Abstract
Background
Age-related macular degeneration (AMD) is one of the most common causes of adult blindness in developed countries. However, the role of long noncoding RNAs (lncRNAs) in the development and progression of early AMD is unclear.
Methods
We established the lncRNA profile of early AMD by reannotation of microarrays from the gene expression omnibus database. Quantitative real-time polymerase chain reaction was used to determine the expression of selected lncRNAs.
Results
The expression profiles of 9 cases of AMD and 7 controls were studied. A total of 266 differentially expressed genes (DEGs) were detected (94 upregulated and 172 downregulated). Among all the DEGs, 64 were lncRNAs. Advanced bioinformatics analyses demonstrated that differentially expressed lncRNAs could play significant roles in visual perception, sensory perception of light stimulus, and cognition. The pathway analyses showed that the two most significantly influenced Kyoto Encyclopedia of Genes and Genomes pathways were those of phototransduction and purine metabolism. By the analyses of the key lncRNAs, it was found that RP11-234O6.2 was downregulated in the aging retinal pigment epithelium (RPE) cellular model. Exogenous RP11-234O6.2 treatment led to increased cell viability and improved apoptosis but it did not affect the cell migration ability of aging RPE cells.
Conclusion
This study indicated that lncRNAs are differentially expressed in early AMD and may produce important regulative effects. An lncRNA, RP11-234O6.2, might be involved in the biological regulation of early AMD and have therapeutic potential.
Keywords: age-related macular degeneration, lncRNAs, microarray, bioinformatics analyses
Introduction
Age-related macular degeneration (AMD) is an age-related multifactorial disease that is regarded as the most common cause of adult blindness in many developed countries.1 It is the primary cause of loss of central vision and significantly decreases the individual’s quality of life. In general, AMD is divided into early and late stages. Late AMD is regarded as neovascular AMD characterized by neovascularization from the choriocapillaris.2 At present, anti-vascular endothelial growth factor is used to treat neovascular AMD and has been shown to benefit patients with AMD. Considering the increasing prevalence of the elderly in the population, the incidence rate of AMD is predicated to be higher in the future.3 Despite the fact that significant efforts have been made to identify risk factors of AMD, currently, there are no recognized methods to prevent it.4 For these reasons, there is an urgent need to develop effective interventions to prevent the progression of early AMD to late AMD. Except for smoking cessation and avoidance of cigarette smoke exposure, effective treatment options for early AMD cases are absent.5 Thus, more knowledge is required on the pathogenesis of AMD; especially key would be insights into the pathophysiology of early AMD.
Given the aging population and the multifactorial nature of AMD, currently, no exact and single etiology of AMD has been identified. However, there are several fairly well-accepted molecular mechanisms that have been reported to be involved in the development and progression of AMD, including inflammation, oxidative damage, and abnormal angiogenesis. Interventions for these potential key factors for its pathogenesis may produce rational treatments for AMD. For instance, polyunsaturated fatty acid, which is both an antioxidant nutritional supplement and an anti-inflammatory agent, has been reported to provide significant benefits in AMD management.6,7 More advanced clinical and laboratory studies are now needed to characterize the detailed pathogenesis and to develop effective therapeutic agents for AMD targeting on the key pathways.
Long noncoding RNAs (lncRNAs), which are a class of noncoding RNAs with more than 200 nucleotides, play critical roles in the development of different kinds of human diseases.8 As reported in the previous studies, lncRNAs can significantly affect epigenetic modification, cell differentiation, transcriptional regulation, immune responses, and other biological processes.9 In a previous study on the murine model of ocular neovascularization by microarray analyses, it was found that lncRNAs were significantly dysregulated.10 The results from advanced bioinformatics analyses suggested that lncRNAs play important roles in ocular neovascularization. However, no previous studies have focused on the role of lncRNAs in early AMD. Considering the extensive effect of lncRNAs on physiological function,11 it is reasonable to think that lncRNAs can significantly influence the development and progression of early AMD. In the present study, we identified a set of lncRNAs that were differentially expressed in early AMD, confirmed the expression patterns of selected lncRNAs in an in vitro model, and performed a preliminary investigation into the biological effects of dysregulated lncRNA on AMD development.
Materials and methods
Data sets
Expression profiles of retinal pigment epithelium (RPE)/choroid from AMD cases and controls were downloaded from the gene expression omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo).12 All of the profiles were from participants from the Department of Ophthalmology and Visual Sciences, the University of Iowa. A total of 9 AMD cases and 7 controls were included in the data analyses. Early AMD in this study was defined as RPE changes (depigmentation or hypo pigmentation) and/or macular drusen without geographic atrophy or choroidal neovascularization. A total of 5 females and 4 males were included in the AMD group, while there were 3 females and 4 males in the control group (P=0.500). The age distribution in the AMD (84.22±6.553) and control (83.00±5.657) groups was matched in this study (P=0.873). As complement factor H (CFH) was regarded as one of the most important genetic risk factors, the single nucleotide polymorphism rs1061170 of CFH was assessed. It was found that the distribution of high-risk genotypes (YH/HH) and low-risk genotypes (YY) was matched between the case and control groups (P=0.596). An exon-based array (the Affymetrix Human Exon ST 1.0 array) was obtained for the microarray analysis.
Data processing and probe reannotation
Microarray re-annotations were divided into two processes: detection of differentially expressed genes (DEGs) and differentially expressed lncRNAs. For the detection of DEGs, the downloaded cell data of GSE50195 based on GPL17629 were added to the information of protein classes (protein_coding, lncRNA, and miRNA). The linear models for microarray data (LIMMA) package was obtained for bioinformatics analyses using R and Bioconductor.13 All the DEGs were presented as both heatmap and volcano plots. In order to extract the lncRNAs from the DEGs, the lncRNA label in protein classes was chosen, and all the differentially expressed lncRNAs were presented in an independent heatmap and were used for advanced functional analyses.
Construction of lncRNA–mRNA coexpression network
To date, the exact biological roles of most lncRNAs are still unclear, and it was reasonable to conduct the bioinformatics functional analyses of the coexpressed mRNAs of related lncRNAs. Functionally related genes generally demonstrated similar expression profiles, and this suggests that these genes may have regulatory relationships. In this study, weighted correlation network analysis14 was used to identify all the DEGs (adjacency threshold =0.6). Among all the coexpression nodes, we selected the nodes that demonstrated a coexpression relationship between lncRNA and mRNA and thus constructed an lncRNA–mRNA coexpression network. In order to verify the lncRNA–mRNA coexpression results, advanced validation analyses by sequencing results were conducted by using RIsearch analysis software;15 this software could detect the interaction between the selected lncRNAs and all coding genes (free energy ≤20). The lncRNA and mRNA pairs, which demonstrated both a coexpression relationship and interactions, were then recorded as an extreme lncRNA–mRNA coexpression network, and a graphical output was obtained by using Cytoscape.16
Bioinformatics analyses
The lncRNA-coexpressed mRNAs were used in the functional enrichment analysis. In order to investigate the potential biological role of related mRNAs, the analyses were conducted by using the Database for Annotation, Visualization and Integrated Discovery (DAVID) for the annotation and functional analysis.17 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were also conducted to show gene set enrichment analysis. In the current study, the clusterProfiler R package was used to conduct the pathway analyses of the DEGs,18 and the Pathview was used to visualize the results of the enrichment pathway analyses.19
Cell culture and the in vitro aging model
Cultured retinal pigmented epithelium (ARPE-19) cells were purchased from American Type Culture Collection (Rockville, MD, USA). The ARPE-19 cells were grown in Dulbecco’s Modified Eagle’s Medium nutrient mixture F-12 (DMEM/F12; Invitrogen Corporation, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. The ARPE-19 cells were used at 80% confluence for each experiment and were passaged every 3 days after being transferred into the culture medium. All the cells were incubated at 37°C in a humidified atmosphere of 5% CO2.
As reported in our previous study, a pulsed H2O2 exposure aging cell model was obtained in this study.20 In general, the cultured ARPE-19 cells were incubated with complete medium containing 800 µM H2O2 for 2 hours every day, and the aging ARPE-19 cells on the 8th day could be used as aging groups.
RNA extraction and quantitative polymerase chain reaction (PCR)
Total RNA was isolated from the ARPE-19 cells by using TRIzol® reagent according to the manufacturer’s instructions (Invitrogen). The concentration and quality of the extracted RNA were measured by using a NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Quantitative PCRs were conducted in a 20 µL reaction volume using an SYBR Green mix. Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control. The fold changes of all RNA expression levels were calculated by using the 2−ΔΔCt method.
Vector construction and transfection
The full-length sequence of RP11-234O6.2 was synthesized and cloned into the pcDNA3.1 (Invitrogen, Shanghai, China) vector according to the manufacturer’s instructions. The transfection of pcDNA3.1–RP11-234O6.2 into ARPE-19 cells was done by using Lipofectamine 2000 (Invitrogen) at a final concentration of 50 nM. The relative expression levels of RP11-234O6.2 were detected by quantitative PCR.
Cell viability assay
Cell viability was monitored by using the Cell Counting Kit 8 (CCK-8; Dojindo, Shanghai, China). The ARPE-19 cells in each group were seeded onto 96-well plates at an initial density of 5,000 cells/well. Then, 10 µL of CCK-8 solution was added to each well, and they were incubated for 2 hours. Cell viability was measured at a wavelength of 450 nm (SpectraMax® Plus 384 Microplate Reader; Molecular Devices, Sunnyvale, CA, USA). The data were calculated using the following formula: relative viability = A450(target) − A450(blank).
Cell migration detection
The impact of lncRNA on aging ARPE-19 cell migration was examined by the cell scratch test. ARPE-19 cells in each group were placed on poly-D-lysine-coated 12-well plates (Corning Inc., Corning, NY, USA) with a cell density of 2.5×105 cells/well in DMEM/F2 1:1 culture media without FBS. A 200-µL pipette tip was used to perform scratches, and all the processed cells were divided into control and lncRNA-treated groups. After 36 hours, distances from the initial scratches were measured. Each detection test was repeated with three independent experiments.
Apoptosis detection
In this study, the apoptosis level was detected by using a flow cytometry method with an annexin V-fluorescein isothiocyanate apoptosis detection kit. The cultured cells in each group were harvested and washed with ice-cold PBS for 2 hours. All the cells were then treated with ice-cold 70% ethanol and then resuspended in 1 mL PBS. Annexin-V fluorescein isothiocyanate and propidium iodide (PI) solutions were added and incubated in the dark for 20 minutes. The results were expressed as the percentage of both early (annexin V+/PI−) and late apoptosis (annexin V+/PI+) cells.
Statistical analysis
The results were expressed as means ± SD of the three repeated experiments. The difference between the means in each index was calculated by using the Student’s t-test. A p-value of <0.05 was considered statistically significant. All the statistical analyses were conducted by using Prism GraphPad 5 software (GraphPad Software Inc., La Jolla, CA, USA).
Results
DEGs and dysregulated lncRNAs
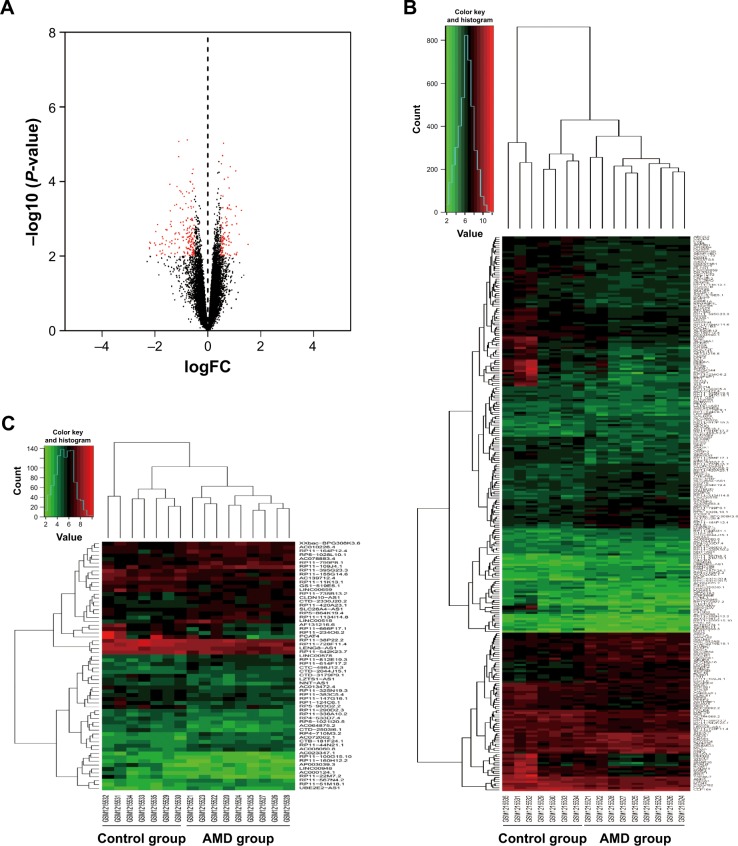
From the GEO database, the cell files of GSE50195 were downloaded for advanced probe reannotation. Using LIMMA with a P-value <0.01 and |logFC| >0.5 as the threshold, a total of 266 DEGs were identified (94 upregulated and 172 downregulated). As shown in Figure 1A, the DEGs were marked red in the volcano plot. Cluster analysis for all the DEGs was conducted, and the expression levels were marked from red to blue (Figure 1B). In advanced analyses, it was found that a total of 64 lncRNAs were differentially expressed when comparing early AMD cases with normal controls. Among the 64 detected lncRNAs, 36 lncRNAs were downregulated and 28 lncRNAs were upregulated. Figure 1C shows the heatmap plot of cluster analysis for the 64 differentially expressed lncRNAs. Table 1 lists the top 20 differentially expressed lncRNAs. The gene ID and symbol as well as P-values were also recorded.
Figure 1.
Differentially expressed genes and lncRNAs of early AMD cases (9 cases versus 8 controls).
Notes: (A) Volcano plot of differentially expressed genes for early AMD. The red points are marked as genes of significance. (B) Clustering heatmap of 266 differentially expressed genes. (C) Clustering heatmap of 64 differentially expressed lncRNAs. AMD group (GSM1215520–GSM1215528) and control group (GSM1215529–GSM1215535) are marked. Red is for high expression, and green is for low expression. Each column represents one sample, and each row represents one gene/lncRNA.
Abbreviations: AMD, age-related macular degeneration; lncRNA, long noncoding RNA.
Table 1.
Top 20 differentially expressed long noncoding RNAs in early AMD
| ID | logFC | t | P-value | Symbol |
|---|---|---|---|---|
| ENSG00000225649 | −0.775314345 | −6.260045806 | 7.65E–06 | AC064875.2 |
| ENSG00000248677 | 0.561298878 | 5.77399922 | 2.03E–05 | CTD-2044J15.1 |
| ENSG00000233705 | 0.514802877 | 5.531509843 | 3.33E–05 | SLC26A4-AS1 |
| ENSG00000205452 | 0.849909105 | 5.441902994 | 4.01E–05 | RP11-812E19.3 |
| ENSG00000258683 | −0.671719098 | −5.007665761 | 9.99E–05 | RP11-11K13.1 |
| ENSG00000257120 | −1.012223924 | −4.705291702 | 0.000190741 | CTD-2503I6.1 |
| ENSG00000229245 | −0.707584769 | −4.673941743 | 0.000204068 | RP11-542K23.7 |
| ENSG00000249797 | −0.954373352 | −4.651608226 | 0.000214138 | CTD-3179P9.1 |
| ENSG00000253898 | 0.572160865 | 4.584293323 | 0.000247655 | RP11-51M18.1 |
| ENSG00000255311 | −0.827712084 | −4.369916079 | 0.000394412 | RP11-160H12.2 |
| ENSG00000255480 | 0.810803659 | 4.330604017 | 0.000429689 | RP4-710M3.2 |
| ENSG00000229267 | 0.748573134 | 4.262839387 | 0.000498169 | AC072062.1 |
| ENSG00000249568 | −0.704593601 | −3.051932202 | 0.005047204 | RP11-234O6.2 |
| ENSG00000259285 | 0.569040604 | 4.256555244 | 0.000505054 | CTD-2330J20.2 |
| ENSG00000240758 | −0.948553638 | −4.207281836 | 0.000562481 | RP11-155G14.6 |
| ENSG00000236837 | −0.600514564 | −3.955002408 | 0.00097778 | AC139712.4 |
| ENSG00000242339 | 0.709975451 | 3.944008193 | 0.00100167 | RP11-735B13.2 |
| ENSG00000246863 | −0.591744536 | −3.921314683 | 0.001052852 | RP11-325N19.3 |
| ENSG00000254610 | −1.025245637 | −3.841380386 | 0.001254953 | RP5-903G2.2 |
| ENSG00000233620 | −1.415375474 | −3.789946295 | 0.00140511 | RP11-22M7.2 |
Abbreviation: AMD, age-related macular degeneration.
LncRNA–mRNA coexpression network analyses
As the detailed functions of lncRNAs were not completely known, the analyses of the coexpressed mRNA would help in the advanced function annotations. In general, it was reasonable to consider that a close expression pattern predicated related gene function. In this study, the lncRNA–mRNA coexpression analysis detected a total of 6 lncRNAs: RP11-234O6.2, RP11-11K13.1, AF131216.6, AC000124.1, LINC00948, and RP11-22M7.2. Figure 2 presents the graphical lncRNA–mRNA coexpression relationship, and it was found that RP11-234O6.2 demonstrated the largest number of coexpression relationship. Considering that RP11-234O6.2 was coexpressed with one lncRNA (LINC00948) and 20 mRNAs, it could be conjectured that it might play an important role in the early AMD development.
Figure 2.
Coexpression of differentially expressed lncRNA and related mRNAs.
Notes: The blue circle represents lncRNAs, the green circle represents protein-coding genes, and the relationship between the two circles represents the coexpression relationship between them.
Abbreviation: lncRNA, long noncoding RNA.
The validation of lncRNA–mRNA relationship was also done using RIsearch by the sequence alignment. Through the interaction relationship analysis, we selected the lncRNA–mRNA pairs, which demonstrated both coexpression and interaction relationships, for advanced analyses. As reported in Table 2, a total of three lncRNAs (ie, RP11-11K13.1, AF131216.6, and RP11-234O6.2) and their related mRNAs were described.
Table 2.
List of lncRNA–mRNA coexpression network after interaction analysis
| lncRNA | Protein coding | Adjacency cor | Free energy |
|---|---|---|---|
| RP11-11K13.1 | MAP2 | 0.618646 | −22.87 |
| RP11-11K13.1 | RDH8 | 0.604374 | −24.17 |
| RP11-11K13.1 | MPP4 | 0.60982 | −33.46 |
| AF131216.6 | SNAP25 | 0.609362 | −20.47 |
| LINC00948 | OPN1MW | 0.673562 | −25.71 |
| RP11-234O6.2 | ROM1 | 0.605834 | −24.38 |
| LINC00948 | GPR98 | 0.664874 | −26.55 |
| AF131216.6 | TMEM244 | 0.617153 | −22.13 |
Abbreviations: cor, correlation; lncRNA, long noncoding RNA.
Functional enrichment analyses
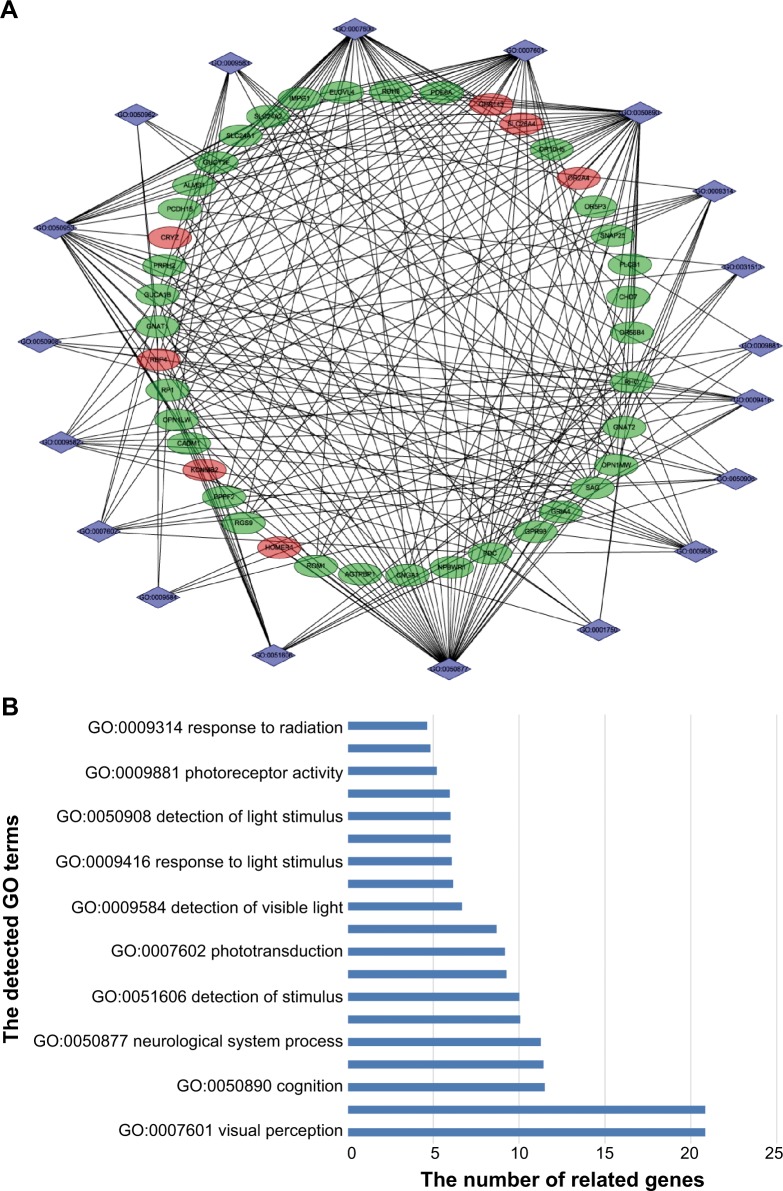
Functional enrichment analyses, which have been widely used in high-throughput data analysis to indicate the function of large-scale genes, would help to understand the role of dysregulated lncRNAs in the development of AMD. Among all the functional enrichment methods, the DAVID is one of the most commonly used integrated data-mining environments, which was used in this study. In this study, a total of 19 gene ontology (GO) terms were detected, and Figure 3A presents these GO terms as well as their related DEGs. Among all the GO terms of significance, visual perception (GO: 0007601, false discovery rate [FDR] =2.38e−18) and sensory perception of light stimuli (GO: 0050953, FDR =2.38e−18) demonstrated the most important effects. Figure 3 presents all the detected GO terms and the significance indexes.
Figure 3.
Functional enrichment of the differentially expressed lncRNA-related mRNAs.
Notes: (A) Graphical presentation of the detected GO terms. Diamonds represent GO term and circles represent genes. The red circles show the upregulated gene and the green circles the downregulated genes. (B) The GO terms of significance are listed with their values of significance.
Abbreviations: GO, gene ontology; lncRNA, long noncoding RNA.
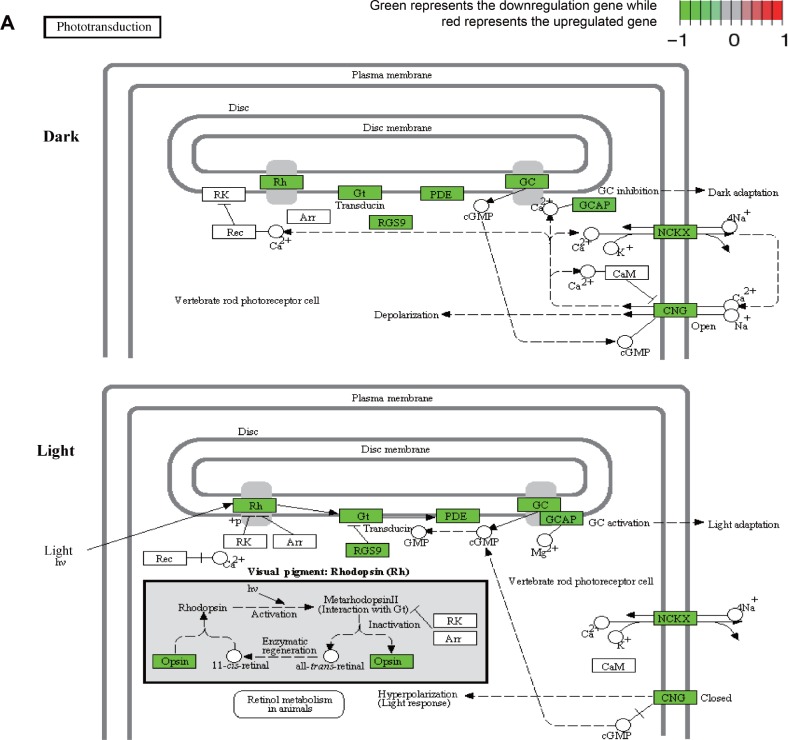
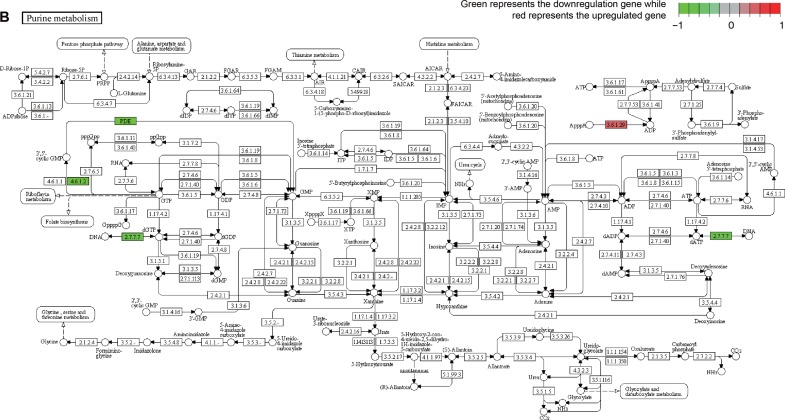
Meanwhile, KEGG pathway analysis was a useful tool in the visualization of the enriched pathways. In this study, two independent pathways, hsa04744 (phototransduction) and hsa00230 (purine metabolism), were associated with the development of early AMD. Figure 4 presents the two KEGG pathways, and the downregulated genes are marked green in the pathways. When the DEGs were considered, there were 9 and 6 gene counts in hsa04744 and hsa00230, respectively.
Figure 4.
The detected KEGG pathway of significance.
Notes: (A) Phototransduction pathway. (B) Purine metabolism pathway. Each panel represents one gene. Red is for high expression, and green is for low expression. Reproduced with permission from Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. © The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.33
Abbreviations: KEGG, Kyoto Encyclopedia of Genes and Genomes; +p, phosphorylation; hv, rovided energy under light conditions; h, Planck constant; v, frequency.
Downregulation of RP11-234O6.2 in early AMD
By the functional enrichment analysis, it was found that RP11-234O6.2 played a key role in the AMD development by both coexpression and interaction analyses. Advanced independent analyses showed that coexpressed mRNAs of RP11-234O6.2 were mainly listed in phototransduction, which was one of the most significant KEGG pathways. These advanced analyses on the expression of RP11-234O6.2 demonstrated an important role. RP11-234O6.2, whose transcript is ENST00000508189.1, was located in Chromosome 4.
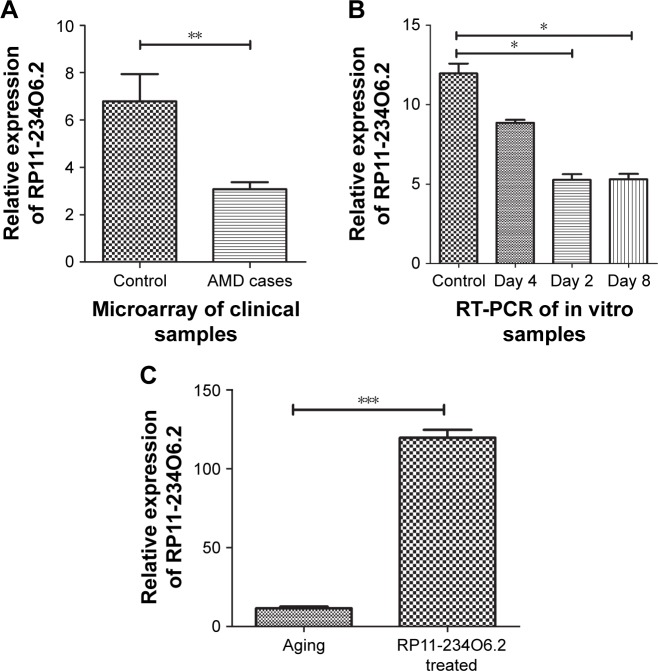
From the microarray analyses of RPE/choroid samples from early AMD cases, it was found that the relative expression of RP11-234O6.2 was significantly decreased compared with controls. In order to detect the expression pattern of RP11-234O6.2 in the aging RPE in vitro model, a real-time PCR method was used to detect the content of RP11-234O6.2. Compared with the normally cultured ARPE-19, it was found that RP11-234O6.2 was significantly downregulated in the aging RPE model. Figure 5A and B show the detailed expression data of RP11-234O6.2 in both tissue samples and in vitro models.
Figure 5.
RP11-234O6.2 was differentially expressed in early AMD.
Notes: (A) RP11-234O6.2 was downexpressed in early AMD cases by microarray analyses; (B) RP11-234O6.2 was downexpressed during aging RPE cell model by RT-PCR; (C) RP11-234O6.2 vector treatment could significantly increase the expression of RP11-234O6.2 in aging ARPE-19 cells. *P<0.05, **P<0.01, ***P<0.0001.
Abbreviations: AMD, age-related macular degeneration; RPE, retinal pigment epithelium; RT-PCR, real-time polymerase chain reaction.
RP11-234O6.2 regulates biological function of aging RPE cells
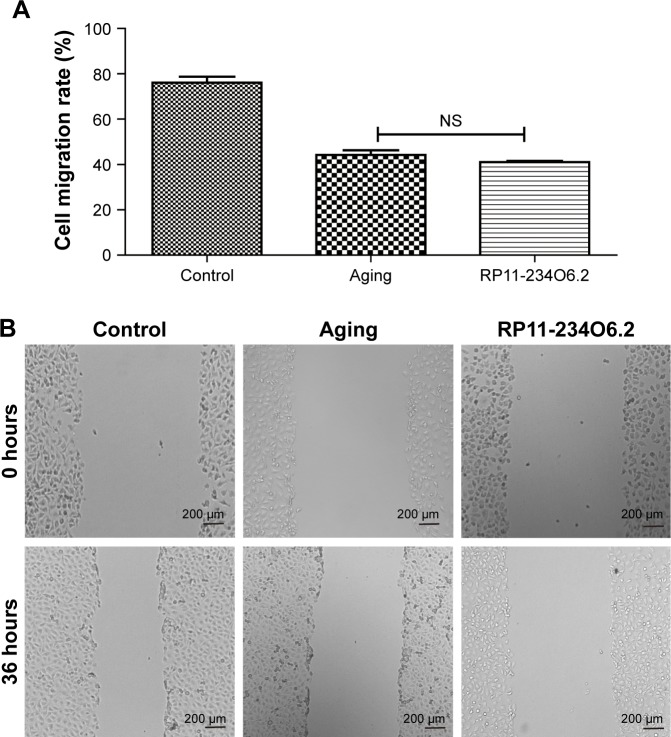
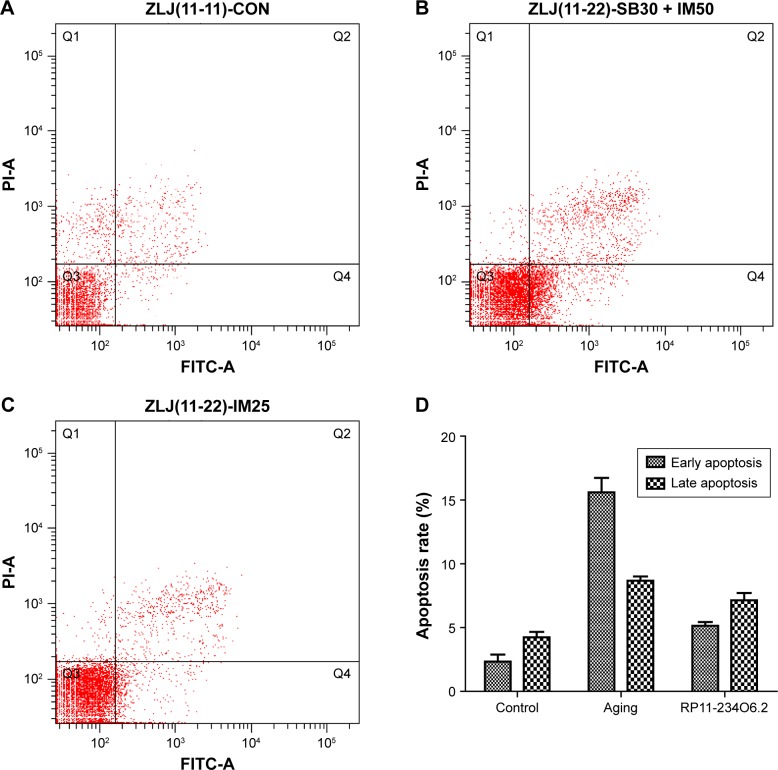
In order to detect the biological effects of RP11-234O6.2 on the RPE in vitro aging model, we conducted a series of experiments, including cell viability, migration, and apoptosis. In this study, it was found that RP11-234O6.2 treatment would upregulate the expression level of RP11-234O6.2 in the aging model (Figure 5C). When considering the general function, it was found that cell viability was significantly decreased in the aging model. After exogenous RP11-234O6.2 treatment, cell viability was significantly improved compared with the aging group (Figure 6; P<0.01). By the cell scratch tests, it was found that the migration rate of the aging RPE cell group was significantly decreased compared with the normal group, whereas it was found that RP11-234O6.2 treatment did not influence the migration effects of aging RPE cells (Figure 7). Through flow cytometry detection, it was found that both the early and late apoptosis rates were significantly increased in the aging group. After RP11-234O6.2 treatment, early apoptosis (P=0.0008), but not late apoptosis (P=0.0848), was significantly improved (Figure 8).
Figure 6.
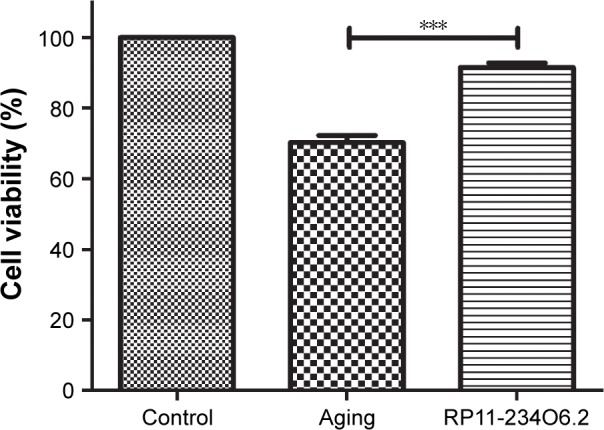
Cell viability of aging RPE cells influenced in RP11-234O6.2 in vitro.
Notes: Cell viability was significantly decreased in the aging group, and RP11-234O6.2 could improve the cell viability significantly. ***P<0.001.
Abbreviation: RPE, retinal pigment epithelium.
Figure 7.
Cell migration ability of aging RPE cells influenced in RP11-234O6.2 in vitro.
Notes: (A) The graphs of each scratch test for 0 and 36 hours. (B) Results for cell migration test. It was found that RP11-234O6.2 treatment did not influence the migration effects of aging RPE cells.
Abbreviations: NS, not significant; RPE, retinal pigment epithelium.
Figure 8.
The apoptosis changes in each group by flow cytometry detection.
Notes: (A) The normal group; (B) the aging group; (C) aging RPE cells after RP11-234O6.2 treatment; (D) results for apoptosis test; early apoptosis (P=0.0008), but not late apoptosis (P=0.0848), was significantly improved by RP11-234O6.2.
Abbreviations: FITC, fluorescein isothiocyanate; PI, propidium iodide; RPE, retinal pigment epithelium.
Discussion
LncRNAs demonstrate important effects and participate in different biological processes. AMD is a multifactorial disease and might also be associated with the dysregulation of lncRNAs. The primary purpose of this study was to detect their global expression patterns in the early AMD. A total of 64 differentially expressed lncRNAs were identified in this study. The advanced bioinformatics analyses of the coexpressed mRNAs showed that a total of 19 GO terms were detected. KEGG pathway analyses showed that two independent pathways, phototransduction and purine metabolism, were associated with the development of early AMD. Based on the functional analyses, one of the differentially expressed lncRNAs, RP11-234O6.2, demonstrated important potential effects. Advanced in vitro studies based on the aging RPE model showed that RP11-234O6.2 could improve cell viability and early apoptosis.
Early AMD has been reported to be influenced by oxidative stress, cell apoptosis, abnormal lipid metabolism, and dysfunction of the immune system,21 whereas lncRNAs might also be associated with these effects in different disorders. In a previous study on RNA-Seq data sets, dysregulated lncRNAs were detected which were associated with oxidative stress in cholangiocarcinoma.22 It was reported that H19 and HULC were upregulated by oxidative stress and regulated cholangiocarcinoma cell migration and invasion by targeting interlukin-6 and CXCR4. In a study on diabetic retinopathy (DR), the results indicated that lncRNA MIAT regulated apoptosis in DR.23 Lipid metabolism was also regarded as a key process in AMD incidence, and a previous study demonstrated that 13 potential lncRNA–mRNA gene pairs were associated with the dysregulation of lipid metabolism in thyroid gland.24 In a study on myocardial infarction and lncRNA expression profiles, the functional analysis demonstrated that a total of 9 lncRNAs were involved in myocardial innate immune responses, and the dysregulation of these lncRNAs led to abnormalities of the immune system.25
Knowledge on the regulatory effects of lncRNAs on AMD progression, and the detailed functions of lncRNAs on the incidence of early AMD, is quite limited. In a previous study, the detection of dysregulated lncRNAs in pathological neovascularization provided evidences of late AMD development.10 However, it focused on late AMD, and most attention was paid to neovascularization, which was quite different during early AMD. Several previous studies reported that lncRNAs were associated with epithelial–mesenchymal transition of RPE cells,26 maintenance of retinal architecture,27 and regulation of microvascular dysfunction in DR.28 In this study, we conducted an overall review of the expression patterns of lncRNAs in early AMD. It was found that a total of 64 lncRNAs were dysregulated in early AMD. Through advanced functional analyses, it was found that lncRNAs may play a key role in the early AMD development. The mapping of lncRNA-related dysregulated mRNAs showed that most DEGs were located in phototransduction and purine metabolism KEGG pathways. As reported in a previous study, the downregulation of phototransduction was a key process in blue light damage of retinas.29 Considering that light damage is regarded as a risk factor for AMD and the light damage model was used in the early AMD research, it was quite reasonable to regard phototransduction as the most significantly influenced pathway. The other pathway addressed in the analyses of microarray data was purine metabolism. Purinergic signaling has been reported to be involved in the degeneration of the injured retina.30 Based on previous research, purine metabolism was associated with the proliferation of retinal glial cells and demonstrated protective effects on retinal degeneration. The finding of this study provided improved understanding of the role of lncRNAs on AMD development by both phototransduction and purine metabolism. Further studies on lncRNA regulation of these two processes may help elucidate the development of AMD.
In this research, we evaluated the expression level of RP11-234O6.2 in vitro and conducted a series of laboratory studies addressing the biological effects of RP11-234O6.2 on the aging model. As far as we know, there are no previous studies on the biological effects of RP11-234O6.2 in other disorders. In this study, the initial data showed that RP11-234O6.2 was downregulated in early AMD. Advanced exogenous RP11-234O6.2 applications showed protective effects by improving cell viability and decreasing early apoptosis rate. The data in this study expand the repertoire of lncRNAs and mRNAs and contribute to our understanding of the molecular mechanisms involved in the development of early AMD. However, one limitation of this study should be recognized. It was difficult to conduct a functional bioinformatics analysis on lncRNAs at this time. The strategy used for analysis, obtained from a previous work,32 was to examine the functions of coexpressed mRNAs. The basic idea was that the relationship between lncRNAs and the coexpressed mRNAs would reflect a potential regulatory relationship. However, the effects of lncRNAs were highly variable and were usually demonstrated directly on DNA and proteins.31 The bioinformatics analyses in this study mainly focused on lncRNA–mRNA regulation, and this certainly could lead to misperceptions of the role of lncRNA on AMD development. Thus, more regulated methods will be required, and the conclusions based on the results of these bioinformatics analyses should be considered with caution.
Conclusion
This study was designed to detect expressed lncRNAs in early AMD by integrating public microarray data. Based on the reannotations of microarray data and functional analyses, it was found that lncRNA–mRNA coexpression appears to play an important role in early AMD development. Among the 64 detected lncRNAs, RP11-234O6.2 was one of the key lncRNAs and is predicated to exert significant regulatory effects. The pattern of expression of RP11-234O6.2 was validated in an in vitro model, and RP11-234O6.2 demonstrated protective effects on the aging RPE model. Considering that preliminary data were obtained in this study, further experiments will be needed to verify the expression and detailed mechanisms of dysregulated lncRNAs in early AMD.
Acknowledgments
This work was supported in whole or in part by the Foundation for Young Scholars of Suzhou, China (grant number kjxw2015044) and the Foundation for Young Medical Talents of Jiangsu Province, 2016 (grant number QNRC2016211). The sponsor and funding organizations had no role in the design or conduct of this research.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Deemer AD, Massof RW, Rovner BW, Casten RJ, Piersol CV. Functional outcomes of the low vision depression prevention trial in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2017;58(3):1514–1520. doi: 10.1167/iovs.16-20001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaiya S, Abu-Amero KK, Kondkar AA, Chalam KV. Sirtuins expression and their role in retinal diseases. Oxid Med Cell Longev. 2017;2017:3187594. doi: 10.1155/2017/3187594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGuinness MB, Karahalios A, Finger RP, Guymer RH, Simpson JA. Age-related macular degeneration and mortality: a systematic review and meta-analysis. Ophthalmic Epidemiol. 2017;24:141–152. doi: 10.1080/09286586.2016.1259422. [DOI] [PubMed] [Google Scholar]
- 4.Pennington KL, DeAngelis MM. Epidemiology of age-related macular degeneration (AMD): associations with cardiovascular disease phenotypes and lipid factors. Eye Vis (Lond) 2016;3:34. doi: 10.1186/s40662-016-0063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garg A, Yang J, Lee W, Tsang SH. Stem cell therapies in retinal disorders. Cells. 2017;6(1) doi: 10.3390/cells6010004. pii: E4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koskela A, Reinisalo M, Petrovski G, et al. Nutraceutical with resveratrol and omega-3 fatty acids induces autophagy in ARPE-19 cells. Nutrients. 2016;8(5) doi: 10.3390/nu8050284. pii: E284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu W, Wu Y, Meng YF, Xing Q, Tao JJ, Lu J. Fish consumption and age-related macular degeneration incidence: a meta-analysis and systematic review of prospective cohort studies. Nutrients. 2016;8(11) doi: 10.3390/nu8110743. pii: E743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao MX, Jiang YP, Tang YL, Liang XH. The crosstalk between lncRNA and microRNA in cancer metastasis: orchestrating the epithelial-mesenchymal plasticity. Oncotarget. 2016;8(7):12472–12483. doi: 10.18632/oncotarget.13957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan P, Su W, Zhuo Y. Precise long non-coding RNA modulation in visual maintenance and impairment. J Med Genet. 2016;54(7):450–459. doi: 10.1136/jmedgenet-2016-104266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu XD, Li KR, Li XM, Yao J, Qin J, Yan B. Long non-coding RNAs: new players in ocular neovascularization. Mol Biol Rep. 2014;41(7):4493–4505. doi: 10.1007/s11033-014-3320-5. [DOI] [PubMed] [Google Scholar]
- 11.Degirmenci U, Lei S. Role of lncRNAs in cellular aging. Front Endocrinol (Lausanne) 2016;7:151. doi: 10.3389/fendo.2016.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitmore SS, Braun TA, Skeie JM, et al. Altered gene expression in dry age-related macular degeneration suggests early loss of choroidal endothelial cells. Mol Vis. 2013;19:2274–2297. [PMC free article] [PubMed] [Google Scholar]
- 13.Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7:252. doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wenzel A, Akbasli E, Gorodkin J. RIsearch: fast RNA-RNA interaction search using a simplified nearest-neighbor energy model. Bioinformatics. 2012;28(21):2738–2746. doi: 10.1093/bioinformatics/bts519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 18.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29(14):1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu W, Wu Y, Meng YF, et al. Effect of curcumin on aging retinal pigment epithelial cells. Drug Des Devel Ther. 2015;9:5337–5344. doi: 10.2147/DDDT.S84979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Handa JT, Cano M, Wang L, Datta S, Liu T. Lipids, oxidized lipids, oxidation-specific epitopes, and age-related macular degeneration. Biochim Biophys Acta. 2017;1862(4):430–440. doi: 10.1016/j.bbalip.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang WT, Ye H, Wei PP, et al. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol. 2016;9(1):117. doi: 10.1186/s13045-016-0348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Chen M, Chen J, et al. Long noncoding RNA MIAT acts as a biomarker in diabetic retinopathy by absorbing of miR-29b and regulating cell apoptosis. Biosci Rep. 2017;37(2) doi: 10.1042/BSR20170036. pii: BSR20170036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen Y, Mao H, Huang M, et al. Long noncoding RNA and mRNA expression profiles in the thyroid gland of two phenotypically extreme pig breeds using Ribo-Zero RNA sequencing. Genes (Basel) 2016;7(7) doi: 10.3390/genes7070034. pii: E34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun C, Jiang H, Sun Z, Gui Y, Xia H. Identification of long non-coding RNAs biomarkers for early diagnosis of myocardial infarction from the dysregulated coding-non-coding co-expression network. Oncotarget. 2016;7(45):73541–73551. doi: 10.18632/oncotarget.11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang S, Yao H, Li M, Li H, Wang F. Long non-coding RNA MALAT1 mediates transforming growth factor beta1-induced epithelial-mesenchymal transition of retinal pigment epithelial cells. PLoS One. 2016;11(3):e0152687. doi: 10.1371/journal.pone.0152687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krol J, Krol I, Alvarez CP, et al. A network comprising short and long noncoding RNAs and RNA helicase controls mouse retina architecture. Nat Commun. 2015;6:7305. doi: 10.1038/ncomms8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan B, Yao J, Liu JY, et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015;116(7):1143–1156. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 29.Calzia D, Panfoli I, Heinig N, et al. Impairment of extramitochondrial oxidative phosphorylation in mouse rod outer segments by blue light irradiation. Biochimie. 2016;125:171–178. doi: 10.1016/j.biochi.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 30.Reichenbach A, Bringmann A. Purinergic signaling in retinal degeneration and regeneration. Neuropharmacology. 2016;104:194–211. doi: 10.1016/j.neuropharm.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, Ge H, Jiang Y, Cheng B, Zhou D, Xu N. Microarray profiling analysis of long non-coding RNAs expression in tendinopathy: identification for potential biomarkers and mechanisms. Int J Experi Pathol. 2015;96(6):387–394. doi: 10.1111/iep.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]