Abstract
Increasing evidence indicates that periodontitis affects non-alcoholic fatty liver disease (NAFLD). We examined the relationship between periodontal bacterial infection and clinical/biochemical parameters in 52 NAFLD patients. Anti-Aggregatibacter actinomycetemcomitans (Aa) antibody titers correlated positively with visceral fat, fasting plasma insulin, and HOMA-IR; and negatively with the liver/spleen ratio. C57BL/6J mice (8-weeks-old) were given Aa or saline (control) for 6 weeks, and were fed either normal chow (NCAa, NCco) or high-fat diet (HFAa and HFco). NCAa and HFAa mice presented impaired glucose tolerance and insulin resistance compared to control mice. HFAa mice showed higher hepatic steatosis than HFco animals. Liver microarray analysis revealed that 266 genes were differentially expressed between NCAa and NCco mice. Upregulated genes in Aa-administrated mice were enriched for glucagon signaling pathway, adipocytokine signaling pathway and insulin resistance. Consistently, plasma glucagon concentration was higher in NCAa mice. In addition, Akt phosphorylation was lower in the liver of NCAa/HFAa than in NCco/HFco mice. Based on 16S rRNA sequencing, Aa administration changed composition of the gut microbiota. Metagenome prediction in gut microbiota showed upregulation of fatty acid biosynthesis and downregulation of fatty acid degradation in Aa-administered mice. Thus, infection with Aa affects NAFLD by altering the gut microbiota and glucose metabolism.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver disease. NAFLD includes a wide spectrum of conditions ranging from non-alcoholic fatty liver (NAFL) to non-alcoholic steatohepatitis (NASH). Generally, NAFL shows a non-progressive clinical course, whereas NASH is a more serious form of NAFLD and can progress to cirrhosis or hepatocellular carcinoma1,2. Many risk factors related to the development of NAFLD have been proposed, such as obesity, diabetes, and insulin resistance3,4.
Periodontal disease is an inflammatory disorder caused by pathogenic oral microorganisms that can lead to the destruction of alveolar bone and connective tissues around the teeth5,6. Periodontal bacteria present in dental plaque possess various virulence factors, such as lipopolysaccharide (LPS), fimbriae, and enzymes, which can trigger inflammation in periodontal tissues7. Considering that the bacterial flora of the oral cavity differs from that of the gut8, there is a possibility that swallowed bacteria could affect the composition of gut microbiome.
Given the infectious nature of periodontal disease, patients with the disease show elevated IgG antibody titers against periodontopathic bacteria. These antibody titers correlate with severity of periodontal disease9. Periodontal infection has long been associated with an increased risk of various diseases, such as atherosclerotic vascular disease10 or type 2 diabetes11. Recently, several studies have indicated that periodontitis might influence NAFLD12,13. In addition, gut microbiota appear to mediate development and progression of NAFLD14–16.
In the present study, we first examined the relationship between periodontal disease and NAFLD by measuring IgG antibody titers to periodontopathic bacteria in NAFLD patients. Based on the results, we then investigated the influence of Aggregatibacter actinomycetemcomitans infection on gut microbiota, glucose/lipid metabolism, and liver steatosis in mice.
Results
Correlation between IgG antibody titers against periodontal pathogens and clinical/biochemical parameters in NAFLD patients
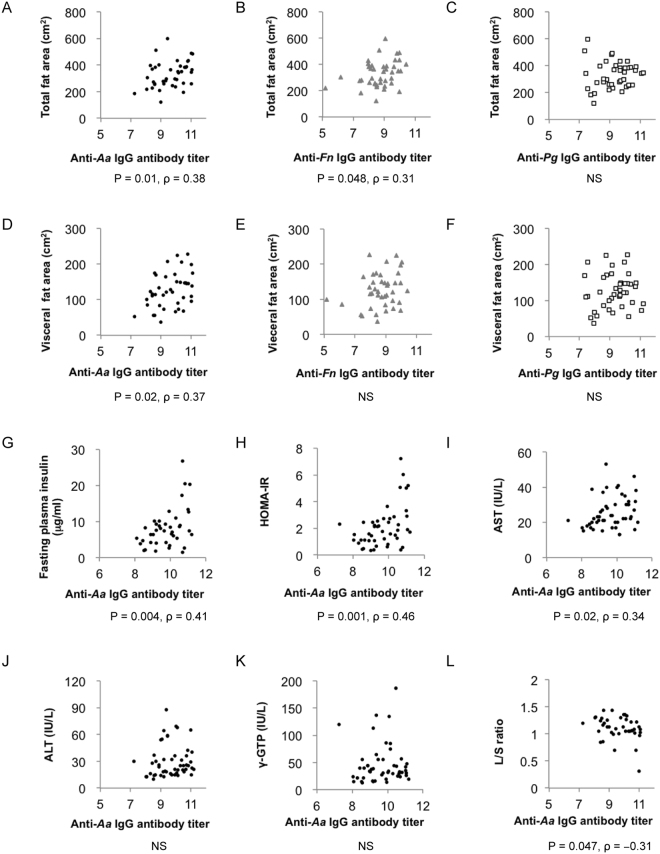
Clinical and biochemical characteristics of the subjects enrolled in this study are summarised in Table 1. We evaluated the correlation between IgG antibody titers to three major periodontopathic bacteria, Aggregatibacter actinomycetemcomitans ATCC 43718 (Aa), Fusobacterium nucleatum (Fn), Porphyromonas gingivalis ATCC 33277 (Pg), and clinical/biochemical parameters in NAFLD patients. Although there was no correlation between IgG antibody titers and body mass index (BMI), anti-Aa (P = 0.01, ρ = 0.38) and anti-Fn (P = 0.048, ρ = 0.31) antibody titers correlated significantly with total fat area evaluated by abdominal computed tomography (CT) scans (Fig. 1A and B). In contrast, no such correlation was observed for anti-Pg IgG antibody titer (Fig. 1C). Moreover, only anti-Aa IgG antibody titer showed a positive correlation with visceral fat area (Fig. 1D, P = 0.02, ρ = 0.37), whereas anti-Fn and anti-Pg titers did not (Fig. 1E and F). Anti-Aa IgG antibody titer correlated positively also with fasting plasma insulin (Fig. 1G, P = 0.004, ρ = 0.41) and the homeostasis model of assessment of insulin resistance (HOMA-IR) (Fig. 1H, P = 0.001, ρ = 0.46). A positive correlation was observed between anti-Aa IgG antibody titer and AST (Fig. 1I, P = 0.02, ρ = 0.34), but not ALT (Fig. 1J) or γ-GTP (Fig. 1K). Interestingly, anti-Aa IgG antibody titer showed a negative correlation with the liver/spleen (L/S) ratio (Fig. 1L, P = 0.047, ρ = −0.31). Correlations between anti-Fn IgG antibody titer/anti-Pg IgG antibody titer and several biochemical parameters are shown in Supplementary Figs S1 and S2.
Table 1.
Characteristics of the patients with NAFLD.
| Variables | All patients | Males | Females |
|---|---|---|---|
| (n = 52) | (n = 27) | (n = 25) | |
| Age (years) | 55 ± 13.8 | 48 ± 14.6 | 63 ± 7.5 |
| Weight (kg) | 68 ± 12.0 | 75 ± 11.0 | 59 ± 6.2 |
| BMI (kg/m2) | 26 ± 2.8 | 27 ± 3.2 | 26 ± 2.2 |
| Blood platelet count (×104/μL) | 22 ± 5.5 | 22 ± 5.3 | 22 ± 4.7 |
| AST (IU/L) | 26 ± 9.1 | 28 ± 9.5 | 23 ± 8.1 |
| ALT (IU/L) | 30 ± 18.2 | 37 ± 20.0 | 22 ± 12.2 |
| ALP (IU/L) | 243 ± 69.8 | 206 ± 63.2 | 284 ± 52.1 |
| γ-GTP (IU/L) | 57 ± 74.4 | 80 ± 95.8 | 31 ± 18.6 |
| Fasting plasma glucose (mg/dL) | 103 ± 19 | 106 ± 16 | 104 ± 11 |
| HOMA-IR | 2.1 ± 1.5 | 1.8 ± 1.1 | 2.4 ± 1.9 |
| Total cholesterol (mg/dL) | 219 ± 48 | 227 ± 46 | 218 ± 25 |
| High density lipoprotein (mg/dL) | 57 ± 14 | 54 ± 14 | 62 ± 12 |
| Triglyceride (mg/dL) | 145 ± 100 | 165 ± 128 | 124 ± 58 |
| L/S ratio | 1.00 ± 0.21 | 1.00 ± 0.26 | 1.00 ± 0.15 |
Data are shown as mean ± SD.
Figure 1.
Correlations between IgG antibody titers to periodontal pathogen and clinical/biochemical parameters in NAFLD patients (n = 52). Correlation between total fat area and (A) anti-Aa IgG antibody titer, (B) anti-Fn IgG antibody titer, (C) anti-Pg IgG antibody titer. Correlation between visceral fat area and (D) anti-Aa IgG antibody titer, (E) anti-Fn IgG antibody titer, (F) anti-Pg IgG antibody titer. Correlation between anti-Aa IgG antibody titer and (G) fasting plasma insulin, (H) HOMA-IR, (I) AST, (J) ALT, (K) γ-GTP, (L) L/S ratio.
A. actinomycetemcomitans administration causes increased body weight, impaired glucose tolerance, and insulin resistance
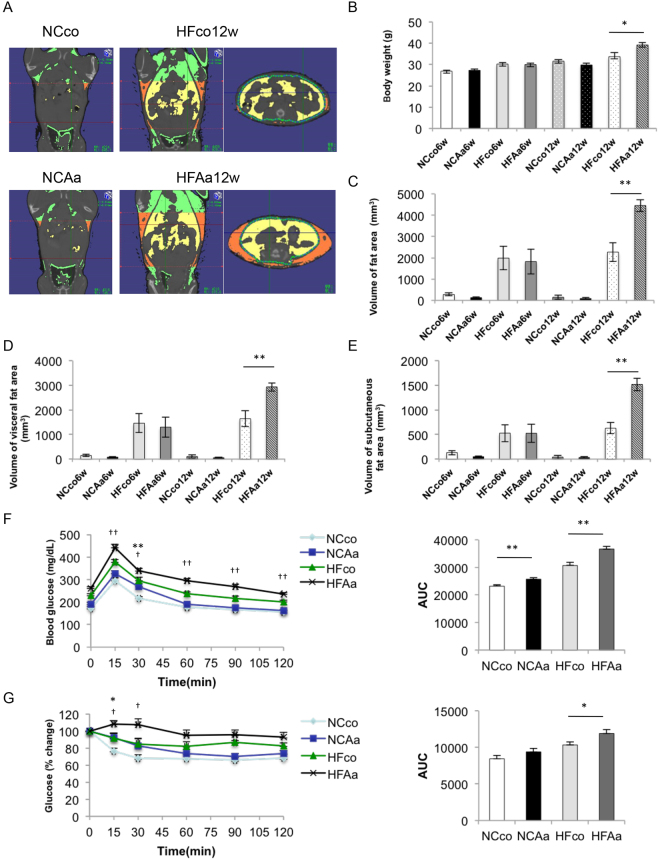
Based on the significant correlation between A. actinomycetemcomitans, as opposed to other bacteria, and clinical/biochemical parameters of NAFLD patients, we conducted a 3D micro-CT analysis to quantify total and separated (visceral and subcutaneous) fat volumes (Fig. 2A). After 12 weeks, body weight was significantly higher in mice given a high-fat diet with A. actinomycetemcomitans administration (HFAa) than in high-fat diet control (HFco) animals. No significant differences could be detected between normal chow diet control (NCco) mice and normal chow diet with A. actinomycetemcomitans administration (NCAa) mice at 6 and 12 weeks, or between HFco and HFAa mice at 6 weeks (Fig. 2B). Total body fat (Fig. 2C), visceral fat (Fig. 2D), and subcutaneous fat (Fig. 2E) volumes were significantly higher in HFAa mice compared to HFco mice at 12 weeks. To determine whether administration of A. actinomycetemcomitans induced impaired glucose tolerance and insulin resistance, we performed a glucose tolerance test (GTT) (Fig. 2F) and an insulin tolerance test (ITT) (Fig. 2G) on both dietary groups at 6 weeks. Accordingly, administration of A. actinomycetemcomitans caused impaired glucose tolerance and insulin resistance in both dietary groups.
Figure 2.
Comparison of body weight, body fat, glucose tolerance and insulin resistance among NCco, NCAa, HFco and HFAa mice. (A) Photographs of Micro-CT imaging. Yellow region represents visceral fat area and orange region represents subcutaneous fat area; (B) Body weight. (C) The volume of fat area, (D) the volume of visceral fat area and (E) the volume of subcutaneous fat area evaluated by Micro-CT imaging (n = 5). *P < 0.05, **P < 0.01. (F) OGTT (1 g/kg) and (G) ITT (1 U/kg) performed 6 h fasting at 6 weeks (n = 9–12). *P < 0.05, **P < 0.01 NCco vs NCAa, †P < 0.05, ††P < 0.01 HFco vs HFAa.
A. actinomycetemcomitans administration increases liver steatosis
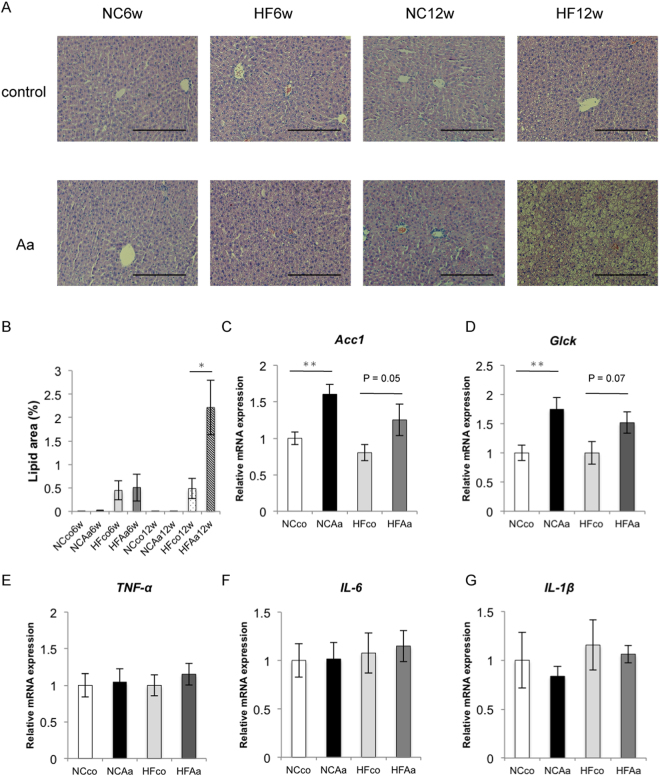
Histological analysis showed marked lipid accumulation in HFAa compared to HFco mice after 12 weeks (Fig. 3A and B). Oral administration of A. actinomycetemcomitans led to increased mRNA expression of Acetyl-CoA carboxylase (Acc1), an enzyme involved in lipid metabolism in the liver, and glucokinase (Glck) in mice fed a normal diet. Acc1 and Glck mRNA expression in the liver of HFAa mice was also generally higher than in that of HFco mice (P = 0.05 and 0.07, respectively) as well as normal chow diet mice (Fig. 3C and D). Moreover, mRNA expression of tumour necrosis factor-alpha (Tnfα), interleukin (Il)-6, and Il1β in the liver did not differ significantly between NCco and NCAa mice, or HFco and HFAa mice at 6 weeks (Fig. 3E–G).
Figure 3.
Evaluation of liver steatosis. (A) HE staining of liver tissue from NCco6w, NCAa6w, HFco6w, HFAa6w, NCco12w, NCAa12w, HFco12w and HFAa12w mice (row magnification × 200, Black bar = 100 μm), and (B) lipid area (%). (C) Acc1 mRNA expressions; (D) Glck mRNA expressions; (E) Tnfα mRNA expressions; (F) Il6 mRNA expressions; (G) Il1β mRNA expressions among NCco, NCAa, HFco and HFAa mice at 6 weeks (n = 9–12). *P < 0.05, **P < 0.01.
Liver microarray analysis with or without A. actinomycetemcomitans administration
To obtain a comprehensive overview of gene expression in the liver following administration of A. actinomycetemcomitans, microarray analysis was performed in NCco and NCAa mice.
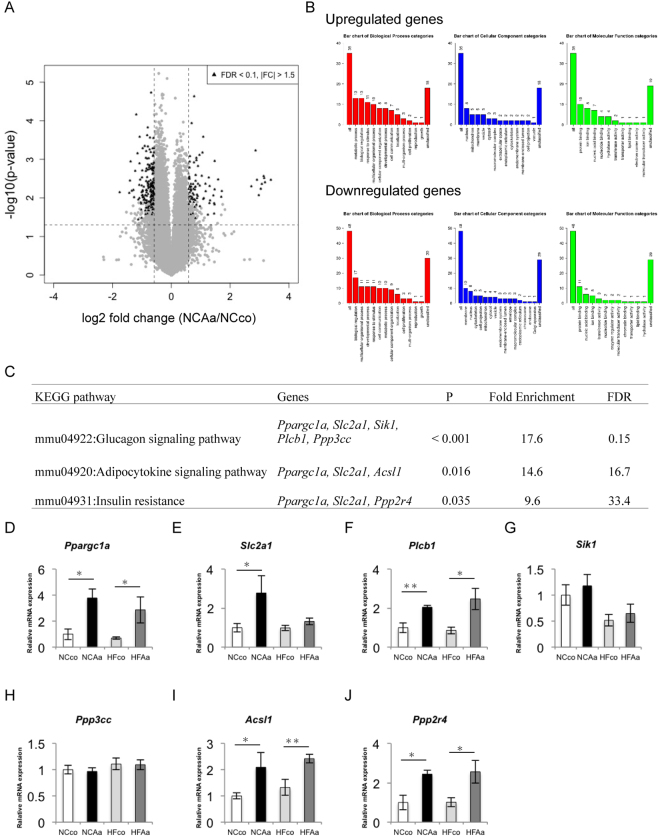
As shown in Fig. 4A, a total of 266 differentially expressed genes (DEGs) were identified, 71 of which were upregulated. All DEGs and corresponding Gene Ontology (GO) terms are listed in Supplementary Table S1, and those with higher expression (average normalised expression levels >first quantile) are clustered in the heatmap (Supplementary Fig. S3). Pathway analysis was used to uncover the significant pathways within differentially expressed gene sets according to the GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Of the data sets with DEGs, 58 upregulated and 128 downregulated Entrez genes were identified in NCAa compared to NCco animals. GO slim overviewed the ontology content in upregulated and downregulated DEGs, respectively (Fig. 4B). Pathway analysis showed that the glucagon signaling pathway was significantly enriched in upregulated DEGs. In addition, adipocytokine signaling pathway and insulin resistance showed P < 0.05 (Fig. 4C). Quantitative PCR validated significant hepatic upregulation of peroxisome proliferative activated receptor, gamma, coactivator 1 mRNA (Ppargc1a), solute carrier family 2 (facilitated glucose transporter), member 1 mRNA (Slc2a1), phospholipase C, beta 1 mRNA (Plcb1), acyl-CoA synthetase long-chain family member 1 mRNA (Acsl1), and protein phosphatase 2A activator, regulatory subunit B mRNA (Ppp2r4) in animals treated with A. actinomycetemcomitans and fed a normal chow diet and/or high-fat diet (Fig. 4D–J).
Figure 4.
Microarray analysis in liver between NCco and NCAa mice (n = 4). (A) Volcano plots. (B) Gene Ontology in DEGs. (C) KEGG pathway (P < 0.05) in upregulated DEGs. Analysis of gene expressions in liver in (D) Ppargc1a mRNA expressions; (E) Slc2a1 mRNA expressions; (F) Plcb1 mRNA expressions; (G) Sik1 mRNA; (H) Ppp3cc mRNA expressions; (I) Acsl1 mRNA expressions; (J) Ppp2r4 mRNA expressions among NCco, NCAa, HFco and HFAa mice at 6 weeks. *P < 0.05, **P < 0.01.
Glucose metabolism in the liver
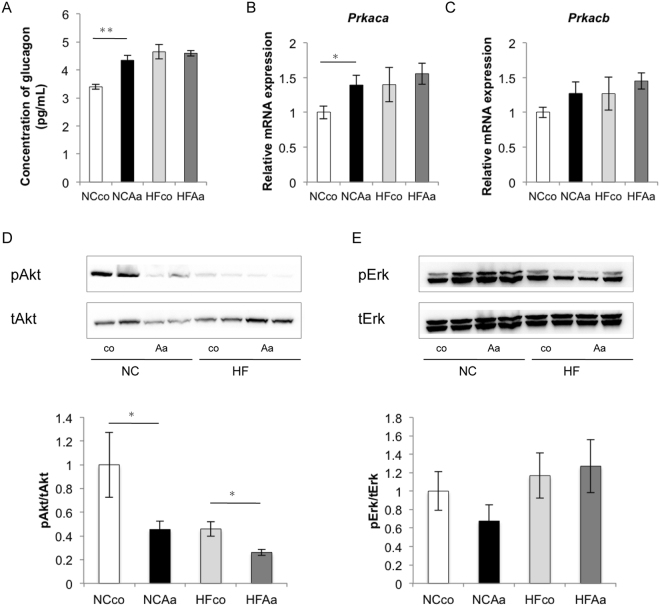
Based on pathway analysis results, we focused on glucose metabolism in the liver. Fasting plasma glucagon was increased in NCAa compared to NCco mice (Fig. 5A). Protein kinase cAMP-activated catalytic subunit alpha (Prkaca) mRNA expression increased significantly after A. actinomycetemcomitans administration in normally-fed animals (Fig. 5B), although no significant differences could be observed for its beta subunit (Prkacb) (Fig. 5C). Furthermore, Akt phosphorylation (pAkt) was significantly lower (by 54%) in the liver of NCAa compared to NCco mice, and in that of HFAa compared to HFco mice (by 43%) (Fig. 5D) without a difference in insulin’s activation of Erk (pErk) (Fig. 5E).
Figure 5.
Analysis of glucose metabolism and insulin resistance among NCco, NCAa, HFco and HFAa mice (n = 6–8). (A) Concentration of fasting plasma glucagon. Analysis of gene expressions in liver in (B) Prkaca mRNA expressions, (C) Prkacb mRNA expressions. (D) Total Akt and pAkt expressions, (E) total Erk and pErk expressions in liver. *P < 0.05, **P < 0.01.
Evaluation of gut microbiome composition based on 16S rRNA sequences
Rarefaction curves indicated that a sufficient number of reads were obtained for 16S rRNA gene analysis (Fig. 6A and Supplementary Fig. S4B). Principal component analysis (PCoA) showed that microbiome composition differed dramatically between mice fed a high-fat or a normal chow diet (Supplementary Fig. S4A). In addition, the number of operational taxonomic units (OTUs), Shannon index, and Chao1 differed significantly between normal chow and high-fat diet feeding mice (P < 0.01), suggesting diversity in gut microbiota were also significantly different between normal chow and high-fat diet feeding mice. To better understand the effect of A. actinomycetemcomitans administration itself, we evaluated gut microbiota separately according to the diet.
Figure 6.
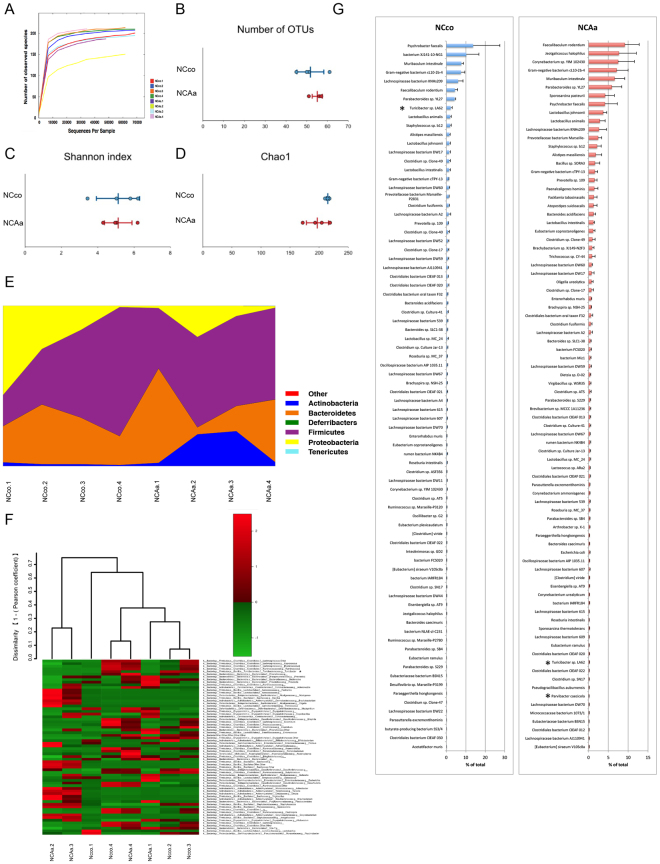
Evaluation of gut microbiome compositions based on 16S rRNA sequences between NCco and NCAa mice (n = 4). (A) rarefaction curve, (B) number of OTUs, (C) Shannon index, (D) Chao1 between NCco and NCAa mice. (E) Microbial composition at a Phylum level. (F) Microbial composition at a Genus level, dendrogram and heatmap constructed based on read abundance. (G) Rank distributions of the spices between NCco and NCAa mice (>0.1% relative abundance). The species name or 16S ribosomal RNA database ID in DDBJ is shown. *P < 0.05 between NCco and NCAa mice.
No significant differences in OTUs (Fig. 6B), Shannon index (Fig. 6C), and Chao1 (Fig. 6D) could be detected between NCco and NCAa mice. Although no significant differences in gut microbiota composition between NCco and NCAa mice were observed at a phylum level (Fig. 6E), the genus Turicibacter from the phylum Firmicutes was significantly underrepresented in NCAa compared to NCco mice (Fig. 6F). It should be noted that three more species showed significantly different abundance between NCco and NCAa mice (Supplementary Table S2). The most abundant species (>0.1%) in NCco and NCAa mice are presented in Fig. 6G.
A comparison of gut microbiota between HFco and HFAa mice is shown in Supplementary Fig. S4. No significant differences in OTUs, Shannon index, and Chao1 could be detected between HFco and HFAa mice. A same as normal chow diet feeding mice, there were no significant differences in gut microbiota composition between HFco and HFAa mice at a phylum level. The genus Turicibacter tended to be underrepresented in HFAa compared to HFco mice.
Metagenome prediction of gut microbiome
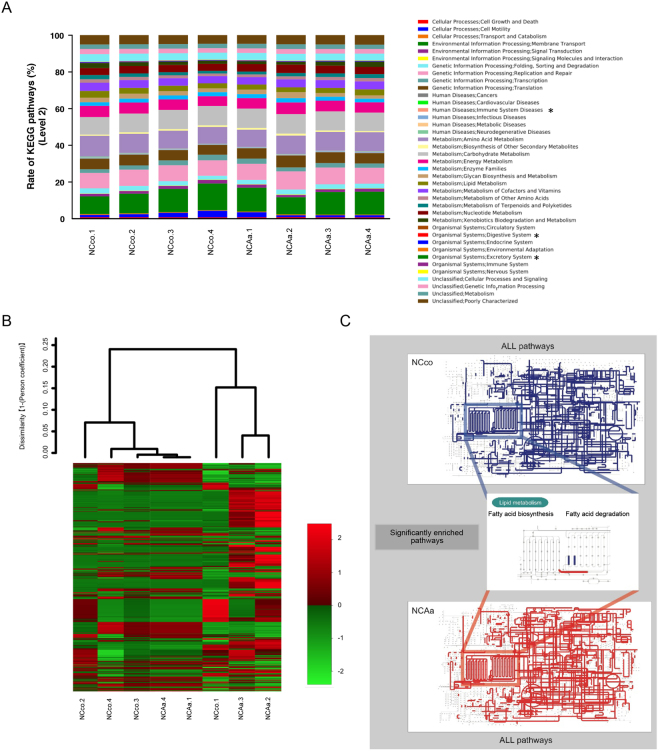
We performed PICRUSt analysis to predict the relative abundance of gene function in gut microbiome. NCco and NCAa mice presented significantly different functional composition at level 2, particularly with respect to excretory system, digestive system, and immune system diseases (Fig. 7A). Although metagenome prediction was not dramatically different between NCco and NCAa mice (Fig. 7B), 130 functional profiles were predicted to exhibit significant differences between NCco and NCAa mice (Supplementary Table S3). Interestingly, fatty acid biosynthesis was significantly increased, but fatty acid degradation was decreased in NCAa compared to in NCco mice (Fig. 7C).
Figure 7.
Metagenome prediction between NCco and NCAa mice. (A) Metagenome prediction of level-2 subsystem. *P < 0.05 between NCco and NCAa mice. (B) Dendrogram and heatmap constructed based on metagenome prediction. (C) Predicted KEGG pathways present in any of samples for NCco (upper figure) and NCAa (lower figure). Middle figure shows significantly enriched pathway. Blue: NCco. Red: NCAa.
Norfloxacin treatment in Aa-administered mice
No significant differences in body weight could be detected between NCAa and NCAa mice treated with norfloxacin (NCAaNor), or between HFAa and HFAa mice treated with norfloxacin (HFAaNor) at 12 weeks. Total body fat, visceral fat, and subcutaneous fat volumes tended to be lower in HFAaNor mice compared to HFAa mice at 12 weeks. In addition, norfloxacin treatment partially improved glucose tolerance and insulin resistance in Aa-administered mice in both dietary groups (Supplementary Fig. S5). Furthermore, norfloxacin treatment could partially supress lipid accumulation in the liver (Supplementary Fig. S6).
Discussion
The “Multiple hit” hypothesis postulates that multiple insults, such as insulin resistance, LPS, nutritional factors, gut microbiota, and genetic and epigenetic factors, act together to induce NAFLD17.
Periodontal diseases are caused predominantly by gram-negative bacteria present in the dental plaque18. A only one gram of dental plaque contains more than 1011 bacteria19. In this study, we measured IgG antibody titers against three major periodontopathic bacteria in NAFLD patients. Detection of oral bacteria in the saliva represents a single-point read-out, whereas the IgG antibody titer is an indicator of chronic infectious status caused by periodontal bacteria. Thus, in this study we focused on IgG antibody titers.
A. actinomycetemcomitans is frequently detected in severe periodontitis20 and is associated with aggressive periodontitis21. Among the three kinds of bacteria, only A. actinomycetemcomitans possess not only endotoxin, but also exotoxin22. Leukotoxin, the exotoxin of A. actinomycetemcomitans, renders this bacterium different from other periodontal pathogens. F. nucleatum has coaggregration properties, which favour the transport of periodontal bacteria23. P. gingivalis belongs to the “red complex”, a group of bacteria associated with periodontal disease24, and is the most common bacterium in periodontal infections.
The association of periodontal disease with NAFLD was first reported by Yoneda et al., who suggested that infection with P. gingivalis might be a risk factor for the development and progression of NAFLD12. A recent study using a mouse model showed that administration of P. gingivalis altered the gut microbiota25. In the present study, a positive correlation could be established only between anti-Aa IgG antibody titer and visceral body fat, whereas a negative correlation was seen with the L/S ratio. The mean value of the anti-Aa IgG antibody titer in 52 NAFLD patients was 9.7. We previously reported that the mean value of the anti-Aa IgG antibody titer after periodontal treatment in systemically healthy subjects was 6.99. Therefore, the anti-Aa IgG antibody titer appears higher in the 52 NAFLD patients than in systemically healthy subjects. Hence, we focused on A. actinomycetemcomitans as a risk factor of NAFLD.
KEGG pathway analysis in liver microarrays suggested the induction of metabolic disorders, such as enrichment of glucagon signaling pathway, insulin resistance and adipocytokine signaling pathway, following A. actinomycetemcomitans administration. What is more, fasting plasma glucagon was increased and Prkaca mRNA expression was significantly increased after A. actinomycetemcomitans administration in animals fed a normal chow diet. Binding of glucagon to its receptor on the hepatocyte plasma membrane leads to activation of adenylyl cyclase, production of the second messenger cyclic AMP, and stimulation of protein kinase A, which phosphorylates protein targets that work in concert to increase hepatic glucose output26. Moreover, compared to NCco and HFco animals, and also NCAa and HFAa mice showed lower pAkt/Akt but equal pErk/Erk levels in the liver. This finding is in line with the concept of “selective insulin resistance” by King et al.27. They reported that elevated levels of glucose, free fatty acids, and inflammatory cytokines caused by diabetes and insulin resistance selectively inhibited insulin’s antiatherogenic actions via the IRS/PI3K/Akt pathway, but not the Grb/Shc/MAPK pathway. In addition, HFAa mice showed enhanced hepatic steatosis by histological analysis.
The gut microbiota has been implicated in NAFLD through its effect on caloric salvage, host energy metabolism, proinflammatory signaling, and via direct hepatotoxicity of bacterial products28,29. Furthermore, metagenomic analysis of both microbiome composition and functional capacity correlated with host clinical parameters, energy homeostasis, and proinflammatory signaling in obese and NAFLD patients30. There are many reports showing that a high-fat diet dramatically changes gut microbiota in mice31,32. Our study also showed remarkable differences in microbiome composition and diversity between normal chow and high-fat diet feeding mice. OTUs of many bacterial species differed significantly between NCco and NCAa mice, and also HFco and HFAa mice, following administration of A. actinomycetemcomitans. Specifically, the genus Turicibacter was underrepresented in A. actinomycetemcomitans administrated mice. This genus reportedly correlates with production of butyric acid33. An increase in butyrate has been associated with improved insulin sensitivity34. Therefore, administration of A. actinomycetemcomitans may affect insulin resistance by altering the gut microbiome and lowering butyrate levels. We also showed that mRNA expression of Tnfα, Il6, and Il1β in the liver did not differ significantly in response to A. actinomycetemcomitans administration at 6 weeks in mice fed a normal chow diet or a high-fat diet. Therefore, insulin resistance is due to alteration of gut microbiota by A. actinomycetemcomitans rather than endotoxin-induced inflammation in the liver at 6 weeks.
There are no antibiotics specific to A. actinomycetemcomitans, so it is impossible to selectively target and disable any function of only A. actinomycetemcomitans. There are several reports about the effect of antibiotic treatment on glucose metabolism and gut microbiota. Antibiotic treatment improved glucose tolerance and liver lipid accumulation35, and also led to changes in gut microbiota36,37. Accordingly, we treated Aa-administered mice with the antibiotic norfloxacin. Norfloxacin treatment improved glucose tolerance, and partially suppressed fat volume and lipid accumulation in the liver. We hypothesised that even if bacterial activity was lost due to antibiotic treatment, norfloxacin could not completely block the effect of A. actinomycetemcomitans because of the bacterium’s endotoxin/exotoxin.
Intestinal mucosal permeability is greater in NAFLD patients than in healthy subjects15. Modulation the gut microbiota is associated with an increased intestinal permeability that precedes the development of metabolic endotoxemia, inflammation, and associated disorders36. Gut dysbiosis might cause translocation of bacteria from the gut to the bloodstream38. Interestingly, metagenome prediction of the gut microbiota showed increased fatty acid biosynthesis and reduced fatty acid degradation. At level 2, KEGG pathway analysis indicated that oral administration of A. actinomycetemcomitans altered gut microbiota, causing the upregulation of functional bacterial genes related to immune system disease, in addition, the digestive and excretory systems were capable of processing higher amounts of fatty acid. Accumulating fatty acid in the gut might be transported to the liver via the portal vein, and exacerbate liver steatosis. Moreover, a high concentration of free fatty acids has other effects on the liver, resulting in dyslipidaemia, hyperinsulinaemia, hyperglycaemia, and hepatic insulin resistance39. Furthermore, increasing the concentration of fatty acids in the gut may stimulate glucagon secretion by pancreatic alpha cells40.
Marked lipid accumulation in the liver was observed in mice fed a high-fat diet, but not in normal chow controls. However, a high-fat diet may in itself alter mRNA expression in the liver. In this study, we intended to evaluate how A. actinomycetemcomitans administration affected gene expression in the liver. Therefore, we performed microarray analysis to compare normal chow diet control and Aa-administered mice. Based on KEGG pathway analysis for differentially expressed genes in the liver of normal chow diet feeding mice, we focused on glucagon signaling pathway, adipocytokine signaling pathway, and insulin resistance. We also confirmed a significant hepatic upregulation of Ppargc1a, Plcb1, Acsl1, and Ppp2r4 in animals treated with A. actinomycetemcomitans in high-fat diet feeding.
Administration of A. actinomycetemcomitans impaired glucose tolerance and insulin resistance in mice fed a high-fat and normal chow diet. There are several reports linking periodontal disease and diabetes41,42, which can be explained by the above argument. Our results revealed that mRNA expression of Tnfα, Il6, and Il1β in the liver showed no significant differences between normal chow diet control and normal chow diet with A. actinomycetemcomitans, or between high-fat diet control and high-fat diet with A. actinomycetemcomitans. Therefore, insulin resistance is due to the alteration of gut microbiota by A. actinomycetemcomitans rather than endotoxin-induced inflammation. This study suggests that periodontal disease influences diabetes not only via inflammation, but also by altering gut microbiota.
In conclusion, A. actinomycetemcomitans might be a risk factor for NAFLD because it alters the gut microbiota. This is the first study to comprehensively evaluate gene expression in the liver and gut microbiota composition following A. actinomycetemcomitans administration in mice.
Methods
Patients
A total of 52 consecutive Japanese patients admitted to Eguchi Hospital and Saga Medical School for the treatment of NAFLD were enrolled in the present study. Patients with evidence of excessive alcohol intake (>20 g/day), other causes of liver diseases (e.g., viral hepatitis, autoimmune liver disease, biliary disease, and hepatocellular carcinoma), or use of antibiotic agents were excluded. NAFLD was diagnosed according to the following criteria: (1) a slight diffuse increase in bright homogeneous echoes in the liver parenchyma, with normal visualisation of the diaphragm and portal and hepatic vein borders, and normal hepatorenal contrast of echogenicity; (2) a diffuse increase in bright echoes in the liver parenchyma, with slightly impaired visualisation of the peripheral portal and hepatic vein borders; and (3) a marked increase in bright echoes at a shallow depth, with deep attenuation and impaired visualisation of the diaphragm and marked vascular blurring43, as previously described44. BMI was calculated as the body weight in kilograms divided by the square of the height in meters (kg/m2). Venous blood samples were taken from all subjects at around 09:00 a.m. following a 12-h overnight fast, and the levels of serum liver enzymes, blood platelet count (×104/μL), total cholesterol (mg/dL), triglycerides (mg/dL), high-density lipoprotein (mg/dL), plasma glucose (mg/dL), and plasma insulin (µg/mL) were determined by standard laboratory techniques. Insulin resistance was calculated by HOMA-IR, using the following formula: HOMA-IR = fasting plasma insulin × fasting plasma glucose/40545. Serum from the patients’ venous blood was collected and stored at −80 °C for the analysis of IgG antibody titers.
The study was performed in accordance with the Ethical Guidelines for Clinical Studies (2008 Notification number 415 of the Ministry of Health, Labor, and Welfare). All patients provided informed consent, and the study protocol was approved by the ethics committees of Saga University and Tokyo Medical and Dental University.
Abdominal computed tomography
Unenhanced spiral acquisition through the liver was obtained during a breath-hold at 5.0-mm collimation, 15.0-mm/rotation table speed (HQ mode, pitch 1:3), 120 kV (p), and auto mA (Light speed QXi; GE Healthcare, Little Chalfont, Buckinghamshire, UK). Images were reconstructed at 10-mm increments. All patients underwent abdominal CT in the morning after a 12-h overnight fast. CT numbers, expressed as Hounsfield units (HU) in a region of interest (ROI) of 40 mm2 in the periphery of the liver and the spleen, away from major vessels, were measured at five points in each organ. Mean numbers were used to determine the L/S ratio, an index of fat accumulation in the liver43,46. In addition, subcutaneous fat area (cm2) and visceral fat area (cm2) were measured at the umbilical level and calculated using FatScan software (N2 System, Osaka, Japan)47.
Cultivation of A.actinomycetemcomitans
A. actinomycetemcomitans ATCC 43718 was inoculated in ATCC Medium 44 (Brain Heart Infusion Broth) and cultured anaerobically (AnaeroPackR-Anaero for Susceptibility, Mitsubishi Gas Chemical Company Inc., Tokyo, Japan) at 37 °C for 24 h.
Serum IgG titer measurement
Specific serum IgG titers were measured by enzyme-linked immunosorbent assay (ELISA) as described previously9,48, using sonicated whole cell extracts of each periodontal bacteria. Briefly, 96-well microplates (EIA plate; Costar, Corning, NY, USA) were coated with sonicated A. actinomycetemcomitans, F. nucleatum, or P. gingivalis at 10 μg/mL in carbonate buffer and incubated for 2 h at 37 °C. After blocking with 2% bovine serum albumin in carbonate buffer, plates were washed with phosphate-buffered saline (PBS)-Tween (1×PBS, 0.05% Tween 20, pH 7.2). Serially diluted reference positive control serum (25–215, 200 µL per well) and single diluted patient serum (210, 200 µL per well) were added into each well, after which the plates were incubated for 1 h at 37 °C and washed again. Subsequently, 200 µL per well of alkaline phosphatase-conjugated goat anti-human IgG (Sigma, St. Louis, MO, USA) was added. Following incubation, plates were washed and developed with phosphatase substrate (Sigma). Optical density at 450 nm was read using a microplate reader (SoftMAX, Molecular Devices, Sunnyvale, CA, USA). Antibody titers were calculated according to the method of Wang et al.48.
Animals
C57BL/6J mice (8-weeks-old; Sankyo Laboratory, Tokyo, Japan) were allowed free access to water and food throughout the experimental period. Mice were randomly divided into four groups: NCco, NCAa, HFco, and HFAa. HFAa and HFco mice were fed High-fat diet 32 (Japan CLEA, Tokyo, Japan); this feed contains 506.8 kcal/100 g (57.5% from fat, 19.7% from protein, and 22.8% from carbohydrate). A total of 108 A. actinomycetemcomitans cells suspended in 100 μL of physiological saline solution were given to NCAa and HFAa mice via a feeding needle. The suspension was given 6 times per week for 6 weeks. NCco and HFco mice were given saline only. To block the effect of A. actinomycetemcomitans, norfloxacin was administered (3 g/mouse) 6 times per week for 6 weeks.
GTT and ITT were performed as previously described49. Briefly, after fasting for 6 h, mice were fed glucose by oral gavage (1 g/kg) or were given an intraperitoneal injection of insulin (1 U/kg) (Humalin R; Eli Lilly and Company, Indianapolis, IN, USA). Glucose concentration was determined with a glucose meter (ACCU-CHEK ST Meter; Roche, Basel, Switzerland). The area under the curve (AUC) (0–120 min) was calculated for each group of mice. Plasma glucagon was measured by the Glucagon ELISA kit (Mercodia, Uppsala, Sweden) after fasting for 6 h.
To evaluate insulin signaling, we fasted the mice for 6 h, injected them with insulin at 1.5 U/kg, and waited 20 min before sacrificing them and freezing the liver at −80 °C.
All experiments with mice were conducted according to the Guidelines for Proper Conduct of Animal Experiments, Science Council of Japan, and protocols were approved by the Animal Care Committee of the Experimental Animal Center at Tokyo Medical and Dental University (0170225 A).
Evaluation of visceral and subcutaneous fat in vivo
Micro-CT imaging was performed with a micro-CT unit (RmCT2; Rigaku Corporation, Tokyo, Japan) with FOV60 for 2 min, on mice anesthetised with isoflurane (induction 2%, maintenance 1.5%). The CT images were visualised and analysed using CTAtlas Metabolic Analysis (ver. 2.03) software (Rigaku Corporation). The fat tissue HU value was adjusted to between −350.0 and −145.0 in accordance with the manufacturer’s instructions. Measurement of body fat volume was limited to the abdominal region, with the initial point set at the diaphragm. Body fat was then divided into visceral fat and subcutaneous fat along the ribs50.
RNA preparation and quantitative real-time PCR
Total RNA was extracted from liver tissue using the NucleoSpin® RNA kit (TaKaRa Bio, Shiga, Japan) and quantified by measuring absorbance at 260 and 280 nm. Next, 500 ng total RNA was reverse-transcribed to cDNA using the PrimeScriptTM RT Master Mix (TaKaRa Bio), and real-time PCR was performed using the Thermal Cycler Dice® Real Time System II (TaKaRa Bio). PCR mixtures were prepared by SYBR® Premix Ex Taq™ II (TaKaRa Bio), and PCR conditions were determined according to the manufacturer’s protocol. Gene expression levels were normalised to those of the reference gene, 36b4. PCR primers used in the study are listed in Supplementary Table S4.
Liver lysate preparation and western blotting
Lysates from frozen livers were extracted with RIPA buffer (150 mM NaCl, 50 mM Tris, pH 7.6, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, supplemented with protease and phosphatase inhibitors), cleared, and used for western blotting51. Antibodies against Erk, pErk, Akt, and pAkt were purchased from Cell Signaling Technology (Danvers, MA, USA); goat anti-rabbit IgG was purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA).
Liver histological analysis
Liver samples were fixed with 4% paraformaldehyde in PBS for 24 h and embedded in paraffin. Next, 5-μm-thick sections were processed with haematoxylin and eosin (HE); the lipid area was quantified using ImageJ software52.
Microarray and data analysis
Total RNA was extracted from the liver of NCco and NCAa mice, and run on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) to check sample quality. The Agilent Low Input Quick Amp Labeling kit was used to generate cRNA with a sample input of 200 ng total RNA for single-color microarray (Cy3) analysis according to the manufacturer’s instructions. Then, cRNA was analysed by hybridisation onto an Agilent SurePrint G3 Unrestricted Gene Expression 8 × 60 K Microarray. Fluorescence signal from the hybridised microarrays was detected using the Agilent Microarray Scanner System. Raw microarray data were extracted using Feature Extraction Software (ver. 11.0.1.1; Agilent Technologies).
Microarray data were 75% tile-normalised and log2-transformed according to the manufacturer’s recommendation53 using R software (ver. 3.3.2). The Limma Bioconductor package (ver. 3.30.4)54 was used to identify DEGs. Statistical significance was measured by Benjamin and Hochberg’s false discovery rate (FDR) to account for multiple testing. DEGs were selected according to FDR < 0.1 and |fold-change| > 1.5. Overrepresentation enrichment analyses for DEGs were performed with the WEB-based Gene SeT AnaLysis Toolkit (http://www.webgestalt.org)55 using GO and KEGG pathway databases.
16S rRNA gene sequencing and Illumina sequence data processing
DNA was extracted from mouse faeces using the NucleoSpin Microbial DNA kit (Machery Nagel, Düren, Germany) and purified with the Agencourt AMPure XP PCR system (Beckman Coulter, Beverly, MA, USA). A multiplexed amplicon library covering the 16S rDNA (V3-V4) region was generated from extracted DNA samples using the 16S (V3-V4) Metagenomic Library Construction Kit for NGS (TaKaRa Bio) following the manufacturer’s protocol and purified again with the AMPure XP system. The Illumina Miseq platform (Illumina Inc., San Diego, CA, USA) was used to generate 250-bp paired-end sequences. The obtained sequence data are available in the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/) under accession number DRA005604 and DRA006060. A total of 2,031,033 sequence reads were generated, corresponding to an average of 126939.6 (range, 90,723–180,451) reads per sample. Noise, low quality sequences, pyrosequencing errors, and chimeras were removed from the data set. The preprocessed reads were clustered into OTUs at 97% identity using the CD-HIT-OTU pipeline (http://weizhongli-lab.org/cd-hit-otu/, ver. 0.01)56.
Taxonomic assignment and metagenome prediction
OTUs were processed and analysed with the Quantitative Insights into Microbial Ecology (QIIME, ver. 1.8) software package57. Taxonomic classification of the sequences at the phylum or genus level was determined using the RDP classifier (ver. 2.2) with default parameters against the GreenGenes database (ver.:gg_13_8). Taxonomic assignment was refined at the species level based on the 16S rRNA database (DNA Data Bank of Japan, as of July 18, 2017) using BLASTN. Hits with E values ≤ 1e-5 were considered significant. Alpha-diversity indices were estimated from the number of observed OTUs, and the Shannon diversity index58 was used as a measure of species richness and evenness. To calculate species richness based on OTUs, rarefaction curves were generated using QIIME with default parameters. At the same time, the PICRUSt (ver. 1.0.0) bioinformatics software package59 was employed to generate metabolic predictions based on closed OTUs at a 97% similarity level. Samples were normalised by randomly resampling the sequences to 10,000 reads per sample using the Seqtk application (https://github.com/lh3/seqtk). The analysed OTUs were normalized to 16S rRNA copy number. Functional composition of the data was predicted based on the KEGG database60. Dendrograms with heatmaps were visualised using R software (ver. 3.3.2). Dissimilarity values (1 – Pearson correlation) were clustered using average linkage methods.
Statistical analysis
Data distribution was assessed using the Shapiro-Wilk test. Correlation between anti-periodontal bacteria IgG antibody titers and clinical/biochemical parameters was evaluated by Spearman’s rank correlation coefficient. In animal experiments, Student’s t-test was applied to compare two groups. One-way analysis of variance, followed by a t-test with Bonferroni connection, was performed for multiple groups’ comparisons using SPSS 22.0 software (SPSS Inc., Chicago, IL, USA). A value of P < 0.05 was considered statistically significant.
Electronic supplementary material
Supplementary Dataset S1, Supplementary Dataset S2, Supplementary Dataset S3.
Acknowledgements
This work was supported by the Japan Society for the Promotion of Science (26463128 to S.K. and 26463129 to Y.T). S.K. is recipient of a grant from the 8020 Promotion Foundation and the Public Health Research Foundation. The authors would like to appreciate Takashi Kohda, Department of Epigenetics, Medical Research Institute, Tokyo Medical and Dental University (TMDU) for his support for experiments. This study was supported by Nanken-Kyoten, TMDU.
Author Contributions
R.K. performed most of the experiments and wrote the 1st draft of the manuscript. S.K., S.M., A.O., S.U., Naoki S., K.W., Y.T., and Y.I. assisted in some studies and reviewed the manuscript. H.T., Y.K., Y.E., and K.A. examined subjects and evaluated clinical and biochemical parameters. Noriko S. and N.M. provided expertise on microarray analysis. Y.T. and T.S. provided advice on 16S rRNA sequencing analysis. S.K. and H.T. supervised all the studies and the writing of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
A correction to this article is available online at https://doi.org/10.1038/s41598-018-23000-6.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14260-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sayaka Katagiri, Email: katagiri.peri@tmd.ac.jp.
Hirokazu Takahashi, Email: takahas2@cc.saga-u.ac.jp.
References
- 1.Liou I, Kowdley KV. Natural history of nonalcoholic steatohepatitis. Journal of clinical gastroenterology. 2006;40(Suppl 1):S11–16. doi: 10.1097/01.mcg.0000168644.23697.31. [DOI] [PubMed] [Google Scholar]
- 2.Torres DM, Williams CD, Harrison SA. Features, diagnosis, and treatment of nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2012;10:837–858. doi: 10.1016/j.cgh.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Marchesini G, et al. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatology. 2003;37:917–923. doi: 10.1053/jhep.2003.50161. [DOI] [PubMed] [Google Scholar]
- 4.Bril F, et al. Relationship between disease severity, hyperinsulinemia, and impaired insulin clearance in patients with nonalcoholic steatohepatitis. Hepatology. 2014;59:2178–2187. doi: 10.1002/hep.26988. [DOI] [PubMed] [Google Scholar]
- 5.Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 6.Nassar H, Kantarci A, van Dyke TE. Diabetic periodontitis: a model for activated innate immunity and impaired resolution of inflammation. Periodontol 2000. 2007;43:233–244. doi: 10.1111/j.1600-0757.2006.00168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolenbrander, P. E. et al. Communication among oral bacteria. Microbiol Mol Biol Rev66, 486–505, table of contents (2002). [DOI] [PMC free article] [PubMed]
- 8.Koren O, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, et al. Antibody response after single-visit full-mouth ultrasonic debridement versus quadrant-wise therapy. J Clin Periodontol. 2006;33:632–638. doi: 10.1111/j.1600-051X.2006.00963.x. [DOI] [PubMed] [Google Scholar]
- 10.Bahekar AA, Singh S, Saha S, Molnar J, Arora R. The prevalence and incidence of coronary heart disease is significantly increased in periodontitis: a meta-analysis. Am Heart J. 2007;154:830–837. doi: 10.1016/j.ahj.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 11.Salvi GE, Carollo-Bittel B, Lang NP. Effects of diabetes mellitus on periodontal and peri-implant conditions: update on associations and risks. Journal of clinical periodontology. 2008;35:398–409. doi: 10.1111/j.1600-051X.2008.01282.x. [DOI] [PubMed] [Google Scholar]
- 12.Yoneda M, et al. Involvement of a periodontal pathogen, Porphyromonas gingivalis on the pathogenesis of non-alcoholic fatty liver disease. BMC gastroenterology. 2012;12:16. doi: 10.1186/1471-230X-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arimatsu K, et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci Rep. 2014;4:4828. doi: 10.1038/srep04828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abu-Shanab A, Quigley EM. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2010;7:691–701. doi: 10.1038/nrgastro.2010.172. [DOI] [PubMed] [Google Scholar]
- 15.Jiang W, et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. 2015;5:8096. doi: 10.1038/srep08096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tilg H, Cani PD, Mayer EA. Gut microbiome and liver diseases. Gut. 2016;65:2035–2044. doi: 10.1136/gutjnl-2016-312729. [DOI] [PubMed] [Google Scholar]
- 17.Buzzetti E, Pinzani M, Tsochatzis EA. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metabolism. 2016;65:1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Listgarten MA. Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J Periodontol. 1976;47:1–18. doi: 10.1902/jop.1976.47.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Jan Lindhe, N. P. L. Thorkild Karring. Clinical Periodontology and Implant Dentistry (2008).
- 20.Mandell RL, Socransky SS. A selective medium for Actinobacillus actinomycetemcomitans and the incidence of the organism in juvenile periodontitis. J Periodontol. 1981;52:593–598. doi: 10.1902/jop.1981.52.10.593. [DOI] [PubMed] [Google Scholar]
- 21.Fine DH, et al. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: longitudinal cohort study of initially healthy adolescents. J Clin Microbiol. 2007;45:3859–3869. doi: 10.1128/JCM.00653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baehni P, Tsai CC, McArthur WP, Hammond BF, Taichman NS. Interaction of inflammatory cells and oral microorganisms. VIII. Detection of leukotoxic activity of a plaque-derived gram-negative microorganism. Infect Immun. 1979;24:233–243. doi: 10.1128/iai.24.1.233-243.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Signat B, Roques C, Poulet P, Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol. 2011;13:25–36. [PubMed] [Google Scholar]
- 24.Rôças IN, Siqueira JF, Santos KR, Coelho AM. “Red complex” (Bacteroides forsythus, Porphyromonas gingivalis, and Treponema denticola) in endodontic infections: a molecular approach. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2001;91:468–471. doi: 10.1067/moe.2001.114379. [DOI] [PubMed] [Google Scholar]
- 25.Nakajima M, et al. Oral Administration of P. gingivalis Induces Dysbiosis of Gut Microbiota and Impaired Barrier Function Leading to Dissemination of Enterobacteria to the Liver. PLoS One. 2015;10:e0134234. doi: 10.1371/journal.pone.0134234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang G, Zhang BB. Glucagon and regulation of glucose metabolism. Am J Physiol Endocrinol Metab. 2003;284:E671–678. doi: 10.1152/ajpendo.00492.2002. [DOI] [PubMed] [Google Scholar]
- 27.King GL, Park K, Li Q. Selective Insulin Resistance and the Development of Cardiovascular Diseases in Diabetes: The 2015 Edwin Bierman Award Lecture. Diabetes. 2016;65:1462–1471. doi: 10.2337/db16-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parnell JA, Raman M, Rioux KP, Reimer RA. The potential role of prebiotic fibre for treatment and management of non-alcoholic fatty liver disease and associated obesity and insulin resistance. Liver Int. 2012;32:701–711. doi: 10.1111/j.1478-3231.2011.02730.x. [DOI] [PubMed] [Google Scholar]
- 29.Le Roy T, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62:1787–1794. doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- 30.Raman M, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11:868–875.e861–863. doi: 10.1016/j.cgh.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 31.Hildebrandt MA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724.e1711–1712. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cani PD, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 33.Zhong Y, Nyman M, Fåk F. Modulation of gut microbiota in rats fed high-fat diets by processing whole-grain barley to barley malt. Mol Nutr Food Res. 2015;59:2066–2076. doi: 10.1002/mnfr.201500187. [DOI] [PubMed] [Google Scholar]
- 34.Vrieze A, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–916.e917. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 35.Membrez M, et al. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J. 2008;22:2416–2426. doi: 10.1096/fj.07-102723. [DOI] [PubMed] [Google Scholar]
- 36.Cani PD, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 37.Fujisaka S, et al. Antibiotic effects on gut microbiota and metabolism are host dependent. J Clin Invest. 2016;126:4430–4443. doi: 10.1172/JCI86674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato J, et al. Gut dysbiosis and detection of “live gut bacteria” in blood of Japanese patients with type 2 diabetes. Diabetes care. 2014;37:2343–2350. doi: 10.2337/dc13-2817. [DOI] [PubMed] [Google Scholar]
- 39.Arner P. Insulin resistance in type 2 diabetes: role of fatty acids. Diabetes Metab Res Rev. 2002;18(Suppl 2):S5–9. doi: 10.1002/dmrr.254. [DOI] [PubMed] [Google Scholar]
- 40.Hong J, Jeppesen PB, Nordentoft I, Hermansen K. Fatty acid-induced effect on glucagon secretion is mediated via fatty acid oxidation. Diabetes Metab Res Rev. 2007;23:202–210. doi: 10.1002/dmrr.663. [DOI] [PubMed] [Google Scholar]
- 41.Khader YS, Dauod AS, El-Qaderi SS, Alkafajei A, Batayha WQ. Periodontal status of diabetics compared with nondiabetics: a meta-analysis. J Diabetes Complications. 2006;20:59–68. doi: 10.1016/j.jdiacomp.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Lalla E, Papapanou PN. Diabetes mellitus and periodontitis: a tale of two common interrelated diseases. Nat Rev Endocrinol. 2011;7:738–748. doi: 10.1038/nrendo.2011.106. [DOI] [PubMed] [Google Scholar]
- 43.Saadeh S, et al. The utility of radiological imaging in nonalcoholic fatty liver disease. Gastroenterology. 2002;123:745–750. doi: 10.1053/gast.2002.35354. [DOI] [PubMed] [Google Scholar]
- 44.Eguchi Y, et al. Visceral fat accumulation and insulin resistance are important factors in nonalcoholic fatty liver disease. J Gastroenterol. 2006;41:462–469. doi: 10.1007/s00535-006-1790-5. [DOI] [PubMed] [Google Scholar]
- 45.Haffner SM, Kennedy E, Gonzalez C, Stern MP, Miettinen H. A prospective analysis of the HOMA model. The Mexico City Diabetes Study. Diabetes Care. 1996;19:1138–1141. doi: 10.2337/diacare.19.10.1138. [DOI] [PubMed] [Google Scholar]
- 46.Piekarski J, Goldberg HI, Royal SA, Axel L, Moss AA. Difference between liver and spleen CT numbers in the normal adult: its usefulness in predicting the presence of diffuse liver disease. Radiology. 1980;137:727–729. doi: 10.1148/radiology.137.3.6934563. [DOI] [PubMed] [Google Scholar]
- 47.Yoshizumi T, et al. Abdominal fat: standardized technique for measurement at CT. Radiology. 1999;211:283–286. doi: 10.1148/radiology.211.1.r99ap15283. [DOI] [PubMed] [Google Scholar]
- 48.Wang D, et al. Elevated serum IgG titer and avidity to Actinobacillus actinomycetemcomitans serotype c in Japanese periodontitis patients. Oral Microbiol Immunol. 2005;20:172–179. doi: 10.1111/j.1399-302X.2005.00208.x. [DOI] [PubMed] [Google Scholar]
- 49.Rask-Madsen C, et al. Loss of insulin signaling in vascular endothelial cells accelerates atherosclerosis in apolipoprotein E null mice. Cell Metab. 2010;11:379–389. doi: 10.1016/j.cmet.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kina-Tanada M, et al. Long-term dietary nitrite and nitrate deficiency causes the metabolic syndrome, endothelial dysfunction and cardiovascular death in mice. Diabetologia. 2017;60:1138–1151. doi: 10.1007/s00125-017-4259-6. [DOI] [PubMed] [Google Scholar]
- 51.Lu M, et al. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat Med. 2012;18:388–395. doi: 10.1038/nm.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shippy R, et al. Using RNA sample titrations to assess microarray platform performance and normalization techniques. Nat Biotechnol. 2006;24:1123–1131. doi: 10.1038/nbt1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li W, Fu L, Niu B, Wu S, Wooley J. Ultrafast clustering algorithms for metagenomic sequence analysis. Brief Bioinform. 2012;13:656–668. doi: 10.1093/bib/bbs035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shannon CE. A Mathematical Theory of Communication. The Bell System Technical Journal. 1948;27:45. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 59.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Dataset S1, Supplementary Dataset S2, Supplementary Dataset S3.