ABSTRACT
Multidrug-resistant tuberculosis (TB) presents a major public health dilemma. Heteroresistance, the coexistence of drug-resistant and drug-susceptible strains or of multiple drug-resistant strains with discrete haplotypes, may affect accurate diagnosis and the institution of effective treatment. Subculture, or passage of cells onto fresh growth medium, is utilized to preserve Mycobacterium tuberculosis cell lines and is universally employed in TB diagnostics. The impact of such passages, typically performed in the absence of drug, on drug-resistant subpopulations is hypothesized to vary according to the competitive costs of genotypic resistance-associated variants. We applied ultradeep next-generation sequencing to 61 phenotypically rifampin-monoresistant (n = 17) and preextensively (n = 41) and extensively (n = 3) drug-resistant isolates with presumptive heteroresistance at two time points in serial subculture. We found significant dynamic loss of minor-variant resistant subpopulations across all analyzed resistance-determining regions, including eight isolates (13%) whose antibiogram data would have transitioned from resistant to susceptible for at least one drug through subculture. Surprisingly, some resistance-associated variants appeared to be selected for in subculture.
KEYWORDS: heteroresistance, rpoB, gyrA, rrs, drug-resistant tuberculosis
INTRODUCTION
An estimated 480,000 new cases of multidrug-resistant tuberculosis (MDR TB) occur each year. Mycobacterium tuberculosis heteroresistance (i.e., the coexistence of drug-resistant and drug-susceptible strains or of multiple drug-resistant strains with discrete haplotypes) may be associated with two or more drug susceptibility profiles within a single specimen and be potentially masked by a majority drug-susceptible population. This can prevent detection by phenotypic and genotypic tests, prolong infectiousness and time to effective treatment initiation, and, potentially, presage treatment failure.
Subculture, where the original culture isolate is reinoculated onto fresh media prior to further genotypic (e.g., MTBDRsl) and phenotypic (e.g., agar proportion method) drug susceptibility testing (DST), is a cornerstone of TB diagnostics. However, subculture can take up to 42 days (e.g., Bactec MGIT 960 system) and is typically performed in the absence of drug. Given that growth on drug-containing agar increases recovery of resistant M. tuberculosis subpopulations up to 100-fold (1) and that the competitive costs of resistance-associated variants (RAV) are diverse (2, 3), we hypothesized that drug-resistant subpopulations might exhibit various levels of decline during culture in the absence of selection pressure. Despite ubiquitous use of subculture, we are aware of no data describing changes in M. tuberculosis heteroresistance with repeated culture passage. This is particularly true at the subphenotypic levels detectable by novel next-generation sequencing approaches (e.g., single-molecule overlapping read [SMOR] analysis) (4), which can lower the sequencing error rate by multiple orders of magnitude (5).
RESULTS
We performed targeted ultradeep sequencing utilizing the SMOR approach at two time points during laboratory processing (primary DST culture and quaternary MGIT subculture) (Fig. 1) for 61 clinical isolates with phenotypic rifampin (RIF) monoresistance (n = 17) or MDR (isoniazid [INH] resistance and RIF resistance) with additional fluoroquinolone (FQ) resistance (n = 28), second-line injectable (SLI) drug resistance (n = 13), or both FQ resistance and SLI resistance (n = 3). The isolates were selected based on Sanger sequencing-determined presumptive heteroresistance, due to either (i) the presence of multiple colocalizing chromatogram peaks within respective resistance-determining regions (RDRs) or (ii) a lack of an identified genotypic correlate (i.e., wild-type RDR), positing the presence of a resistant subpopulation at a level below the resolution of Sanger sequencing (6).
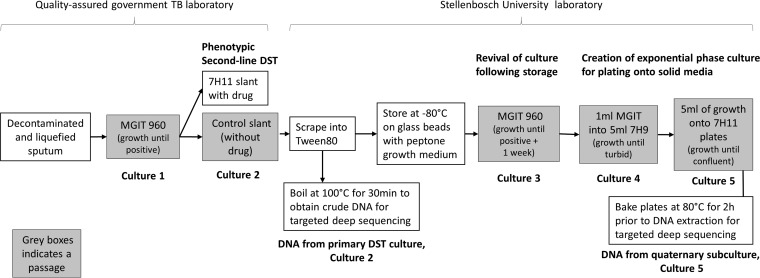
FIG 1.
Derivation of study samples. M. tuberculosis laboratory procedures and the rationale employed in derivation of analyzed specimens were as follows: primary drug susceptibility testing (DST) culture (Culture 2) and quaternary subculture (Culture 5). MGIT, Mycobacteria Growth Indicator Tube.
In primary DST culture (Culture 2 in Fig. 1), SMOR detected 316 resistant variants, of which 125 (40%) represented fixed resistance (≥95% of total M. tuberculosis population), 74 (23%) were macroheteroresistant (5% to 95% of the total population), and 117 (37%) were microheteroresistant (<5% of the total population) across all RDRs of the 61 analyzed isolates. The proportion of microheteroresistant subpopulations declined significantly from culture 2 to culture 5, representing 117/316 (37%) to 48/237 (20%) of the total resistant populations (P < 0.001), respectively. The largest contribution to the decline in microheteroresistance was accounted for by the disappearance (i.e., the lack of detection in culture 5) of 79 subpopulations (median size, 0.2%; interquartile range [IQR], 0.14% to 0.35%) with subculture. In addition, two microheteroresistant subpopulations grew to macroheteroresistance in subculture, and 12 new microheteroresistant subpopulations were detected (2 appearing de novo and 10 resulting from the decline of previously macroheteroresistant subpopulations) (Fig. 2). The proportions of an additional 8 (6%) microheteroresistant subpopulations declined, but the data could not be verified due to fewer than 20 matched reads being available. The median relative decline in heteroresistant subpopulation size across all RDRs within an individual was −68% (IQR, −100% to +4%), and loss of microheteroresistance was pronounced across all RDRs as follows (gene name, number of respective microheteroresistant subpopulations/total number of resistant populations): inhA, 9/41 (22%) to 1/33 (3%) (P = 0.02); katG, 8/35 (23%) to 0/27 (0%) (P < 0.01); rpoB, 42/94 (45%) to 11/60 (18%) (P < 0.001); gyrA, 41/94 (44%) to 26/74 (34%) (P = 0.19); rrs, 10/41 (24%) to 5/33 (15%) (P = 0.34). The decline in the RAV subpopulation size was a function of the original subpopulation size, with 79% (95% confidence interval [CI], 69% to 86%) of the microheteroresistant subpopulations (<1%) declining, compared with 63% (95% CI, 42% to 81%) of the microheteroresistant populations (1% to 5%), 18% (95% CI, 10% to 28%) of the macroheteroresistant populations (5% to 95%), and 0% (95% CI, 0% to 3%) of the fixed populations (≥95%). Assuming that a threshold of a 1% resistant proportion as measured by deep sequencing reliably predicts phenotypic drug susceptibility (7), eight (13%) isolates would have altered their antibiogram profiles through subculture from resistant to susceptible for RIF (n = 3), for aminoglycosides (n = 2), for FQ (n = 1), and for both FQ and aminoglycosides (n = 1) and for both RIF and INH (n = 1) (Fig. 2). Clinical records from 20 (71%) patients with fluoroquinolone-resistant TB were available, 10 (50%) of whom were receiving ofloxacin (OFX) at the time of the original sputum collection. Among these isolates, neither median declines in the subpopulation proportion nor changes in subpopulation diversity differed significantly from those seen with patients not currently on treatment.
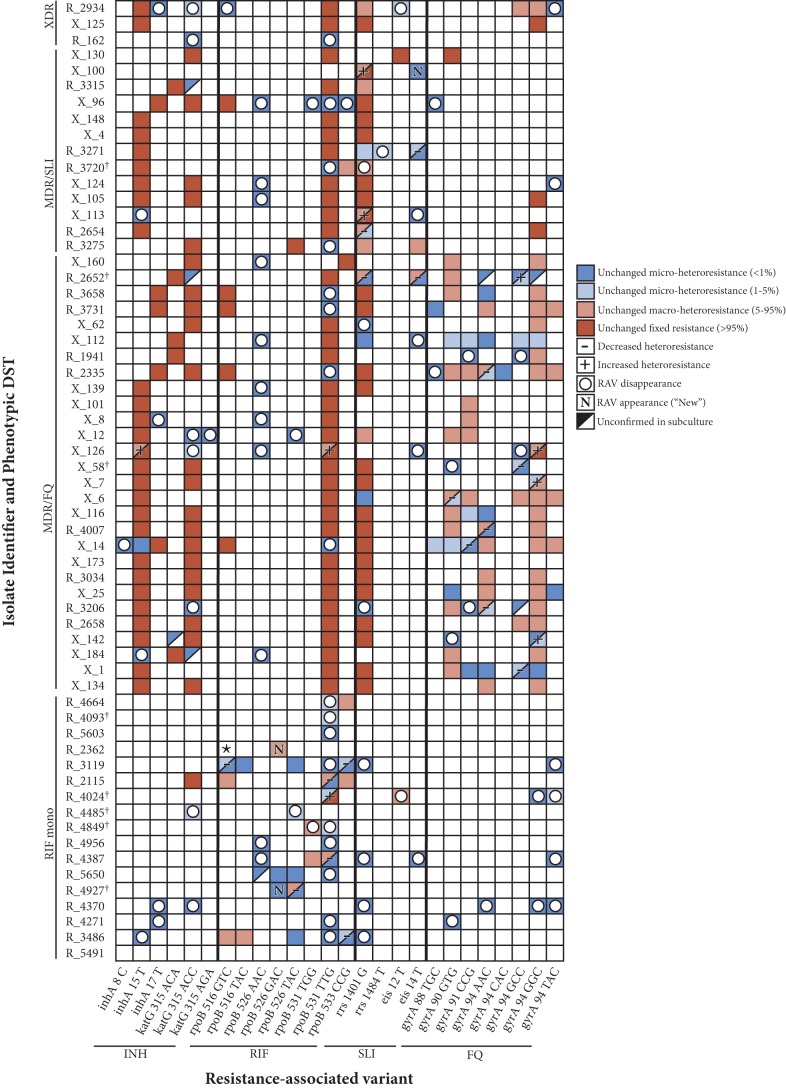
FIG 2.
Dynamic changes in heteroresistant M. tuberculosis profiles with serial subculture. The color-coded map illustrates the overall loss of M. tuberculosis resistance-associated variant (RAV) subpopulations (empty circles or “−” label), while some RAVs (e.g., gyrA 94GGC) appear to be selected for in subculture (“N” or “+” label). Red boxes indicate fixed-resistance mutations (≥95% of the total M. tuberculosis population) in both samples (cultures 2 and 5); salmon-colored boxes indicate macroheteroresistance (5% to 95% of the total M. tuberculosis population) in both samples; light blue boxes indicate microheteroresistance (<5% of the total M. tuberculosis population) in both samples; dark blue boxes indicate microheteroresistance (<1% of the total M. tuberculosis population) in both samples. Also illustrated is RAV disappearance (empty circles, with the background color illustrating the subpopulation size in the original culture), appearance (“New”), and categorical decline or growth (− or +, respectively). Please note that data corresponding to growth or decline within categories (e.g., R_3206 gyrA 90GTG increasing from 25% to 44%, with both samples being “macroheteroresistant”) are not shown. Sample R_5491 was demonstrated to be wild type at all RDRs analyzed. ★, all early reads for sample R_2362 identified the 9-bp deletion at rpoB 516–525 in the original culture, though in subculture this deletion was strongly offset by a minority strain harboring the rpoB 526GAC mutation (please see Discussion for details). †, isolates transitioning from resistant to susceptible for at least one drug. XDR, extensively drug resistant (MDR, with additional resistance to both FQ and SLI).
In conjunction with the overall decline in resistant M. tuberculosis subpopulations, a small subset (13/74; 18%) of macroheteroresistant subpopulations, and 2/117 (1.7%) of the microheteroresistant subpopulations, had a relative increase in size of 83% (IQR, 30% to 606%; absolute change, +26%; IQR, +17% to 60%). The mutations 94GGC (n = 4) and 90GTG (n = 2) within gyrA and the mutation 531TTG (n = 2) within rpoB comprised the majority of strongly increasing RAVs; other subpopulations coupled to those respective RAVs tended to decline proportionally (Fig. 2).
DISCUSSION
Subculture is a standard microbiological laboratory practice for clinical diagnostics (8, 9), and DST results are assumed to be agnostic to subculture passages. However, among other heteroresistant human pathogens, multiple passages on antibiotic-free media do cause reversion to drug susceptibility (10, 11), and M. tuberculosis subjected to serial in vitro passage demonstrates important phenotypic changes, including irreversible attenuation of H37Rv pathogenicity due to loss of virulence (12). We found that M. tuberculosis population dynamics continue through serial culture, with resistant subpopulations large enough to confer phenotypic resistance often being outcompeted by both wild-type strains and alternative resistant variants.
In our study, heteroresistant subpopulation diversity declined with serial culture due to disappearance of individual RAVs, in particular, those which were originally subphenotypic in nature (<1% of the total population). In a separate work, we demonstrated these minor resistant variants to be clinically relevant in conferring phenotypic resistance and further corroborated their presence through single-colony selection on drug-containing media (4). Rifampin-monoresistant isolates appeared especially susceptible to loss of microheteroresistant subpopulations; this may have been due to their higher likelihood of being pretreatment specimens.
The overall loss of minor variants was typically accompanied by a transition to a more homogenous RAV-specific population of mycobacteria, hypothetically one with no evolutionary disadvantage in harboring the resistant codon (13). Moreover, and surprisingly, certain RAVs appeared to be selected for in subculture despite the absence of drug pressure. These RAVs were often, though not always, those most frequently noted among clinical resistant strains. For example, the gyrA 94GAC→GGC (D94G) mutation is associated with high-level resistance to both OFX and newer-generation fluoroquinolones (14), and rpoB 531TCG→TTG (S531L) is the most common rifampin-conferring mutation globally (15). Equivalent or enhanced levels of fitness of these circulating strains (in contrast to laboratory-derived strains) vis-à-vis their drug-susceptible counterparts has been demonstrated in competition assays (2). Haplotype analysis, the characterization of which of multiple mutations coexist on individual sequencing reads thereby differentiating the respective strains, is further illustrative of this process. For sample R2362 (Fig. 2), the 9-bp deletion at rpoB 515–518 dominated the original culture, occurring in all but 31 of >262,000 total reads. Of those 31 original sequencing reads, 16 (<0.01% of the total) had the rpoB 526CAC→GAC mutation. In subculture, rpoB 526GAC grew to represent 33% of the culturable population, replacing the less-fit rpoB 515–518 deletion that had presumably been clinically selected for under drug pressure.
Our study was limited in that it tested dynamic changes across only two time points, both of which occurred following at least two rounds of purification. The loss of M. tuberculosis-resistant variant diversity that we describe, and the potential for discrepant results among molecular TB DST assays, may be conservative relative to that experienced with earlier passages, such as those from direct clinical sputum specimens to primary culture. Second, while we sampled the majority of each slant, it is conceivable that additional minor subpopulations of mycobacteria with rare variants might have existed in the unsampled portion. Third, although positive selection in strains exhibiting increasing levels of drug resistance mapped almost entirely to resistance-associated genes (16), additional adaptive changes occurring outside RDRs were likely cooccurring. Further, within RDRs, we targeted deep sequencing at those codons accounting most often for drug resistance and therefore cannot comment on the behavior of less-prevalent and, theoretically, less-fit RAVs. Lastly, our study revealed many instances of false-negative phenotypic DST reporting, in particular, for SLIs, among preextensively drug-resistant isolates with known FQ resistance. Though this may not be unusual for ultra-high-throughout laboratories, additional caution seems warranted in interpreting phenotypic DST results for this group.
In conclusion, we describe significant dynamic loss of minor-variant resistant subpopulations through serial subculture, with potential consequences for molecular TB assays performed on subculture as well as phenotypic DST antibiograms. This report further documents the previously underdescribed nature of M. tuberculosis microheteroresistance and further emphasizes the need for additional study of the effects of this phenomenon on both clinical diagnostics and patient outcomes.
MATERIALS AND METHODS
M. tuberculosis culture and DNA extraction.
Decontaminated and liquefied sputum was first cultured within the South African National Health Laboratory Service (NHLS) by the use of an MGIT 960 (Becton Dickinson, Sparks, MD) system until positive (Culture 1 in Fig. 1), after which DST was done on Middlebrook 7H11 slants (Becton Dickinson, Sparks, MD) at WHO-recommended critical concentrations (OFX, 2 μg/ml; amikacin [AMK], 4 μg/ml; RIF, 1 μg/ml) (17). M. tuberculosis growth on control slants was then processed within the Stellenbosch University laboratory to obtain a crude DNA lysate (primary DST culture; culture 2). Briefly, a scrape from the control slant was added to 400 μl Tween 80, and 200 μl of the cells was inactivated by incubation at 100°C for 30 min. The other half was stored at −80°C on glass beads with peptone growth medium for future use. In order to obtain high-quality purified DNA, an aliquot from the stored culture was inoculated in MGIT 960 for 1 week after reading positive (culture 3), after which it was subcultured into fresh 7H9 liquid media supplemented with oleic acid-albumin-dextrose-catalase (OADC) until it reached turbidity (culture 4) before being plated on supplemented 7H11 plates (culture 5). These plates were incubated at 80°C for 2 h prior to a phenol-chloroform DNA extraction.
Targeted deep sequencing.
DNA specimens were coded, processed under blind conditions, amplified, and prepared for targeted SMOR sequencing to allow high-resolution detection of low-level (<5%) minor populations of RAVs (i.e., “microheteroresistance”), as previously described (5), with the following modifications. Following the gene-specific multiplex PCR, primer-dimer artifacts were removed using a single 0.8×, Agencourt AMPure XP bead (Beckman Coulter, Brea, CA) cleanup, instead of two sequential bead cleanups, eluting the amplicons in 15 μl of a 10 mM Tris-HCl–0.05% Tween 20 solution. SMOR primers are designed to have complete overlap of amplicon reads, allowing double coverage (confirmatory sequencing) of each locus of interest on every DNA molecule. All loci were covered with ≥10,000 total sequencing reads, and all RAV subpopulations within the inhA, katG, rpoB, gyrA, rrs, and the eis promoter gene regions were counted if the resistant allele was covered by 10 or more SMOR reads (i.e., ≥20 matched reads, or ≥10 pairs of reads for each sequenced amplicon molecule), equivalent to a 0.1% minor population. SMOR analysis has been shown to lower the intrinsic sequencing error rate in high-GC amplicons from ∼1% to ∼0.03% (5); a cutoff of 0.1% detection was employed to conservatively ensure high confidence in minor variant calling. Further, the SMOR process uses a high fidelity polymerase with an associated error rate of 10−7, many orders of magnitude below the sequencing error rate. No-template controls were included throughout the process to ensure the absence of well-to-well sample or amplicon contamination. DNA from a confirmed pan-susceptible M. tuberculosis H37Rv strain was used as a sequencing error control throughout the SMOR assay (5). Targeted sequencing additionally allows the detection of multiple RDR-associated RAVs within individual amplicons (i.e., haplotype analysis). ASAP software detects and quantifies the presence of multiple RAV haplotypes among the amplicons to allow further analysis of the nature of heteroresistance within resistant subpopulations.
Accession number(s).
All sequencing read files were deposited in the NIH Sequence Read Archive (SRP105767).
ACKNOWLEDGMENTS
We thank the National Health Laboratory Service for provision of the Mycobacterium tuberculosis cultures.
This work was supported by the Doris Duke Charitable Foundation (J.Z.M.), the National Institute of Allergy and Infectious Diseases (NIAID) (ACTG Supplement to J.Z.M.), the South African Medical Research Council (SAMRC), and a National Research Foundation (NRF) Research Career Advancement Award (E.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NRF or the SAMRC.
J.Z.M., E.S., G.T., R.M.W., and D.M.E. were responsible for either the conception or the design of the study. Acquisition of data was performed by E.S. and R.M.W. Analysis and interpretation of data were performed by J.Z.M., E.S., C.A., D.L., and D.M.E. J.Z.M., E.S., and D.M.E. drafted the work. Revision of the data for important intellectual content was performed by G.T., R.E.C., and R.M.W.
REFERENCES
- 1.Mitchison DA. 1949. Tests for streptomycin sensitivity of tubercle bacilli in tween 80 albumin liquid medium. Lancet ii:694–696. [DOI] [PubMed] [Google Scholar]
- 2.Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJ. 2006. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312:1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]
- 3.Rigouts L, Gumusboga M, de Rijk WB, Nduwamahoro E, Uwizeye C, de Jong B, Van Deun A. 2013. Rifampin resistance missed in automated liquid culture system for Mycobacterium tuberculosis isolates with specific rpoB mutations. J Clin Microbiol 51:2641–2645. doi: 10.1128/JCM.02741-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Metcalfe JZ, Streicher E, Theron G, Colman RE, Allender C, Lemmer D, Warren R, Engelthaler DM. 14 June 2017. Cryptic micro-heteroresistance explains M. tuberculosis phenotypic resistance. Am J Respir Crit Care Med doi: 10.1164/rccm.201703-0556OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colman RE, Schupp JM, Hicks ND, Smith DE, Buchhagen JL, Valafar F, Crudu V, Romancenco E, Noroc E, Jackson L, Catanzaro DG, Rodwell TC, Catanzaro A, Keim P, Engelthaler DM. 2015. Detection of low-level mixed-population drug resistance in Mycobacterium tuberculosis using high fidelity amplicon sequencing. PLoS One 10:e0126626. doi: 10.1371/journal.pone.0126626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Streicher EM, Bergval I, Dheda K, Bottger EC, Gey van Pittius NC, Bosman M, Coetzee G, Anthony RM, van Helden PD, Victor TC, Warren RM. 2012. Mycobacterium tuberculosis population structure determines the outcome of genetics-based second-line drug resistance testing. Antimicrob Agents Chemother 56:2420–2427. doi: 10.1128/AAC.05905-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canetti G, Fox W, Khomenko A, Mahler HT, Menon NK, Mitchison DA, Rist N, Smelev NA. 1969. Advances in techniques of testing mycobacterial drug sensitivity, and the use of sensitivity tests in tuberculosis control programmes. Bull World Health Organ 41:21–43. [PMC free article] [PubMed] [Google Scholar]
- 8.Adami AG, Gallo JF, Pinhata JM, Martins MC, Giampaglia CM, de Oliveira RS. 2017. Modified protocol for drug susceptibility testing of MGIT cultures of Mycobacterium tuberculosis by the MGIT 960. Diagn Microbiol Infect Dis 87:108–111. doi: 10.1016/j.diagmicrobio.2016.10.023. [DOI] [PubMed] [Google Scholar]
- 9.Anonymous. 2014. Mycobacteriology laboratory manual. World Health Organization. http://www.who.int/tb/laboratory/mycobacteriology-laboratory-manual.pdf Accessed 30 July 2017.
- 10.Morand B, Muhlemann K. 2007. Heteroresistance to penicillin in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 104:14098–14103. doi: 10.1073/pnas.0702377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mondon P, Petter R, Amalfitano G, Luzzati R, Concia E, Polacheck I, Kwon-Chung KJ. 1999. Heteroresistance to fluconazole and voriconazole in Cryptococcus neoformans. Antimicrob Agents Chemother 43:1856–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domenech P, Reed MB. 2009. Rapid and spontaneous loss of phthiocerol dimycocerosate (PDIM) from Mycobacterium tuberculosis grown in vitro: implications for virulence studies. Microbiology 155:3532–3543. doi: 10.1099/mic.0.029199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillespie SH. 2001. Antibiotic resistance in the absence of selective pressure. Int J Antimicrob Agents 17:171–176. doi: 10.1016/S0924-8579(00)00340-X. [DOI] [PubMed] [Google Scholar]
- 14.Farhat MR, Jacobson KR, Franke MF, Kaur D, Sloutsky A, Mitnick CD, Murray M. 2016. Gyrase Mutations are associated with variable levels of fluoroquinolone resistance in Mycobacterium tuberculosis. J Clin Microbiol 54:727–733. doi: 10.1128/JCM.02775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Gan X, Li N, Wang J, Li K, Zhang H. 2010. rpoB gene mutation profile in rifampicin-resistant Mycobacterium tuberculosis clinical isolates from Guizhou, one of the highest incidence rate regions in China. J Antimicrob Chemother 65:1299–1301. doi: 10.1093/jac/dkq102. [DOI] [PubMed] [Google Scholar]
- 16.Farhat MR, Shapiro BJ, Kieser KJ, Sultana R, Jacobson KR, Victor TC, Warren RM, Streicher EM, Calver A, Sloutsky A, Kaur D, Posey JE, Plikaytis B, Oggioni MR, Gardy JL, Johnston JC, Rodrigues M, Tang PK, Kato-Maeda M, Borowsky ML, Muddukrishna B, Kreiswirth BN, Kurepina N, Galagan J, Gagneux S, Birren B, Rubin EJ, Lander ES, Sabeti PC, Murray M. 2013. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet 45:1183–1189. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anonymous. World Health Organization Global TB Programme. Updated interim critical concentrations for first-line and second-line DST. http://www.stoptb.org/wg/gli/assets/documents/Updated%20critical%20concentration%20table_1st%20and%202nd%20line%20drugs.pdf Accessed 1 March 2017.