Abstract
Silicon and glass were the main fabrication materials of microfluidic devices, however, plastics are on the rise in the past few years. Thermoplastic materials have recently been used to fabricate microfluidic platforms to perform experiments on cellular studies or environmental monitoring, with low cost disposable devices. This review describes the present state of the development and applications of microfluidic systems used in cell biology and analyses since the year 2000. Cultivation, separation/isolation, detection and analysis, and reaction studies are extensively discussed, considering only microorganisms (bacteria, yeast, fungi, zebra fish, etc.) and mammalian cell related studies in the microfluidic platforms. The advantages/disadvantages, fabrication methods, dimensions, and the purpose of creating the desired system are explained in detail. An important conclusion of this review is that these microfluidic platforms are still open for research and development, and solutions need to be found for each case separately.
I. INTRODUCTION
Miniaturization of devices and systems by means of microfabrication technologies has become very popular in scientific advances, and the micro-nanofluidics field has thus emerged. Particularly, the electronics and chemical, biological, and medical fields have benefited from developing microscale technologies. Conventional laboratory handling, processing, and analytical techniques have been revolutionized with the help of microfluidics. In the field of microfluidics, there has been much valuable work with materials such as glass, polydimethylsiloxane (PDMS) or thermoplastics, and living cells, but this review deals only with devices made of thermoplastics used in cell biology.
A. Choice of material
In the biochemical and biomedical fields, polymer-based materials are primarily used since their surface can be easily modified.1 Being composed of linear and branched molecules, the thermoplastic materials are durable against temperature and pressure changes and they do not suffer from any structural breakdown. The properties/characteristics of thermoplastics used to fabricate chips are summarized in Table I. Thermoplastic-based materials have good physical and chemical characteristics such as low electrical conductivity and high chemical stability, and they are suitable for mass production at low cost. Thermoplastics can be softened and made to flow by applying heat and pressure. During cooling, the softened polymer hardens and it takes the shape of the container or mold without any chemical change.2,3 Low-cost fabrication methods for high-throughput production can be successfully used in thermoplastics made with microfluidic systems.
TABLE I.
Summary of properties for thermoplastics.5–11
| Thermoplastics | Thermal expansion coefficient [m/(m K)] 10−6 | Young's modulus (GPa) | Tg () | Tm () | Solubility parameter (MPa)1/2 | Water absorption (%) | O2 permeability (x10−13 cm3. cm cm−2 s−1 Pa−1) | Biocompatibility | Transparency | Auto-fluorescence |
|---|---|---|---|---|---|---|---|---|---|---|
| Cyclo olefin (co) polymer (COC/COP) | 60–70 | 1.7–3.2 | 70–180 | 190–320 | 17.7 | 0.01 | NA | Biocompatible | Transparent | Low |
| Polymethyl methacrylate (PMMA) | 7077 | 2.4–3.4 | 105 | 250–260 | 20.1 | 0.1–0.4 | 0.1 | Biocompatible | Transparent | Low |
| Polyethylene terephthalate (PET) | 59.4 | 2–2.7 | 70 | 255 | 20.5 | 0.16 | 0.03 | Biocompatible | Transparent | Medium |
| Polyethylene-low density (LDPE) | 100–200 | 0.11–0.45 | −125 | 105–115 | 17.6 | 0.005–0.015 | 2 | Biocompatible | Both opaque and transparent | Medium |
| Polyethylene-high density (HDPE) | 120 | 0.8 | −80 | 120–180 | 18.2 | 0.005–0.01 | 0.4 | Biocompatible | Both opaque and transparent | Medium |
| Polypropylene (PP) | 72–90 | 1.5–2 | −20 | 160 | 16.3 | 0.01–0.1 | 1.7 | Biocompatible | Both opaque and transparent | Medium |
| Polystyrene (PS) | 70 | 3–3.5 | 95 | 240 | 18.7 | 0.02–0.15 | 2 | Biocompatible | Transparent | High |
| Polycarbonate (PC) | 65–70 | 2.6 | 145 | 260–270 | 19.4 | 0.23 | 1 | Biocompatible | Transparent | High |
| Polyvinyl chloride (PVC) | 54–110 | 2.4–4.1 | 80 | 100–260 | 19.4 | 0.04–0.4 | 0.04 | Biocompatible | Transparent | High |
| Polyamide (Nylon) | 110 | 2.5 | 47–60 | 190–350 | 28 | 1.6–1.9 | 0.03 | Biocompatible | Transparent | High |
| Polysulfone(PSU) | 55–60 | 2.48 | 185 | 180–190 | 18.7 | 0.2–0.8 | NA | Biocompatible | Translucent | High |
| Polylactic acid (PLA) | 740 | 3.5 | 60–65 | 150–160 | 0.68 | NA | Biocompatible (problematic) | Transparent | High | |
| Polytetrafluoroethylene (PTFE) | 112–135 | 0.4 | 115 | 326 | 12.6 | 0.005–0.01 | 3 | Biocompatible | Translucent | High |
| Polyetheretherketone (PEEK) | 26 | 4–24 | 143 | 343 | 21.9 | 0.1–0.5 | 0.1 | Biocompatible | Opaque | NA |
| Acrylonitrile butadiene styrene (ABS) | 72–108 | 1.4–3.1 | 105 | Amorphous | 18.8 | 0.05–1.8 | 0.5 | Not suitable | Both opaque and transparent | High |
It is very important to select the material type by taking the design of the device into consideration, the compatibility of the material with the chemicals, as well as the applied temperature and pressure. Thus, microfabrication techniques and functional components involved in the miniaturized systems should be planned in detail before constructing the microfluidic system.4
The mechanical, chemical, and optical properties of the polymers are listed in Table I. According to this table, it is not convenient to use LDPE or HDPE as a construction material of microfluidic devices due to their high thermal expansion coefficients and low melting point that make the bonding process difficult. Moreover, they are susceptible to stress cracking.12 PVC is largely employed in medical products but its usage is still open to discussion.13 Automotive, electrical/electronic, and industrial applications have benefited from the advantages of polyamide polymers. However, polyamide has disadvantages such as high moisture pick-up with related dimensional instability and high shrinkage in molded sections. Therefore it is not convenient to use this polymer in microfluidic applications.14 PMMA, PET, and PS offer good thermal stability and insulation properties. High mechanical strength, hardness, and rigidity are the advantageous characteristics of the PMMA substrate but COP/COC is better than these polymers.15–18 The COP/COC is a fully saturated olefin polymer and it shows very little interaction with proteins. The material's ring structure provides high stability. The COP/COC polymer has a smaller Young's modulus than PMMA, and its rigidity and strength is higher. This also makes the dimensional stability property of the COP/COC polymer better.19
When the chemical properties are considered, the material used in cell biology must be biocompatible. According to Table I, most of the thermoplastics are biocompatible except ABS and PLA. The oxidative stability of PET might cause problems during the long experiments.20 PCs are classified as linear polymers that include two germinal ether bonds and a carbonyl bond. While this bond is hydrolytically stable, surface erosion might occur during the in vivo applications. In addition, the release of Bisphenol A (BPA), which is very hazardous in food contact situations, might show up during hydrolysis.21 PVC and nylon are also known as biocompatible materials but, PVC can release toxic gases during manufacturing and nylon is a heat sensitive material.13,14 PS is widely used in molecular and cell biology studies due to its biocompatibility. Petri dishes, test tubes, microplates, and other laboratory containers are all made of PS and this material has high resistivity against alcohols, polar solvents, and alkalis.18,22 PMMA has chemical inertness to many solutions and solvents; however, unfortunately it is affected by ethanol, isopropyl alcohol (IPA), acetone, and other important solvents used in microfabrication and sterilization. The polishing of PMMA is also easy and it displays low water absorption and excellent water resistance.15,16 Most importantly, PMMA is a biocompatible polymer, except when its surface is treated with ozone or O2 plasma.17 The COP/COC polymer shows resistance against acids, bases, and almost all solvents including ethanol, IPA, and acetone, and it has the lowest water absorbency among all plastics. This polymer's stability does not change under moisture conditions, exhibiting a good structure for microfluidic device fabrication.1 The high moisture barrier of COP/COC is beneficial; when working with cell cultures, the cells consume more oxygen from water, instead of its absorption onto the surface of the polymers, and the COP/COC material provides an inert low binding surface property.23
In order to follow the changes inside the microfluidic devices under a microscope, the optical properties of the selected material (e.g., transparency) are very important. Therefore PEEK, PP, PSU, and PTFE are not suitable to work with. In addition to transparency, the auto-fluorescence characteristic of a substrate determines the natural fluorescence that appears in it. Although the auto-fluorescent characteristics of LDPE and HDPE are better than most of the other thermoplastics, their bonding is difficult.12 PET also shows medium auto-fluorescence characteristics, but using PET as a fabrication material of a microfluidic device for biological applications is questionable. There are many microfluidic devices made of PC, but its auto-fluorescence is high. Thus, this makes PC difficult to use when working with fluorescently labeled cells or materials. PS has high transparency and the surface of PS is suitable for long-term cell studies.18 Despite all these appealing characteristics, the thermo-compression bonding of PS pieces has rarely been utilized, and not enough attention has been given to directing the research further into such device applications.24 However, compared to COP/COC and PMMA it has higher background auto-fluorescence. PMMA has a low auto-fluorescence background and this material exhibits excellent transparency.15,16 On average, 92% of light in the visible range can pass through a typical PMMA grade and PMMA materials can be found easily at a low-cost.17 COC/COP materials are the best among other thermoplastics in terms of auto-fluorescence.4
Glass is an extensively used material in microfluidic practices. Glass has a good surface stability and solvent compatibility. In addition, glass is also biocompatible, chemically inert, and hydrophilic. It provides superior optical transparency and high-pressure resistance. However, glass microfluidic devices are relatively expensive and they are prone to breaking. PDMS is a widely used soft-elastomer in microfluidic systems. Soft-lithography techniques can be employed easily to create complex fluidic circuits by PDMS. This material is optically transparent and flexible. Furthermore, it is inert, non-toxic, and biocompatible. Unfortunately, organic solvents can be absorbed by PDMS and it is gas permeable. Proteins or small hydrophobic solvents can also be absorbed and this may create cell adhesion problems and clogging in the microfluidic devices. Since PDMS is an inherently hydrophobic material, it requires surface treatments to transform its surface into a hydrophilic surface. Moreover, PDMS is not a durable material for lengthy experiments. Consequently, it is not possible to commercialize the microfluidic systems made of PDMS as it is not a rigid material.25–29
In this review, PMMA, COP/COC, PC or PS made microfluidic devices, which are fabricated mainly to work with living organisms and tissues, will be focused on, and among them PMMA is the most extensively used one for cell biology applications. COP/COC and PS are promising substrates. When publications from the year 2000 are examined, more than 70% of the research belongs to PMMA, and there are only 12 publications (9%) on COP/COC made devices used in work related to cells (WOS). Nevertheless, COP/COC is a rising substrate for the commercialization of microfluidic devices made of thermoplastics.
B. Fabrication
Photolithography, oxidation, e-beam evaporation, wet etching, sputtering, injection molding, micromilling, laser ablation, computer numerical control (CNC) machining, hot embossing, and CO2 laser engraving are some of the methods applied to create the patterns of microfluidic devices. Double sided adhesives, UV/ozone assisted thermal bonding, solvent assisted thermal bonding, pressure sensitive adhesive tapes, laminating adhesives or thermal fusion bonding were some of the common methods to seal these devices. So far, microfluidic devices made from thermoplastics are simple systems lacking active components such as micropumps, microvalves, and sensors.
Interfacial interactions play an important role for the performance of the microfluidic devices. Several surface modification techniques, such as plasma or UV treatment, can be applied to the polymers to control the surface properties.30 A hydrophilic surface is required to have a smooth and consistent flow of analytes within the microchannels to conduct and to monitor the experiments properly. Moreover, cell or tissue interactions with biocompatible materials can be managed by biological recognition. In order to increase this biological recognition, most often the surface of the device is coated with protiens.31 For example, PC12 cell adhesion, proliferation, and differentiation into a microfluidic neural interface platform were enhanced via polypeptide surface treatment.30 In another example, biotinylated- Bovine serum albumin (BSA) was used to treat the surface of the microfluidic device to immobilize the mother yeast cells.32 We have been fabricating microfluidic devices using thermoplastic substrates like PMMA, COP/COC, and PS by hot embossing, chemical etching, and thermal bonding methods. These 1 nl microbioreactors are successfully used for yeast culturing. Fortunately, for yeast cell cultivation, COP/COC, PMMA, and PS made microfluidic devices do not require any modifications on the surface.25
C. Applications and commercialization
Cell or tissue culturing, separation, detection, analysis or reaction-production studies can be conducted in microfluidic devices by using small sample volumes. In addition, high-throughput drug screening, single cell or molecule analysis and manipulation, drug delivery and therapeutics, biosensing, and point-of-care diagnostics are some of the biological applications that can be accomplished via these devices.33–36
Although microfluidic devices are capable of making high-throughput screening by using a small amount of consumables, commercialization of these devices is not at the desired level. This might be the result of challenges encountered such as finding adequate funding for product development and manufacturing during the stages of commercialization. However, a few companies have overcome these steps, and they have introduced their products into the market. Pico-Gen Picodroplet Formation Chips by Sphere Fluidics Ltd., Multiflux by Dolomite Microfluidics or Chips by Microfluidic ChipShop are the examples of commercialized products. Microfluidic products in the market can be increased if industrial partners and academic partners can come together and reach an agreement. Moreover, as the developments increase in both bioMEMS technologies and commercialization of microfluidic equipment, real-world problems can be solved by researchers effectively at high throughput and low cost. At the end, point-of-care diagnostics, rapid, quantitative, and multiplexed immunoassays, biosensors or instruments for rapid detection of pathogens can be more widely produced and individuals suffering from diseases can be treated more quickly and easily.37,38
In this review paper, microfluidic devices made from thermoplastics, that have been used to study living-organisms or tissues are described. This paper includes 4 main titles, which are related to Cultivation: Organism-on-a-chip, Separation/Isolation, Detection and Analysis and Reaction: Microbial Fuel Cells in thermoplastic made microfluidic devices.
II. CULTIVATION: ORGANISM-ON-A-CHIP
Cells, capable of dividing and increasing in size, continue to grow when the appropriate media and conditions are available. It is generally accepted that, a single cell is the building block for human life. Every cell includes a genetic material that holds the secret to inherited diseases. Scientists have improved the methods of studying the behavior of single cells. The effects of the amount and type of the nutrients, temperature, humidity, and gaseous atmosphere on cells can be investigated to provide optimum cultivation conditions. The cell's response in a culture is measured to reveal the relation between other kinds of cells, carcinogenic agents or, eventually, drugs. Nowadays, these experiments can be conducted in precisely controlled micro-environments, where a low volume of sample and energy are needed. High-throughput screening can also be conducted in these microfluidic devices.39,40
A. Culturing of bacterial cells
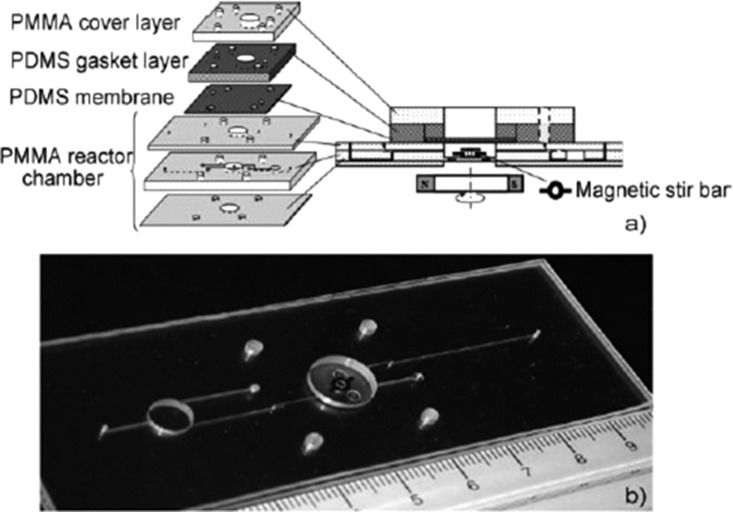
Cell culturing experiments have been conducted with several living organisms. In 2005, Szita and his colleagues fabricated PMMA and PDMS made microbioreactors to monitor bacteria cells, Escherichia coli (E. coli). A multiplexed microbioreactor system including magnetic motors for magnetic stirring and optics to observe the optical density (OD), were used to measure the parameters, dissolved oxygen (DO), and pH, during fermentation. The system can make four parallel microbial fermentations.41 Another study with bacteria cells in a polymer-based microbioreactor system is capable of measuring optical density (OD), pH, and dissolved oxygen (DO) in real-time. Continuous cultivation of E. coli was done in a microbioreactor of 150 μl volume with membrane aeration. The device consists of PMMA and PDMS layers with three connecting microchannels of 250 μm height and 250 μm width (Fig. 1). The surface modification of the PMMA and PDMS parts of the device was conducted via poly(ethylene glycol)-grafted poly(acrylic acid) (PAA) copolymer films to obtain bio-inert surfaces resistant to non-specific protein adsorption and cell adhesion. This surface modification reinforced the cultivation time and prevented wall growth of the cells.42
FIG. 1.
(a) Schematic view of the microfluidic system. (b) Photograph of the PMMA chamber. Reproduced with permission from Zhang et al., Lab Chip 6, 906 (2006). Copyright 2006 Royal Society of Chemistry.
In 2010, glass slides and polymer films were used to create low-cost and detachable microfluidic chips by applying adhesive wax as a bonding material. This biocompatible wax-based microfluidic chips were used to perform PCR tests and to culture GFP-tagged (Green fluorescence protein) E. coli to see the effect of the antibiotic ciprofloxacin concentration on the E. coli migration. The fluorescence expression was found to decrease as the ciprofloxacin concentration increased in the reagent cell. Otherwise, E. coli cells survived 15 days in the chip without any leakage.43 In the same year, Skolimowski and his colleagues built a five layer microfluidic chip allowing gas transition and examined the active oxygen depletion. Simulations were performed via COMSOL Multiphysics 3.5a to follow the relation between O2 generation and measured oxygen concentration. Pseudomonas aeruginosa bacterium was used in the microchip to examine the growth patterns under different oxygen concentrations. A GFP-tagged bacterium was allowed to grow in the flow chambers of the device. The oxygen concentration affected the attachment of bacterium to the substrate.44 The drug resistance of several bacterial strains E. coli, Shigella flexneri, Shigella boydii, Shigella sonnei, and Uropathogenic E. coli was tested using the Resazurin dye reduction method (RRM) as the colorimetric antibiogram in the PMMA microfluidic system and 96-well microtiter plates. Bacteria suspensions were inoculated into the wells of both the 96-well plates and the microfluidic device. Visual and OD620 results of the microfluidic system and 96-well microtiter plates were compared with the standard turbidity tests. The data obtained from both systems were compatible with each other, so the developed microfluidic device can be used in the determination of the antibiogram of the drug-resistant bacteria.45 Recently, the determination of E. coli K12 concentration was done by using positively and negatively charged electrospun poly(vinyl alcohol) (PVA) nanofibers in PMMA made microchannels. Fiber distribution and fiber mat height on analyte retention were also examined. In order to have a large surface area for analyte concentration and to obstruct size-related retention of the E. coli cells, the 3 D morphology of the mats was improved. Positively charged nanofibers showed better performance than the negatively charged ones. Then, the negatively charged nanofibers were customized with anti-E.coli antibodies, and consequently they became capable of specific capturing the bacterial cells.46
B. Fungi cultivation
PMMA made microfluidic devices were also employed for fungi cultivation. As a new approach for the cultivation, monodispersed agar beads were produced by using a temperature-controlled microfluidic device with 5 layers. The dimensions of the agar beads were determined by the flow, which can be dispersed or continuous (Fig. 2). These agar beads were used as the substrate for the Cordyceps militaris cells. This technology is seen as a promising one because several experiments can be performed, like encapsulating biomaterials, enzymes, and drugs into agar beads, for biomedical applications.47 In another study on fungal and bacterial cells, Bolic et al. developed a milliliter-scale bioreactor (0.5–2 ml volume) consisting of gas connections, heater, temperature sensor, optical fibers, magnetic stirrer, and optical sensors. pH, dissolved oxygen, and optical density can be measured and the system is capable of aeration and mixing. The performance of the device was evaluated by considering mixing time, residence time distribution, and oxygen transfer rates in several conditions. The mixing time was determined as 0.4 s - 2 s, and the oxygen transfer rate was 1000 h−1. Bacterial and yeast cells (Lactobacillus paracasei and Saccharomyces cerevisiae (S. cerevisiae) cells) were successfully cultivated in the device.48
FIG. 2.
(a) The 5 layers of the microfluidic device. (b) Assembled view of the temperature-controlled chip. Reproduced with permission from Lin et al., Electrophoresis 32, 3157 (2011). Copyright 2011 Wiley Online Library.
By using ultrasonic hot embossing and welding techniques, microfluidic devices made from PC were fabricated and the functionality of these devices was shown via yeast cultivation experiments. Cells survived at least 22 h in the device and enhanced green fluorescent protein (eGFP) expression was observed with a supply of the inducer galactose. It was proven that, ultrasonic processing can be used for microfluidic device fabrication in the future to conduct microbial analysis.49
C. Zebra fish cultivation
The zebrafish is the most used vertebrate model organism in scientific studies. Zebrafish (Danio rerio), which can be used for drug research and environmental toxicology studies, was investigated for its developmental analysis in a 3 D multilayer microfluidic system. By using this system, one embryo can be kept in one trap and each of these embryos can be encoded. On a large scale, it is possible to make high-throughput docking and recovery of single embryos. The device included conical traps of 2 mm in diameter at the top plane and 1.6 mm in diameter at the bottom plane. Wild type and GFP tagged Tg(fli1a:EGFP) zebrafish embryos were trapped, and kept for 3 days via active suction-based immobilization and then analyzed. 100% trapping of the cells was attained successfully, and these cells kept their positions during the 72 h experiments. This system can also be adapted to the kinetic analysis of pharmacological agents prohibiting blood vessel growth (angiogenesis) in zebrafish.50,51 In 2013, another PMMA made microfluidic device for zebrafish was developed, and Environmental Scanning Electron Microscope (ESEM) imaging was used to observe the zebrafish larvae. There were an engraved reservoir, 36 circular microwells, and 6 microchannels in the chip. The reservoir with the multiple semispherical microwells, located in the device, was to keep the larvae and to drain the excess medium. In order to activate the device, a paper filter was used, and the trapping of the larvae was achieved by the suction of the cells due to water drainage. This microfluidic system was important for the ESEM imaging for prospective laser microsurgery and tissue regeneration. However, the experiments were not successful due to the damaged tissues under a low vacuum environment, and more optimization was needed to make ESEM imaging without tissue damage.52 In 2014, Akagi et al. developed another microfluidic system to trap and immobilize the transgenic zebrafish embryos with the help of low-pressure suction. The PMMA made microchip platform consisted of piezoelectric microdiaphram pumps, embryo-trapping suction manifold, drug delivery manifold, and tin oxide heating element. Gambit 2.3 and Finite-volume-based Fluent 6.3 softwares were used for computational fluid dynamics (CFD) simulations. Embryo loading and recovery were done in the main channel, single embryo trapping and immobilization were conducted in an array of 16 traps, and the drug delivery was done via a drug delivery channel. This system was capable of making rapid and automated manipulation of zebrafish cells for drug discovery.53 A year later, Akagi and his group improved their microfluidic system and this system was able to reveal the morphological features of the zebrafish larvae by employing ESEM imaging technology. Microwells were used to keep the yolk of the zebrafish larvae and microchannels were used to provide immobilization of the larvae.54 In another study on zebrafish, a continuous flow embryo sorter device capable of analyzing, sorting, and dispensing the zebrafish embryos was developed. In order to actuate the system, DC gearmotors with a D-shaped output shaft (3 mm diameter) were integrated into the system. In the main body of the device, a rectangular channel for embryo loading, sorter wheel and suction manifold for keeping the embryos in position during the rotation were located, whereas in the 3 D printed part, a DC motor and stainless steel ball bearings were present. Through this study, a new concept for rapid and automated zebrafish embryo sorting, and a device to do so, was introduced.55 An electronic interface integrated Lab-on-a-chip biomicrofluidic device was used to make automatic immobilization, cultivation, and treatment of zebrafish embryos. The design and optimization of the microfluidic device were done with the Gambit 2.3 CFD simulation program. The main channel for embryo loading, 20 traps for embryo trapping and immobilization, and a plenum suction manifold of 0.7 mm height for creating the drag force to immobilize the cells were located within the device. The Field Programmable Gate Array (FPGA) hardware/software controlled the cell loading and immobilization, flow dynamics, temperature, and image acquisition of zebrafish embryos.56 Furthermore, Zhu et al. fabricated a minimized high-throughput Lab-on-a-Chip microfluidic device to perform a fish embryo toxicity (FET) assay. The device had a 96-well microtiter plate, a main loading channel, and 21 miniaturized embryo traps. Gambit 2.3 and Finite-volume based Fluent 6.3 softwares were used to run CFD simulations. Rapid loading, separating, and immobilizing of the zebrafish cells in the traps, providing continuous perfusion and live imaging were done with this 3 D device. The off-chip interface carried peristaltic pumps, USB-imaging station, and ITO heaters. In addition, anti-angiogenesis drug tests were performed in this device, and the image acquisition was done with an imaging cytometer.57 Fluorescence Ratiometric Imaging (FRIM) technology was employed to develop the zebrafish embryos in a microfluidic device. FRIM technology can make the kinetic quantification of the aqueous oxygen gradients, and the oxygen consumption of the cells can be measured. The device included a main loading channel for embryo loading and toxicant transportation, 18 embryo traps of 1.5 mm × 1 mm sizes for trapping and immobilization, a suction manifold with the interconnecting channels and a sensing manifold with Presens Sensor Foils. This microfluidic system can be used to reveal the metabolism and physiology of the cells in the future.58
D. Fruit fly (drosophila) cultivation
In 2016, a PMMA made microfluidic device for Drosophila was fabricated to study the actions of flies in the system. In the device, the chambers with visual and auditory stimuli were used to manage the liquid food presentations. A behavior chamber and a feeding alcove were placed in the chip for flies to feed from a microchannel. The behavior of the flies and microfluidic food channel were video recorded. According to the repeated experiments, the flies learned to access the food in a more direct way.59
E. Mammalian cell cultivation
For human cell and tissue culturing experiments, PMMA, PS, PC, and COC substrates were investigated as the biocompatible materials alternative to PDMS. In order to decrease the adsorption of hydrophobic compounds, UV-generated ozone or oxygen plasma surface treatments were done on the polymer substrates. The validity of the surface treatment was evaluated via the contact angle of water on the surface. In order to reveal the biocompatibility, human hepatoma (HepG2) cells were used on the treated surfaces of the microfluidic chips. After cultivation of the cells on well-plates, they were exposed to acridine orange and propidium iodide solutions for viability tests. PDMS and PMMA showed a lower recovery of the hydrophobic compounds. After surface treatment, HepG2 cells were stuck to the PMMA substrate due to the unstable peroxides on the PMMA surface after the treatment. PC and COC chips showed good performance about gas permeability and COC is better than PC due to its lower auto-fluorescence. Overall, PC and COC chips were mostly suitable for incorporation of cells and tissues.60 To study the co-cultured cell behavior, human U937 and MG-63 cell lines were employed where monitoring of real-time cytokine release, generation of a linear cytokine gradient for drug discovery, and non-contact co-culturing processes were accomplished.61 Shear-stress acting on the cells may give information about the dissolved oxygen (DO) level. Using HT1080 cells, the transportation of the oxygen through the fabricated microbioreactor was first investigated, then the low-shear stress at the cell level and oxygen tension of the materials and dimensions were regulated. There were two channels inside the microbioreactors, one of them is the culture channel placed in the bottom sheet, and the other one is the oxygen supply channel on the top plate. The number of cells and their viability (fluorescent live/dead staining), the cell density, and the circularity of the cells were investigated.62 HEK-293 T cells were cultivated in PDMS and PMMA microfluidic devices to reveal the optimum conditions for growth under several experimental conditions such as with or without the cell adhesion agent poly-D-lysine. Microchannel geometries, thicknesses, and flow rates in the devices were also evaluated. The PMMA chip had three distinct microchannel structures (linear, zigzag, and square waves) of 40 μm height × 0.4 mm width and 3.68 μl volume. The other chip had 40 μm height with serpentine structures of 100 μm width. The length changed between 10 and 80 mm and the volume was 17.8 μl. Poly-D-lysine increased the cell adhesion and viability under continuous or discontinuous flow. Cell adhesion was mostly seen in the corners of the microchannels and in large channels due to lower flow rate. This recent study provides an insight into the future studies on the microbioreactor design.63 In 2009, the hard top-soft bottom microfluidic devices were fabricated. Polyethylene terephthalate glycol (PETG) and cyclic olefin copolymer (COC) or PS were used as hard tops and the channels were imprinted by hot embossing. The hard tops were bonded to elastomeric PDMS or polyurethane (PU). The device included an X region and the height of this area was 200 μm while the height of other channels was 30 μm. HepG2 cells and C2C12 cells were cultured in the devices, and the cell survival was ∼100%.64 A microfluidic device made of polytetrafluoroethylene (PTFE) was fabricated to conduct the encapsulation of living, therapeutically active cells within monodisperse alginate microspheres. HEK293, U-2 OS, and PC12 cell lines (GFP tagged) were successfully encapsulated and loss of cell viability was minimum. High and medium guluronic acid concentrated alginate samples were more applicable to micro-reaction processes.30 Due to the limited functions of the PDMS polymer, conventional plastics are used to rapidly prototype microfluidic systems. Thermal scribing, a one-step fabrication method, was used to produce the PS made microfluidic devices. The applicability of the system was shown via induction of functional neutrophil extracellular traps (NETs). The experiments were helpful to understand the mechanism of neutrophil culture systems.65
1. 3 D culturing
Nowadays, cell cultures are created in a (3-dimensional) 3 D environment to imitate the in vivo conditions. 3 D cultured cells are more reminiscent of the native tissue from which they originated. They can conceive a more elaborate extracellular matrix and better intercellular communication.66,67 3 D cultured cells are independent of the cell density in contrast to 2 D cultured cells. A 3 D cell culture-based chemosensitivity assay was performed in the microfluidic cell culture chip, which contained 36 microbioreactors. Human colorectal adenocarcinoma cells were loaded to the microfluidic device to obtain information about the micro-scale perfusion of the 3 D cell culture and chemosensitivity assay. This platform provides stable, well-defined, and a biologically more relevant culture environment to perform high-precision and high-throughput 3 D cell-culture based assays.68 Huang et al. developed another high-throughput 3 D microfluidic cell culture system including 30 microbioreactors. The system was capable of implementing durable thermal conditions for cell culturing and efficient sample loading, and contained multiple medium perfusion mechanisms with a waste medium collector for bioassays. The microchannels were used to deliver the medium into the microbioreactors or to the waste reservoirs. In the fabricated device, the chemosensitivity assay, the DNA content detection (via fluorescence labeling) and viability experiments were conducted by using a human oral cancer cell line (OEC-M1).69 High-throughput 3 D cell culture, drug administration, and quantitative in situ assays were conducted in a micro-scaffold array system. The microfluidic device included sponge-like micro-scaffolds for absorption of cell or drug loading as well as for avoiding cell loss during medium exchange. The device had a 96 microwell array with 2 mm diameter on the top layer for culture and drug medium, and had a smaller 96 microwell array with 1.5 mm diameter on the bottom layer for the 3 D cell culture. RFP-labeled NIH3T3 fibroblasts, human fibrosarcoma cells (HT1080), human hepatocellular carcinoma cells (HepG2), and human non-small lung cancer cells (NCI-H460) were cultured in this device, and the cells were stained with fluorescence for imaging. In this system, cancer cells showed higher drug resistance than those on the planar high-density multiwall plates (2 D). These 3 D cultured cells were independent of the cell density in contrast to 2 D cultured cells.70 Nery et al. developed a flow-through sensor array to perform cell viability and cell toxicity tests in a microbioreactor system. Cell toxicity experiments were conducted with A549 cells treated with 1,4-dioxane and 5-fluorouracil. In order to enhance the functionality of the device, rectangular obstacles of 0.35 mm × 1.2 mm dimensions were placed in the microchannel. The comparison of the system with the standard methods indicated that this system can be successfully used to carry out cell culture monitoring and drug testing experiments after toxic treatment.71
2. Nanoparticles (NPs) in microfluidic platforms
Nanoparticle (NP) study is very popular in scientific research. NPs find a place in potential applications of biomedical, optical, and electronic fields. NPs build a bridge between the bulk materials and atomic or molecular structures, and they provide a high specific surface area, high reactivity, and rapid diffusion.72 Koh et al. worked with NPs to examine the Bcl-2 down-regulation at the mRNA and protein levels with cellular uptake and apoptosis by using K562 human erythroleukemia cells and G3139 as the drug, and they produced a multi-inlet microfluidic hydrodynamic focusing (MF) system. The lipopolyplex (LP) nanoparticles produced via the MF method were smaller than those produced by the bulk mixing (BM) method. In addition, Bcl-2 antisense uptake was higher in MF LP nanoparticles and these nanoparticles were more influential in the down-regulation of the Bcl-2 protein level than the BM LP nanoparticles.73 In order to increase the transmission of exogenous oligonucleotides (ODN) in vitro, the semi-continuous flow electroporation (SFE) chip was used with the liposome nanoparticles (LNs) containing the target ligand. K562 cells and transferrin-targeted lipoplex encapsulating ODN G3139 nanoparticles were mixed and incubated to increase the nanoparticle binding. In the fabricated microchip, electric pulses were applied during the mixture flow through the channel and Al pieces were used as both electrodes and channel walls in SFE. Since electroporation application makes cell membranes permeable, ODN transmission efficiency was higher when the non-targeted LNs and SFE were combined rather than utilizing targeted LNs alone.74
3. Electric field (EF)-based microfluidic systems
Electrophoresis, electroosmosis or electroporation are generally applied to manipulate the biological cells and record their responses.75 An electrotaxis study was conducted in a microfluidic cell culture chip. Two types of chips were used: the single-field chip (SFC) consisted of only one microchannel of 3000 μm width, 70 μm height and 15 mm length. The multi-field electrostatic chip (MFC) included a cell culture microchannel of 24 mm length, having three segments of 5000, 1667, and 1000 μm dimensions. In SFC, three distinct electric fields (EFs) were applied and the cellular response was recorded. Numerical simulation was done via the CFD-ACE+ software and the results were compared with those of the measured ones. In the MFC chip, CL1–5 and CL1–0 lung cancer cell lines were used, and these two cancer cell lines gave distinct answers under different Efs.76 A multilayer contactless dielectrophoresis (cDEP) system was developed, where sample and electrode channels were placed on different layers. Several simulations were performed via COMSOL and the results were validated experimentally. The developed device had identical characteristics with the other cDEP devices and was capable of increasing fluid throughput. The limiting elements in these devices were the breakdown voltage of the barrier material and the capability of producing high-voltage/high-frequency signals. The ultimate aim was to immobilize the cells in the saw tooth structure with the application of the electric field. When the electric field was applied, individual cells (MDA-MB-231 human breast cells) began to create the pearl chains, and they travelled through the saw tooth structure for trapping. The difference of this device from other devices was that the fluid electrodes and the sample channels were separated from each other by a thin film.77 For cell trapping, a stable electrode with a salt bridge was combined with the microfluidic sensor chip. HEK293 cells were used in this system and the PMMA device was fabricated. By using FIB milling, the micropores were created at the middle of the cell trapping area to form the incubation-type planar patch clamp. The laser-induced channel currents of channel rhodopsin wide receiver (ChRWR) expressing the HEK 293 sensor cell were measured. The results were compared with the pipette patch clamp design, and they were found to be compatible with each other.78 In another study, a PMMA cell culture microchip and a multichannel lens-free CMOS (complementary metal-oxide semiconductor)/LED imaging system were combined to monitor the cell growth. The LabVIEW program was used to manage the CMOD/LED imaging system. HepG2 cancer cells were tested in this system, and cytotoxicity experiments were conducted by using cyclophosphamide solution at several concentrations. Cell proliferation in the device was monitored, and cell growth was successfully obtained under different environmental conditions.79 In the same year, a 1 D scanning detector and a parallel array of flow channels were coupled to create the parallel microfluidic cytometer (PMC) for cell screening assays. A live-cell translocation assay using a NF-kB/GFP fusion protein in Chinese hamster ovary cells, CD3/CD28 receptor capping in Jurkat cells, and nuclear translocation of NF-kB in Jurkat T-cells employing an antibody label were successfully cultured in this system.80
Micro-optical tweezers (μOTs) were combined with the microfluidic device for trapping as well as for accomplishing the mechanical and chemical spectroscopic analyses of the cells. The microfluidic device was made of hybrid PMMA and glass, and μOT achieved the trapping and exciting of the Raman and fluorescence response of the cells. Microprism reflectors were created by Two Photon Lithography (TPL) on the fiber facets to create optical trapping inside the microfluidic device. Red blood cells (RBCs) and tumor cells were used in the system, and the device was capable of trapping cells by setting the power output at each prism at 5 mW. In addition, monitoring of the single cell response with different environmental stress can be conducted.81
There are several advantages of using microfluidic devices for culturing experiments. Most importantly, scientists can make high-throughput analysis by using a single device. These devices allow one to conduct parallel experiments. Experimental conditions such as temperature, cultivation time, and oxygen or pH level settings can be arranged more precisely. In order to monitor the cell growth during cultivation, several imaging technologies, i.e., ESEM or FRIM, can be integrated into the microfluidic devices. Rapid loading, separating, immobilizing of cells, and continuous perfusion through the device can be realized fairly easily. Cell manipulation and cell response observations can be made instantly via these microfluidic devices. In vivo conditions can be created in the 3 D culturing chips, and experiments can be done in a stable, well defined, and biologically relevant culture environment to obtain high-precision and high-throughput.
F. Tissue engineering (TE)
Since the beginning of the 21st century, bio-based materials have been used in many research fields. Healthcare facilities need continuous innovations and the engineering applications in new materials contribute to the development of such innovations. According to the statistics, 100 000 people are on the donor waiting list and 22 people on an average die every day due to insufficiency in organs or tissues. Over the past decade, tissue engineering has made subtle progress in finding solutions to these problems. The field of TE combines notions of materials science, engineering, medicine, and biology to improve cell, tissue, and organ performance.82–84
The first tissue engineering study involving PMMA in the microfield was developed by Dalby and his colleagues. The reaction between primary human osteoprogenitor cell populations and nanotopographies of 10 nm in size was examined. Colloidal lithography and polymer demixing on silicon was used to generate the topographies and then they were hot embossed on PMMA. The cell morphology, cell cytoskeleton, adhesion formation, cell growth, and differentiation were studied using human bone marrow cells.85 The microfabrication and microcontact printing methods were combined to create a spherical organoid (spheroid) microarray culture system. Cylindrical cavities of 300 μm diameter were placed in the chip and these cavities were characterized as being either a supporter or inhibitor of the cell adhesion. Collagen (Col) and polyethylene glycol were used to generate the adhesive and non-adhesive regions in the chip, respectively. Primary hepatocytes were produced as identical spheroids in the middle of the cavities, and hepatocytes were produced as the cuboidal shaped organoids, similar to the in vivo experiments.86 The chitosan microfibers coated with collagen are necessary for cell cultivation, and can be produced in PMMA made microfluidic devices. One of these devices was fabricated with a 45° cross-junction microchannel to pass the chitosan solution and sodium tripolyphosphate (STPP). Hydrodynamic focusing was used to create the laminar flow. Schwann cells and fibroblast cells were cultured in these chitosan microfibers, which created a satisfying environment for the cells. In tissue engineering applications, they can be used as the scaffold for cell cultures.87 In the study on prosthetic cornea matter, PMMA was treated with the help of polydopamine-based adhesive surface chemistry for the improvement of the biointegration of soft tissues. Polydopamine (PDA) treatment with cell adhesive peptide RGD (PDA-PEG-RGD) increased the corneal epithelial cell proliferation and keratocytes. Adhesion to the collagen gels was achieved by PDA but was not achieved by PDA-PEG-RGD and untreated PMMA. When the subcutaneous implantation was applied, tissue reaction to polydopamine-coated surfaces was benevolent even after 45 days. Tissue integration of implants with soft tissues can be operated with polydopamine-based surface chemistries.88 Microengineering of vascular structures was also investigated in microfield studies by the combination of self-assembled monolayer (SAM)-based cell transfer and gelatin methacrylate hydrogel photopatterning methods. Two SAM desorption tools, which are photoinduced and electrochemically triggered, were displayed during the transfer of human umbilical vein cells (HUVECs) from oligopeptide SAM-coated surfaces to the hydrogel. In order to generate the microvascular structure, a perfusion culture chamber made of PMMA was used. This study can be seen as a good start for more complex, vascularized tissue constructs for regenerative medicine and tissue engineering applications with the combination of SAM-based cell transfer and hydrogel photocrosslinking.89 Microfluidic hydrogels with helical microchannels were produced and their perfusion features were evaluated both experimentally and numerically (COMSOL Multiphysics V4.2). Helical microchannels and straight microchannels were compared with each other by means of cell viability and post-encapsulation. The cooled agarose solution was injected into the PMMA chamber, and fabricated via the laser etcher. In order to have different sized helical microchannels, several helical springs were employed. Rhodamine B solution and the NIH 3T3 cell line were used to reveal the diffusion property and the cell viability under the perfusion culture of microfluidic hydrogels, respectively. Helical microchannels were better than straight microchannels in perfusion ability and oxygen and nutrient delivery to cells.90 Two different kinds of nanoengineered polystyrene surfaces (NPS), including nanopillar (NPS-Pi) or nanopore (NPS-Po) were fabricated to study the topographical effects of surfaces on MC3T3-E1 cells. When compared with the flat substrates, NPS has serious effects on cells in terms of the cell morphology, attachment, proliferation, and osteogenic differentiation. In addition, cell proliferation and osteogenesis differentiation were better in NPS-Po.91 By using the microinjection molding technique, osteoinductive micro-pillared PS surfaces were fabricated for bone replacement operations. Micro topography parameters, pillars diameter, aspect ratio, and spacing, were evaluated according to MC3T3-E1 cell adhesion and proliferation criteria after 1, 3, and 7 days from seeding. It is observed that, micro-pillared surfaces were better than flat surfaces.92 In 2013, another study on tissue engineering was done by Sivashankar et al. A 3 D microfluidic system capable of continuous perfusion was established to monitor liver tissue cultures. The microfluidic device included PMMA microbioreactors which were connected with each other, and poly(ethylene glycol) diacrylate (PEG-DA) microstructures with mesothelial cells were placed in these microbioreactors. The major roles of the mesothelial cells were to contribute to the adhesive surface and help tissue repair. The hematoxylin and eosin (H&E) staining method was used to stain the tissue parts, and the terminal deoxynucleotide transferase (dUTP) nick end labeling (TUNEL) assay was utilized to monitor the DNA fragments. The liver tissue conserved its viability after twelve days of culture.93 Primary human alveolar bone osteoblast (PHABO) morphogenesis was investigated in both microchip-based 3 D-static conditions and 3 D-fluid flow-mediated biomechanical stimulation in perfusion bioreactors. The morphogenesis of the PHABO was evaluated by respective imaging, fluorescence based live/dead staining, and SEM and time-lapse imaging techniques. Cubic microcavities of 300 μm length were placed in the microstructured area in the PMMA made microfluidic device. In static cultures and fluid-flow mediated cultures, PHABO showed different morphogenesis, and mechanobiological studies under hard tissue-specific environments stimulate the osteoblasts to the bone phenotype.94 In 2013, a mechanical microconnector system (mMS) was created to regulate the retracted spinal cord stumps. This system was made of PMMA to fill the spinal cord tissue gap after transection. The two discs consisted of 55 small and 15 large honeycombs. The spinal cord stumps were sent through the honeycomb-structured holes by the negative pressure utilization at the outlet tubing system of the mMS and these stumps kept their location in the mMS walls. Axonal regrowth was achieved after 2, 5, and 19 weeks with the mMS, and in-bleeding or cyst was not observed.95
In 2015, a new cell line was created by combining spheroids and tissues in a microfluidic device without the need for scaffolds or metabolic biosynthesis. Liposome fusion, bio-orthogonal chemistry, and cell surface engineering were used in the device by taking advantage of click chemistry. The device has a Y-shaped channel to mix the cell suspensions including C3H10T1/2 stem cells. By using this system, bio-orthogonal chemical groups with the help of click chemistry were created in a rapid, straightforward, and flexible way.96 In another bone-related study, several parameter effects on the vascularization of bone-mimicking tissues were investigated. The design aims to create a link between the macroscale and microscale tissue engineering studies. Here, the effect of endothelial cell (EC) density, cell ratio among ECs, mesenchymal stem cells (MSCs) and osteo-differentiated MSCs, culture medium, hydrogel type, and tissue geometry parameters were researched. The geometry and oxygen gradient of the hydrogels were optimized by using computational simulations (Rhinoceros and COMSOL), and the analyses of microvascular network features were done. Mcells/ml ECs, 10:1:0 cell ratio, osteo-medium, 2 × 2 × 5 mm3 cage, and 2.5 mg/ml fibrin (60%) + collagen (40%) hydrogels were the best choices to produce bone-mimicking pre-vascularized matrices. Isolation of specific cellular populations and genetic analyses can be conducted in this system.97 Although PDMS is a preferred polymer for cell-based research, it has adverse effects on cells due to high gas permeability and surface hydrophobicity of the material. Therefore, the performance of a PS made microfluidic device was tested and compared to that of PDMS. In this process, COP (at the bottom) and cellulose acetate (CA) were used to flatten the surface and prevent adhesion between PS and COP, respectively. Human umbilical vein endothelial cell (HUVEC) culturing in one application and blood neutrophils under chemoattractant exposure observation in another application were conducted. The results showed that, PS made microfluidic devices can be used for long-term cell studies.98 A microfluidic platform with improved 3 D gel capabilities, controlled surface properties, and better high-volume functions was produced in COC via commercially viable fabrication methods. Human microvascular endothelial cells (hMVECs) were used in the experiments and the results showed that COC has no negative effect on cells like PDMS devices.99 A multi-organ-tissue-flow (MOTiF) biochip including a perfusable membrane was constructed in COC to be used in cell culture experiments. Nutrition medium supply, catabolic cell metabolites removal, and shear stress application on endothelial cells (ECs) can be achieved. The results obtained on cell viability, EC marker protein expression, and adhesion of ECs under low and high shear stress environments were compared with the two-dimensionally perfused flow chambers under a stable environment. The MOTiF biochip provided higher cellular density in monolayer with increased cell layer thickness.100 A body-on-a-chip device was also produced to imitate the drug distribution and metabolism processes in the body. A pumpless 14 chamber (chambers were considered as distinct organs) microfluidic cell culture device was fabricated to conduct the separation between the barrier and nonbarrier cell cultures as well as to reveal the interactive responses between the cell lines. A549 (liver) and Caco2 (GI) represented the barrier lines and HepG2 C3A (liver), Meg01 (bone marrow) and HK2 (kidney) represented the nonbarrier lines. The device, comprised of polycarbonate frame, 0.5 mm thick two layer silicon gaskets for cell/organ chambers, 0.25 mm thick two layers of PMMA gaskets for channels, a porous PC membrane, and 1.6 mm thick silicone gaskets. Cell lines lived 7 days in the device. By using this system, the usefulness of building, managing, and cultivating a multi-organ microphysiological system was proved.101
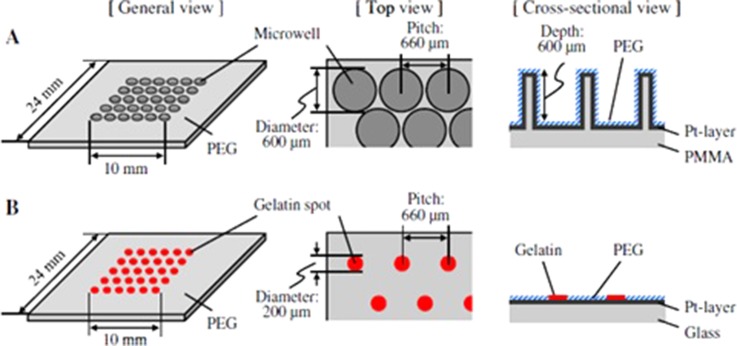
Microwell chip and micropatterned chip were operated and compared by monitoring the proliferation and differentiation characteristics of the embryoid bodies (EB) obtained from the mouse embryonic stem (ES) cells (Fig. 3). The microwell chip had 270 microwells (each 600 μm in diameter and depth). In order to coat the surface with PEG, the chip was immersed into the 50% ethanol solution of 2.5 mM PEG with a thiol group. For the micropatterned chip, 270 gelatin spots (200 μm in diameter) were created on the glass substrate via microcontact printing. The remaining area was again coated with PEG to increase the nonadhesive property. A high cell growth rate and expression of the endotherm as well as an increment in the mesoderm markers were seen in the EBs in the micropatterned chip. The proliferation and differentiation of the EBs may vary from design to design.102 Another microdevice was developed to produce spheroids with almost the same size, and this device was capable of transferring these spheroids from a floating situation to a micropatterned adherent culture. 270 microwells, which were PEG treated to obtain the nonadhesive surface, were located on the PMMA frame and PDMS sheet. On the PMMA frame, the microwells were created via programmable micro-milling technology. On the bottom of the PMMA frame, PDMS was bonded. In this study, first mouse ES cells and then 3T3 cells, HepG2 cells, and primary rat hepatocytes were sequentially produced as spheroids in each microwell chip. This technology can be auspicious in spheroid studies.103 The polymer surfaces, which have changeable cell-attractive and cell-repellent characteristics, were used for local lift-off of the mammalian cells. In order to implement the cell adhesion, colloidal microgels were used to create micropatterned thermoresponsive polymer coatings. The microgels of 200 μm were fabricated through automated nanodispensing or microcontact printing (μCP). The microfluidic device was made of a PMMA plate, a double sided sticky pressure-sensitive adhesive (PSA) foil, and a glass with microgel patterns at the bottom. The microchannel was created on the PSA foil of 86 μm height, 500 μm width, and 1 cm length. L929 mouse fibroblasts were cultivated and the cell detachment was accomplished via both of the coating strategies depending on the temperature shift. The lower limit of the surface coverage was determined by changing the average microgel distance and the cell detachment efficiency.104
FIG. 3.
(a) Microwell designed chip. (b) Micropatterned chip. Reproduced with permission from Sakai et al., J. Biosci. Bioeng. 111, 85 (2011). Copyright 2011 Elsevier.
In another study on cell attachment and alignment using physically microstripped-nanoengineered polystyrene surfaces (PMS-NPS), the PMS-NPS was produced via nano-injection molding, UV-photolithography, and electroforming methods. Controlled MG-63 cell attachment and alignment were conducted by biophysical cue (without biochemical cue) and PMS-NPSs were found to be better than flat PS surfaces.105 In order to study the cell-nanoengineered surface (NES) interactions, PS nano Petri dishes were fabricated. During mass fabrication via nano-injection, the Taguchi method was used to evaluate the fabrication parameters as well as to optimize and conduct efficient and reliable cell culture studies. MG-63 cell attachment and proliferation was examined in nano Petri dishes including nanopore arrays with and without the oxygen plasma treatment. The nanopore array surface gave better performance than the flat surfaces for cell attachment and proliferation.106
The advantages of the microfluidic platforms employed for tissue engineering studies can be summarized as the effective management of multi-organ microphysiological systems, biointegration of soft tissues, tissue integration of implants, microengineering of vascular structures, monitoring parameter effects on the vascularization of bone-mimicking tissues, isolation of specific cellular populations, and imitating the drug distribution and metabolism processes in the body. This part is summarized in Table II.
TABLE II.
Summary table of CULTIVATION: Organism-on-a-Chip part.
| Format | Microorganism | Material used | Fabrication techniques | Dimensions | Comment | Reference |
|---|---|---|---|---|---|---|
| Multiplexed microbioreactor | E. coli | PMMAPDMS | Milling Hermetical sealing | 150 μl reactor chamber | OD, DO and pH measuring 4 parallel microbial fermentation | 41 |
| Microbioreactor | E. coli | PMMAPDMS PAA | CNC milling Thermal bonding | 150 μl reactor chamber | OD, DO, pH measuring Reinforced cultivation time and prevented wall growth of the cells | 42 |
| 3D microfluidic chip | HeLa E. coli | PMMAPC | Laser cutter Wax bonding | 2 mm width and 500 μm height of chamber | PCR, HeLa cell EP and E. coli culturing Effect of ciprofloxacin concentration on E. coli | 43 |
| Microfluidic chip | Pseudomonas aeruginosa | PMMAPDMS | Laser ablation Adhesive tape for sealing | 150 μm thick cell culture chamber | Chip allows the gas transition Active oxygen depletion examination | 44 |
| Microfluidic chip | E. coli Shigella flexneri Shigella boydii Shigella sonnei Uropathogenic E. coli | PMMA | CO2 laser cutter glue bonding | 5 mm in diameter and 800 μm in the height of reservoir | Drug resistance testing of several bacterial strains Comparison of the microfluidic system and 96-well microtiter plates | 45 |
| PMMA made microchannels | E. coli K12 | PMMA | Hot embossing UVO-assisted thermal bonding | 42 μm height, 1 mm width and 20 mm length of microchannels | Positively charged nanofibers were better than negatively charged ones | 46 |
| Microfluidic chip | Cordyceps militaris | PMMA | CO2 laser machine Screw binding | NA | Monodispersed agar beads for cultivation | 47 |
| Milliliter-scale bioreactor | Lactobacillus paracasei S. cerevisiae | PMMAPDMS | CNC machining PDMS for sealing glue for bonding | 0.5–2 ml bioreactor | OD, DO, pH measuring. Aeration and mixing capability | 48 |
| Microfluidic chip | S. cerevisiae | PC | Ultrasonic hot embossing Ultrasonic welding | Microchannels with a depth between 50 μm and 1 mm and a width between 100 μm and 3 mm | eGFP tagged protein monitoring with a supply of the inducer galactose 22 h operation in the device | 49 |
| 3D multilayer microfluidic system | Zebrafish (Danio rerio) | PMMA | Infrared laser machine Thermal bonding | Conical traps of 2 mm in diameter at the top and 1.6 mm in diameter at the bottom planes | One embryo in one trap 100% trapping of the cells 72 h experiments | 50, 51 |
| Microfluidic chip | Zebrafish (Danio rerio) | PMMA | High-speed infrared CO2 laser cutting Thermal bonding | 0.75 mm in diameter and 0.5 mm in height of microwells | ESEM imaging Damaged tissues under a low vacuum environment | 52 |
| 3D microfluidic chip | Zebrafish (Danio rerio) | PMMA | Infrared laser machine Thermal bonding | 1.7 mm × 1.5 mm × 55 mm of main channel | Rapid and automated manipulation of zebrafish cells for drug discovery | 53 |
| Microfluidic chip | Zebrafish (Danio rerio) | PMMA | CO2 laser cutting | 36 circular microwells (0.75 mm in diameter and 0.5 mm in depth | Microwells for keeping zebrafish yolk revealing morphological features of zebrafish via ESEM imaging | 54 |
| 3D multilayer microfluidic chip | Zebrafish (Danio rerio) | PMMA | Infrared laser micromachining Thermal bonding | Suction manifold of 0.5 mm width and 1.8 mm height | Analyzing, sorting and dispensing of cells | 55 |
| 3D multilayer microfluidic chip | Zebrafish (Danio rerio) | PMMA | Laser micromachining Thermal bonding | 20 miniaturized traps and 1.7 mm × 1.5 mm × 55 mm of main channel | Making automatic immobilization, culture and treatment of cells | 56 |
| 3D multilayer microfluidic chip | Zebrafish (Danio rerio) | PMMA | Infrared laser micromachining Thermal bonding | 96-well microtiter plate 21 miniaturized traps of 1.5 mm × 1 mm | FET assay performing Rapid loading separating immobilizing of cells and continuous perfusion Anti-angiogenesis drug tests | 57 |
| 3D multilayer microfluidic chip | Zebrafish | PMMA | Infrared laser micromachining | 18 embryo traps of 1.5 mm × 1 mm | FRIM technology for kinetic quantification of the aqueous oxygen gradients | 58 |
| Small-animal Nutritional Access Control (SNAC) chip | Drosophila melanogaster | PMMA | CNC machining Thermal fusion bonding | Behavior chamber of 20 mm × 15 mm × 2 mm Feeding alcove of 400 μm width | Studying the actions of flies | 59 |
| Microfluidic chip | HepG2 | PMMAPS PC COC | Hot embossing Thermal bonding | 3.5 μl culture chambers | Investigation of biocompatible materials | 60 |
| Microfluidic coculture chip | Human U937 and MG-63 cell lines | PMMA | CO2 laser scriber Thermal bonding | Trenches of 200 μm in height and width | Studying coculture behavior | 61 |
| Microbioreactor | HT 1080 | PMMA | Milling Double sided adhesive tape | Culture channel of 4 cm × 300 μm × 300 μm | DO level and shear-stress acting on the cells investigation | 62 |
| Microfluidic chip | HEK-293T | PDMS PMMA | PDMS: silicon wafer mold + high frequency generator PMMA: CO2 laser etching | Microchannels of 40 μm H × 0.4 mm W Microchannels of 40 μm H × 100 μm W | Revealing optimum conditions for growth under several experimental conditions | 63 |
| Hard-soft hybrid material microfluidic chip | HepG2 C2C12 | PETG COC PS | Hot embossing Soft lithography Argon and oxygen plasma | X region of 200 μm height | Combination of hard and soft materials Cell culturing ∼100% cell survival rate | 64 |
| Microfluidic chip | HEK293 U-2 OS PC12 | PTFE | CNC machining Stainless steel clamping | Fluidic input channels of 500 μm2, opening out of 1000 μm2 | Encapsulation of living, therapeutically active cells within monodisperse alginate microspheres | 30 |
| Microfluidic chip | IMR-90 fetal lung fibroblast Human neutrophils | PS PDMS | Thermal scribing | Microchannel of 500 μm width, 250 μm height and 25 mm length | Understanding the mechanism of neutrophil culture systems | 65 |
| Microfluidic chip | Human colorectal adenocarcinoma cells | PDMS PMMA | CNC machining Replica molding Oxygen plasma | 36 microbioreactors of 1.5 mm in diameter and 1 mm in height | High-throughput 3D cell culture and chemosensitivity assay | 68 |
| Microfluidic chip | Human oral cancer cell line | PDMS PMMA | CNC machining Replica molding Oxygen plasma | 30 microbioreactors of 3 mm in diameter and 2.5 in height | High-throughput 3D cell culture system Chemosensitivity assay DNA content detection Viability experiments | 69 |
| Micro-scaffold array chip and Drug laden chip | NIH3T3 fibroblasts Human fibrosarcoma cells Human hepatocellular carcinoma cells Human non-small lung cancer cells | PMMA | CO2 laser system | 96 microwell array of 2 mm in diameter | 3D cell culture Drug administration Quantitative in situ assays | 70 |
| Microbioreactor | Human lung adenocarcinoma epithelial cell line (A549) | PMMA | Micromilling Screw binding | Rectangle obstacles of 0.35 mm × 1.2 mm in microchannel of 0.8 mm × 0.35 mm | Cell viability and cell toxicity tests Drug testing experiments | 71 |
| Microfluidic chip | K562 human erythroleukemia cells | PMMA | machining Thermal bonding | Microchannel of 254 μm W × 150 μm H | NP study to examine the Bcl-2 down-regulation at the mRNA and protein levels | 73 |
| Semi-continuous flow electroporation (SFE) chip | K562 cells | PMMA | Hot embossing Lamination | Channels of 2 mm width | Increasing the transmission of exogenous oligonucleotides in vitro | 74 |
| Single field chip (SFC) and multi-field electrostatic chip (MFC) | CL1-5 and CL1-0 lung cancer cell lines | PMMA | CO2 laser ablation Double sided adhesive tape | SFC: microchannel of 3000 μm × 70 μm × 15 mm MFC: microchannel of 24 mm in length with 5000, 1667, 1000 μm in width segments | Electrotaxis study Distinct electric fields applications for cellular response | 76 |
| Microfluidic chip | MDA-MB-231 human breast cells | PMMA | NA | Barrier thickness of 60 μm ± 40 μm | Multilayer contactless dielectrophoresis system | 77 |
| Sensor chip | HEK293 cells | PMMA | Hot embossing FIB milling | Micropore of 1.5–2 μm in diameter | Cell trapping | 78 |
| Microfluidic chip | HepG2 cells | PMMA | CO2 laser ablation Double-sided adhesive tape Hot-press bonding | Rectangular cell culture chamber of 8 mm × 6 mm | Monitoring cell growth Cytotoxicity experiments | 79 |
| Parallel microfluidic cytometer (PMC) | Chinese hamster ovary cells Jurkat cells Jurkat T-cells | PMMA | Deep reactive-ion etching | Microchannel of 40 μm × 150 μm | Cell screening assays | 80 |
| Micro-optical tweezers (μOT) | RBC Tumor cells | Glass PMMA | Micromilling Solvent assisted bonding | Channels of 60 μm × 1 mm × 2 cm | Mechanical and chemical spectroscopic analysis | 81 |
| Nanotopography | Primary human osteoprogenitor cells | PMMA | Colloidal lithography Polymer demixing Embossing | 10 nm size topographies | Cell morphology, cell cytoskeleton, adhesion formation, cell growth and differentiation studies | 85 |
| Spheroid microarray chip (SM chip) | Hepatocytes | PMMA | Micromilling Press bonding | Cylindrical cavities of 300 μm | Production of spheroids by hepatocytes | 86 |
| Microfluidic chip | Schwann cells Fibroblast cells | PMMA | CO2 laser machining Screw bonding | Microchannel width of 200 μm, depth of the chip is 1.5 mm | Can be used as scaffold for cell cultures | 87 |
| Surface modification | Human corneal limbal epithelial cells (HCLEs) Human keratocytes | PMMA | Surface modifications with chemicals | NA | Adhesion of collagen gel studies | 88 |
| Microengineering vascular structures | Human umbilical vein cell (HUVEC) | PMMA | Gelatin methacrylate hydrogel photopatterning | NA | More complex, vascularized tissue constructs for regenerative medicine and tissue engineering applications with the combination of SAM-based cell transfer and hydrogel photocrosslinking | 89 |
| Helical and straight microchannels | NIH 3T3 | PMMA | Agarose solution for hydrogel fabrication Laser etcher | Chamber: 24 mm × 12 mm × 5 mm Wire diameter: 300 μm | Helical and straight microchannels comparison Better perfusion ability and oxygen and nutrient delivery to cells in helical microchannels | 90 |
| Surface modification | MC3T3-E1 | PS | Anodization Hot embossing Nickel electroforming | Nanopore structure with a diameter of 200 nm, depth of 500 nm | Studying the topographical effects of surfaces on MC3T3-E1 cells | 91 |
| Surface modification | MC3T3-E1 | PS | Injection molding | Several patterns with 2,3,4 μm diameter holes | Bone replacement operations Microtopography, pillar diameter, aspect ratio and spacing evaluations Micro-pillared surfaces are better than flat surfaces | 92 |
| 3D microfluidic structures | Liver tissue cultures Mesothelial cells | PMMA | Engraving machine UV photo polymerization | PEGDA structures of 60 μm height | PEGDA microstructures with mesothelial cells for contribution of adhesive surface and tissue repair | 93 |
| Microfluidic chip | Primary human alveolar bone osteoblast (PHABO) | PMMA | NA | 300 μm cubic cavities 10 mm × 10 mm device | PHABO morphogenesis investigation in both microchip-based 3D-static conditions and 3D-fluid flow-mediated biomechanical stimulation in perfusion bioreactors | 94 |
| Mechanical microconnector system (mMS) | Spinal cord stumps | PMMA | Vacuum application | Thickness of 350 μm and outer diameters of 1.7 mm and 2.7 mm | Regulation of retracted spinal cord stumps Axonal regrowth was achieved after 2, 5 and 19 weeks with the mMS. | 95 |
| Microfluidic chip | C3H10T1/2 stem cells | PMMA | CO2 laser etching Thermal bonding | 170 μm width, 200 μm height and 1.5 cm length | Application of click chemistry Bio-orthogonal chemical group generation in a rapid, straightforward and flexible way | 96 |
| Microvascular network | Endothelial cell Mesenchymal stem cells | PMMA | Laser cut Silicone glue bonding | 2 × 2 × 2 mm3 and 2 × 2 × 5 mm3 masks | Parameter effects on the vascularization of bone-mimicking tissues Creation a link in between the macroscale and microscale tissue engineering studies | 97 |
| Microfluidic chip | Human umbilical vein endothelial cells (HUVEC) Blood neutrophil | PS COP | Hot embossing Thermal bonding | 3 different aspect ratios: 10, 20 and 50 Height: 10 μm | HUVEC culturing in one application and blood neutrophil culturing under chemoattactant in another PS can be used for long term studies | 98 |
| Microfluidic chip | Human microvascular endothelial cells (hMVECs) | COC | Hot embossing Oxygen plasma Roller-lamination | 50 mm × 4 mm × 100 μm (L×W×H) | No adverse effect of COC material on cells | 99 |
| MOTiF biochip | Endothelial cells | COC | Injection molding Surface oxidation | 140 μm thick | Nutrient medium supply, catabolic cell metabolites removal and shear stress applications | 100 |
| Body-on-a-chip | A549 Caco2 HepG2 C3A Meg01 HK2 | PC Silicone PMMA | Milling Screwing | 2.5 μl volume | Imitation the drug distribution and metabolism processes in the body Building, managing and cultivating of multi-organ microphysiological system | 101 |
| Microwell and patterned chip | Embryoid bodies from mouse embryonic stem cells | PMMA | Micromilling Microcontact printing | 270 microwells of 600 μm in diameter and depth, 270 gelatin spots of 200 μm in diameter | The proliferation and differentiation of EB variation from design to design | 102 |
| Spheroid transfer chip (ST chip) | Mouse ES cells, 3T3 cells, HepG2 cells and primary hepatocytes | PMMA PDMS | Micromilling | 270 microwells of 600 μm in diameter, depth and pitch | Spheroid production Cell proliferation and cell viability in the spheroid and spheroid size monitoring | 103 |
| Microfluidic chip | L929 mouse fibroblast | PMMA | Automated nanodispensing or microcontact printing Double side sticky pressure-sensitive adhesive | Microgel coatings of 200 μm | Cultivation of L929 mouse fibroblast Studying cell detachment | 104 |
| Surface modification | MG-63 | PS | Nano-injection molding UV photolithography electroforming | The width and spacing of microgroove patterns: 50 μm | Cell attachment and alignment using PMS-NPS PMS-NPS are better than flat PS surfaces | 105 |
| Surface modification | MG-63 | PS | Nano-injection | Nanopore array: 20 mm × 20 mm | Cell attachment and proliferation study on NES Nanopore surface is better than flat surface | 106 |
III. SEPARATION/ISOLATION
In biological and biomedical research as well as in clinical therapy, cell separation or cell sorting methods are mostly used to perform analyses. For example; in some cases, circulating tumor cells (CTCs), red blood cells (RBCs), and white blood cells (WBCs) are held together in blood samples and CTCs should be separated to work with. Cell separation with a low risk of contamination is very important to further study a targeted cell or an individual cell in an isolated area. Heterogeneous cell populations can only give lumped averaged data about the populations, but one can obtain an important result from an isolated subpopulation via cell sorting. There are many technologies for cell isolation such as flow cytometry, laser capture microdissection, limiting dilution, manual cell picking or microfluidic devices. In the following parts, studies on cell isolation in microfluidic devices will be discussed in detail.107,108
A. DNA-RNA extraction/isolation
In the prokaryotic cell domain, bacteria cells occupy a large area, and E. coli cells are the most widely used pathogens among all, in many experiments. In 2004, Chung et al. employed immobilized beads in their microfluidic device to extract DNA from lysed cells. The solution in the device flowed back and forth, and the DNA was successfully isolated in the device. The experiments conducted with serum had higher efficiency.109 In 2007, an optimal fuzzy sliding-mode control (OFSMC) bio-microfluidic device was fabricated with 8051 microprocessors. In this system, the collision of molecules was increased via a back and forth process to enhance the biochemical reaction efficiency, and DNA extraction experiment was performed. When the beads were immobilized, the extraction efficiency is higher than that of the free beads.110 Geissler and his group employed Bacillus atrophaeus subsp. globigii spores for the nucleic acid separation. A PMMA made microfluidic device was used to manage the bead-based mechanical cell lysis of the bacteria. The chip was composed of the mechanical slides of PMMA and a metal disk with microbeads in the lysis chamber. Cell destruction was handled via the collisions and frictional forces were created by the metal disk magnetic actuation. Gathered DNA molecules were counted via the PCR method. The composition of the lysis matrix such as the size or amount of the microbeads and the instrumental parameters, like duration and frequency of the agitation, affected the yield of the experiment.111 The isolation and amplification of eukaryotic mRNA from Cryptosporidium parvum (C. parvum) cells were performed in another type of microfluidic device. The surface of the microfluidic channels of the device was carboxylated via the UV/ozone and coated with polyamidoamine (PAMAM) dendrimers to enhance the binding of the thymidine oligonucleotide (oligo(dT)25) for the nucleic acid sequence-based amplification (NASBA) reaction. The device includes 6 microchannels to perform mRNA isolation and amplification. mRNA was successfully separated from C. parvum oocysts.112
Viruses are neither prokaryotic nor eukaryotic pathogens, and duplicate their DNA or RNA only inside the living cells of other organisms. They cause infectious diseases and can affect all type of living forms.113 In 2008, viral RNA extraction from mammalian cells infected with influenza A (H1N1) virus experiments were conducted in a cyclic polyolefin made microfluidic device. A solid-phase extraction system was created for isolation of RNA and separation was done via reversible binding of nucleic acids to the silica particles in the monolith.114 In order to detect the norovirus (NoV) in oysters, cell concentration, lysis (RNA extraction), nucleic acid amplification, and detection operations were assembled in one microfluidic device. The virus concentration and lysis events were conducted by charge switchable microbeads in a shape changeable microchamber. The murine NoVs were adsorbed on the microbeads and their RNA was extracted via bead beating. Then, the extracted RNA was sent to the amplification chamber and finally murine NoVs in the oyster were detected. The microfluidic device was produced from PMMA and PDMS.115
B. Separation/isolation of microorganisms
Cell separation has been conducted using several organisms and systems. The trapping and qualification of neutral particles such as polystyrene beads (representing non-living organisms) and living cells (E. coli as gram-negative bacteria and Enterococcus faecalis as gram-positive bacteria), were considered in a microfluidic study. Dielectrophoretic trapping ability of these different particles was tested in this platform. PMMA and glass wafers were used to create the microfluidic device. Bacteria and polystyrene beads were separated easily when the optimum frequency was set. The voltage amplitude affected the trapping event.116 The dielectric behavior of different bacterial species under different conditions can be used for the separation and detection of pathogens. Not only E. coli but also other types of bacterial cells have been studied in microfluidic devices. In 2016, multiplex sorting and detection of Salmonella typhimurium (S. typhimurium) and E. coli 0157 from several cultures were done in a magnetophoresis-based microfluidic chip. Dynabeads anti-salmonella and Hyglos-Streptavidin magnetic beads of various sizes with conforming pathogen-specific biotinylated recombinant phages were used to arrest the cells. The PMMA device comprised of a separation chamber and a buffer inlet channel divided into 3 similar channels. The depth of the device was 100 μm and a biaxially oriented polypropylene (BOPP) tape was used to seal the device. 72% S. typhimurium-bound Dynabeads and 67% E. coli 0157-bound Hyglos beads were recovered from a 10 μl mixture in 1.2 min. It was proved that, more than one pathogen containing culture can be sorted and isolated.117 An integrated microfluidic electrostatic sampler (IMES) was developed including a unipolar charging chamber, a half cylinder precipitation electrode and a collection chip, which had a half-open microchannel for air transport. During the experiments, airborne molecules were sent through the charging chamber; then with the help of the electrostatic field, precipitation occurred in the half-open microchannel. The collection liquid was fed into the microchannel, which then transported the molecules to the collection reservoir. In order to produce the half-open microchannel, the hydrophobic mesh was bonded via an adhesive tape. Bacillus subtilis cells were used and the collection efficiency was around 16%. The particle loss on the hydrophobic mesh affected the collection efficiency of the system.118
Fungi, especially S. cerevisiae, are the model organisms of eukaryotic cells. Cell guiding experiments were done in the microfluidic device by applying dielectrophoresis (DEP) to the S. cerevisiae cells. It was monitored that, as the flow rate increased, guiding efficiency decreased in the device. Moreover, several medium conductivities and frequencies were tested, and the DEP behavior of cells was monitored. The microfluidic device included glass, polyethylene (PET), and ITO covered PMMA layers (being used as the counter electrode) and a microchannel. By using this system, pre-treatment steps of PCR amplification of DNA can be conducted via dielectrophoresis guiding of the cells to gather and isolate them from a complex sample.119 In another study, a capillary-driven, self-propelled PCR was developed for the quantitative real-time detection of pathogenic microorganisms in less than 18 minutes. The new autonomous disposable plastic, COP, was used for the fabrication of the device, which consisted of two microchannels; one for PCR and the other for controlling the capillary flow of the solution (a driving microchannel). The walls of the microchannels were coated with a non-ionic surfactant to create a hydrophilic surface and the system also had high and low temperature aluminum heater blocks. The chip was integrated with a fluorescence detection system to analyze the amplified product (human β-actin, E.coli DNA and E.coli O157) on the chip. This chip and detection system were successfully applied for highly sensitive reproducible quantitative DNA analysis.120 In a study conducted with C. parvum cells (parasite), the separation of cells from polystyrene beads (analogous to pathogens) was performed in spiral microchannels. An external force or an additional buffer was not used in the system, and the particle isolation was done by the application of high flow rates. The channel length, flow rate, particle size, and particle concentration parameters were investigated to optimize the separation. The device was made of PMMA, and included a focusing channel with 6 loops. Inertial focusing was used for the first time and polystyrene beads of 4 to 7.5 μm, similar in size to pathogens, were successfully examined at channel Reynolds numbers of about 100.121
C. Separation of blood cells
An adult human has an average of 5 l of blood in the body. Delivering of oxygen and other nutrients to living cells and removing waste products, fighting diseases, and plug forming in a damaged blood vessel are the roles of the red blood cells, white blood cells, and platelets in the blood, respectively.122 In some analyses, it is necessary to separate these cell types from blood. Quantitative information can be obtained by using buffers with several conductivities and with the application of a microfluidic system. Particle retention was accomplished by dielectrophoresis (DEP) employing polystyrene beads of 500 nm, 2 and 6 μm in diameter and erythrocytes of around 6 μm in diameter. Cross-over frequency, i.e., transition from the negative to positive DEP frequency, was determined, and the separation of beads of several sizes were accomplished in the microfluidic device, which included platinum electrodes with interdigitated or intercastellated structures deposited on glass or nitride. Significant quantitative data about the retention of beads and blood cells were gathered and active DEP forces were roughly calculated in this study.123 The human arteriolar system, including the circular channel cross-section, network asymmetry, bifurcation, and side channels, was modeled in the microfluidic device, which had a channel diameter ranging from 200 μm to 100 μm. The μPIV (micro-Particle Image Velocimetry) method was used to reveal the flow field within the device by using fluorescently labeled tracers. Red blood cells (RBCs) were separated from the blood sample and used in the device for the analysis of the cell-deplation layer. Finally, the quantification of haematocrit distribution was made through the Neubauer haemocytometer with the RBS collected from the outlet of the device.124 In another study, white blood and red blood cells were separated from each other by using the density difference feature of the cells in the microfluidic device. The device separation technique was based on magnetic-levitation of the cells in a magnetic field. In order to monitor the cells, a smartphone was used. The microcapillary channel, N52 grade neodymium magnets, and side mirrors were located in the magnetic levitation chip. FITC and DAPI stained blood cells were used in the device and the images taken via the smartphone were analyzed by the ImageJ program. In this system, the white and red blood cells were isolated and quantified easily by using imaging with the magnetic levitation (i-LEV) platform.125 Immunomagnetic-based cell separation was managed by designing a high throughput microfluidic device. The CD45-conjugated magnetic particles were used to tag the white blood cells (WBCs). WBCs were captured with the help of the magnetic field, which was created via magnets in the chip. The immunomagnetic-based system depends on the magnetic and fluid dynamic forces under laminar flow in the device. Before the experiments, a finite element model (FEA) via COMSOL Multiphysics was created to optimize the cell separation mechanism. When the separation was completed, the captured WBCs were counted, and 99.9% WBCs were isolated from the blood.126 The cell separation, cell lysis, and DNA purification experiments by using rat blood samples were carried out in a microfluidic device, which included a microfilter, a micromixer, a micropillar array, a microweir, a microchannel, a microchamber and a porous matrix. Continuous flow operation was conducted in the microfluidic device and crossflow filtration was used to isolate the blood cells. Cell lysis was done via guanidine buffer and genomic DNA was kept by the porous matrix. Fluent software was employed for flow characterization. Two main parts located in the microfluidic platform were the microfilter for cell isolation and the microchannel for cell lysis and DNA purification. 37 ng DNA was isolated and separated from the 1 μl blood, and it only took 50 min to complete the experiment.127 A cost effective calorimetric COP made diagnostic device was fabricated for ABO and Rh blood typing. The microfluidic device consisted of a screw pump, serpentine reaction channels, chaotic micromixers, and low aspect ratio filters (Fig. 4). The maximum loading volume was determined as 6, and 1 μl of blood sample collected via the finger prick method was loaded into the loading reservoir and 5 μl of phosphate buffered saline (PBS) buffer was introduced into all channels. The pressure difference occurring between the interactive (matched) and non-interactive (mismatched) blood types caused the PBS buffer solution to separate the non-aggregated RBCs out of the outlet reservoir in just 1 min. When the blood sample contained the related antigen, then an agglutination reaction occurred and agglutinated RBCs blocked the filters near the outlet and the reaction channel turned red. The device was also used for testing thalassemia (a type of anemia) blood samples with smaller RBCs.128
FIG. 4.
Chip test results for (a) A Rh+, (b) B Rh+, (c) AB Rh+, and (d) and (e) O Rh+ blood types, respectively. (e) and (f) Thalassemia samples with smaller RBCs and lower hematocrit. (d) Healthy blood sample and (f) (i) Normal blood sample (age 23; male) (ii) thalassemia blood sample (age 37; male). Reproduced with permission from Chen et al., Lab Chip 15, 4533 (2015). Copyright 2015 Royal Society of Chemistry.
D. Separation/isolation of tumor cells
In cancer, unhealthy cells divide continuously and they can spread throughout the body. Therefore, it is very important to identify and perform several analyses on cancer cells. Circulating tumor cells (CTCs) in whole blood were isolated with the help of monoclonal antibodies (mABs) in a high throughput microsampling unit (HTMSU). CTCs were not labeled, but they were detected via conductivity sensor after capturing. The microfluidic device included 51 high-aspect ratio linear or sinusoidally constructed microchannels. The effectiveness of capturing CTCs from the whole blood was found to be >97%. Trypsin was used to release the captured CTCs from mABs, and the enumeration was done by detection electrodes at 100% detection efficiency.129 In 2011, high recovery rate was achieved in capturing the CTCs. Several simulations were performed parametrically to enable the capturing of rare target cells in the microfluidic device. A high flow rate device (HFRD) was produced according to the results of the simulations. The simulated CTCs (MCF-7 cells) were gathered and spiked in 40% hematocrit solutions of human red blood cells. On average, an 80% recovery rate was attained in capturing the CTCs in the HFRD.130 In another study, separation and detection of the rare cells existing in the peripheral blood mononuclear cells were investigated in a disk shaped microfluidic device. A magnetic field was created to increase the trapping efficiency. The inlet reservoir, which can hold 300 μl cell solution, was used for introducing the cell culture and trapping. The disk had 12 cm diameter and 4 compartments placed on it were used to perform 4 experiments. Here, MCF7 cells attached to magnetic beads were employed to represent CTCs as the target and Jurkat clone E6–1 was employed to represent the leukocytes. Positive selection of rare cells from the abundant ones was obtained via the magnetic field. Cell separation, autoMACS (magnetic activated cell sorting) comparison, and cell viability analyses were managed in this microfluidic system. 80% of rare cells (MCF7) were detected, and this result was 20% higher than those obtained by autoMACS. However, the viability of the cells was around 90% ± 20%, which may have resulted from the damage of the trapped cells.131 Another high-throughput separation of CTCs from the blood was conducted in a microfluidic system integrating 3 modules. The first system was made of COC for CTC selection. The CTC selection bed included anti-EpCAM antibodies for blood processing. The second system was an impedance module and made of PMMA. This module included two perpendicular microchannnels. The first microchannel was used to carry the cells through this module, and the second one was used to make single cell impedance measurements (CTC counting). The final module, also made of PMMA, was used for staining and imaging. There were two sets of microchannels; 8 μm base and either 6 μm or 50 μm height. Blood samples with local resectable and metastatic pancreatic ductal adenocarcinoma (PDAC) were analysed via this modular device for phenotypic identification. EpCAM positive CTCs from PDAC patients were successfully detected, and with this system CTC assay time was reduced to 1.5 h instead of 8 h of conventional methods.132 In 2014, Jackson et al. fabricated surface treated (UV-induced) microfluidic devices having different sized microfluidic channels to isolate and analyse CTCs. For colorimetric assay and imaging, the fluorescent dye-labeled oligonucleotides were immobilized on the surface of the channels. PMMA, COC and PC made channels were used but PC was eliminated due to its high auto-fluorescence characteristics, which interfered with the fluorescence imaging of CTCs. In order to select the CTCs from the blood sample, anti-EpCAM was coated on the surface of the devices. There were 50-curvilinear channels in the device to create the cell selection bed, which had a 596 mm2 surface area. COC made microfluidic devices achieved higher clinical CTC yield and better purity of the selected fractions than the PMMA made devices.133 A two-stage microfluidic device was developed for sorting and isolation of the CTCs. The first stage of the chip enabled the separation of the white blood cells (WBC) via microfluidic magnetic activated cell sorting (μ-MACS), and the second stage accomplished the CTC isolation via a geometrically activated surface interaction (GASI) chip. In order to create the device, PMMA and polyester films were used (Fig. 5). The cancer cell isolation ability of the system ranged from 10.19% to 22.91%. The system was capable of identifying the heterogeneous CTCs according to their features.134 The tumor cell isolation was alternatively achieved via a negative selection method, which did not require any biomarker expression. CTC separation from the blood sample proceeded in a PMMA made microfluidic chip without any tumor specific antigen. In the microfluidic system, first white blood cells (WBCs) were separated via magnetophoresis, then red blood cells (RBCs) were isolated by a micro-slit membrane, and only tumor cells remained. The CTC separation chip contained two main modules, the first one included microfluidic chamber with a magnetic array around it for WBC separation, and the second one included a circular parylene-C membrane for RBC isolation. In the experiments, WBCs and RBCs were successfully separated from the whole blood and more than 80% CTC recovery was accomplished.135
FIG. 5.
The schematic illustration of the two-stage microfluidic system. Reproduced with permission from Hyun et al., Biosens. Bioelectron. 67, 86 (2015). Copyright 2015 Elsevier.
Other than CTCs, different tumor cell lines like human histiocytic leukemia cells were investigated via the application of dielectrophoresis (DEP) combined with the environmental scanning electron microscopy (ESEM). The designed microfluidic device had curved microelectrodes, which were produced via chrome/gold deposition on the glass piece. The PMMA polymer was used to create the separable microculture chamber of 2 mm height on the glass substrate along with the electrode arrays. 3 D surface topographical information can be obtained via ESEM images instead of flat or reconstructed images produced by confocal imaging. Since a very low number of cells were used in this system, rare cell studies can be conducted via this system due to its good performance.136 The amount of ovarian cancer cells (SKOV3) in the blood was determined by an electrochemical Lab-on-a-Disc (eLoaD) system. The device sensitively allowed blood decomposition and cancer cell separation from plasma by applying label-free electrochemical impedance. In order to allow detection, gold electrodes were coated with anti-EpCAM. The device consisted of 3 PMMA and 2 adhesive layers. The capture efficiency of the device was 87% at the AC amplitude of 50 mV. Five different assays can be done in parallel within the device. By using gold electrodes, SKOV3 cells were detected and separated successfully.137 A centrifugal-force-based size-selective CTC isolation microfluidic device was produced to separate the CTCs from blood and to count them. The device includes PC membranes of 8 μm pore sizes. The MCF-7 breast cancer cell line was used and 61% capture efficiency was obtained depending on several dilution factors and flow conditions.138
E. Separation/isolation of other cells
Male reproductive cell (sperm) isolation was conducted in a PMMA made microfluidic device. This device was capable of separating the healthy, motile, and morphologically normal sperms without the centrifugation of unprocessed semen. A microfluidic sperm sorter (MSS) had macroreservoirs with micropores which allowed most motile and functional sperms. The bottom chamber was bonded to a glass slide with a double sided adhesive (DSA), and PC membrane filters were placed on the bottom chamber. The top chamber was attached to the bottom chamber. By using this chip, lesser ROS and DNA fragmentation were observed than that of the conventional swim-up method.139 A lensless charge-coupled device (CCD) that facilitated the monitoring of a large field of view (FOV) and made the recording automatically, was integrated into a microfluidic platform to sort and track the sperm inside the channel. The sperm cell solution was loaded through the inlet via pipetting; the images were taken with CCD in both the horizontal and vertical configuration. Tracing the shadow paths of the individual sperm was used to reveal the sperm motilities. This technique will be useful while working with the oligozoospermic and oligospermaesthenic samples in which the most motile sperms need to be isolated.140 A microfluidic sperm-sorting (MFSS) device was fabricated using the COP polymer instead of the commonly used PDMS or quartz. Two separate inlets were designed for channels (medium inlet and semen inlet) to separate the motile and nonmotile sperms. Two microfluidic channels of different dimensions (chip A: 0.3 × 0.5 mm and chip B: 0.1 × 0.6 mm) were fabricated and the fluids moving through the outlets should move parallel to each other. The sperm separation efficiency changed depending on the position (bottom-center-top) of the sperm and the height of the channel. In chip A, the linear velocity distribution was higher than that in chip B and the highest amount of motile spermatozoa was monitored at the bottom of the channel. In order to increase the recovery of spermatozoa at higher velocity, the width of the channels should be increased.141
In 2002, isotachophoresis (ITP) and zone electrophoresis (ZE) were coupled to increase the concentration limit of detection in PMMA made multi-channel chips. THP-1 cells (human monocytşc cells) were used in the experiments and the evaluation of sample injections from buffers with various ionic strengths was done. Efficient stacking and separations were achieved in both low and high conductivity buffers. Moreover, it was found that the stacking process by ITP was consistent with cell based assays and assays for surface proteins with live intact cells.142
By using microfluidic devices, separation/isolation of living and non-living cells can be conducted in a more controlled manner with increased efficiency. Several processes such as lysis, amplification and detection can be carried out in a single device without any need for conventional methods. Beads are generally placed in the devices to facilitate the separation. Complex samples (including more than one species) can be sorted and isolated without damaging them. A higher clinical yield and better purity can be attained and sequential operations such as isolation of the cell and further analysis can be done successfully. This part is tabulated and given as the supplementary material.
IV. DETECTION AND ANALYSIS
Cell division, differentiation, maturation, and death are the parts of the cell cycle of living organisms. Detection and characterization of a cell are among the most popular areas in scientific research for diagnosis and therapy. There are several methods to detect the cells, and Raman spectroscopy, biosensing, and electro-based detection are some of the examples.143,144
A. Electro-based detection
Detection of bacterial cells via electro-based detection systems is widely used in microfluidic devices. In 2002, Yang et al. operated a stacked microfluidic chip capable of making electric-driven immunoassay and DNA hybridization. The device consisted of a patterned polyimide layer with a flip-chip bonded CMOS chip, a pressure sensitive acrylic adhesive (PSA) layer, a PMMA layer, and a glass cover layer. The detection of E. coli bacteria and Alexa-labeled protein toxin staphylococcal enterotoxin B (SEB) was conducted via electric-field-driven immunoassays. The Shiga-like toxin gene (SLT1) from E. coli cells was also identified by using the strand displacement amplification (SDA) module.145 In another microfluidic device, E. coli and S. cerevisiae cells were used to study the electrokinetic transport. The devices were made of UV-modified PMMA and PC. The apparent mobility (μapp) of the cells showed around 10% differences from chip to chip. Since yeast cells have z smaller electrophoretic mobility than the electroosmotic flow (EOF), they moved towards the cathode of the devices. In contrast, E. coli cells went towards the anode in 0.5 and 1 mM PBS due to their higher mobility than EOF. In 20 mM PBS, E. coli cells migrated to the cathode due to higher ionic strength.146 Jezierski and his group fabricated a microfluidic free-flow electrophoresis (μFFE) chip to perform separation and biosensing experiments with adherent HEK cells. Fluorescent calcium indicators were loaded with HEK 293 cells to detect the ATP, which is a nonfluorescent active molecule with high electrophoretic mobility. The chips were made of either PMMA or COP or bought from ChipShop. Electrophoretic separation of the ATP stream in the μFFE chips loaded with HEK 293 cells was visualized by the fluorescence imaging of sensing cells. This system was capable of on-line detection of native and unlabeled compounds.147 In another study, the identification and quantification processes of bacterial cells and spores were adapted into an electronic microfluidic device. The device was capable of immune-localization of spores, which were captured by the membrane filter within the chip. The analyte signal was recorded, and collective response or detection and counting of individual spores and particles were done. The device was made of PMMA and nuclepore membrane, and stainless steel tubing was inserted via drilling into the PMMA for fluid passage. The limit of detection of the chip was around 500 Bacillus globigii (Bg), which is a stimulant for Bacillus anthracis (Ba).148 In 2008, an electrochemical impedance spectroscopy (EIS)-based microfluidic device was developed to study the adhesion of bacteria cells onto a semiconducting indium tin oxide (ITO) plate with the aim of electrochemical detection and characterization. Pseudomonas stutzeri (PS) and Staphylococcus epidermidis (SE) bacterial strains were used in this system to compare the adhesion behavior and charge transporting property of these cells. Impedance variations were measured both at low and high frequencies for electrical detection and electrical characterization, respectively. The flow chamber was made of two parallel PMMA sheets, and a silicone membrane was put in between these sheets. The ITO plate was located at the bottom sheet, and a circular chamber was placed on the upper sheet for bacterial suspensions. As the number of adhered cells increased, the intenseness of the impedance decreased exponentially. First PS cells, then SE cells were identified electrically and PS cells were more prone to make charge transfer to the electrode than SE cells.149 Similarly, protein digestions was performed in an electrokinetically driven solid-phase trypsin microfluidic device integrated with matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (MALDI-TOF MS). The microbioreactor included a microchannel and the micropost system was located in that channel. Several proteins were employed in the system and cytochrome c digestion was the most efficient one with 97% sequence coverage for protein identification. Bovine serum albumin (BSA) with 46%, phosphorylase b with 63% and β-casein with 79% sequence coverages were detected in the device. E.coli cells were also used for fingerprint analysis of intact cells.150 Vila and his colleagues operated an optical analysis for the determination of environmental pollution. The system included a PMMA optofluidic arrangement, comprised of light emitting diodes (LEDs) and detectors, and they managed optical measurement and elimination of ambient light interference via an electronic circuit. The device included 4 layers of PMMA with distinct patterns and optical and fluidic aspects. The system was applied to the detection of water toxicity. In these toxicity tests, bacterial reduction kinetics of ferricyanide was used, where ferricyanide collaborated with the membrane proteins of E. coli, and simple, precise, and reliable water toxicity determination was enabled.151 Recently, electrical sensing of the bacterial lysate was used to detect the Pseudomonas aeruginosa and Staphylococcus bacteria cells that cause keratitis. The device included PMMA wells and electrodes of 10 μm width and 20 μm spacing. A statistically remarkable impedance change was obtained for 10 CFU/ml diluted bacteria samples.152
The electroporation chips have been produced to eliminate the limit in the amount of target cells to be detected, potential risk of using high voltage, and the undesired effect of temperature rising encountered in conventional systems. Electrical square pulses were applied through the channel, and the mixture of Huh-7 cells lines and reporter genes were sent into the channel. In order to see the cells under the fluorescent microscope (BX60, Olympus, Japan), trypan blue was used for staining. Transfection was successfully accomplished in the device and the cell survival rate was increased when the low pulse frequency and high flow speed were employed.153 Optical fibers, mirrors and electrodes were used to make cytometric analysis of blood cells in three-dimensional microfluidic devices. In the study of Kummrov et al. erythrocytes and thrombocytes were distinguished via forward light scatter. The fluorescence imaging was used to monitor the two dimensional focusing of sample flow and the results were confirmed by finite element calculations. In this study, T-helper cells labeled by fluorescent monoclonal antibodies were also determined.154
B. Magnetic field based detection
Nowadays, the creation of a magnetic field in microfluidic devices for cell detection is heavily used. For E. coli detection, Laczka et al. benefited from immunomagnetic capture and ampertometric detection techniques. The combination of horseradish peroxidase (HRP), hydrogen peroxide (H2O2), and hydroquinone (HQ) was used for electrochemical monitoring. An incubation micro-chamber, which had magnetic particles (MPs), was employed for the enzymatic reactions. The microelectrodes were used to monitor the enzyme product, which flowed in the microchannels. The PC polymer formed the bottom of the chip, where two magnets, necessary for keeping the MP upstream from the electrode, and a pocket for housing the chip were located. The experiment took 1 h and the bacterial cells were identified with a limit of detection of 55 cell ml−1.155 In another study, the polymerase chain reaction (PCR) of bacterial cell culture was conducted in a magnetically functionalized closed-loop PCR microfluidic device. A ferrofluid plug was employed as the valve and actuator to prevent liquid flow in the device. The loaded sample visited all the sections of the device to accomplish the PCR amplification, the thermal lysis, and the denaturation of the PCR mixture. The limit of detection was found as four bacterial cells. This device was capable of shortening the reaction time compared to conventional thermocycler systems, and the results were comparable.156 Spinning magnets were employed to handle the sample manipulation, mixing, and controlled target release in the microfluidic device, called MagTrap, which was used for detection of E. coli cells. MagTrap consisted of magnets, which were placed in a rotating wheel and microfluidic channels. In this system, the strip magnet design was used, and the detection procedure was first the binding of E. coli to antibody coated superparamagnetic fluorescently coated beads, and then releasing them into the microchannel. As the wheel rotated, the beads were trapped and separated. MagTrap was successful in detecting the bacterial cells.157 In 2012, Verbarg and his group have combined their MagTrap device with another device to do cytometry experiments using E. coli, Salmonella, and Shigella bacteria cells. Again, target-specific magnetic beads were used in the MagTrap and rotational movements of the device (forward and backward) conducted reagent processing in MagTrap. The MagTrap device was made of PMMA and consisted of a trapezoidal channel. In order to activate the magnetic wheel for bead manipulation, a motor was used. A PDMS microflow cytometer was connected to the outlet of the MagTrap to accomplish the cytometry experiments.158 In 2013, the combined MagTrap and cytometer system was used to identify the particles in the microfluidic system as well as to obtain sample-to-answer diagnosis. Fluorescently stained microspheres grabbed the target molecules, and these molecules were taken out of the sample matrix. These grabbed molecules were exposed to biotinylated tracer molecules and streptavidin-labeled phycoerythrin, respectively. Finally, these molecules passed through the microflow cytometer for color analysis. In order to detect the E. coli cells, three sets of MagPlex microspheres were prepared, and the concentration of 1 × 104 cells/ml could be recognized via MagTrap.159
C. Chemical detection
Daunert and her group produced a microfluidic device having a chemical detection system. Arsenite and antimonite were used as the target analytes to be sensed by the E. coli cells, which included plasmid pSD10. The microfluidic platform was able to recognize the regulatory protein ArsR fused with GFPuv, which is a reporter protein. The GFPuv in the cells was capable of fluorescence emission, enabling arsenite and antimone detection. The device consisted of 2 reagent reservoirs, a mixing channel and a detection reservoir. Whole-cell biosensing system was successfully accomplished, and the response time was shortened compared to conventional systems.160 Later, the same group improved their design, and they introduced two different spore-based whole-cell sensing portable microfluidic systems to detect arsenite and zinc. The detection limit of the system in both serum and water media was 1 × 10−7 M for arsenite and 1 × 10−6 M for zinc, respectively. The first platform (CD 1) was used for zinc detection (left) and the second platform (CD 2) was employed to detect the arsenic (right). During the experiments, all the luminescence measurements were conducted via a spectrophotometer that had a fiber optic probe. This probe was placed 2 mm above the detection chamber perpendicularly, and all the measurements were taken at 20 min intervals. In this sensing systems, the Bacilus subtilis strain was used for arsenic detection and Bacilus megaterium cells was used for the zinc detection.161
Artemia franciscana (Artoxkit MTM) cells were used to test the aquatic toxicity in a microfluidic system. In order to determine the outcomes of the reference toxicant on the identified behavioral parameters, fully automated time-resolved video data analysis was carried out. A continuously microperfused microfluidic system was used to monitor the free-swimming actions of Artemia sp. nauplii cells. CorelDraw X3 CAD and SolidWorks programs were used to model the device in 2 and 3 D, respectively. The device included a circular caging chamber for free swimming larvae and 10 microchannels were placed in between the inlet/outlet and caging chamber. Several chemical stressors were applied through the system and the behavior of the cells was followed. Hyperactivity and hypoactivity syndromes were analyzed in response to chemical stressors.162 In another study, Allorchestes compresa marine amphipod was used to monitor sublethal behavioral toxicity by employing a microfluidic device capable of continuous perfusion. The toxicants were perfused through the device, and automated behavioral tests were done. 2 and 3 D versions of the device were designed via CorelDraw × 3 and SolidWorks 2015 software, respectively. COMSOL Multiphysics 4.4 was used to study the fluid dynamics inside the device (Fig. 6). Detection and automatic analysis of swimming behavior of the A. compresa cells were done successfully.163 In a study on drug delivery, lectins were attached on the inner surface of the PMMA made microfluidic device to target cells in the gastrointestinal tract. Aminolysis was used to obtain amine-terminated surfaces. The device contained reservoirs and Caco-2 cell lines were employed in the experiments. Avidin molecules were bound to the PMMA surfaces rendering the PMMA cytoadhesive. This system demonstrated strong potential for use in drug delivery.164
FIG. 6.
(a) The layers of the microfluidic system. (b) Assembled chip microphotograph. Reproduced with permission from Cartlidge et al., Sens. Actuators, B 239, 660 (2017). Copyright 2017 Elsevier.
D. Biosensing/biosensor based detection
Enteropathogenic E. Coli (EPEC) cells, which cause several gastroenteric illnesses, were investigated in a special microfluidic system. A Fabry–Perot (FP) cavity-based biosensing platform was employed to detect the EPEC in 5 min. A micro-thin double sided adhesive tape and two semi-transparent FP mirror plates were used for the construction. In order to measure the spectral changes, an optofluidic FP cavity was employed, and the channel of 2 mm width was cut via a CO2 laser. Capillary force was applied through the system to fill the channel, and then the transmission spectra was detected in order to arrange the adhesion time of cells on the functionalized surfaces. Surface functionalization was conducted by a translocated intimin receptor (TIR). Four taxonomically or distantly related bacterial strains were specifically recognized by TIR surfaces. The efficiency of trapping on the functionalized gold mirrors was determined by confocal optical imaging via SEM.165 In another study, electrokinetic capture was employed to detect E. coli cells via a cantilever biosensor combined with electrodes for piezoelectric actuation. The PiezoMUMPs process was used to create a biosensor consisting of four layers; a silicon structural layer, a silicon oxide insulating layer, an aluminum nitride piezoelectric layer, and a gold electrode layer. The microfluidic platform responsible for sample/solution delivery to biosensor was made of PMMA and PDMS. When the sample concentration was 107 cells/ml, the signal to noise ratio was 82 within 10 min; and when the sample concentration was 105 cells/ml, this ratio was 26. Therefore, E. coli cells were successfully detected via the cantilever biosensor.166 A high-throughput centrifugal microfluidic device was designed to make colorimetric analysis of the foodborne pathogen. A sample reservoir, a spiral shaped sample injection microchannel, 24 aliqouting chambers, cross capillary valves, and 24 reaction chambers were placed in the device (Fig. 7). The microfluidic system was made of PMMA using a pressure sensitive adhesive (PSA). Loop mediated isothermal amplification (LAMP) was employed to identify E. coli, Salmonella typhimurium and Vibrio parahaemolyticus pathogens and all of the genetic analytical process was finalized within 1 h.167 Gold nanorods (AuNRs) were used to detect the orchid viruses in the fiber optic particle plasmon resonance (FOPBR) immunosensor system. A near infrared sensing window was generated by the AuNRs to make direct sensing. Cymbidium mosaic virus (CymMV) and Odontoglossum ringspot virus (ORSV) were identified by the antibodies present on the AuNRs by the unclad fiber core surface. The AuNR-FOPPR sensing window was established in the microfluidic channel, which was made of PMMA. The limits of detection (LOD) of the microfluidic system were compared with those of the ELISA test and the results confirmed that this microfluidic system has better performance. The immunosensor achieved faster analysis, better reproducibility, and lower detection limits compared to ELISA. In addition, this sensor was capable of categorizing the healthy and infected orchids, and also displayed the infection level.168 A micromixer, including a microballoon, was fabricated for creating 3 D reciprocating flow and to detect dengue virus. The system was capable of expanding and contracting, and hence could change the flow dynamics. Two different CD-like designs were constituted, where Design A was used to evaluate the mixing in the microballoon, to make comparison with the stopped-flow mixing method and to theoretically analyze the rotational frequencies. Design B was used to see the effects of the microballoon as well as the stopped-flow mixing technique on the ELISA results. Two loading reservoirs, a mixing chamber and two microchannels were located on both designs A and B. The biomolecule reaction rate and detection sensitivity of the dengue viruses were increased compared to the conventional methods.169 In the study of Morant-Minana et al. the detection of C. Camplyobacter spp. in real food samples was performed by using a biodevice integrated with a DNA sensor. For the detection, an electrochemical geosensor based on thin-film gold electrodes was deposited onto the COP surface. The feasibility and sensitivity of the biosensors developed in this study were tested by Atomic Force Microscopy (AFM), X-ray diffraction (XRD), Fourier Transform Infrared spectroscopy (FTIR), Cyclic Voltammetry (CV) and Square Wave Voltammetry (SWV), and the detection of PCR amplicon of C.Camplyobacter spp. concentration was successfully performed. Photolithography and sputtering were used to create the electrodes on COP. The device presents an integration of electrochemistry and microfluidics for real time detection and analysis of microorganisms in food. The amplification of bacterial genomic DNA in a short time by using electrochemical biosensor technology on the COP polymer made the microfluidic device showed that it can be used for further diagnostic studies in the food sector.170
FIG. 7.
The design of the centrifugal LAMP microdevice. Reproduced with permission from Seo et al., Sens. Actuators, B 246, 146 (2017). Copyright 2017 Elsevier.
Dong and Zhao designed a PS made biochip; (i) to determine the specific pathogen among 13 types of uropathogenic microbes, that cause urinary tract infections (UTIs) by employing the immunosorbent ATP-bioluminescence assay (IATP-BLA) and (ii) to find the relevant antibiotic for patient according to detected microbial organisms. The biochip consisted of 5 layers, two glasses as cover, two PS sheets with 384 vertical reaction chambers as the sample layer (urine and oxygen/air) or culture layer (microbes and other reagents), and a fiberglass membrane, in which 8 different types of antibodies were immobilized, fixed between two PS sheets to target and capture microbes. The immobilized antibodies captured the specific microbes in a loaded urine sample. Calcium alginate gel was used to encapsulate the captured cells and these encapsulated cells were evaluated by ATP-BLA in a microplate reader by using different methods for different channels. As a result, the biochip decreased the detection time of the pathogens from days to minutes and can be a model for the diagnosis of other diseases.171
The human embryonic kidney cell line (HEK-293) is widely used in cell biology studies. Since many neuronal proteins can be expressed by this cell line, it is accepted that this cell line was generated from embryonic neural cells.172 A portable ion channel biosensor (4 channel incubation type planar patch clamp biosensor) was developed, and the channelrhodopsin wide receiver (ChRWR) expressed by HEK293 cells was used to evaluate the performance of the microfluidic system. The system consisted of a cell trapping area and a pipette solution well. A micropore was created in the middle of the round area to achieve the cell trapping. The biosensor's surface was coated with ECMs of poly-L-lysine and the cells were fed through this coated chip. During the experiments, a laser-evoked channel current of the cells was recorded via a patch-clamp amplifier. This system can safely be used for high throughput screening of neural cells.173 In order to conduct immunostaining-based cytometry, a microfluidic device was developed, where antibodies were placed in a gelatin layer. The chip included flow chambers and counting chambers on a laminating adhesive, and was sandwiched in between the PMMA layers. Inkjet printing was used to establish the gelatin/antibody layer. A homogeneous antibody distribution was accomplished in the device due to the long maturation process of gelatin, and the antibody release was done by heating up the gelatin layer. In the experiments, CD4 positive (CD4+) T-lymphocytes were used to perform on-chip immunostaining to whole blood, and cell counting was done effectively. The temperature-switch antibody release from gelatin/antibody layers was successfully completed.174
E. PCR based detection systems based on genetic material
Creating a new strand of DNA complementary to a template strand is called the polymerase chain reaction. Several organisms' DNA can be copied or reproduced by using this technique.175 Tumor virus identification via DNA manipulation was conducted in a microfluidic device, in which the micro-reverse transcription polymerase chain reaction (μRT-PCR) was carried out. PMMA with SU-8 was used as the construction material of the device that integrated sample reservoirs, RT-PCR meanders, and capillary electrophoresis (CE). The device design was simulated via ANSYS, CFD-RC and IntelliSuite softwares, and then validated by the experiments. Heat transfer, reaction temperature control, and fluid flow clogging preclusion were successfully accomplished in this device.176 In another study, μRT-PCR, CE, and on-line detection with buried optical fibers were combined in the microfluidic platform to detect Piscine nodavirus cells in a fast, sensitive and automated way. PDMS, PMMA and soda-lime glass were used to fabricate the device. By using this system, virus cells were successfully detected from fish samples and the detection limit was 12.5 copies/μl.177 In 2015, the detection of Human papilloma virus (HPV) 16 was accomplished by a biomicrosystem employing PCR based detection system. The device included 98 biosensors, and the monoclonal antibody (mAb) 5051 immobilization was done on the PMMA substrate with a gold layer. Electrochemical impedance spectroscopy, cyclic voltammetry, impedance measurements, AFM and SEM were used to define the system. The wells in diameter were located on the PMMA substrate for detection. The comparison of the results of HPV 16 detection with those of the standard PCR tests confirmed the effectiveness of this biomicrosystem.178 A COP made microfluidic device was developed to achieve a rapid genetic testing system, such as the detection of the point mutation of FGFR3 gene. This microfluidic device had a serpentine channel with 80 loops, was covered with a thin pressure sensitive adhesive transparent film, and consisted of two temperature zones for denaturation and annealing reactions. The rapid thermal cycling was performed by a continuous flow of PCR, the solution was thermally cycled through the created multiple local temperature zones in the microfluidic channels. Due to the high surface area to volume ratio, the fast heat conduction to the solution was obtained via channel walls as an advantage of this process. A point mutation of FGFR3 gene was detected in 6.5 min by using Bacilus subtilis spores. As the genetic mutation of the FGFR3 gene is related to bladder cancer, this study is a promising one for rapid genetic testing of various diseases.179 In another study, poly(cyclic olefin) microfluidic devices were fabricated to detect and identify the bacteria cells. The device was suitable to perform DNA amplification, microfluidic valving, sample injection, on-column labeling, and separation. There was a channel reactor of 29 nl to accomplish the PCR. E. coli O157 and S. typhimurium cells were used in the study and the limit of detection was found to be around 6 copies of target DNA.180
F. Miscellaneous detection systems for diseases
1. Fluorescence based detection for Human Immunodeficiency Virus (HIV) point of care testing systems
Acquired Immunodeficiency Syndrome (AIDS) disease is caused by the Human Immunodeficiency Virus (HIV). This disease can be spread via sexual contact, infected blood or mother-to-child interaction.181 HIV and AIDS studies have also been conducted in microfluidic platforms. For example, the anti-gp120 antibody was immobilized into the microfluidic device for capturing and imaging of HIV from an untreated HIV-infected blood sample. The quantum dots (Qdot525 and Qdot655) were employed to label the envelope of gp120 glycoprotein and high-mannose glycans of captured HIV, respectively. The virus images were counted via a fluorescence microscope. PMMA, a double sided adhesive film (DSA), and glass were used to build the microfluidic device. The device was capable of identifying HIV in less than 10 min from a fingerprick volume of blood. Here, Qdots implemented a new and effective tool for analyzing the HIV molecules without any need for pre-processing.182 Detection of CD4+ T-lymphocytes was done in a microfluidic device with lensless imaging for HIV point-of-care testing. Anti-CD4 antibody was immobilized in the device for capturing of CD4+ T-lympohocytes from the blood. A charge coupled device (CCD) sensor was employed to make the identification of the captured cells, and the gray scale images were obtained via this lensless imaging CCD sensor. An automatic cell counting software was used to enumerate the cells, and a fluorescence microscope was also used for manual counting. This microfluidic system had 70.2 ± 6.5% capture efficiency and 88.8% ± 5.4% capture specificity for CD4+ T-lymphocytes.183
2. Sensors for detection and analysis of cancer
In recent years, it was observed that cancer risk is increasing in industrialized communities/societies.184 There are many studies on cancer treatments, and the detection of cancer cells can be done in microfluidic devices. Weigum et al. produced a cell-based sensor for the detection of oral cancer biomarkers via the epidermal growth factor receptor (EGFR). The device included a track-etched membrane served as a micro-sieve for capturing and enriching the cells from the biological sample. Automated microscopy and fluorescent image analysis were used to monitor the presence of the isotype of the captured cells on membrane, and the EGFR tests were conducted in less than 10 min via this biosensor. The cell-based lab-on-a-chip (LOC) sensor was made of PMMA and a PC membrane of 0.4 μm pores. The device consisted of a microchannel and a circular membrane capture area with an imaging window of 200 μm in diameter, resulting in a volume of 3.9 μl. EGFR expression in cancer cells was found to be higher than that of the control cells, which had EGFR expression similar to the normal squamous epithelium. The LOC sensor can thus be used to detect the oral cancer biomarkers as well as to characterize the EGFR over-expression in oral malignancies reliably.185 A chemotaxis assay study was done in a microfluidic chamber by using breast cancer cells. In order to fabricate the microchannels for chemoattractant transportation and barriers/conduits for flow diversion, the photolithography method was employed. The device consisted of three layers; the bottom layer was glass including a microchannel, the middle layer was made of PDMS to seal the reservoir, and the top layer was made of PMMA connected to a pressure regulator. The directionality and instantaneous speed of cell protrusion were studied in this platform. In the presence of yjr epidermal growth factor receptor, breast cancer cells answered more directionally and quickly than the uninduced cells.186 A prostate-specific membrane antigen (PSMA) was used as an indicator of the prostate tumor cells. The antibodies and aptamers can bind to PSMA. A high-throughput micro-sampling unit (HTMSU) was made of PMMA, and it consisted of immobilized anti-PSMA aptamers for capturing the circulating prostate tumor cells from the peripheral blood. The device included 51 curvilinear microchannels and the reason for using a curvilinear channel was to increase the cell capture efficiency. During the experiments, 90% of the LNCaP cells (prostate cancer cell line) were captured. In order to reinforce the detection and efficiency, a conductivity sensor was combined with the HTMSU for cell counting.187 Microfluidic flow cytometry (μFCM) technology was developed to observe the caspase-dependent cell death in a user-friendly chip-based system. This system included an off-chip electronic interface and was capable of making multivariate analysis using a small amount of sample (10 μL). Human leukemia U937 and THP1α cells were used to conduct 2 D hydrodynamic cell focusing of cells in the PMMA made microfluidic device. Two solid-state lasers (blue: 473 nm and red: 640 nm) and four photomultiplier tube-based (PMT) detection channels with band-pass filters were placed on the off-chip hardware system in order to catch the fluorescent signals. The chip included a sheath fluid reservoir, a microchannel for cell focusing and sheath fluid gathering chambers. This device enabled the analysis of the DNA, the recording of the pharmacologically induced activation of caspases, and the dissipation of mitochondrial inner membrane potential in living cells.188
Point mutations can be detected with the application of the capillary electrophoresis assay, polymerase chain reaction (PCR) or ligase detection reaction (LDR). A microfluidic device for the detection of CTCs was developed. This system had electrokinetic enrichment ability to reveal the mutations within the DNA of the CTCs. SW620 and HT 29 cells in the blood samples were used because these cells can overexpress the integral membrane protein EpCAM. The wall of the microfluidic device was coated with the anti-EpCAM antibodies to detect the cells. In the electromanipulation unit, 125 μm diameter Pt wires were used as the electrodes to perform the conductivity-based enumeration, following the release of the CTCs from the antibody selection surface. 96% ± 4% efficiency was attained during the CTC selection.189 The detection of CTCs was further conducted in the label-free reflectometric interference spectroscopy (RIF) functionalized microfluidic biosensor system. The RIF sensing structure was created by anodic aluminium oxide (AAO), which was produced by the electrochemical anodization. This AAO surface was coated with a biotinylated anti-EpCAM antibody to bind the human cancer cells of epithelial origin. The device had two microfluidic channels with simple mixers for sending the fluid evenly through the AAO sensing platform. This system did not require any bonding procedure, the PMMA layers were just clamped to each other. In addition, this detection method did not need any fluorescence tagging or pre-enhancement processes; the samples were simply sent through the system, and the cells were detected via the AAO RIF sensor. The device successfully detected around 1000–100 000 cells/ml in a 5 min response time for 50 μl sample.190 In order to fight against cancer cells, many scientists have been struggling to develop therapies that strengthen the immune system and cure the disease.191 In order to get insight into the biological behavior of the myeloma tumor cells after mechanical stress, a microfluidic device was developed and optimized. A microgear pump was fabricated via the deep X-ray lithography (DXRL) method by using PMMA sheets of 500 μm thickness. A 1 × 1 mm2 magnet was placed in the device for actuation and the solvent bonding technique was used for sealing. The gear included 20 teeth. The cell suspension was fed to the system, pumped for a determined time, and the local head pressure difference was recorded. As the stress increased, the cell viability was found to decrease.192 Mechanical dissociations of the digested tumor tissue and cell aggregates into single cell experiments were done in polyethylene terephthalate (PET) made microfluidic devices to enhance the quality of the results obtained. Several sized (millimeters to microns) channels were placed in the device to create well defined high shear force regions to increase the dissociation of clusters into single cells. HCT 116 colon cancer cells were used in the experiments and by using this microfluidic device, cell recovery, cell viability, and process time were improved.31 3 D micro-chambers for cell capturing and analysis were fabricated by COC. The device was capable of creating high flow velocity in 3 D microchannels to provide selective cell immobilization. The performance of the device was tested via immune-fluorescence labeling and Fluorescence in Situ Hybridization analysis on cancer cell lines and on a patient pleural effusion sample. This device, fabricated at a low cost, can be used for medical diagnostics.193
3. Morphology based detection and analysis systems for diagnostic purposes
Several studies on red blood cell detection have been conducted in microfluidic devices. The quantification of blood was handled in the microfluidic device using the impedance analysis technique. Ansys Fluent 12.1.4 simulations were performed to evaluate the diffusive mixing practices. The microfluidic system used in this study had the microfluidic lysis block, the impedance analysis chip, the electronic detection, and data collection processing parts. The microfluidic system was made of PMMA and the height and channel widths were 100 and 200 μm, respectively. As the cells flowed in this impedance chip, the electrical cell volume and cell membrane capacitance were measured by applying two different sinusoidal voltages through the electrodes. The results obtained from the red blood cell (RBC) lysis experiments conformed to those in the conventional hematology devices.194 The red blood cells, which have concave surfaces, have been used to reveal their shapes for diagnostic purposes. The particular microfluidic device used for RBC analysis was made of PMMA, and it has 1000 μm × 200 μm (width × height) dimensions. In order to hold the RBCs in the channel, optical traps were used, and by using laser power the cells were rotated. During the rotation, the images of the RBCs were taken from several viewpoints and recorded. It was proven that the system was capable of revealing the holographic images of the RBCs. Therefore, it is possible to apply this system for diagnostic analysis without any labeling.195 Alapan et al. developed a biochip for Sickle cell disease (SDC) and made quantitative evolution of red blood cells (RBCs) in a microfluidic system with the help of protein coated microchannels. By using the SDC biochip, the relation between the RBC adhesion to fibronectin (FN) and laminin (LN) proteins as well as the features of RBCs were revealed. The SDC chip contained FN or LN coated glass slide at the bottom, and PMMA substrate including inlets and outlets at the top. The adhered RBCs were monitored and classified as deformable and nondeformable by considering their morphology. The SDC biochip is a promising tool for quantitative assessment of RBCs and for observing disease progression and vaso-occlusion.196
By using microfluidic devices, several detection experiments such as pathogen detection or disease detection can be done. Micro-nano particles or magnets can be used as assistant elements in these devices to enhance the quality of detection. Two microfluidic devices can be combined to make consecutive experiments. For example, first detection of cells in one device then enumeration of detected cells can be done in another device. Moreover, environmental stress is an important factor for living organisms. Microfluidic devices give opportunities to monitor the changes in cells instantaneously. Taxonomically or distantly related cells can be observed in one single device. In developing countries, disease diagnosis is very difficult. Healthcare services are deprived of advanced medical devices. Therefore, microfluidic devices are seen as a savior for disease identification in these countries, enabling the detection in a short and easy way. This section is tabulated and given as the supplementary material.
V. REACTION: MICROBIAL FUEL CELLS
Fossil fuels, renewable sources and nuclear sources are the energy sources of life. Fossil fuels satisfy a considerable amount of the energy need in the world. In this regard, microbial fuel cell (MFC) studies have gained great attention since the early 20th century. The MFC field combines different scientific and engineering knowledge, and MFCs were produced to benefit from the small volume and high output power density.197–199
A mini-MFC including one chamber, and a plate-shaped (instead of stripe-shaped) gold anodic electrode were operated to obtain higher electrochemical activity. Shewanella oneidensis MR-1 cells were used to create the biofilm on the gold electrode. The gold anodic electrode layer was fabricated on the glass and 8 wells were located on the PMMA made anode well layer. The mini-MFCs contained a 2 mm thick PMMA supporting layer, and a proton exchange membrane (PEM) layer carried the nafion membrane (proton-conductive polymer film) with 8 Pt/C electrodes (Fig. 8). The biofilm formation on gold electrodes was attained after one day of operation that increased the electricity production. 29 mW/m2 power density and 2148 mA/m2 current density were collected via this mini-MFC.200 Two years later, monitoring of electrochemically active bacteria was conducted in five layer microbial fuel cells (MFCs). An anode electrode layer made of Cr/Au on PMMA, an anode chamber layer (gasket), a proton exchange membrane (PEM), a cathode chamber layer (gasket), and a cathode electrode layer made of Cr/Au on PMMA layers constituted the MFC from the bottom to the top. The electricity generation capacities of bacterial cells, wild-type Shewanella oneidensis (S. oneidensis) and Pseudomonas aeruginosa (P. aeruginosa) along with the isogenic nirS, lasl, bdlA, and PilT mutants were compared, and they were found to perform different efficiencies of extracellular electron transfer. 6 different MFCs showed only 1.4% difference among each other.201 In 2016, Mardanpour and Yaghmaei investigated nickel based microfluidic microbial fuel cells (MFCs) and E. coli cells that were used as the biocatalyst. Bioelectricity generation from glucose and urea of human blood and urine, respectively, was accomplished by using MFC as a power production. The MFC can be operated in both fed-batch and continuous modes. Glucose-fed and urea-fed MFCs can have 5.2 μW cm−2 and 14 Wm−3 power densities, respectively, providing high power density, self-regeneration, waste management, and low production cost instead of conventional methods.202 Cyanobacterial photosynthetic and respiratory processes are capable of electricity production all day. Nine bio-solar panel microsystems, having a PMMA energy capturing layer, an anodic PMMA chamber layer, a proton exchange membrane (PEM), and a bottom PMMA layer for exposing an air-cathode to oxygen, were fabricated. Synechocystis sp. PCC 6803 cells produced 5.59 μW maximum power and 1.28 V energy. This study can be a progression in bio-solar cells to promote high energy production.203 Very recently, a microbial electrolysis cell (MEC) was fabricated to produce biohydrogen for medical applications. E. coli, non-pathogenic bacteria cells, were used to bioxidate the human excreta in the MEC to generate the biohydrogen. 0.94 μl hydrogen (μl urea)−1 day−1 from urea and 0.84 μl hydrogen (μl glucose)−1 day−1 from glucose were attained. Chemotaxis phenomena were also investigated in an MEC device and the results were found to be compatible with real conditions.204 An integrable and scalable power source of MFC, based on laminar-flow was produced, where the device was operated without any physical membrane in it, and the virtual membrane was created to harvest the electricity. Wild type Pseudomonas aeruginosa PAO1 cells were used in the system, and a power density of 60.5 μW/cm2 was obtained using a 100 kΩ load power output.205
FIG. 8.
(a) Mini-MFC schematic illustration. (b) Gold anodic layer on glass and PMMA anodic well layer. (c) Image of the bonded PMMA anodic well layer on glass substrate. (d) Metal straw sealed by silicon rubber to the anodic layer. (e) Assembled mini-MFC. Reproduced with permission from Chen et al., Biosens. Bioelectron. 26, 2841 (2011). Copyright 2011 Elsevier.
The manufacturing of products having importance on human life like biological hydrogen production from renewable resources can be accomplished in thermoplastics made devices, as they are hard and durable to certain reactions.
VI. CONCLUSION/FUTURE DIRECTIONS
This review provides an overview of recent advances in microfluidic devices made from PMMA, PS, and COP that have been used in biological applications. Different aspects related to the cultivation, separation/isolation, detection, and reaction experiments in microfluidic devices have been discussed and an overview of these studies carried out since 2000 till today is given. The main conclusions are as follows: (1) high-throughput analysis at low cost can be done in a single device that is capable of conducting parallel experiments, (2) more detailed results can be obtained by integrating imaging technologies, (3) experiments can be conducted in stable, well defined, and biologically relevant culture environments, (4) multi-organ microphysiological systems can be constituted by mimicking the real conditions, and (5) more than one process such as lysis, amplification, and separation can be accomplished in a single device.
Microfluidic system technology continues to evolve and progress in every field. However, more economical, environmentally friendly, and portable devices should be fabricated. Scientists should be more interested in real-world problems, such as epidemic illnesses etc., and serious steps should be taken to solve these problems. Many developing countries are short of healthcare facilities and medicines. Devices for point-of-care diagnostics can be fabricated for pathogen or disease detection and this can be a life saver in these countries. With many devices in or under development, innovative designs, improvements in processes and growth in consumer applications will lead mems products to have a market share of 24 billion US dollars in 2020.206 The breakthrough technologies in bio-mems will overcome the challenges encountered so far and make room for new mems applications in multiple markets.
VII. SUPPLEMENTARY MATERIAL
See supplementary material for the tabulated versions of the Separation/Isolation and Detection and Analysis sections.
The Separation/Isolation summary table includes the device format, microorganisms, materials used for device fabrication, fabrication techniques, separation criteria, remarks on the study, and reference parts.
The Detection and Analysis table includes the device format, microorganisms, materials used for device fabrication, fabrication techniques, detection criteria, remarks on the study, and reference parts.
ACKNOWLEDGMENTS
The financial support of the Boğaziçi University Research Fund through project Nos. R9701 and M11141 is gratefully acknowledged.
References
- 1. Yi L., Xiaodong W., and Fan Y., J. Mater. Process. Technol. 208, 63 (2008). 10.1016/j.jmatprotec.2007.12.146 [DOI] [Google Scholar]
- 2. Kricka L. J., Fortina P., Panaro N. J., Wilding P., Alonso-Amigo G., and Becker H., Lab Chip 2, 1 (2002). 10.1039/b109775j [DOI] [PubMed] [Google Scholar]
- 3. Ebewele R. O., “ Condensation (step-reaction) polymerization,” in Polymer Science and Technology ( CRC Press, 2000). [Google Scholar]
- 4. Nunes P. S., Ohlsson P. D., Ordeig O., and Kutter J. P., Microfluid. Nanofluid. 9, 145 (2010). 10.1007/s10404-010-0605-4 [DOI] [Google Scholar]
- 5. Shadpour H., Musyimi H., Chen J., and Soper S. A., J. Chromatogr. A. 1111, 238 (2006). 10.1016/j.chroma.2005.08.083 [DOI] [PubMed] [Google Scholar]
- 6. de Tayrac R., Chentouf S., Garreau H., Braud C., Guiraud I., Boudeville P., and Vert M., J. Biomed. Mater. Res. B: Appl. Biomater. 85, 529 (2007) 10.1002/jbm.b.30976. [DOI] [PubMed] [Google Scholar]
- 7. Young E. W. K., Berthier E., and Beebe D. J., Anal. Chem. 85, 44 (2013). 10.1021/ac3034773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rosenzweig D. H., Carelli E., Steffen T., and Jarzem P., Int. J. Mol. Sci. 16, 15118 (2015). 10.3390/ijms160715118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heckmann W., Microsc. Microanal. 11, 2036 (2015) 10.1017/S1431927605501508. [DOI] [Google Scholar]
- 10. Chang T. Y., Pardo-Martin C., Allalou A., Wahlby C., and Yanik M. F., Lab Chip 12, 711 (2012). 10.1039/C1LC20849G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crawford L., Higgins J., and Putnam D., Sci. Rep. 5, 13177 (2015). 10.1038/srep13177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhengzhou Aroent Power Co. Ltd. Properties Compare for LDPE and HDPE, 2012.
- 13. Goodman D., J. Vinyl Addit. Technol. 16, 156 (1994). 10.1002/vnl.730160312 [DOI] [Google Scholar]
- 14.C&J Industries, Generic Material Polyamide-Nylon.
- 15. Piruska A., Nikcevic I., Lee S. H., Ahn C., Heineman W. R., Limbach P. A., and Seliskar C. J., Lab Chip 5, 1348 (2005). 10.1039/b508288a [DOI] [PubMed] [Google Scholar]
- 16. Wright W. W., Br. Polym. J. 21, 525 (1989) 10.1002/pi.4980210617. [DOI] [Google Scholar]
- 17. Abdel-Wahab A. A., Ataya S., and Silberschmidt V. V., Polym. Test. 58, 86 (2017). 10.1016/j.polymertesting.2016.12.016 [DOI] [Google Scholar]
- 18. Becker H. and Gärtner C., Electrophoresis 21, 12 (2000). [DOI] [PubMed] [Google Scholar]
- 19. Nevitt M., Microfluid., BioMEMS, Med. Microsystems XI 8615, 86150F (2013). 10.1117/12.2002122 [DOI] [Google Scholar]
- 20. Jabarin S. A. and Lofgren E. A., Polym. Eng. Sci. 24, 1056 (1984). 10.1002/pen.760241311 [DOI] [Google Scholar]
- 21. Ulery B. D., Nair L. S., and Laurencin C. T., J. Polym. Sci. B: Polym. Phys. 49, 832 (2011). 10.1002/polb.22259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kodzius R., Xiao K., Wu J., Yi X., Gong X., Foulds I. G., and Wen W., Sens. Actuators, B 161, 349 (2012). 10.1016/j.snb.2011.10.044 [DOI] [Google Scholar]
- 23. McCann R., Bagga K., Stalcup A., Vázquez M., and Brabazon D., Proc. SPIE 9351, 93511N (2015) 10.1117/12.2076916. [DOI] [PubMed] [Google Scholar]
- 24. Cao Y., Bontrager-Singer J., and Zhu L., J. Micromech. Microeng. 25, 65005 (2015). 10.1088/0960-1317/25/6/065005 [DOI] [Google Scholar]
- 25. Puza S., Gencturk E., Odabasi I. E., Iseri E., Mutlu S., and Ulgen K. O., Biomed. Microdevices 19, 40 (2017). 10.1007/s10544-017-0182-3 [DOI] [PubMed] [Google Scholar]
- 26. Lee J. N., Park C., and Whitesides G. M., Anal. Chem. 75, 6544 (2003). 10.1021/ac0346712 [DOI] [PubMed] [Google Scholar]
- 27. Whitesides G. M., Nature 442, 368 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- 28. Tan S. H., Nguyen N. T., Chua Y. C., and Kang T. G., Biomicrofluidics 4, 32204 (2010). 10.1063/1.3466882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Halldorsson S., Lucumi E., Gómez-sjöberg R., and Fleming R. M. T., Biosens. Bioelectron. 63, 218 (2015). 10.1016/j.bios.2014.07.029 [DOI] [PubMed] [Google Scholar]
- 30. Workman V. L., Dunnett S. B., Kille P., and Palmer D. D., Macromol. Rapid Commun. 29, 165 (2008). 10.1002/marc.200700641 [DOI] [Google Scholar]
- 31. Qiu X., De Jesus J., Pennell M., Troiani M., and Haun J. B., Lab Chip 15, 339 (2015). 10.1039/C4LC01126K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xie Z., Zhang Y., Zou K., Brandman O., Luo C., Ouyang Q., and Li H., Aging Cell 11, 599 (2012). 10.1111/j.1474-9726.2012.00821.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yeo L. Y., Chang H. C., Chan P. P. Y., and Friend J. R., Small 7, 12 (2011). 10.1002/smll.201000946 [DOI] [PubMed] [Google Scholar]
- 34. Becker H. and Heim U., Sens. Actuators A 83, 130 (2000). 10.1016/S0924-4247(00)00296-X [DOI] [Google Scholar]
- 35. Mark D., Haeberle S., Roth G., von Stetten F., and Zengerle R., Chem. Soc. Rev. 39, 1153 (2010). 10.1039/b820557b [DOI] [PubMed] [Google Scholar]
- 36. Mauk M. G., Chiou R., Genis V., and Carr M. E., in ASEE Annual Conference and Exposition, Atlanta, Georgia (2013). [Google Scholar]
- 37. Volpatti L. R. and Yetisen A. K., Trends Biotechnol. 32, 347 (2014). 10.1016/j.tibtech.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 38. Chin C. D., Linder V., and Sia S. K., Lab Chip 12, 2118 (2012). 10.1039/c2lc21204h [DOI] [PubMed] [Google Scholar]
- 39.“What is cell culture?,” Coriell Institute for Medical Research 403 Haddon Avenue Camden, New Jersey 08103, see https://www.coriell.org/research-services/cell-culture/what-is-cell-culture.
- 40. Chaudry A., Sci. Creat. Q. 2, 1 (2004), see http://www.scq.ubc.ca/cell-culture/. [Google Scholar]
- 41. Szita N., Boccazzi P., Zhang Z., Boyle P., Sinskey A. J., and Jensen K. F., Lab Chip 5, 819 (2005). 10.1039/b504243g [DOI] [PubMed] [Google Scholar]
- 42. Zhang Z., Boccazzi P., Choi H.-G., Perozziello G., Sinskey A. J., and Jensen K. F., Lab Chip 6, 906 (2006). 10.1039/B518396K [DOI] [PubMed] [Google Scholar]
- 43. Gong X., Yi X., Xiao K., Li S., Kodzius R., Qin J., and Wen W., Lab Chip 10, 2622 (2010). 10.1039/c004744a [DOI] [PubMed] [Google Scholar]
- 44. Skolimowski M., Nielsen M. W., Emnéus J., Molin S., Taboryski R., Sternberg C., Dufva M., and Geschke O., Lab Chip 10, 2162 (2010). 10.1039/c003558k [DOI] [PubMed] [Google Scholar]
- 45. Elavarasan T., Chhina S. K., Parameswaran M., and Sankaran K., Sens. Actuators, B 176, 174 (2013). 10.1016/j.snb.2012.10.011 [DOI] [Google Scholar]
- 46. Matlock-Colangelo L., Coon B., Pitner C. L., Frey M. W., and Baeumner A. J., Anal. Bioanal. Chem. 408, 1327 (2016). 10.1007/s00216-015-9112-5 [DOI] [PubMed] [Google Scholar]
- 47. Lin Y. S., Yang C. H., Lu K., Huang K. S., and Zheng Y. Z., Electrophoresis 32, 3157 (2011). 10.1002/elps.201100343 [DOI] [PubMed] [Google Scholar]
- 48. Bolic A., Larsson H., Hugelier S., Eliasson Lantz A., Krühne U., and Gernaey K. V., Chem. Eng. J. 303, 655 (2016). 10.1016/j.cej.2016.05.117 [DOI] [Google Scholar]
- 49. Runge T., Sackmann J., Karl W., Lars S., and Blank M., Microsyst. Technol. 23, 2139 (2017). 10.1007/s00542-016-3007-z [DOI] [Google Scholar]
- 50. Akagi J., Khoshmanesh K., Hall C. J., Crosier K. E., Crosier P. S., Cooper J. M., and Wlodkowic D., Procedia Eng. 47, 84 (2012). 10.1016/j.proeng.2012.09.090 [DOI] [Google Scholar]
- 51. Akagi J., Khoshmanesh K., Hall C. J., Cooper J. M., Crosier K. E., Crosier P. S., and Wlodkowic D., Sens. Actuators, B 189, 11 (2013). 10.1016/j.snb.2012.11.036 [DOI] [Google Scholar]
- 52. Akagi J., Hall C. J., Crosier K. E., Crosier P. S., and Wlodkowic D., Proc. SPIE 8923, 892346 (2013) 10.1117/12.2033405. [DOI] [Google Scholar]
- 53. Akagi J., Zhu F., Hall C. J., Crosier K. E., Crosier P. S., and Wlodkowic D., Cytometry Part A 85, 537 (2014). 10.1002/cyto.a.22464 [DOI] [PubMed] [Google Scholar]
- 54. Akagi J., Zhu F., Skommer J., Hall C. J., Crosier P. S., Cialkowski M., and Wlodkowic D., Cytometry Part A 87, 190 (2015). 10.1002/cyto.a.22603 [DOI] [PubMed] [Google Scholar]
- 55. Fuad N. M. and Wlodkowic D., Proc. SPIE 8923, 892347 (2013) 10.1117/12.2033406. [DOI] [Google Scholar]
- 56. Wang K. I. K., Salcic Z., Yeh J., Akagi J., Zhu F., Hall C. J., Crosier K. E., Crosier P. S., and Wlodkowic D., Biosens. Bioelectron. 48, 188 (2013). 10.1016/j.bios.2013.04.033 [DOI] [PubMed] [Google Scholar]
- 57. Zhu F., Akagi J., Hall C. J., Crosier K. E., Crosier P. S., Delaage P., and Wlodkowic D., Proc. SPIE 8923, 892345 (2013). 10.1117/12.2033404 [DOI] [Google Scholar]
- 58. Zhu F., Baker D., Skommer J., Sewell M., and Wlodkowic D., Cytometry Part A 87, 446 (2015). 10.1002/cyto.a.22662 [DOI] [PubMed] [Google Scholar]
- 59. Navawongse R., Choudhury D., Raczkowska M., Stewart J. C., Lim T., Rahman M., Toh A. G. G., Wang Z., and Claridge-Chang A., Neurobiol. Learn. Mem. 131, 176 (2016). 10.1016/j.nlm.2016.03.019 [DOI] [PubMed] [Google Scholar]
- 60. van Midwoud P. M., Janse A., Merema M. T., Groothuis G. M., and Verpoorte E., Anal. Chem. 84, 3938 (2012). 10.1021/ac300771z [DOI] [PubMed] [Google Scholar]
- 61. Wei C. W., Cheng J. Y., and Young T. H., Biomed. Microdevices 8, 65 (2006). 10.1007/s10544-006-6384-8 [DOI] [PubMed] [Google Scholar]
- 62. Abaci H. E., Devendra R., Smith Q., Gerecht S., and Drazer G., Biomed. Microdevices 14, 145 (2012). 10.1007/s10544-011-9592-9 [DOI] [PubMed] [Google Scholar]
- 63. Peñaherrera A., Payés C., Sierra-Rodero M., Vega M., Rosero G., Lerner B., Helguera G., and Pérez M. S., Microelectron. Eng. 158, 126 (2016). 10.1016/j.mee.2016.03.059 [DOI] [Google Scholar]
- 64. Mehta G., Lee J., Cha W., Tung Y. C., Linderman J. J., and Takayama S., Anal. Chem. 81, 3714 (2009). 10.1021/ac802178u [DOI] [PubMed] [Google Scholar]
- 65. Chandrasekaran A., Kalashnikov N., Rayes R., Wang C., Spicer J., and Moraes C., Lab Chip 17, 2003 (2017). 10.1039/C7LC00356K [DOI] [PubMed] [Google Scholar]
- 66. Ravi M., Paramesh V., Kaviya S. R., Anuradha E., and Paul Solomon F. D., J. Cell. Physiol. 230, 16 (2015). 10.1002/jcp.24683 [DOI] [PubMed] [Google Scholar]
- 67. Fey S. J. and Wrzesinski K., “ Determination of Acute Lethal and Chronic Lethal Dose Thresholds of Valporic Acid using 3D Spheroids Constructed from the Immortal Human Hepatocyte Cell Line HepG2-C3A,” in Valproic Acid: Pharmacology, Mechanisms of Action and Clinical Implications Chapters ( Nova Science Publishers, 2013), p. 141. [Google Scholar]
- 68. Wu M. H., Chang Y. H., Liu Y. T., Chen Y. M., Wang S. S., Wang H. Y., Lai C. S., and Pan T. M., Sens. Actuators, B 155, 397 (2011). 10.1016/j.snb.2010.11.027 [DOI] [Google Scholar]
- 69. Huang S.-B., Wang S.-S., Hsieh C.-H., Lin Y. C., Lai C.-S., and Wu M.-H., Lab Chip 13, 1133 (2013). 10.1039/c2lc41264k [DOI] [PubMed] [Google Scholar]
- 70. Li X., Zhang X., Zhao S., Wang J., Liu G., and Du Y., Lab Chip 14, 471 (2014). 10.1039/C3LC51103K [DOI] [PubMed] [Google Scholar]
- 71. Witkowska Nery E., Jastrzebska E., Zukowski K., Wróblewski W., Chudy M., and Ciosek P., Biosens. Bioelectron. 51, 55 (2014). 10.1016/j.bios.2013.07.023 [DOI] [PubMed] [Google Scholar]
- 72. Nomura T., Miyazaki J., Miyamoto A., Kuriyama Y., Tokumoto H., and Konishi Y., Environ. Sci. Technol. 47, 3417 (2013) 10.1021/es400053x. [DOI] [PubMed] [Google Scholar]
- 73. Koh C. G., Zhang X., Liu S., Golan S., Yu B., Yang X., Guan J., Jin Y., Talmon Y., Muthusamy N., Chan K. K., Byrd J. C., Lee R. J., Marcucci G., and Lee L. J., J. Controlled Release 141, 62 (2010). 10.1016/j.jconrel.2009.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang S., Zhang X., Yu B., Lee R. J., and Lee L. J., Biosens. Bioelectron. 26, 778 (2010). 10.1016/j.bios.2010.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cemazar J., Miklavcic D., and Kotnik T., Inf. MIDEM 43, 143 (2013). [Google Scholar]
- 76. Huang C. W., Cheng J. Y., Yen M. H., and Young T. H., Biosens. Bioelectron. 24, 3510 (2009). 10.1016/j.bios.2009.05.001 [DOI] [PubMed] [Google Scholar]
- 77. Sano M. B., Salmanzadeh A., and Davalos R. V., Electrophoresis 33, 1938 (2012). 10.1002/elps.201100677 [DOI] [PubMed] [Google Scholar]
- 78. Uno H., Wang Z. H., Nagaoka Y., Takada N., Obuliraj S., Kobayashi K., Ishizuka T., Yawo H., Komatsu Y., and Urisu T., Sens. Actuators, B 193, 660 (2014). 10.1016/j.snb.2013.12.019 [DOI] [Google Scholar]
- 79. Chang K. T., Chang Y. J., Chen C. L., and Wang Y. N., Electrophoresis 36, 413 (2015). 10.1002/elps.201400272 [DOI] [PubMed] [Google Scholar]
- 80. Cheung M. C., McKenna B., Wang S. S., Wolf D., and Ehrlich D. J., Cytometry Part A 87, 541 (2015). 10.1002/cyto.a.22609 [DOI] [PubMed] [Google Scholar]
- 81. Liberale C., Cojoc G., Bragheri F., Minzioni P., Perozziello G., La Rocca R., Ferrara L., Rajamanickam V., Di Fabrizio E., and Cristiani I., Sci. Rep. 3, 1258 (2013). 10.1038/srep01258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bedian L., Rodríguez A. M. V., Vargas G. H., Parra-Saldivar R., and Iqbal H. M. N., Int. J. Biol. Macromol. 98, 837 (2017). 10.1016/j.ijbiomac.2017.02.048 [DOI] [PubMed] [Google Scholar]
- 83. Gershlak J., Hernandez S., Fontana G., Perreault L., Hansen K., Larson S., Binder B., Dolivo D., Yang T., Dominko T., Rolle M., Weathers P., Medina-Bolivar F., Cramer C., Murphy W., and Gaudette G., Biomaterials 125, 13 (2017). 10.1016/j.biomaterials.2017.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Limongi T., Lizzul L., Giugni A., Tirinato L., Pagliari F., Tan H., Das G., Moretti M., Marini M., Brusatin G., Falqui A., and Torre B., Microelectron. Eng. 175, 12 (2017). 10.1016/j.mee.2016.12.014 [DOI] [Google Scholar]
- 85. Dalby M. J., McCloy D., Robertson M., Agheli H., Sutherland D., Affrossman S., and Oreffo R. O. C., Biomaterials 27, 2980 (2006). 10.1016/j.biomaterials.2006.01.010 [DOI] [PubMed] [Google Scholar]
- 86. Fukuda J., Sakai Y., and Nakazawa K., Biomaterials 27, 1061 (2006). 10.1016/j.biomaterials.2005.07.031 [DOI] [PubMed] [Google Scholar]
- 87. Yeh C.-H., Lin P.-W., and Lin Y.-C., Microfluid. Nanofluid. 8, 115 (2010). 10.1007/s10404-009-0485-7 [DOI] [Google Scholar]
- 88. Jeong K. J., Wang L., Stefanescu C. F., Lawlor M. W., Polat J., Dohlman C. H., Langer R. S., and Kohane D. S., Soft Matter 7, 8305 (2011). 10.1039/c1sm05918a [DOI] [Google Scholar]
- 89. Sadr N., Zhu M., Osaki T., Kakegawa T., Yang Y., Moretti M., Fukuda J., and Khademhosseini A., Biomaterials 32, 7479 (2011). 10.1016/j.biomaterials.2011.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Huang G., Wang S., He X., Zhang X., Lu T. J., and Xu F., Biotechnol. Bioeng. 110, 980 (2013). 10.1002/bit.24764 [DOI] [PubMed] [Google Scholar]
- 91. Cha K. J., Hong J. M., Cho D., and Kim D. S., Biofabrication 5, 025007 (2013). 10.1088/1758-5082/5/2/025007 [DOI] [PubMed] [Google Scholar]
- 92. Lucchetta G., Sorgato M., Zanchetta E., Brusatin G., Guidi E., Di Liddo R., and Conconi M. T., Express Polym. Lett. 9, 354 (2015). 10.3144/expresspolymlett.2015.33 [DOI] [Google Scholar]
- 93. Sivashankar S., Puttaswamy S. V., Lin L. H., Dai T. S., Yeh C. T., and Liu C. H., Sens. Actuators, B 176, 1081 (2013). 10.1016/j.snb.2012.09.087 [DOI] [Google Scholar]
- 94. Altmann B., Löchner A., Swain M., Kohal R., Giselbrecht S., Gottwald E., Steinberg T., and Tomakidi P., Biomaterials 35, 3208 (2014). 10.1016/j.biomaterials.2013.12.088 [DOI] [PubMed] [Google Scholar]
- 95. Brazda N., Voss C., Estrada V., Lodin H., Weinrich N., Seide K., Müller J., and Müller H. W., Biomaterials 34, 10056 (2013). 10.1016/j.biomaterials.2013.09.057 [DOI] [PubMed] [Google Scholar]
- 96. O'Brien P. J., Luo W., Rogozhnikov D., Chen J., and Yousaf M. N., Bioconjugate Chem. 26, 1939 (2015). 10.1021/acs.bioconjchem.5b00376 [DOI] [PubMed] [Google Scholar]
- 97. Bersini S., Gilardi M., Arrigoni C., Tal?? G., Zamai M., Zagra L., Caiolfa V., and Moretti M., Biomaterials 76, 157 (2016). 10.1016/j.biomaterials.2015.10.057 [DOI] [PubMed] [Google Scholar]
- 98. Young E. W. K., Berthier E., Guckenberger D. J., Sackmann E., Lamers C., Meyvantsson I., Huttenlocher A., and Beebe D. J., Anal. Chem. 83, 1408 (2011). 10.1021/ac102897h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jeon J. S., Chung S., Kamm R. D., and Charest J. L., Biomed. Microdevices 13, 325 (2011). 10.1007/s10544-010-9496-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Raasch M., Rennert K., Jahn T., Peters S., Henkel T., Huber O., Schulz I., Becker H., Lorkowski S., Funke H., and Mosig A., Biofabrication 7, 015013 (2015). 10.1088/1758-5090/7/1/015013 [DOI] [PubMed] [Google Scholar]
- 101. Miller P. G. and Shuler M. L., Biotechnol. Bioeng. 113, 2213 (2016). 10.1002/bit.25989 [DOI] [PubMed] [Google Scholar]
- 102. Sakai Y., Yoshiura Y., and Nakazawa K., J. Biosci. Bioeng. 111, 85 (2011). 10.1016/j.jbiosc.2010.08.014 [DOI] [PubMed] [Google Scholar]
- 103. Sakai Y. and Nakazawa K., J. Biochip Tissue Chip S4, 3 (2011) 10.4172/2153-0777.S4-001. [DOI] [Google Scholar]
- 104. Uhlig K., Wegener T., He J., Zeiser M., Bookhold J., Dewald I., Godino N., Jaeger M., Hellweg T., Fery A., and Duschl C., Biomacromolecules 17, 1110 (2016). 10.1021/acs.biomac.5b01728 [DOI] [PubMed] [Google Scholar]
- 105. Cha K. J., Lim J., Na M., and Sung D., Microelectron. Eng. 158, 11 (2016). 10.1016/j.mee.2016.02.034 [DOI] [Google Scholar]
- 106. Cha K. J., Na M., Kim H. W., and Kim D. S., J. Micromech. Microeng. 24, 055002 (2014). 10.1088/0960-1317/24/5/055002 [DOI] [Google Scholar]
- 107. Tomlinson M. J., Tomlinson S., Yang X. B., and Kirkham J., J. Tissue Eng. 4 (2013) 10.1177/2041731412472690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gross A., Schoendube J., Zimmermann S., Steeb M., Zengerle R., and Koltay P., Int. J. Mol. Sci. 16, 16897 (2015). 10.3390/ijms160816897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chung Y.-C., Jan M.-S., Lin Y.-C., Lin J.-H., Cheng W.-C., and Fan C.-Y., Lab Chip 4, 141 (2004). 10.1039/b310849j [DOI] [PubMed] [Google Scholar]
- 110. Chung Y. C., Wen B. J., and Lin Y. C., Control Eng. Pract. 15, 1093 (2007). 10.1016/j.conengprac.2007.01.005 [DOI] [Google Scholar]
- 111. Geissler M., Beauregard J. A., Charlebois I., Isabel S., Normandin F., Voisin B., Boissinot M., Bergeron M. G., and Veres T., Eng. Life Sci. 11, 174 (2011). 10.1002/elsc.201000132 [DOI] [Google Scholar]
- 112. Reinholt S. J., Behrent A., Greene C., Kalfe A., and Baeumner A. J., Anal. Chem. 86, 849 (2014). 10.1021/ac403417z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Koonin E. V., Senkevich T. G., and Dolja V. V., Biol. Direct 1:29 (2006) 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Bhattacharyya A. and Klapperich C. M., Sens. Actuators, B 129, 693 (2008). 10.1016/j.snb.2007.09.057 [DOI] [Google Scholar]
- 115. Chung S. H., Baek C., Cong V. T., and Min J., Biosens. Bioelectron. 67, 625 (2015). 10.1016/j.bios.2014.09.083 [DOI] [PubMed] [Google Scholar]
- 116. Chow K. S. and Du H., Sens. Actuators, A 170, 24 (2011). 10.1016/j.sna.2011.03.053 [DOI] [Google Scholar]
- 117. Ngamsom B., Esfahani M. M. N., Phurimsak C., Lopez-Martinez M. J., Raymond J. C., Broyer P., Patel P., and Pamme N., Anal. Chim. Acta 918, 69 (2016). 10.1016/j.aca.2016.03.014 [DOI] [PubMed] [Google Scholar]
- 118. Ma Z., Zheng Y., Cheng Y., Xie S., Ye X., and Yao M., J. Aerosol Sci. 95, 84 (2016). 10.1016/j.jaerosci.2016.01.003 [DOI] [Google Scholar]
- 119. Christensen T. B., Pedersen C. M., Bang D. D., and Wolff A., Microelectron. Eng. 84, 1690 (2007). 10.1016/j.mee.2007.01.236 [DOI] [Google Scholar]
- 120. Tachibana H., Saito M., Shibuya S., Tsuji K., Miyagawa N., Yamanaka K., and Tamiya E., Biosens. Bioelectron. 74, 725 (2015). 10.1016/j.bios.2015.07.009 [DOI] [PubMed] [Google Scholar]
- 121. Jimenez M., Miller B., and Bridle H. L., Chem. Eng. Sci. 157, 247 (2017). 10.1016/j.ces.2015.08.042 [DOI] [Google Scholar]
- 122. Dean L., Blood Groups Red Cell Antigens ( National Center for Biotechnology Information, 2005), Vol. 1, Chap. 1, see https://www.ncbi.nlm.nih.gov/books/NBK2261/. [Google Scholar]
- 123. Auerswald J. and Knapp H. F., Microelectron. Eng. 67–68, 879 (2003). 10.1016/S0167-9317(03)00150-3 [DOI] [Google Scholar]
- 124. Carugo D., Capretto L., Nehru E., Mansour M., Smyth N., Bressloff N., and Zhang X., Curr. Anal. Chem. 9, 47 (2013). 10.2174/157341113804486437 [DOI] [Google Scholar]
- 125. Baday M., Calamak S., Durmus N. G., Davis R. W., Steinmetz L. M., and Demirci U., Small 12, 1222 (2016). 10.1002/smll.201501845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Bhuvanendran Nair Gourikutty S., Chang C. P., and Puiu P. D., J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 1011, 77 (2016). 10.1016/j.jchromb.2015.12.016 [DOI] [PubMed] [Google Scholar]
- 127. Chen X., Cui D., Liu C., Li H., and Chen J., Anal. Chim. Acta 584, 237 (2007). 10.1016/j.aca.2006.11.057 [DOI] [PubMed] [Google Scholar]
- 128. Chen J.-Y., Huang Y.-T., Chou H.-H., Wang C.-P., and Chen C.-F., Lab Chip 15, 4533 (2015). 10.1039/C5LC01172H [DOI] [PubMed] [Google Scholar]
- 129. Adams A., Okagbare P. I., Feng J., Hupert M. L., Patterson D., Göttert J., McCarley R. L., Nikitopoulos D., Murphy M. C., and Soper S. A., J. Am. Chem. Soc. 130, 8633 (2008). 10.1021/ja8015022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Park T., Park D. S., and Murphy M. C., in ASME 2011 International Mechanical Engineering Congress and Exposition (2011), Vol. 2, p. 1185. [Google Scholar]
- 131. Chen K. C., Pan Y. C., Chen C. L., Lin C. H., Huang C. S., and Wo A. M., Anal. Biochem. 429, 116 (2012). 10.1016/j.ab.2012.07.007 [DOI] [PubMed] [Google Scholar]
- 132. Kamande J. W., Hupert M. L., Witek M. A., Wang H., Torphy R. J., Dharmasiri U., Njoroge S. K., Jackson J. M., Aufforth R. D., Snavely A., Yeh J. J., and Soper S. A., Anal. Chem. 85, 9092 (2013). 10.1021/ac401720k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Jackson J. M., Witek M. A., Hupert M. L., Brady C., Pullagurla S., Kamande J., Aufforth R. D., Tignanelli C. J., Torphy R. J., Yeh J. J., and Soper S. A., Lab Chip 14, 106 (2014). 10.1039/C3LC50618E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hyun K., Yoon T., Hyun S., and Jung H., Biosens. Bioelectron. 67, 86 (2015). 10.1016/j.bios.2014.07.019 [DOI] [PubMed] [Google Scholar]
- 135. Bhuvanendran Nair Gourikutty S., Chang C. P., and Poenar D. P., J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 1028, 153 (2016). 10.1016/j.jchromb.2016.06.016 [DOI] [PubMed] [Google Scholar]
- 136. Khoshmanesh K., Akagi J., Nahavandi S., Kalantar-Zadeh K., Baratchi S., Williams D. E., Cooper J. M., and Wlodkowic D., Anal. Chem. 83, 3217 (2011). 10.1021/ac2002142 [DOI] [PubMed] [Google Scholar]
- 137. Nwankire C. E., Venkatanarayanan A., Glennon T., Keyes T. E., Forster R. J., and Ducrée J., Biosens. Bioelectron. 68, 382 (2015). 10.1016/j.bios.2014.12.049 [DOI] [PubMed] [Google Scholar]
- 138. Lee A., Park J., Lim M., Sunkara V., Kim S. Y., Kim G. H., Kim M. H., and Cho Y. K., Anal. Chem. 86, 11349 (2014). 10.1021/ac5035049 [DOI] [PubMed] [Google Scholar]
- 139. Asghar W., Velasco V., Kingsley J. L., Shoukat M. S., Shafiee H., Anchan R. M., Mutter G. L., Tüzel E., and Demirci U., Adv. Healthcare Mater. 3, 1671 (2014). 10.1002/adhm.201400058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zhang X., Khimji I., Gurkan U. A., Safaee H., Catalano P. N., Keles H. O., Kayaalp E., and Demirci U., Lab Chip 11, 2535 (2011). 10.1039/c1lc20236g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Matsuura K., Takenami M., Kuroda Y., Hyakutake T., Yanase S., and Naruse K., Reprod. Biomed. Online 24, 109 (2012). 10.1016/j.rbmo.2011.09.005 [DOI] [PubMed] [Google Scholar]
- 142. Wainright A., Williams S. J., Ciambrone G., Xue Q., Wei J., and Harris D., J. Chromatogr. A 979, 69 (2002). 10.1016/S0021-9673(02)01246-3 [DOI] [PubMed] [Google Scholar]
- 143. Archana M., Yogesh T. L., and Kumaraswamy K. L., Indian J. Cancer 50, 274 (2013). 10.4103/0019-509X.118720 [DOI] [PubMed] [Google Scholar]
- 144. Pantel K. and Alix-Panabières C., J. Thorac. Dis. 4, 446 (2012) 10.3978/j.issn.2072-1439.2012.08.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Yang J. M., Bell J., Huang Y., Tirado M., Thomas D., Forster A. H., Haigis R. W., Swanson P. D., Wallace R. B., Martinsons B., and Krihak M., Biosens. Bioelectron. 17, 605 (2002). 10.1016/S0956-5663(02)00023-4 [DOI] [PubMed] [Google Scholar]
- 146. Witek M. A., Wei S., Vaidya B., Adams A. A., Zhu L., Stryjewski W., McCarley R. L., and Soper S. A., Lab Chip 4, 464 (2004). 10.1039/b317093d [DOI] [PubMed] [Google Scholar]
- 147. Jezierski S., Klein A. S., Benz C., Schaefer M., Nagl S., and Belder D., Anal. Bioanal. Chem. 405, 5381 (2013). 10.1007/s00216-013-6945-7 [DOI] [PubMed] [Google Scholar]
- 148. Floriano P. N., Christodoulides N., Romanovicz D., Bernard B., Simmons G. W., Cavell M., and McDevitt J. T., Biosens. Bioelectron. 20, 2079 (2005). 10.1016/j.bios.2004.08.046 [DOI] [PubMed] [Google Scholar]
- 149. Bayoudh S., Othmane A., Ponsonnet L., and Ben Ouada H., Colloids Surf. A 318, 291 (2008). 10.1016/j.colsurfa.2008.01.005 [DOI] [PubMed] [Google Scholar]
- 150. Lee J., Soper S. a., and Murray K. K., Analyst 134, 2426 (2009). 10.1039/b916556h [DOI] [PubMed] [Google Scholar]
- 151. Pujol-Vila F., Giménez-Gómez P., Santamaria N., Antúnez B., Vigués N., Díaz-González M., Jiménez-Jorquera C., Mas J., Sacristán J., and Muñoz-Berbel X., Sens. Actuators, B 222, 55 (2016). 10.1016/j.snb.2015.07.095 [DOI] [Google Scholar]
- 152. Pandya H. J., Kanakasabapathy M. K., Verma S., Chug M. K., Memic A., Gadjeva M., and Shafiee H., Biosens. Bioelectron. 91, 32 (2017). 10.1016/j.bios.2016.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Lin Y.-C. and Huang M.-Y., J. Micromech. Microeng. 11, 542 (2001). 10.1088/0960-1317/11/5/315 [DOI] [Google Scholar]
- 154. Kummrow A., Theisen J., Frankowski M., Tuchscheerer A., Yildirim H., Brattke K., Schmidt M., and Neukammer J., Lab Chip 9, 972 (2009). 10.1039/b808336c [DOI] [PubMed] [Google Scholar]
- 155. Laczka O., Maesa J. M., Godino N., del Campo J., Fougt-Hansen M., Kutter J. P., Snakenborg D., Muñoz-Pascual F. X., and Baldrich E., Biosens. Bioelectron. 26, 3633 (2011). 10.1016/j.bios.2011.02.019 [DOI] [PubMed] [Google Scholar]
- 156. Lok K. S., Kwok Y. C., Lee P. P. F., and Nguyen N. T., Sens. Actuators, B 166–167, 893 (2012). 10.1016/j.snb.2012.03.001 [DOI] [Google Scholar]
- 157. Verbarg J., Kamgar-Parsi K., Shields A. R., Howell P. B., and Ligler F. S., Lab Chip 12, 1793 (2012). 10.1039/c2lc21189k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Verbarg J., Plath W. D., Shriver-Lake L. C., Howell P. B., Erickson J. S., Golden J. P., and Ligler F. S., Anal. Chem. 85, 4944 (2013). 10.1021/ac303801v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Golden J. P., Verbarg J., Howell P. B., Shriver-Lake L. C., and Ligler F. S., Biosens. Bioelectron. 40, 10 (2013). 10.1016/j.bios.2012.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Rothert A., Deo S. K., Millner L., Puckett L. G., Madou M. J., and Daunert S., Anal. Biochem. 342, 11 (2005). 10.1016/j.ab.2004.10.048 [DOI] [PubMed] [Google Scholar]
- 161. Date A., Pasini P., and Daunert S., Anal. Bioanal. Chem. 398, 349 (2010). 10.1007/s00216-010-3930-2 [DOI] [PubMed] [Google Scholar]
- 162. Huang Y., Persoone G., Nugegoda D., and Wlodkowic D., Sens. Actuators, B 226, 289 (2016). 10.1016/j.snb.2015.11.128 [DOI] [Google Scholar]
- 163. Cartlidge R., Nugegoda D., and Wlodkowic D., Sens. Actuators, B 239, 660 (2017). 10.1016/j.snb.2016.08.058 [DOI] [Google Scholar]
- 164. Tao S. L., Lubeley M. W., Desai T. A., and Contorl J., Release 88, 215 (2003). 10.1016/S0168-3659(03)00005-1 [DOI] [PubMed] [Google Scholar]
- 165. Ivanova E. P., Truong V. K., Gervinskas G., Mitik-Dineva N., Day D., Jones R. T., Crawford R. J., and Juodkazis S., Biosens. Bioelectron. 35, 369 (2012). 10.1016/j.bios.2012.03.019 [DOI] [PubMed] [Google Scholar]
- 166. Leahy S. and Lai Y., Sens. Actuators, B 238, 292 (2017). 10.1016/j.snb.2016.07.069 [DOI] [Google Scholar]
- 167. Seo J. H., Park B. H., Oh S. J., Choi G., Kim D. H., Lee E. Y., and Seo T. S., Sens. Actuators, B 246, 146 (2017). 10.1016/j.snb.2017.02.051 [DOI] [Google Scholar]
- 168. Lin H. Y., Huang C. H., Lu S. H., Kuo I. T., and Chau L. K., Biosens. Bioelectron. 51, 371 (2014). 10.1016/j.bios.2013.08.009 [DOI] [PubMed] [Google Scholar]
- 169. Aeinehvand M. M., Ibrahim F., Harun S. W., Djordjevic I., Hosseini S., Rothan H. A., Yusof R., and Madou M. J., Biosens. Bioelectron. 67, 424 (2015). 10.1016/j.bios.2014.08.076 [DOI] [PubMed] [Google Scholar]
- 170. Morant-Miñana M. C. and Elizalde J., Biosens. Bioelectron. 70, 491 (2015). 10.1016/j.bios.2015.03.063 [DOI] [PubMed] [Google Scholar]
- 171. Dong T. and Zhao X., Anal. Chem. 87, 2410 (2015). 10.1021/ac504428t [DOI] [PubMed] [Google Scholar]
- 172. Madhusudana S. N., Sundaramoorthy S., and Ullas P. T., Int. J. Infect. Dis. 14, e1067 (2010). 10.1016/j.ijid.2010.07.004 [DOI] [PubMed] [Google Scholar]
- 173. Wang Z., Uno H., Takada N., and Kumar O., 16th International Conference on Miniaturized Systems for Chemistry and Life Sciences 28 October–1 November, Okinawa, Japan (2012), pp. 1033. [Google Scholar]
- 174. Zhang X., Wasserberg D., Breukers C., Terstappen L. W. M. M., and Beck M., ACS Appl. Mater. Interfaces 8, 27539 (2016). 10.1021/acsami.6b09206 [DOI] [PubMed] [Google Scholar]
- 175. Works H. I., Natl. Cent. Biotechnol. Inf. (2017), see https://www.ncbi.nlm.nih.gov/probe/docs/techpcr/.
- 176. Tsai N. C. and Sue C. Y., Biosens. Bioelectron. 22, 313 (2006). 10.1016/j.bios.2005.12.023 [DOI] [PubMed] [Google Scholar]
- 177. Lee S. H., Ou M. C., Chen T. Y., and Lee G. B., Solid-State Sensors, Actuators and Microsystems Conference (2007), p. 943. [Google Scholar]
- 178. Urrego L. F., Lopez D. I., Ramirez K. A., Ramirez C., and Osma J. F., Sens. Actuators, B 207, 97 (2015). 10.1016/j.snb.2014.10.036 [DOI] [Google Scholar]
- 179. Nagai H. and Fuchiwaki Y., Electron. Commun. Jpn. 98, 1 (2015). 10.1002/ecj.11753 [DOI] [Google Scholar]
- 180. Koh C. G., Tan W., Zhao M., Ricco A. J., and Fan Z. H., Anal. Chem. 75, 4591 (2003). 10.1021/ac0343836 [DOI] [PubMed] [Google Scholar]
- 181. Girard M. P., Osmanov S., Assossou O. M., and Kieny M.-P., Vaccine 29, 6191 (2011). 10.1016/j.vaccine.2011.06.085 [DOI] [PubMed] [Google Scholar]
- 182. Kim Y. G., Moon S., Kuritzkes D. R., and Demirci U., Biosens. Bioelectron. 25, 253 (2009). 10.1016/j.bios.2009.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Moon S. and Keles H., Biosens. Bioelectron. 24, 3208 (2009). 10.1016/j.bios.2009.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Climent M. Á., Torregrosa M. D., Vázquez S., Gironés R., and Arranz J. A., Cancer Treat. Rev. 55, 173 (2017). 10.1016/j.ctrv.2017.03.007 [DOI] [PubMed] [Google Scholar]
- 185. Weigum S. E., Floriano P. N., Christodoulides N., and McDevitt J. T., Lab Chip 7, 995 (2007). 10.1039/b703918b [DOI] [PubMed] [Google Scholar]
- 186. Fok S., Domachuk P., Rosengarten G., Krause N., Braet F., Eggleton B. J., and Soon L. L., Biophys. J. 95, 1523 (2008). 10.1529/biophysj.107.115246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Dharmasiri U., Balamurugan S., Adams A. A., Okagbare P. I., Obubuafo A., and Soper S. A., Electrophoresis 30, 3289 (2009). 10.1002/elps.200900141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Akagi J., Takeda K., Fujimura Y., Matuszek A., Khoshmanesh K., and Wlodkowic D., Procedia Eng. 47, 88 (2012). 10.1016/j.proeng.2012.09.091 [DOI] [Google Scholar]
- 189. Dharmasiri U., Njoroge S. K., Witek M. A., Adebiyi M. G., Kamande J. W., Hupert M. L., and Barany F., Anal. Chem. 83, 2301 (2011). 10.1021/ac103172y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Kumeria T., Kurkuri M. D., Diener K. R., Parkinson L., and Losic D., Biosens. Bioelectron. 35, 167 (2012). 10.1016/j.bios.2012.02.038 [DOI] [PubMed] [Google Scholar]
- 191. Minn A. J. and Wherry E. J., Cell 165, 272 (2016). 10.1016/j.cell.2016.03.031 [DOI] [PubMed] [Google Scholar]
- 192. Matteucci M., Lakshmikanth T., Shibu K., De Angelis F., Schadendorf D., Venuta S., Carbone E., and Di Fabrizio E., Microelectron. Eng. 84, 1729 (2007). 10.1016/j.mee.2007.01.242 [DOI] [Google Scholar]
- 193. Mottet G., Perez-Toralla K., Tulukcuoglu E., Bidard F. C., Pierga J. Y., Londono-Vallejo A., Descroix S., Malaquin L., and Viovy J. L., Biomicrofluidics 8, 024109 (2014). 10.1063/1.4871035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Han X., Van Berkel C., Gwyer J., Capretto L., and Morgan H., Anal. Chem. 84, 1070 (2012). 10.1021/ac202700x [DOI] [PubMed] [Google Scholar]
- 195. Memmolo P., Miccio L., Merola F., Gennari O., Netti P. A., and Ferraro P., Cytometry Part A 85, 1030 (2014). 10.1002/cyto.a.22570 [DOI] [PubMed] [Google Scholar]
- 196. Alapan Y., Kim C., Adhikari A., Gray K. E., Gurkan-Cavusoglu E., Little J. A., and Gurkan U. A., Transl. Res. 173, 74 (2016). 10.1016/j.trsl.2016.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Logan B. E., Hamelers B., Rozendal R., Schröder U., Keller J., Freguia S., Aelterman P., Versraete W., and Rabaey K., Environ. Sci. Technol. 40, 5181 (2006). 10.1021/es0605016 [DOI] [PubMed] [Google Scholar]
- 198. Rahimnejad M., Adhami A., Darvari S., Zirepour A., and Oh S.-E., Alexandria Eng. J. 54, 745 (2015). 10.1016/j.aej.2015.03.031 [DOI] [Google Scholar]
- 199. Chaturvedi V. and Verma P., Bioresour. Bioprocess. 3, 38 (2016). 10.1186/s40643-016-0116-6 [DOI] [Google Scholar]
- 200. Chen Y. P., Zhao Y., Qiu K. Q., Chu J., Lu R., Sun M., Liu X. W., Sheng G. P., Yu H. Q., Chen J., Li W. J., Liu G., Tian Y. C., and Xiong Y., Biosens. Bioelectron. 26, 2841 (2011). 10.1016/j.bios.2010.11.016 [DOI] [PubMed] [Google Scholar]
- 201. Mukherjee S., Su S., Panmanee W., Irvin R. T., Hassett D. J., and Choi S., Sens. Actuators, A 201, 532 (2013). 10.1016/j.sna.2012.10.025 [DOI] [Google Scholar]
- 202. Mardanpour M. M. and Yaghmaei S., Biosens. Bioelectron. 79, 327 (2016). 10.1016/j.bios.2015.12.022 [DOI] [PubMed] [Google Scholar]
- 203. Wei X., Lee H., and Choi S., Sens. Actuators, B 228, 151 (2016). 10.1016/j.snb.2015.12.103 [DOI] [Google Scholar]
- 204. Mardanpour M. M. and Yaghmaei S., Electrochim. Acta 227, 317 (2017). 10.1016/j.electacta.2017.01.041 [DOI] [Google Scholar]
- 205. Yang W., Lee K. K., and Choi S., Sens. Actuators, B 243, 292 (2017). 10.1016/j.snb.2016.11.155 [DOI] [Google Scholar]
- 206.Yole Development Status of the Microfluidic Industry, Market & Technology Report No. May, 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for the tabulated versions of the Separation/Isolation and Detection and Analysis sections.
The Separation/Isolation summary table includes the device format, microorganisms, materials used for device fabrication, fabrication techniques, separation criteria, remarks on the study, and reference parts.
The Detection and Analysis table includes the device format, microorganisms, materials used for device fabrication, fabrication techniques, detection criteria, remarks on the study, and reference parts.