify S. lugdunensis, among other bacteria, directly from blood cultures (11, 12). This revolution in the usual laboratory workflow might directly impact patient management and prognosis in the context of bacteremia, as recently reported for S. aureus bacteremia (13).
S. LUGDUNENSIS VIRULENCE: GROWING EVIDENCES FROM CLINICAL STUDIES
According to the literature, S. lugdunensis colonizes 30% to 50% of patients (14, 15). Three studies precisely analyzed S. lugdunensis colonization and detected inguinal colonization in 22% to 39% of patients, followed by axillary colonization in 19.8% to 20% of patients and nasal colonization in 9.3% to 17.9% cases (14–16).
Retrospective clinical studies.
Until recently, most studies that emphasized the role of S. lugdunensis in clinical settings were retrospective analyses that mainly described its role in skin and soft tissue infections. Some reports also described the occurrence of bacteremia and endocarditis owing to this bacterium. Liu et al. reviewed the literature regarding endocarditis and showed that S. lugdunensis was absent at the portal of entry in 45% of cases and occurred on native valves in 80% of cases, with a global mortality rate of 39% (17). Overall, even if mortality rates vary between studies, severe valvular lesions are a common finding. Although retrospective data have to be carefully interpreted, they intrigue clinicians, because the results reported in the literature suggest that the mortality rate of S. aureus endocarditis is approximately 20%, whereas the global mortality rate of CoNS endocarditis is approximately 12% (18, 19). The occurrence of bone and joint infections appeared recently in retrospective studies describing prosthetic joint infections, particularly knee joint infections (20). Argemi et al. (3) and Douiri et al. (21) showed in two recently published studies that 40% of all clinical samples that tested positive for S. lugdunensis were obtained from patients with proven infections, particularly bone and joint infections. This infection rate confirmed the status of this Staphylococcus species retrospectively, and we further confirmed these results through a prospective clinical trial.
Prospective clinical studies.
We recently published the first and currently only prospective study (named VISLISI) (5). In this monocentric clinical trial, all bacteriological samples that yielded positive results for S. lugdunensis were systematically screened during a 3-year period. We provided evidence that 37.2% of the 347 strains isolated originated from infected patients, particularly from those with bone and joint infections (34.6%). We also showed that the inoculation of pediatric blood culture bottles with joint fluids, tissue specimens, and sonicated prosthetic materials could significantly improve the diagnostic rate of these infections, as previously shown in prospective cohort studies (22). This prospective clinical trial once again confirmed, but with a strong methodological background, the virulence of S. lugdunensis at the clinical level. At the microbiological level, it appears that this bacterium might also produce a range of putative virulence factors that could explain these clinical findings.
MICROBIOLOGICAL STUDIES THAT STRENGTHEN CLINICAL EVIDENCE
Several studies described the occurrence of putative virulence factors in S. lugdunensis. In 1997, Donvito et al. described the hemolytic properties of this bacterium that were then linked to the presence of a delta-like hemolysin encoded by a gene that was found in the non-agr locus named slush (23). Since the publication of this study, several other virulence factors have been characterized.
Adhesion factors.
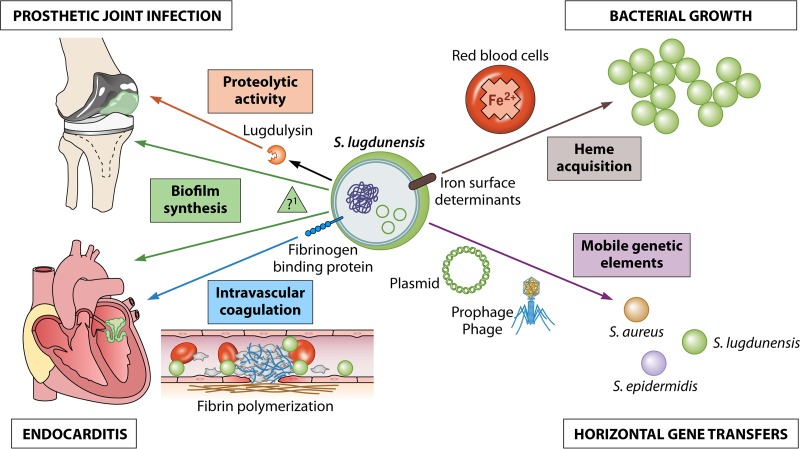
Similar to S. aureus, S. lugdunensis produces a fibrinogen-binding protein linked to the bacterial cell wall that acts as a clumping factor (24). Fibrinogen-binding proteins have been involved in vitro and in animal models in the occurrence of S. aureus endocarditis and persistent bacteremia (25). We also showed in the VISLISI clinical trial that the production of a clumping factor was strongly associated with the occurrence of bacteremia. Staphylococcus lugdunensis produces various other adhesion proteins that belong to a group of molecules, called microbial surface components recognizing adhesive matrix molecules, that covalently link the bacterial membrane through LPXTG motifs and the action of a sortase enzyme (26). This bacterium also produces a von Willebrand factor-binding protein and functionally displays high binding capacities to various extracellular matrix components, such as fibronectin, collagen, vitronectin, laminin, and human IgG (27). The functions of these adhesion factors are not limited to extracellular matrix molecule binding; these factors have numerous other functions, such as immune evasion and biofilm formation (25).
Biofilm.
Staphylococcus lugdunensis is a biofilm-producing bacterium with some specific properties. Frank and Patel showed that it frequently forms biofilms, but unlike the matrix of biofilms formed by other CoNSs, that of biofilms formed by S. lugdunensis is not made of poly-N-acetylglucosamine and is instead mainly proteinaceous, even when an ica locus has been identified in this species (28). Recent reports have also described the role of the autolysin atlL in biofilm formation and the role of a novel locus, comEB, in DNA-dependent biofilm formation (29). In a secondary analysis of the VISLISI clinical trial, we showed evidence using the BioFilm ring test (Biofilm Control, Saint-Beauzire, France), which is a new test to evaluate the kinetics of biofilm production, with respect to all the 28 strains found in osteoarticular infections producing biofilms within 6 h after culturing (30). The relationship between osteoarticular infections and biofilm formation has been described previously for S. aureus and might also be linked to S. lugdunensis as a causative agent (31). Perhaps this could explain why all patients infected with S. lugdunensis had previously undergone surgical interventions. Antibiotic susceptibility of biofilm-embedded bacteria has not been studied with S. lugdunensis, but several in vitro studies have demonstrated that S. aureus and Gram-negative bacteria display lower antibiotic susceptibility when they are grown in a biofilm than that when grown in a plankton, although the potency of some antibiotics, such as rifampin and linezolid, appear to be less impaired (32). New pharmacodynamic parameters, for example biofilm bactericidal concentration or minimal biofilm inhibitory concentration, are probably needed to more accurately evaluate the antibiotic susceptibility of the bacteria in a biofilm (33).
Proteolytic activity.
We recently described a previously unknown novel protease, named lugdulysin, that might also be implicated in osteoarticular infections (5). This Zn2+-dependent protease is similar to hyicolysin, another metalloprotease found in S. hyicus, a CoNS isolated from pigs presenting with exudative epidermatitis. In addition to hyicolysin, lugdulysin is possibly a member of the M30 family of proteases (according to MEROPS database). Lugdulysin still needs further chemical and structural characterization, but its implication in pathogenicity remains coherent with previous reports regarding metalloproteases (34). Cassat et al. characterized the role of aureolysin, another metalloprotease secreted by S. aureus strains that might play a role in osteomyelitis (35). Aureolysin acts as a conductor for the virulence repertoire of S. aureus to modulate bone remodeling. The involvement of metalloproteases in human diseases relies on their capacity to remodel the extracellular matrix, as observed in osteomyelitis, tumor invasion and metastasis, and inflammatory vascular diseases.
Iron metabolism.
Iron plays a crucial role in bacterial metabolism and growth. Staphylococcus lugdunensis is the only CoNS that carries a complete operon dedicated to iron capture and metabolism that encodes the iron-regulated proteins, IsdB, IsdC, IsdJ, and IsdK (36). It is the only CoNS that has a captation system similar to that of S. aureus.
Virulence factor regulation.
Staphylococcus lugdunensis also bears an agr locus and produces an RNAIII-like RNA molecule that is distinct from the delta-like hemolysin activity that relies on a different locus (slush). The role of agr is crucial in S. aureus, and a similar locus has been described in other CoNSs, such as S. epidermidis, that also bear different regulation loci such as the LuxS/AI-2 system that contributes to the quorum sensing machinery. These systems are now well understood in S. aureus; however, the agr locus still needs further characterization in S. lugdunensis.
Clinical manifestations and their putative correlations with virulence factors are illustrated in Fig. 1.
FIG 1.
Clinical and bacteriological roles of the main putative virulence factors identified in S. lugdunensis.
Antibiotic susceptibility.
Staphylococcus lugdunensis remains remarkably sensitive to most antibiotics, particularly beta-lactams, contrary to other CoNSs. Fosfomycin is the only antimicrobial with highly variable results and a resistance level of >50% depending on the study. This resistance is mostly because of the presence of the gene fusB. Beta-lactamase production is also variable depending on the study, with production reported in anywhere from 0% to 70% of the strains. However, this phenotype is not unusual among staphylococci, and methicillin resistance still appears to be marginal in S. lugdunensis, although only some strains have the mecA gene associated with methicillin resistance. In the largest collection of bacteria tested for the presence of methicillin resistance and the mecA gene, Kleiner et al. found that 3% of 36 strains tested were oxacillin resistant and displayed the mecA gene (37). According to American (Clinical and Laboratory Standards Institute) and European (European Committee on Antimicrobial Susceptibility Testing) guidelines, S. aureus and S. lugdunensis have identical clinical breakpoints, higher than those of other CoNSs (38). These two species with oxacillin MIC values of >2 mg/liter are mostly methicillin resistant due to the presence of the mecA gene. The corresponding MIC for other CoNSs is >0.25 mg/liter, because a lower breakpoint correctly classifies most CoNSs with the mecA gene whereas it overcalls resistance for S. lugdunensis (39). It is of interest to note that cefoxitin has appeared as a more reliable indicator to detect methicillin resistance, and once again, S. aureus and S. lugdunensis share similar breakpoints. Cefoxitin MICs of >4 mg/liter predict methicillin resistance and the presence of the mecA gene.
Thus, despite its occurrence in nosocomial infections and its colocalization on the skin with other CoNSs and S. aureus that is commonly methicillin resistant, S. lugdunensis does not seem to share resistance genes through horizontal genetic transfer. Nevertheless, recent genetic reports have emphasized the occurrence of various mobile genetic elements.
SPECIFIC GENETIC FEATURES
Staphylococcus lugdunensis was fully sequenced in 2010 by Tse et al. (40). Since then, 19 genome assembly and annotation projects have become currently available. The genome lengths from the various strains of this bacterium range from 2.5 to 2.6 Mb with a GC content of 33.7% to 33.9%. The closest related species is S. haemolyticus, with 78.3% homology of the coding sequences, followed by S. aureus with 77.8% homology (41). The genes encoding virulence factors that we cited previously have been identified in all strains. One distinctive feature of this CoNS is the presence of several mobile genetic elements (MGE).
Identification of MGE.
We recently identified several plasmid and prophage sequences through the whole-genome sequencing of seven strains obtained from the VISLISI clinical trial (42). We did not identify any virulence or resistance genes in these MGE, but we did find several homologies to some previously described prophages and plasmids, particularly pVISLISI_3 that has 100% homology to pRIVM6519_1, a plasmid first identified in S. aureus. This result suggests the occurrence of horizontal genetic exchange between these two species, characterized by their clinical virulence. We also found full nucleotide similarities between the plasmids pVISLISI_1, pVISLISI_2, and pLUG_10 from S. lugdunensis and SAP108B from S. epidermidis. With respect to the four prophage sequences, we showed that ϕSL2 to ϕSL5 shared 25% to 44% of the putative encoded proteins with stB12 from S. hominis and PH15 from S. epidermidis.
TIME TO CHANGE MICROBIOLOGICAL AND CLINICAL CRITERIA OF INTERPRETATION
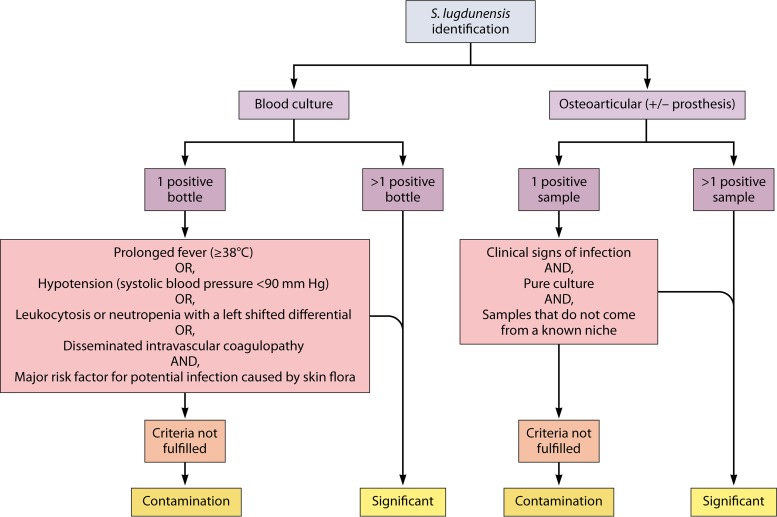
The Infectious Diseases Society of America (IDSA) was the first to consider S. lugdunensis as a different CoNS in its 2015 guidelines for the diagnosis management of osteomyelitis in adults (43). IDSA does not recommend a bone biopsy in patients with suspected osteomyelitis when S. aureus or S. lugdunensis infection has been established based on positive blood cultures. In the same year, the European Society of Cardiology published guidelines for the management of endocarditis and emphasized the role of S. lugdunensis in destructive infectious endocarditis, unlike other CoNSs. These results are clearly supported by growing clinical evidence, and the causative role of the virulence factors described in this review is likely, even if in vitro and animal models are lacking to model this relationship. At the same time, the European Manual of Clinical Microbiology still advises that two positive samples for CoNS are required in a clinical sample to consider it significant, because contamination and colonization remain frequent (44). In contrast, only one S. aureus-positive sample in blood cultures or a bone sample is enough to be considered pathological. Clinical and microbiological evidences are now concordant enough to consider that S. lugdunensis cannot be regarded as a regular CoNS. Fadel et al. demonstrated in a retrospective analysis that 45% of 29 patients with a single S. lugdunensis-positive blood culture did indeed have bacteremia (45). The authors used the criteria published by Souvenir et al. (46), which have proven useful to determine the clinical significance of blood culture positivity for CoNS. Regarding bone, joint, and prosthetic joint infections, the most informative data emerged from the VISLISI clinical trial (5). Among 28 patients with proven infections, 25% had only one positive sample. In this trial, infection was considered to have been proven in cases with only a single positive sample if three criteria were fulfilled, namely, there were clinical signs of infection, a pure culture, and a sample that did not come from a known niche for this organism.
Thus, we propose that S. lugdunensis be considered pathogenic in deep clinical samples, such as blood cultures or bone and articular samples, even if only one sample is positive but in pure culture, at least until any other diagnosis has been proven. However, we would advise caution when interpreting single positive samples in skin and soft-tissues or within a known niche of this bacterium (inguinal, axillary, and nasal). In addition, the positivity of two deep samples is sufficient to consider the bacterium clinically significant. Those aspects are summarized in Fig. 2.
FIG 2.
Clinical significance of microbiological samples with S. lugdunensis identification in blood cultures and osteoarticular samples. Major risk factors for potential infection caused by skin flora are: long-term intravascular catheterization, peritoneal dialysis, hemodialysis, or extensive postsurgical infections with CoNS. S. lugdunensis known niches are inguinal and axillary.
CONCLUSION
Frank et al. were the first to show that S. lugdunensis appeared to be different from other CoNS; since then, clinical, microbiological, and genetic evidences continue to distinguish S. lugdunensis from other CoNSs (6). It is now time to change how clinicians and microbiologists interpret the positivity of clinical samples of S. lugdunensis, particularly blood cultures and osteoarticular samples. This bacterium may not show a high virulence level similar to S. aureus, but its virulence is higher than that of all other CoNSs. The mechanisms linking the identified virulence factors with the clinical observations remain to be elucidated.
ACKNOWLEDGMENTS
We thank the Collège des Universitaires des Maladies Infectieuses et Tropicales (CMIT), research laboratory EA7290 Virulence Bactérienne Précoce from Faculté de Médecine, Strasbourg, that supported all the published studies from our research team regarding S. lugdunensis virulence. We also thank Enago for the English language review and Patrick Lane for art enhancement services.
X.A., Y.H., P.R., and G.P. wrote and corrected the review. The authors declare no conflicts of interest.
There was no financial support for this work.
REFERENCES
- 1.Becker K, Heilmann C, Peters G. 2014. Coagulase-negative staphylococci. Clin Microbiol Rev 27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel R. 2013. Matrix-assisted laser desorption ionization-time of flight mass spectrometry in clinical microbiology. Clin Infect Dis 57:564–572. doi: 10.1093/cid/cit247. [DOI] [PubMed] [Google Scholar]
- 3.Argemi X, Riegel P, Lavigne T, Lefebvre N, Grandpré N, Hansmann Y, Jaulhac B, Prévost G, Schramm F. 2015. Implementation of matrix-assisted laser desorption ionization–time of flight mass spectrometry in routine clinical laboratories improves identification of coagulase-negative staphylococci and reveals the pathogenic role of Staphylococcus lugdunensis. J Clin Microbiol 53:2030–2036. doi: 10.1128/JCM.00177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto M. 2009. Staphylococcus epidermidis–the “accidental” pathogen. Nat Rev Microbiol 7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argemi X, Prévost G, Riegel P, Keller D, Meyer N, Baldeyrou M, Douiri N, Lefebvre N, Meghit K, Ronde Oustau C, Christmann D, Cianférani S, Strub JM, Hansmann Y. 2017. VISLISI trial, a prospective clinical study allowing identification of a new metalloprotease and putative virulence factor from Staphylococcus lugdunensis. Clin Microbiol Infect 23:334.e1–334.e8. doi: 10.1016/j.cmi.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 6.Frank KL, Del Pozo JL, Patel R. 2008. From clinical microbiology to infection pathogenesis: how daring to be different works for Staphylococcus lugdunensis. Clin Microbiol Rev 21:111–133. doi: 10.1128/CMR.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupont C, Sivadon-Tardy V, Bille E, Dauphin B, Beretti JL, Alvarez AS, Degand N, Ferroni A, Rottman M, Herrmann JL, Nassif X, Ronco E, Carbonnelle E. 2010. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin Microbiol Infect 16:998–1004. doi: 10.1111/j.1469-0691.2009.03036.x. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, Shen D, Guo J, Wang K, Wang H, Yan Z, Chen R, Ye L. 2012. Clinical and microbiological characterization of Staphylococcus lugdunensis isolates obtained from clinical specimens in a hospital in China. BMC Microbiol 12:168. doi: 10.1186/1471-2180-12-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinsky BA, Samson D, Ghafghaichi L, Baron EJ, Banaei N. 2009. Comparison of real-time PCR and conventional biochemical methods for identification of Staphylococcus lugdunensis. J Clin Microbiol 47:3472–3477. doi: 10.1128/JCM.00342-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elamin WF, Ball D, Millar M. 2015. Unbiased species-level identification of clinical isolates of coagulase-negative staphylococci: does it change the perspective on Staphylococcus lugdunensis? J Clin Microbiol 53:292–294. doi: 10.1128/JCM.02932-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yonetani S, Ohnishi H, Ohkusu K, Matsumoto T, Watanabe T. 2016. Direct identification of microorganisms from positive blood cultures by MALDI-TOF MS using an in-house saponin method. Int J Infect Dis 52:37–42. doi: 10.1016/j.ijid.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Chen JHK, Ho P-L, Kwan GSW, She KKK, Siu GKH, Cheng VCC, Yuen K-Y, Yam W-C. 2013. Direct bacterial identification in positive blood cultures by use of two commercial matrix-assisted laser desorption ionization–time of flight mass spectrometry systems. J Clin Microbiol 51:1733–1739. doi: 10.1128/JCM.03259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verroken A, Defourny L, le Polain de Waroux O, Belkhir L, Laterre P-F, Delmée M, Glupczynski Y. 2016. Clinical impact of MALDI-TOF MS identification and rapid susceptibility testing on adequate antimicrobial treatment in sepsis with positive blood cultures. PLoS One 11:e0156299. doi: 10.1371/journal.pone.0156299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho P-L, Leung SM-H, Chow K-H, Tse CW-S, Cheng VC-C, Tse H, Mak S-K, Lo W-K. 2015. Carriage niches and molecular epidemiology of Staphylococcus lugdunensis and methicillin-resistant S. lugdunensis among patients undergoing long-term renal replacement therapy. Diagn Microbiol Infect Dis 81:141–144. doi: 10.1016/j.diagmicrobio.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 15.van der Mee-Marquet N, Achard A, Mereghetti L, Danton A, Minier M, Quentin R. 2003. Staphylococcus lugdunensis infections: high frequency of inguinal area carriage. J Clin Microbiol 41:1404–1409. doi: 10.1128/JCM.41.4.1404-1409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bieber L, Kahlmeter G. 2010. Staphylococcus lugdunensis in several niches of the normal skin flora. Clin Microbiol Infect 16:385–388. doi: 10.1111/j.1469-0691.2009.02813.x. [DOI] [PubMed] [Google Scholar]
- 17.Liu P-Y, Huang Y-F, Tang C-W, Chen Y-Y, Hsieh K-S, Ger L-P, Chen Y-S, Liu Y-C. 2010. Staphylococcus lugdunensis infective endocarditis: a literature review and analysis of risk factors. J Microbiol Immunol Infect 43:478–484. doi: 10.1016/S1684-1182(10)60074-6. [DOI] [PubMed] [Google Scholar]
- 18.Petti CA, Simmon KE, Miro JM, Hoen B, Marco F, Chu VH, Athan E, Bukovski S, Bouza E, Bradley S, Fowler VG, Giannitsioti E, Gordon D, Reinbott P, Korman T, Lang S, Garcia-de la-Maria C, Raglio A, Morris AJ, Plesiat P, Ryan S, Doco-Lecompte T, Tripodi F, Utili R, Wray D, Federspiel JJ, Boisson K, Reller LB, Murdoch DR, Woods CW, International Collaboration on Endocarditis-Microbiology Investigators . 2008. Genotypic diversity of coagulase-negative staphylococci causing endocarditis: a global perspective. J Clin Microbiol 46:1780–1784. doi: 10.1128/JCM.02405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. 2015. Staphylococcus aureus Infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lourtet-Hascoët J, Bicart-See A, Félicé MP, Giordano G, Bonnet E. 2016. Staphylococcus lugdunensis, a serious pathogen in periprosthetic joint infections: comparison to Staphylococcus aureus and Staphylococcus epidermidis. Int J Infect Dis 51:56–61. doi: 10.1016/j.ijid.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Douiri N, Hansmann Y, Lefebvre N, Riegel P, Martin M, Baldeyrou M, Christmann D, Prevost G, Argemi X. 2016. Staphylococcus lugdunensis: a virulent pathogen causing bone and joint infections. Clin Microbiol Infect 22:747–748. doi: 10.1016/j.cmi.2016.05.031. [DOI] [PubMed] [Google Scholar]
- 22.Velay A, Schramm F, Gaudias J, Jaulhac B, Riegel P. 2010. Culture with BACTEC Peds Plus bottle compared with conventional media for the detection of bacteria in tissue samples from orthopedic surgery. Diagn Microbiol Infect Dis 68:83–85. doi: 10.1016/j.diagmicrobio.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 23.Donvito B, Etienne J, Denoroy L, Greenland T, Benito Y, Vandenesch F. 1997. Synergistic hemolytic activity of Staphylococcus lugdunensis is mediated by three peptides encoded by a non-agr genetic locus. Infect Immun 65:95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell J. 2004. Characterization of the fibrinogen-binding surface protein Fbl of Staphylococcus lugdunensis. Microbiology 150:3831–3841. doi: 10.1099/mic.0.27337-0. [DOI] [PubMed] [Google Scholar]
- 25.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. 2014. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heilbronner S, Hanses F, Monk IR, Speziale P, Foster TJ. 2013. Sortase A promotes virulence in experimental Staphylococcus lugdunensis endocarditis. Microbiology 159:2141–2152. [DOI] [PubMed] [Google Scholar]
- 27.Nilsson M, Bjerketorp J, Wiebensjö A, Ljungh A, Frykberg L, Guss B. 2004. A von Willebrand factor-binding protein from Staphylococcus lugdunensis. FEMS Microbiol Lett 234:155–161. doi: 10.1111/j.1574-6968.2004.tb09527.x. [DOI] [PubMed] [Google Scholar]
- 28.Frank KL, Patel R. 2007. Poly-N-acetylglucosamine is not a major component of the extracellular matrix in biofilms formed by icaADBC-positive Staphylococcus lugdunensis isolates. Infect Immun 75:4728–4742. doi: 10.1128/IAI.00640-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajendran NB, Eikmeier J, Becker K, Hussain M, Peters G, Heilmann C. 2015. Important contribution of the novel locus comeb to extracellular DNA-dependent Staphylococcus lugdunensis biofilm formation. Infect Immun 83:4682–4692. doi: 10.1128/IAI.00775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Argemi X, Prévost G, Riegel P, Provot C, Badel-Berchoux S, Jehl F, Olivares E, Hansmann Y. 2017. Kinetics of biofilm formation by Staphylococcus lugdunensis strains in bone and joint infections. Diagn Microbiol Infect Dis 88:298–304. doi: 10.1016/j.diagmicrobio.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. 2011. Staphylococcus aureus biofilms. Virulence 2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tasse J, Croisier D, Badel-Berchoux S, Chavanet P, Bernardi T, Provot C, Laurent F. 2016. Preliminary results of a new antibiotic susceptibility test against biofilm installation in device-associated infections: the Antibiofilmogram. Pathog Dis 74:ftw057. doi: 10.1093/femspd/ftw057. [DOI] [PubMed] [Google Scholar]
- 33.Macià MD, Rojo-Molinero E, Oliver A. 2014. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin Microbiol Infect 20:981–990. doi: 10.1111/1469-0691.12651. [DOI] [PubMed] [Google Scholar]
- 34.Malemud CJ. 2006. Matrix metalloproteinases (MMPs) in health and disease: an overview. Front Biosci 11:1696–1701. doi: 10.2741/1915. [DOI] [PubMed] [Google Scholar]
- 35.Cassat JE, Hammer ND, Campbell JP, Benson MA, Perrien DS, Mrak LN, Smeltzer MS, Torres VJ, Skaar EP. 2013. A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe 13:759–772. doi: 10.1016/j.chom.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrand AJ, Haley KP, Lareau NM, Heilbronner S, McLean JA, Foster T, Skaar EP. 2015. An iron-regulated autolysin remodels the cell wall to facilitate heme acquisition in Staphylococcus lugdunensis. Infect Immun 83:3578–3589. doi: 10.1128/IAI.00397-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleiner E, Monk AB, Archer GL, Forbes BA. 2010. Clinical significance of Staphylococcus lugdunensis isolated from routine cultures. Clin Infect Dis 51:801–803. doi: 10.1086/656280. [DOI] [PubMed] [Google Scholar]
- 38.CLSI. 2017. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. CLSI document M100–S27 CLSI, Wayne, PA. [Google Scholar]
- 39.Hussain Z, Stoakes L, Massey V, Diagre D, Fitzgerald V, El Sayed S, Lannigan R. 2000. Correlation of oxacillin MIC with mecA gene carriage in coagulase-negative staphylococci. J Clin Microbiol 38:752–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tse H, Tsoi HW, Leung SP, Lau SKP, Woo PCY, Yuen KY. 2010. Complete genome sequence of Staphylococcus lugdunensis strain HKU09-01. J Bacteriol 192:1471–1472. doi: 10.1128/JB.01627-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heilbronner S, Holden MTG, van Tonder A, Geoghegan JA, Foster TJ, Parkhill J, Bentley SD. 2011. Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors: Staphylococcus lugdunensis genome sequence. FEMS Microbiol Lett 322:60–67. doi: 10.1111/j.1574-6968.2011.02339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Argemi X, Martin V, Loux V, Dahyot S, Lebeurre J, Guffroy A, Martin M, Velay A, Keller D, Riegel P, Hansmann Y, Paul N, Prévost G. 2017. Whole-genome sequencing of seven strains of Staphylococcus lugdunensis allows identification of mobile genetic elements. Genome Biol Evol 9:evx077. doi: 10.1093/gbe/evx077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berbari EF, Kanj SS, Kowalski TJ, Darouiche RO, Widmer AF, Schmitt SK, Hendershot EF, Holtom PD, Huddleston PM, Petermann GW, Osmon DR. 2015. 2015 Infectious Diseases Society of America (IDSA) clinical practice guidelines for the diagnosis and treatment of native vertebral osteomyelitis in adults. Clin Infect Dis 61:e26–e46. doi: 10.1093/cid/civ482. [DOI] [PubMed] [Google Scholar]
- 44.Cornaglia G, Courol R, Herrmann J-L, Kahlmeter G (ed). 2012. European manual of clinical microbiology. European Society of Clinical Microbiology and Infectious Diseases, Basel, Switzerland. [Google Scholar]
- 45.Fadel HJ, Patel R, Vetter EA, Baddour LM. 2011. Clinical significance of a single Staphylococcus lugdunensis-positive blood culture. J Clin Microbiol 49:1697–1699. doi: 10.1128/JCM.02058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Souvenir D, Anderson DE Jr, Palpant S, Mroch H, Askin S, Anderson J, Claridge J, Eiland J, Malone C, Garrison MW, Watson P, Campbell DM. 1998. Blood cultures positive for coagulase-negative staphylococci: antisepsis, pseudobacteremia, and therapy of patients. J Clin Microbiol 36:1923–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]