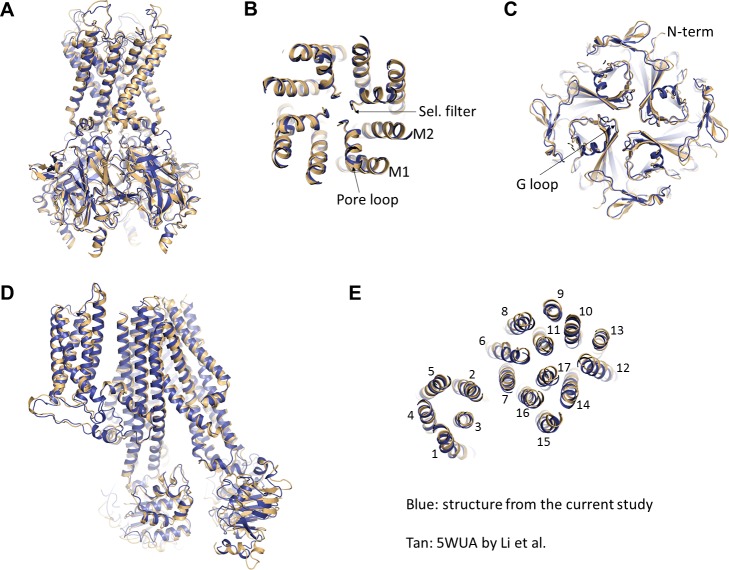
Figure 8. Comparison of the current structure with the GBC-bound, ATP-free structure from Li et al.(PDB ID: 5WUA).
(A) Overlay of the Kir6.2 structure viewed on the side. (B) Overlay of the Kir6.2 membrane helices viewed from the top. (C) Overlay of the Kir6.2 cytoplasmic domain. (D) Side view of the overlay of the SUR1 structure. (E) Overlay of the SUR1 transmembrane helices 1–17 viewed from the top. In all panels, the higher resolution structure from the current study is colored in blue, and the 5WUA structure from Li et al. is colored in tan.
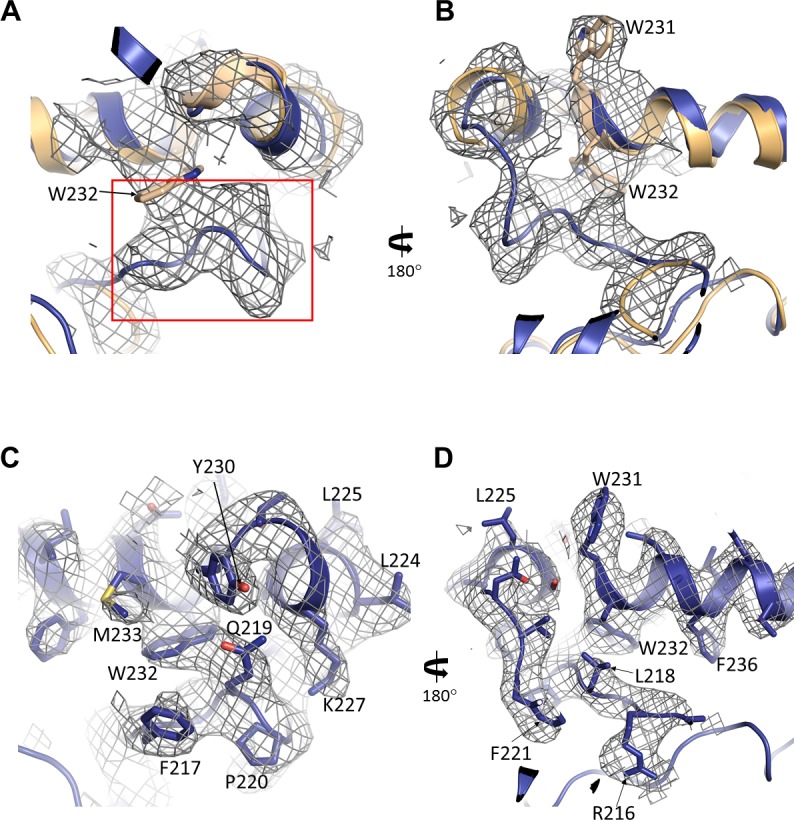
Figure 8—figure supplement 1. Reinterpretation of the GBC cryo-EM density proposed in Li et al.