ABSTRACT
Mutations in crrAB genes encoding a two-component regulator involved in modifications of lipopolysaccharide were searched for among a collection of colistin-resistant Klebsiella pneumoniae isolates. Four isolates, respectively, producing carbapenemases NDM-1, OXA-181, or KPC-2 showed mutated CrrB proteins compared with those in wild-type strains. Complementation assays with a wild-type CrrB protein restored the susceptibility to colistin in all cases, confirming the involvement of the identified substitutions in the resistance phenotype.
KEYWORDS: polymyxin, rapid polymyxin NP test, susceptibility testing, resistance mechanisms, CrrAB, Klebsiella pneumoniae
TEXT
The emergence and spread of carbapenemase-producing Klebsiella pneumoniae isolates worldwide has forced clinicians to reintroduce colistin as last-resort therapy (1). Besides the plasmid-mediated mcr-1 and mcr-2 genes in K. pneumoniae (2, 3), the chromosomally encoded alterations of the mgrB gene and the PmrAB and PhoPQ two-component systems are currently the most commonly reported mechanisms of acquisition of polymyxin resistance in this enterobacterial species (4). Recently, mutations in the crrB gene, belonging to a third two-component system (named CrrAB for colistin resistance regulation) and involved in lipopolysaccharide (LPS) modifications, were associated with colistin resistance (5, 6). Mutations in the crrB gene are responsible for the increase in crrC gene transcription, which in turn regulates the expression of the pmrC gene and the pmrHFIJKLM operon, through the PmrAB two-component system (6). Expression of these genes leads to the addition of cationic groups on the LPS and consequently to colistin resistance.
In previous studies, the colistin resistance mechanisms of a collection of 185 K. pneumoniae isolates recovered from human samples worldwide (Europe, Turkey, Colombia, South Africa) were analyzed. Chromosomally encoded modifications of LPS through alterations (mutation, truncation) of the mgrB gene (7–9), the pmrAB genes (10), or the phoPQ genes (11) were identified in 152 isolates. However, 33 isolates did not present any substitutions in these genes, and they were negative for the plasmid-mediated resistance genes mcr-1 and mcr-2.
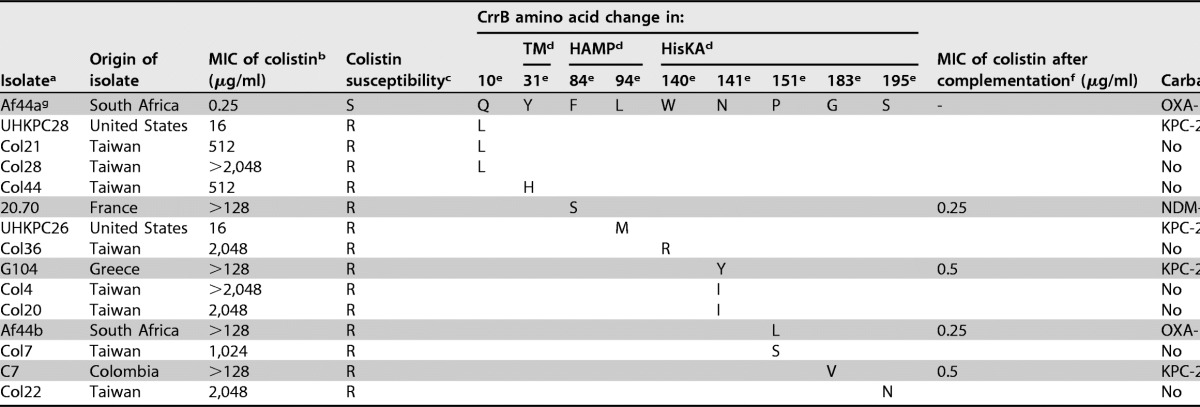
We analyzed the genes encoding the CrrAB system in these 33 isolates. Amplification of the crrA and crrB genes was performed using primers crrAB-extF (5′-GTGAGGCCATCAAATTCTCG-3′) and crrAB-extR (5′-AAGTCCCAAAAGAGGCAAAC-3′) located on each extremity of this operon. No amplification was obtained with the primers for 19 of the 33 isolates. We obtained the same result by using internal primers annealing into the crrB gene, namely, crrB-intF (5′-GTGACTATCTTACGTGGGAG-3′) and crrB-intR (5′-CACTCAGCATCAAGGAGTAC-3′). This absence of amplification suggested an absence of the crrAB operon in these strains, which is in accordance with the variability of the lateral acquisition of the crrAB operon in K. pneumoniae (5). Amplification of the crrAB gene was obtained for the 14 remaining isolates. Subsequent sequencing identified mutations leading to amino acid changes (F84S, N141Y, P151L, and G183V) in the CrrB protein in 4 of the 14 colistin-resistant K. pneumoniae isolates (Table 1).
TABLE 1.
Features of colistin-resistant K. pneumoniae clinical isolates
aIsolates from this study are indicated by shading.
bMICs of colistin were determined using the manual broth microdilution reference method for this study, Etest for the study by Wright et al. (5), and agar dilution for the study by Cheng et al. (6).
cS, susceptible (MIC, ≤2 μg/ml); R, resistant (MIC, >2 μg/ml), according to EUCAST breakpoints (http://www.eucast.org/).
dDomains of the CrrB protein predicted by SMART software are indicated as follows: TM, transmembrane domain (amino acids 12–34); HAMP, histidine kinase, adenylyl cyclase, methyl binding protein, and phosphatase domain (amino acids 81–135); HisKA, histidine kinase A (phosphoacceptor) domain (amino acids 136–200).
eAmino acid positions where mutations have been detected.
fMICs of colistin after complementation with a wild-type CrrB protein (with plasmid crrB-pTRIC).
gThe colistin-susceptible isolate Af44a is the isogenic colistin-susceptible counterpart of Af44b.
Three of the CrrB amino acid substitutions were located in the histidine kinase A (HisKA) phosphoacceptor domain (amino acids 136 to 200) (Table 1). Previously, four mutations in this HisKA domain were found to be involved in colistin resistance (6) (Table 1). Among them, two were at the same position as the mutations observed in our strains (amino acids 141 and 151), but the amino acid changes were different. The fourth strain presented a mutation in the HAMP domain of CrrB, whereas a single mutation in this domain has been shown (5). All of the substitutions in the CrrB protein, known to be responsible for colistin resistance, are shown in Table 1.
Determination of the colistin MICs by use of the reference broth microdilution method showed a high level of colistin resistance (MIC of colistin, >128 μg/ml for all four isolates with mutated crrB genes) (Table 1). The high MICs of colistin in the K. pneumoniae strains are in accordance with results reported by Cheng et al. (6) (Table 1). Furthermore, Wright et al. (5) reported lower MICs of colistin (16 μg/ml) for two isolates exhibiting CrrB mutations. However, MICs were determined by use of the Etest strip technique, which is known to underestimate colistin MIC values (12).
We performed complementation assays to confirm the involvement of the mutated crrB gene in the colistin resistance phenotype. A recombinant plasmid (pTRIC) was built by cloning a triclosan resistance gene (mFabI) (13) into the low-copy-number plasmid pBR322. The wild-type crrB gene from the colistin-susceptible K. pneumoniae strain Af44a (MIC of colistin, 0.25 μg/ml) was amplified by PCR and cloned into this plasmid. The recombinant crrB-pTRIC plasmid and the pTRIC plasmid were separately transformed by electroporation into the four resistant strains presenting the mutations in the crrB gene. Transformants were selected by overnight incubation at 37°C on Mueller-Hinton agar supplemented with triclosan (1 μM), and the recombinant clones were checked by PCR and sequencing. MICs of colistin for the transformants revealed that production of a wild-type CrrB protein (crrB-pTRIC plasmid) restored the susceptibility to colistin in all isolates (MIC, ≤0.5 μg/ml) (Table 1), confirming that the different substitutions in CrrB were, respectively, responsible for the resistant phenotypes. As expected, transformation with the pTRIC plasmid used as a negative control did not restore any susceptibility to colistin.
All isolates presenting a CrrB amino acid change produced a carbapenemase (NDM-1, OXA-181, or KPC-2) (Table 1). They were recovered from patients who had been treated with colistin in Colombia, France, Greece, and South Africa. For the patient harboring the colistin-resistant isolate Af44b, one colistin-susceptible isolate (Af44a) was recovered before the colistin treatment, and pulsed-field gel electrophoresis analysis confirmed the clonal relationship of the two isolates (data not shown). Sequencing of the crrB gene of the Af44a isogenic susceptible strain identified a wild-type CrrB protein, reinforcing the hypothesis that the in vivo emergence of colistin resistance under colistin pressure might be related to CrrB mutations in some instances.
In conclusion, four novel mutations in the CrrB protein were identified at the sources of acquisition of high-level colistin resistance among carbapenemase-producing K. pneumoniae.
ACKNOWLEDGMENTS
This work was funded by the University of Bordeaux and the University of Fribourg and by grants from the ANIWHA ERA-NET project, Switzerland, and the Office Fédéral de la Santé Publique, Bern, Switzerland (grant no. 16009294).
REFERENCES
- 1.Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 2.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. [DOI] [PubMed] [Google Scholar]
- 3.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21:pii=30280 http://www.eurosurveillance.org/images/dynamic/EE/V21N27/art22525.pdf. [DOI] [PubMed] [Google Scholar]
- 4.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright MS, Suzuki Y, Jones MB, Marshall SH, Rudin SD, van Duin D, Kaye K, Jacobs MR, Bonomo RA, Adams MD. 2015. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother 59:536–543. doi: 10.1128/AAC.04037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng YH, Lin TL, Lin YT, Wang JT. 2016. Amino acid substitutions of CrrB responsible for resistance to colistin through CrrC in Klebsiella pneumoniae. Antimicrob Agents Chemother 60:3709–3716. doi: 10.1128/AAC.00009-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Jayol A, Bontron S, Villegas MV, Ozdamar M, Turkoglu S, Nordmann P. 2015. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother 70:75–80. [DOI] [PubMed] [Google Scholar]
- 8.Jayol A, Nordmann P, Desroches M, Decousser JW, Poirel L. 2016. Acquisition of broad-spectrum cephalosporin resistance leading to colistin resistance in Klebsiella pneumoniae. Antimicrob Agents Chemother 60:3199–3201. doi: 10.1128/AAC.00237-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordmann P, Jayol A, Poirel L. 2016. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg Infect Dis 22:1038–1043. doi: 10.3201/eid2206.151840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayol A, Poirel L, Brink A, Villegas MV, Yilmaz M, Nordmann P. 2014. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Chemother 58:4762–4766. doi: 10.1128/AAC.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jayol A, Nordmann P, Brink A, Poirel L. 2015. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother 59:2780–2784. doi: 10.1128/AAC.05055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Humphries RM. 2015. Susceptibility testing of the polymyxins: where are we now? Pharmacotherapy 35:22–27. [DOI] [PubMed] [Google Scholar]
- 13.Jang C-W, Magnuson T. 2013. A novel selection marker for efficient DNA cloning and recombineering in E. coli. PLoS One 8:e57075. doi: 10.1371/journal.pone.0057075. [DOI] [PMC free article] [PubMed] [Google Scholar]