Abstract
This study aimed to investigate genetic polymorphisms of miR-146a, miR-149, miR-196a2, and miR-499 and genetic susceptibility of ischemic stroke in the population of Guangxi in China. A case–control study was used to investigate miRNAs genetic polymorphisms in 298 patients with ischemic stroke and 303 healthy controls. Single-base extension polymerase chain reaction genotyping principle was used to detect genetic polymorphisms of miRNAs,and the relationship of genotype in each group and blood lipid was compared and analyzed. The genetic polymorphism of miR-499A>G (rs3746444) was associated with ischemic stroke (P < 0.05), and the risk of ischemic stroke was high in patients with G allele (OR = 1.455; 95% CI = 0.531–2.381; P = 0.039) and AG (OR = 1.339; 95% CI = 1.126–1.967; P = 0.037) genotype. The levels of low-density lipoprotein cholesterol, very-low-density lipoprotein cholesterol, homocysteine, and lipoprotein in the ischemic stroke group were higher than those in the control group (P < 0.05). The genetic polymorphism of miR-499A>G (rs3746444) was related to ischemic stroke, and G allele and AG genotype may increase the risk of ischemic stroke in the population of Guangxi in China.
Keywords: allele, genotype, ischemic stroke, microRNA, single nucleotide polymorphism
INTRODUCTION
Ischemic stroke (IS) is a cardiovascular disease that seriously threatens the health of middle-aged and elderly people. In China, due to its high morbidity, high disability rate, high mortality, and high recurrence rate, IS has surpassed the morbidity of heart disease and tumors [1], which brings tremendous economic pressure and health burden to the family and the society [2–4]. Multiple factors including smoking, hypertension, diabetes, hyperlipidemia, and high homocysteine content are risk factors for IS [5]. However, these risk factors can only explain a small part of the etiology. IS is a complex disease induced by the combined action of a variety of genetic and environmental factors, and genetic mutations play a crucial role in its development [6–8]. Nevertheless, the exact molecular mechanism underlying the development of IS has not yet been determined.
MicroRNAs (miRNAs) are endogenous noncoding small-molecule RNAs mainly composed of 19–25 nucleotides in eucaryon. Through complementary binding with 3′ untranslated regions in the target gene of mRNA, miRNAs can post-transcriptionally regulate the gene expression. Several studies have shown that miRNAs are involved in a series of important life processes, including cell proliferation, development, differentiation, apoptosis, and metabolism [9, 10]. Genetic mutations existing in miRNA sequences affect the expression of mature miRNAs, thereby influencing the expression of target genes. Moreover, studies have shown that miRNAs are closely associated with growth/development, differentiation, maturation, and lesion of the nervous systems [11].
Single nucleotide polymorphism (SNP) is one of the most common types of genetic variation. SNP that exists in the precursor miRNAs (pre-miRNAs) can affect the mature expression of miRNAs, and then change multiple biological functions mediated by the target genes. Currently, many studies have shown that SNP loci that exist in the four precursor miRNAs, including miR-146aC>G (rs2910164), miR-149T>C (rs2292832),miR-196a2T>C(rs11614913), and miR-499A>G (rs3746444), are closely associated with various diseases that are seriously harmful to human health, such as tumors, hypertension, cardiovascular disease, and diabetes [12–15]. Recently,the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G alleles are possible genetic predisposing factors [16–20]. These 4 miRNAs can affect vascular damage responses [21–24]. Moreover, The miR-146a, miR-149, miR-196a2, and miR-499 alleles are closely associated with regulation of tumor necrosis factor-α(TNF-α) [21], methylenetetrahydrofolate reductasefour [22], and C-reactive protein (CRP) [24], These miRNA targets are related to thrombosis or inflammation pathways in the circulation system. So far, the correlation of genetic polymorphisms of miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G in the population Guangxi in China has not been reported both at domestic and foreign. Therefore, this study aimed to investigate the correlation of IS and SNP of the four miRNAs in the population of Guangxi.
MATERIALS AND METHODS
Subjects of Study
The study population comprised 298 patients with IS and 303 controls. All the population were recruited from the department of neurology, Affiliated Hospital of Youjiang Medical University for Nationalities, Guangxi, China between January 2015 and June 2015. All of them had the first-time IS. The study was approved by the Ethics Committee of You Jiang Medical College and conformed to the guidelines set forth by the Declaration of Helsinki. The diagnosis met the criteria approved at the Fourth National Cerebrovascular Disease Conference in 1995. The patients were diagnosed by at least two experienced clinical neurologists through brain computed tomography (CT) and (or) magnetic resonance imaging (MRI). Patients with IS caused by cardiovascular malformations or tumors, trauma, blood diseases, drugs, and infectious diseases were excluded. The exclusion criteria of the population in the control group were the same as that in the IS group. The patients had no history of stroke or severe genetic diseases in the family.
Research methods
Extraction of genomic DNA: EDTA-K2-mediated anti-coagulated venous blood (3mL) was obtained from people who received physical examination. A whole-blood DNA extraction kit (Shenzhen Yaneng Bioscience Co., Ltd., China) was used to extract the genomic DNA of whole blood according to the manufacturer instruction. The extracted DNA was stored at −70°C.
The design and synthesis of the primers: According to the GeneBank of National Center for Biotechnology Information, the human full-length sequences of the four SNP loci [miR-146aC>G (rs2910164), miR-149T>C(rs2292832), miR-196a2T>C(rs11614913), and miR-499 A>G (rs3746444)] were used. Primer3.0 was used to design the primers, and the primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd. Sequences of each primer are shown in Table 1.
Table 1. Primers sequences of each SNP locus in miR-146a, miR-149, miR-196a2, and miR-499.
| SNP loci | Sequences of PCR primers | Sequences of extension primers | |
|---|---|---|---|
| rs2292832 | Upstream primer | 5′-TCTGGCTCCGTGTCTTCACTCC-3′ | 5′-TTTTTTTTCGGCGACCTGCGTTGTTCC-3′ |
| Downstream primer | 5′-GGCCCGAAACACCCGTAAGATA-3′ | ||
| rs3746444 | Upstream primer | 5′-CTGGGAGACAGACCCTCCCTCT-3 | 5′-TTTTTTTTTTTTTTTTTTTTTTTTTGTTTAACTCCTCTCCACGTGAAC-3′ |
| Downstream primer | 5′-GCCCTGCACTTTTGCTCTTTCA-3 | ||
| rs2910164 | Upstream primer | 5′-AGGAAGCAGCTGCATTGGATTT-3′ | 5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGGGTTGTGTCAGTGTCAGACCT-3′ |
| Downstream primer | 5′-GTCCTCAAGCCCACGATGACAG-3′ | ||
| rs11614913 | Upstream primer | 5′-CCCCTTCCCTTCTCCTCCAGAT-3′ | 5′-TTTTTTTTTTTTTTTTTTTTTTTTTGAACTCGGCAACAAGAAACTG-3′ |
| Downstream primer | 5′-TTGTTCTGCAACCCCACTCACA-3′ | ||
The amplification of polymerase chain reaction (PCR): Four SNP loci, including miR-146aC>G (rs2910164), miR-149T>C (rs2292832), miR-196a2T>C (rs11614913), and miR-499A>G (rs3746444) were used. Reaction system for each PCR amplification contained a total volume of 20 mL, including 2 mL of PCR buffer (1× GC buffer I, Takara), 2.0 mL of 2.5 mmol/L Mg2+, 2.0 mL of 2 mmol/L dNTP, 1 μL of 0.2 mol/L upstream primer, 1 μL of 0.2 mol/L downstream primer, 1.0 U of polymerase, and 1.0 μL of template DNA. If the volume was less than 20 μL, then sterile double-distilled water was used to complement. PCR product was obtained using Multiplex PCR Master Mix with HotStar Taq DNA Polymerase (Qiagen). Then, 8 μL of PCR product was purified with 0.5 U of shrimp alkaline phosphatase (SAP) enzyme (Promega) and 4 U of exonuclease I (Epicentre), and incubated at 37°C for 60 min using the technique of SNaPshot Multiplex Kit (ABI), followed by incubation at 75°C for 15 min for extension reaction. Finally, the extension product was sequenced by electrophoresis using ABI3730XL sequencer after purification with SAP. GeneMapper4.0 (ABS) was used to analyze SNP genotype. The reaction condition of PCR was as follows: 95°C for 2 min; 11-cycle reaction (94°C for 20 s, 65°C for 40 s, 72°C for 1 min); 24-cycle reaction (94°C for 20 s, 59°C for 40 s, 72°C for 1 min); 72°C for 2 min; 4°C.
The measurement of serum homocysteine (Hcy) and blood lipid level
Heparin-anticoagulated blood was collected from patients within 24 h after being hospitalized, and then the blood was centrifuged at 3000 rpm for 5 min to separate the serum to be used in the measurement of blood lipid and Hcy. Enzymatic measurement was used for detecting TC, TG, and Hcy, and the method of masking was used to directly measure high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), very-low-density lipoprotein cholesterol (VLDL-C), and lipoprotein a [Lp(a)] (Hitachi 7600 automatic biochemical analyzer).
The relative expression levels of miR-499 and C-reactive protein(CRP) in patients with IS harboring different alleles
Total RNA was extracted from 1ml of 100 plasma samples(50 AA and 50 AG+GG genetype) using miRCURY Exosome Isolation Kit (Takara, Dalian, China) according to the manufacturer's instructions .5.0μg of total RNA was reverse transcribed into cDNA using poly(A) polymerase and Mir-X miRNA First-Strand Synthesis Kit(Takara, Cat. No. 638313).Real-time reverse transcription PCR(RT-PCR) and Mir-X miRNA qRT-PCR SYBR Kit (Takara, Cat. No. 638314) were performed to validate expression levels of miR-499.Quantitative PCR was performed on ABI 7900HT real-time PCR machine (Applied Biosystems, CA, USA), three shrimps were analyzed for each genetype of plasma samples. U6 RNA was used as an endogenous control,the primers were purchased from Takara., China. The relative serum levels of miR-499 expression were calculated using the delta-delta Ct method (2−ΔΔCt). The levels of serum CRP in 215 AA genotypes were compared with 83 (AG + GG) genotype by immunoturbidimetry Method(Hitachi 7600 series).
Statistical analysis
SPSS13.0 (SPSS, Inc., IL, USA) was used to conduct statistical analysis. Allele and genotype frequencies of the IS and control groups used the χ2 or Fisher's exact test. Measurement data were represented by x ± s, and categorical data were represented by χ2. OR value and 95% confidence intervals were used to evaluate the correlation of each allele and genotype distribution frequencies. Multivariate logistic regression was used to evaluate the relationship between the risk factors (gender, age, smoking situation, diabetes, hypertension, and Hcy) and the disease. χ2 was used to evaluate whether the allele and genotype in the control group met the conditions of Hardy–Weinberg equilibrium. All the tests were bilateral detections, and P < 0.05 was considered to be statistically significant.
RESULTS
Clinical data of the enrolled population
Comparison of clinical indicators between the IS and control groups is shown in Table 2. Patients with IS were predominantly smokers. So, patients with hypertension or diabetes were significantly more than those in the control group (P < 0.05). The levels of LDL-C, VLDL-C, Hcy, and Lp(a) were significantly higher than those in the control group (P < 0.05), but the level of HDL-C in patients with IS was significantly lower than that in the control group (P < 0.05).
Table 2. Comparison of basic clinical data between the IS and control groups.
| Groups | IS group (298 cases) | Control group (303 case) | χ2 | P |
|---|---|---|---|---|
| M/F (cases) | 196/102 | 181/122 | 2.341 | 0.126 |
| Age (year) | 60.70 ± 12.33 | 60.17 ± 10.32 | 1.030 | 0.303 |
| Smoking (cases) | 96 (32.2) | 55 (18.2) | 15.80 | < 0.001 |
| Hypertension (cases) | 204 (68.5) | 75 (24.8) | 115.38 | < 0.001 |
| Diabetes (cases) | 79 (26.5) | 26 (8.6) | 33.50 | < 0.001 |
| TC (mmol/L) | 5.10 ± 1.18 | 5.51 ± 0.32 | 0.087 | 0.930 |
| TG (mmol/L) | 1.85 ± 1.65 | 2.41 ± 0.10 | 0.226 | 0.821 |
| HDL-C (mmol/L) | 1.23 ± 0.31 | 1.73 ± 0.47 | 15.498 | < 0.001 |
| LDL-C (mmol/L) | 2.30 ± 1.91 | 2.36 ± 0.99 | 5.078 | < 0.001 |
| VLDL-C (mmol/L) | 0.86 ± 0.77 | 0.69 ± 0.51 | 3.219 | 0.001 |
| HCY (μmmol/L) | 13.98 ± 7.15 | 8.96 ± 7.02 | 5.084 | < 0.001 |
| LP(a) (mg/dL) | 24.30 ± 20.35 | 12.43 ± 11.51 | 8.099 | < 0.001 |
The genotyping result of miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G
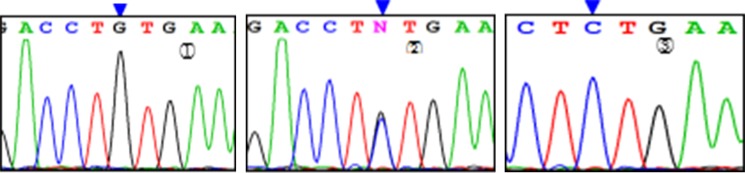
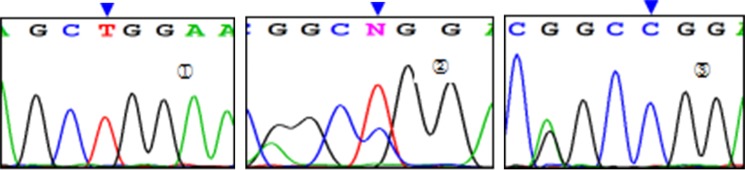
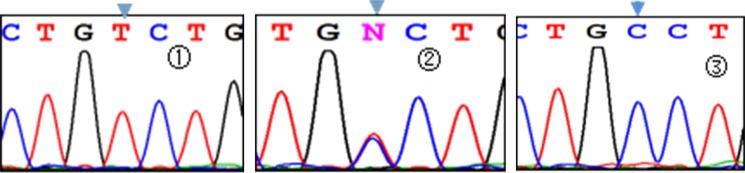
Three genotypes were detected by SNP of miR-146aC>G gene, including GG, GC, and CC (Figure 1); miR-149T>C gene also had three genotypes, including CC, CT, and TT (Figure 2). Moreover, CC, CT, and TT (Figure 3) were the genotypes for miR-196a2T>C, while AA, AG, and GG (Figure 4) were the genotypes for miR-499A>G.
Figure 1. Sequencing diagram of miR-146C>G (rs2910164).
Note:①, ②, and ③ represent genotypes of GG, CG, and CC, respectively; arrows are loci of genetic mutations.
Figure 2. Sequencing diagram of miR-149T>C (rs2292832).
Note: ①, ②, and ③ represent genotypes of TT, CT, and CC, respectively; arrows are loci of genetic mutations.
Figure 3. Sequencing diagram of miR-196a2T>C (rs11614913).
Note: ①, ②, and ③ represent genotypes of TT, CT, and CC, respectively; arrows are loci of genetic mutations.
Figure 4. Sequencing diagram of miR-499A>G (rs3746444).
Note: ①, ②, and ③ represent genotypes of AA, AG, and GG, respectively; arrows are loci of genetic mutations.
Comparison of the allele and genotype distribution frequencies between the IS and control groups
Table 4 shows the allele and genotype distribution of miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G in the IS and control groups. Genotype distribution of the control group met the requirements of the Hardy–Weinberg equilibrium (rs2910164C>G, P = 0.672; rs2292832T>C, P = 0.447; rs11614913T>C, P = 0.385; rs3746444A>G, P = 0.131). After multivariate logistic regression analysis (including gender, age, smoking, hypertension, diabetes, high Hcy, and high lipoprotein), statistical differences were found in the allele and genotype distribution of miR-499A>G in the IS group [comparison of miR-499A> G allele and genotype between the IS and control groups showed significant differences [AG vs AA, adjusted OR value (AOR) = 1.339, 95% CI = 1.126– 1.967, P = 0.037; G vs A, AOR = 1.455, 95% CI = 1.019–2.381, P = 0.037)] compared with the control group. No statistical difference in the genetic polymorphisms of miR-146aC>G, miR-149T>C, and miR-196a2T>C was found between the IS and control groups (P > 0.05).
Table 4. Comparison of genotype distribution frequencies of the allele and genotypes of polymorphic loci in the IS and control groups.
| SNPs | Stroke (298 cases) | Control (303 cases) | AOR (95% CI) | P value |
|---|---|---|---|---|
| miR-149T>C | ||||
| TT | 131 (44.0) | 121 (39.9) | 1.000 (reference) | |
| CT | 127 (42.6) | 136 (44.9) | 1.108 (0.879–1.647) | 0.756 |
| CC | 40 (13.4) | 46 (15.2) | 1.226(0.963–1.726) | 0.301 |
| Dominant model (TT vs CT + CC) | 0.958(0.713–1.295) | 0.363 | ||
| Recessive model(TT + CT vs CC) | 0.776(0.659–1.375) | 0.575 | ||
| T | 389 (65.3) | 378 (62.4) | 1.000 (reference) | |
| C | 207 (34.7) | 228 (37.6) | 1.156 (0.916–1.597) | 0.325 |
| HWE P | 0.300 | 0.447 | ||
| miR-499A>G | ||||
| AA | 215 (72.1) | 244 (80.5) | 1.000 (reference) | |
| AG | 78 (26.2) | 53 (17.5) | 1.339 (1.126–1.967) | 0.037 |
| GG | 5 (1.7) | 6 (2.0) | 1.077 (0.361–2.024) | 0.669 |
| Dominant model (AAvs AG + GG) | 1.621 (1.079–2.516) | 0.027 | ||
| Recessive model (AA+ AG vs GG) | 0.879 (0.531–2.557) | 0.665 | ||
| A | 508 (85.2) | 541 (89.3) | 1.000 (reference) | |
| G | 88 (14.8) | 65 (10.7) | 1.455 (1.019–2.381) | 0.039 |
| HWE P | 0.491 | 0.131 | ||
| miR-146aC>G | ||||
| CC | 129 (43.3) | 119 (39.3) | 1.000 (reference) | |
| CG | 130 (43.6) | 139 (45.9) | 1.163(0.939–1.681) | 0.596 |
| GG | 39 (13.1) | 45 (14.9) | 0.991(0.533–1.475) | 0.557 |
| Dominant model (CC vs CG + GG) | 1.015 (0.623–1.273) | 0.329 | ||
| Recessive model (CC + CG vs GG) | 1.006(0.543–1.319) | 0.317 | ||
| C | 388 (65.1) | 377 (62.2) | 1.000(reference) | |
| G | 208 (34.9) | 229 (37.8) | 1.091 (0.075–1.451) | 0.776 |
| HWE P | 0.490 | 0.672 | ||
| miR-196a2T>C | ||||
| TT | 73 (24.5) | 75 (24.8) | 1.000 (reference) | |
| CT | 138 (46.3) | 159 (52.5) | 1.257 (0.976–2.058) | 0.190 |
| CC | 87 (29.2) | 69 (22.8) | 1.339 (0.894–1.996) | 0.216 |
| Dominant model (TTvsCT + CC) | 0.803 (0.576–1.127) | 0.249 | ||
| Recessive model (CCvsCT +TT) | 1.376 (0.759–1.963) | 0.173 | ||
| T | 284 (47.7) | 309 (51.0) | 1.000 (reference) | |
| C | 312 (52.3) | 297 (49.0) | 1.035 (0.799–1.541) | 0.559 |
| HWE P | 0.215 | 0.385 |
Note: HWE P: Wendy–Harberger's law; miRNA: microRNA; 95% CI: 95% confidence interval; AOR: adjusted OR value.
Corrected factors of the AOR include gender, age, smoking, hypertension, diabetes, high Hcy, and high Lp(a).
Comparison of blood lipid, Lp(a), and Hcy in each genotype locus in the IS group
No statistical difference in blood lipid, lipoprotein (a), and Hcy was observed in each genotype of miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G in the IS group (P > 0.05, Table 3 Comparison of relative expression levels of miR-499 and C-reactive protein(CRP) in patients with IS harboring different alleles
Table 3. Comparison of the levels of blood lipid, Lp(a), and Hcy of each genotype in the IS group .
| Items | rs2292832 T/C | rs3746444 A/G | rs2910164 C/G | rs11614913 T/C | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | CT + CC | P value | AA | AG + GG | P value | CC | CG + GG | P value | TT | CT + CC | P value | |
| 131 cases | 167 cases | 215 cases | 83 cases | 129 cases | 169 cases | 73 cases | 225 cases | |||||
| TC (mmol/L) | 5.17 ± 1.29 | 5.05 ± 1.11 | 0.38 | 5.07 ± 1.21 | 5.21 ± 1.04 | 0.44 | 5.16 ± 1.25 | 5.05 ± 1.14 | 0.46 | 5.00 ± 1.14 | 5.13 ± 1.20 | 0.35 |
| TG (mmol/L) | 1.90 ± 1.50 | 1.81 ± 1.74 | 0.62 | 1.87 ± 1.04 | 1.75 ± 1.18 | 0.63 | 1.95 ± 1.40 | 1.78 ± 1.02 | 0.91 | 1.72 ± 1.04 | 1.88 ± 1.81 | 0.46 |
| HDL-C (mmol/L) | 1.22 ± 0.26 | 1.23 ± 0.33 | 0.71 | 1.22 ± 0.30 | 1.25 ± 0.31 | 0.51 | 1.21 ± 0.32 | 1.24 ± 0.30 | 0.50 | 1.25 ± 0.32 | 1.22 ± 0.30 | 0.42 |
| LDL-C (mmol/L) | 2.94 ± 0.84 | 3.03 ± 2.36 | 0.70 | 2.98 ± 2.08 | 3.03 ± 0.81 | 0.87 | 2.95 ± 0.88 | 3.02 ± 2.35 | 0.72 | 2.81 ± 0.86 | 3.05 ± 2.14 | 0.33 |
| VLDL(mmol/L) | 0.86 ± 0.68 | 0.85 ± 0.83 | 0.93 | 0.80 ± 0.54 | 0.80 ± 0.54 | 0.51 | 0.91 ± 0.61 | 0.83 ± 0.50 | 0.24 | 0.80 ± 0.49 | 0.88 ± 0.84 | 0.40 |
| HC(μmmol/L) | 14.27 ± 7.44 | 13.78 ± 6.98 | 0.56 | 14.22 ± 7.57 | 14.22 ± 7.57 | 0.78 | 13.90 ± 6.77 | 14.02 ± 7.40 | 0.88 | 14.94 ± 7.88 | 13.66 ± 6.88 | 0.23 |
| LP(a)(mg/dL) | 22.90 ± 18.51 | 25.20 ± 21.45 | 0.34 | 23.82 ± 19.52 | 26.47 ± 23.81 | 0.39 | 23.38 ± 18.66 | 24.89 ± 21.40 | 0.48 | 24.65 ± 22.00 | 24.18 ± 20.23 | 0.96 |
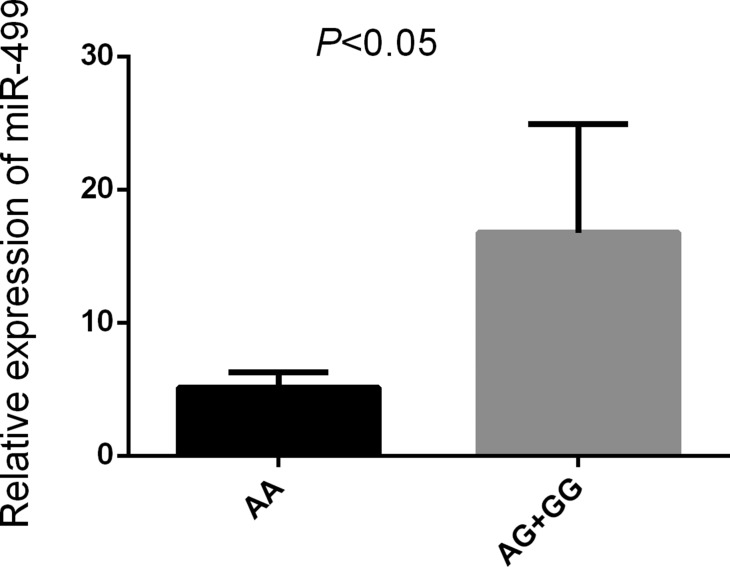
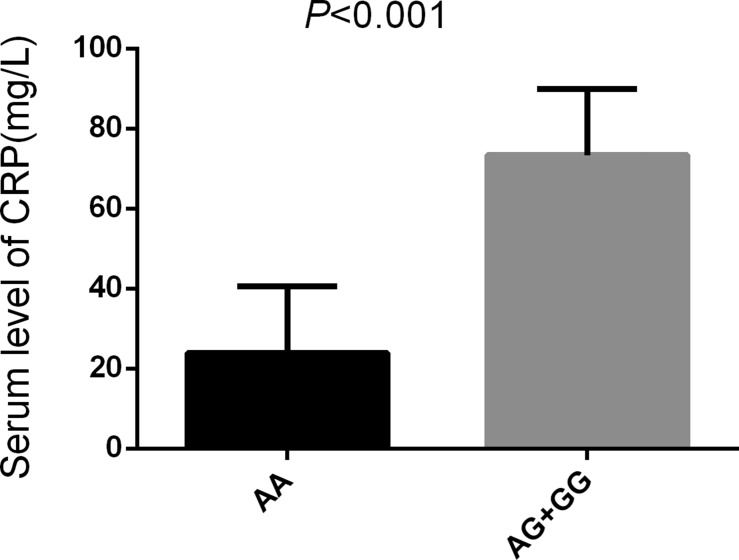
The results show that patients carrying the rs3746444AG/GG genetype had a higer level of miR-499 and CRP compared with those carrying the rs3746444AA genetype (P < 0.05) (Figures 5, 6).
Figure 5. Relative expression of miR-499.
Note:Increased level of miR-499 in IS patients carrying the AG/GG (n = 50) compared with AA (n = 50) (P < 0.05).
Figure 6. Serum level of CRP in different alleles of miR-499.
Note: Significant difference of serum CRP level between AA (n = 215) and AG/GG (n = 83) in IS patients (P < 0.001).
DISCUSSION
As the major type of stroke, IS refers to cerebral necrosis caused by stenosis or occlusion of the cerebral artery, and cerebral insufficiency, which is a complex neurological disease induced by interaction of genes with the environment that causes permanently disabling disease [6, 25–26]. One in six people in the world suffer from stroke [27], and IS seriously increases the burden on economy and medical resource in the world [28]. However, traditional diagnosis relies on CT or MRI, which cannot completely diagnose all types of strokes. Satisfactory clinical results cannot be obtained especially regarding the risk factors for IS. The role of genetic factors is increasingly evident in the state of the disease, especially in stroke [29]. MiRNAs are small-molecule RNAs recently found to be involved in the physiological and pathological processes of various diseases, including tumors [30], functions of immune system [31], cell proliferation [9], cardiovascular diseases [13], and nervous system diseases [32]. MiRNAs have a high degree of stability, tissue specificity, specificity of cell expression, and easy detectability in the human circulation. Therefore, miRNAs have become a molecular marker for the clinical diagnosis in many diseases, especially for early damage of the nervous system, cardiovascular diseases, and early painless diagnosis of IS [25, 33].
MiRNAs can regulate about 30% human gene expression [34]. SNP existing in pre-miRNAs or genetic mutations can affect the expression of mature miRNAs or cause mutative expression [35]. SNP existing in miRNAs are closely related to traditional risk factors for IS, such as atherosclerosis, hypertension, hyperlipidemia (high cholesterol), and diabetes [36–39]. Based on preliminary studies, four miRNAs were selected [miR-146aC>G (rs2910164), miR-149T>C (rs2292832), miR-196a2T>C (rs11614913), and miR-499A>G (rs3746444] to investigate the genetic susceptibility of SNP and IS in the population of Southwest region of Guangxi in China. It was found that miR-499A>G (rs3746444) was correlated with IS in the population Southwest region of Guangxi in China (P < 0.05). The risk of stroke would increase in people with G allele (OR = 1.455; 95% CI = 0.531–2.381) and AG genotype (OR = 1.339; 95% CI = 1.126–1.967; P = 0.037). We found that carrying the rs3746444AG/GG genetype had a higer level expression of miR-499, These findings were similar to the findings of the study by Liu et al. [40] that genetic polymorphism of miR-499A>G (rs3746444) might increase the risk of stroke in the Sichuan Han population. The rs3746444 A/G polymorphism association with ischemic stroke risk might be involved in thrombosis and/or inflammation pathways in the circulation system. The changes in pre-miR-499A: U or G: U base pairing would affect the expression of the target genes of mature miR-499 and biological functions [41, 42], cell hypoxia, or apoptosis caused by overexpression of miR-499 [43], and all these can increase the risk of stroke. The study found that carrying the rs3746444 AA had a lower level of serum CRP compared to those rs3746444AG/GG, miR-499 would increase the concentration of serum or plasma C-reactive protein, causing increased risk factors for stroke, such as inflammation, hypertension, and hyperlipoidemia. In summary, the pathological process of miR-499 in stroke and its potential mechanism are still unclear, but the mechanism that can increase the incidence of stroke through regulating the level of traditional risk factors for IS has been recognized. It is of immense significance in searching new diagnostic markers of stroke through in-depth study of the relationship between genome-wide associative analysis and stroke.
This study showed no correlation between IS and genetic polymorphisms of miR-146a, miR-149, and miR-196a2 in the population of Guangxi in China. However, the study by Jeon et al. [44] suggested that miR-146a G allele was associated with the morbidity of IS in the Korean population. A study by Zhu et al. [45]demonstrated that miR-146a C allele and CC genotype could increase the risk of stroke in the Han population in northern China. Qu et al. [46]reported that miR-146a (rs2910164) can be used as a diagnostic marker for the prognosis of stroke in the Asian population. Huang et al. [47] claimed that miR-146a (rs2910164) was significantly correlated with the stroke in the Han population in Shenzhen, China. Moreover, Chen et al. [48] indicated that miR-149 was associated with the risk of stroke in the Han population in Jiangsu, China. However, no study has elucidated that the genetic polymorphism of miR-196a2 is associated with stroke. The results of the aforementioned studies varied in different ethnic groups in different countries, which might be due to regional differences and different lifestyles, environment, stresses. And it suggested that the polymorphisms might have tissue-specific and different effect on human diseases in different organs, reflecting the diversities of the etiological factors for different diseases.
Ischemic stroke is a complex disease caused by a variety of factors. It can be caused by the interaction among multiple genes or by the interaction between genes, regions, and ethnic groups. Certain limitations of this study were as follows: (1) the subjects of the study were limited to the population of Guangxi in China, and the sample size was small. Thus, studies with varied ethnic groups and regions and large sample size are needed. (2) Selective bias was possible, and significant differences in clinical data between the IS and control groups were found. In addition, potential risk factors for stroke existed in people in the control group. (3) The pathological mechanism underlying the genetic polymorphism of miR-499A>G and ischemic stroke was still unclear. All genetic mutations in miR-499 and other factors might cause stroke, which had an impact on the explanation of the results of this study.
In conclusion, this study suggested that the genetic polymorphism of miR-499A>G was associated with IS in the population of Southwest region of Guangxi in China, and G allele and AG genotype of miR-499 (rs3746444) could increase the risk of IS. The molecular mechanism underlying the pathogenesis of IS and the genetic polymorphism of miR-499A>G in different ethnic groups need further exploration.
Footnotes
CONFLICTS OF INTEREST
The authors declared that they have no competing financial interests.
FUNDING
This study was supported by the National Natural Science Foundation (No. 81260234).
REFERENCES
- 1.Liu L, Wang D, Wong KS, Wang Y. Stroke and stroke care in China: huge burden, significant workload, and a national priority. Stroke. 2011;42:3651–3654. doi: 10.1161/STROKEAHA.111.635755. [DOI] [PubMed] [Google Scholar]
- 2.Krishnamurthi RV, Moran AE, Feigin VL, Barker-Collo S, Norrving B, Mensah GA, Taylor S, Naghavi M, Forouzanfar MH, Nguyen G, Johnson CO, Vos T, Murray CJ, et al. Stroke Prevalence, Mortality and Disability-Adjusted Life Years in Adults Aged 20–64 Years in 1990-2013: Data from the Global Burden of Disease 2013 Study. Neuroepidemiology. 2015;45:190–202. doi: 10.1159/000441098. [DOI] [PubMed] [Google Scholar]
- 3.Murray CJ, Atkinson C, Bhalla K, Birbeck G, Burstein R, Chou D, Dellavalle R, Danaei G, Ezzati M, Fahimi A, Flaxman D, Foreman Gabriel S, et al. The state of US health, 1990-2010: burden of diseases, injuries, and risk factors. JAMA. 2013;310:591–608. doi: 10.1001/jama.2013.13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsieh FI, Chiou HY. Stroke: morbidity, risk factors, and care in taiwan. J Stroke. 2014;16:59–64. doi: 10.5853/jos.2014.16.2.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein LB, Adams R, Becker K, Furberg CD, Gorelick PB, Hademenos G, Hill M, Howard G, Howard VJ, Jacobs B, Levine SR, Mosca L, Sacco RL, et al. Primary prevention of ischemic stroke: A statement for healthcare professionals from the Stroke Council of the American Heart Association. Stroke. 2001;32:280–299. doi: 10.1161/01.str.32.1.280. [DOI] [PubMed] [Google Scholar]
- 6.Titov BV, Matveeva NA, Martynov MY, Favorova OO. [Ischemic stroke as a complex polygenic disease] Mol Biol (Mosk) 2015;49:224–248. [PubMed] [Google Scholar]
- 7.Della-Morte D, Guadagni F, Palmirotta R, Testa G, Caso V, Paciaroni M, Abete P, Rengo F, Ferroni P, Sacco RL, Rundek T. Genetics of ischemic stroke, stroke-related risk factors, stroke precursors and treatments. Pharmacogenomics. 2012;13:595–613. doi: 10.2217/pgs.12.14. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh YC, Jeng JS, Lin HJ, Hu CJ, Yu CC, Lien LM, Peng GS, Chen CI, Tang SC, Chi NF, Tseng HP, Chern CM, Hsieh FI, et al. Epistasis analysis for estrogen metabolic and signaling pathway genes on young ischemic stroke patients. PLoS One. 2012;7:e47773. doi: 10.1371/journal.pone.0047773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia LF, Gan YH, Yu GY. [Relationships between microRNA expressions and prognosis in patients with tongue squamous cell carcinoma and the mechanisms microRNA regulating tongue squamous cell carcinoma biological behavior] Beijing Da Xue Xue Bao. 2016;48:5–9. [PubMed] [Google Scholar]
- 10.Mu W, Yao W. [Research Progress on the Development and Regulation of Embryonic Hematopoietic Stem Cells] Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2015;32:1141–1145. [PubMed] [Google Scholar]
- 11.Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 13.Volny O, Kasickova L, Coufalova D, Cimflova P, Novak J. microRNAs in Cerebrovascular Disease. Adv Exp Med Biol. 2015;888:155–195. doi: 10.1007/978-3-319-22671-2_9. [DOI] [PubMed] [Google Scholar]
- 14.Gomez-Banoy N, Mockus I. [MicroRNAs: circulating biomarkers in type 2 Diabetes Mellitus and physical exercise] Rev Med Chil. 2016;144:355–363. doi: 10.4067/S0034-98872016000300011. [DOI] [PubMed] [Google Scholar]
- 15.Marques FZ, Charchar FJ. microRNAs in Essential Hypertension and Blood Pressure Regulation. Adv Exp Med Biol. 2015;888:215–235. doi: 10.1007/978-3-319-22671-2_11. [DOI] [PubMed] [Google Scholar]
- 16.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeon YJ, Choi YS, Rah H, Kim SY, Choi DH, Cha SH, Shin JE, Shim SH, Lee WS, Kim NK. Association study of microRNA polymorphisms with risk of idiopathic recurrent spontaneous abortion in Korean women. Gene. 2012;494:168–173. doi: 10.1016/j.gene.2011.12.026. [DOI] [PubMed] [Google Scholar]
- 18.Min KT, Kim JW, Jeon YJ, Jang MJ, Chong SY, Oh D, Kim NK. Association of the miR-146aC>G, 149C>T, 196a2C>T, and 499A>G polymorphisms with colorectal cancer in the Korean population. Mol Carcinog. 2012;51:E65–E73. doi: 10.1002/mc.21849. [DOI] [PubMed] [Google Scholar]
- 19.Park YS, Jeon YJ, Lee BE, Kim TG, Choi JU, Kim DS, Kim NK. Association of the miR-146aC>G, miR-196a2C>T, and miR-499A>G polymorphisms with moyamoya disease in the Korean population. Neurosci Lett. 2012;521:71–75. doi: 10.1016/j.neulet.2012.05.062. [DOI] [PubMed] [Google Scholar]
- 20.Wang AX, Xu B, Tong N, Chen SQ, Yang Y, Zhang XW, Jiang H, Liu N, Liu J, Hu XN, Sha GZ, Chen M. Meta-analysis confirms that a common G/C variant in the pre-miR-146a gene contributes to cancer susceptibility and that ethnicity, gender and smoking status are risk factors. Genet Mol Res. 2012;11:3051–3062. doi: 10.4238/2012.August.31.2. [DOI] [PubMed] [Google Scholar]
- 21.El G M, Church A, Liu T, McCall CE. MicroRNA-146a regulates both transcription silencing and translation disruption of TNF-alpha during TLR4-induced gene reprogramming. J Leukoc Biol. 2011;90:509–519. doi: 10.1189/jlb.0211074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu C, Gong Y, Sun A, Zhang Y, Zhang C, Zhang W, Zhao G, Zou Y, Ge J. The human MTHFR rs4846049 polymorphism increases coronary heart disease risk through modifying miRNA binding. Nutr Metab Cardiovasc Dis. 2013;23:693–698. doi: 10.1016/j.numecd.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Luthra R, Singh RR, Luthra MG, Li YX, Hannah C, Romans AM, Barkoh BA, Chen SS, Ensor J, Maru DM, Broaddus RR, Rashid A, Albarracin CT. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27:6667–6678. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- 24.Yang B, Chen J, Li Y, Zhang J, Li D, Huang Z, Cai B, Li L, Shi Y, Ying B, Wang L. Association of polymorphisms in pre-miRNA with inflammatory biomarkers in rheumatoid arthritis in the Chinese Han population. Hum Immunol. 2012;73:101–106. doi: 10.1016/j.humimm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Grasso M, Piscopo P, Crestini A, Confaloni A, Denti MA. Circulating microRNAs in Neurodegenerative Diseases. EXS. 2015;106:151–169. doi: 10.1007/978-3-0348-0955-9_7. [DOI] [PubMed] [Google Scholar]
- 26.Falcone GJ, Malik R, Dichgans M, Rosand J. Current concepts and clinical applications of stroke genetics. Lancet Neurol. 2014;13:405–418. doi: 10.1016/S1474-4422(14)70029-8. [DOI] [PubMed] [Google Scholar]
- 27.Bushnell CD. Stroke and the female brain. Nat Clin Pract Neurol. 2008;4:22–33. doi: 10.1038/ncpneuro0686. [DOI] [PubMed] [Google Scholar]
- 28.Barquera S, Pedroza-Tobias A, Medina C, Hernandez-Barrera L, Bibbins-Domingo K, Lozano R, Moran AE. Global Overview of the Epidemiology of Atherosclerotic Cardiovascular Disease. Arch Med Res. 2015;46:328–338. doi: 10.1016/j.arcmed.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Felling RJ, Song H. Epigenetic mechanisms of neuroplasticity and the implications for stroke recovery. Exp Neurol. 2015;268:37–45. doi: 10.1016/j.expneurol.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 31.Carissimi C, Fulci V, Macino G. MicroRNAs: novel regulators of immunity. Autoimmun Rev. 2009;8:520–524. doi: 10.1016/j.autrev.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Bushati N, Cohen SM. MicroRNAs in neurodegeneration. Curr Opin Neurobiol. 2008;18:292–296. doi: 10.1016/j.conb.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 34.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 35.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schober A, Weber C. Mechanisms of MicroRNAs in Atherosclerosis. Annu Rev Pathol. 2016;11:583–616. doi: 10.1146/annurev-pathol-012615-044135. [DOI] [PubMed] [Google Scholar]
- 37.Boucherat O, Potus F, Bonnet S. microRNA and Pulmonary Hypertension. Adv Exp Med Biol. 2015;888:237–252. doi: 10.1007/978-3-319-22671-2_12. [DOI] [PubMed] [Google Scholar]
- 38.Li T, Yang GM, Zhu Y, Wu Y, Chen XY, Lan D, Tian KL, Liu LM. Diabetes and hyperlipidemia induce dysfunction of VSMCs: contribution of the metabolic inflammation/miRNA pathway. Am J Physiol Endocrinol Metab. 2015;308:E257–E269. doi: 10.1152/ajpendo.00348.2014. [DOI] [PubMed] [Google Scholar]
- 39.Gomez-Banoy N, Mockus I. [MicroRNAs: circulating biomarkers in type 2 Diabetes Mellitus and physical exercise] Rev Med Chil. 2016;144:355–363. doi: 10.4067/S0034-98872016000300011. [DOI] [PubMed] [Google Scholar]
- 40.Hu Z, Liang J, Wang Z, Tian T, Zhou X, Chen J, Miao R, Wang Y, Wang X, Shen H. Genetic polymorphisms in pre-microRNAs and risk of ischemic stroke in a Chinese population. J Mol Neurosci. 2014;52:473–480. doi: 10.1007/s12031-013-0152-z. [DOI] [PubMed] [Google Scholar]
- 41.Xiang Y, Fan S, Cao J, Huang S, Zhang LP. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30:79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 42.Xiang Y, Fan S, Cao J, Huang S, Zhang LP. Association of the microRNA-499 variants with susceptibility to hepatocellular carcinoma in a Chinese population. Mol Biol Rep. 2012;39:7019–7023. doi: 10.1007/s11033-012-1532-0. [DOI] [PubMed] [Google Scholar]
- 43.Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP, Li YR, Li PF. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med. 2011;17:71–78. doi: 10.1038/nm.2282. [DOI] [PubMed] [Google Scholar]
- 44.Jeon YJ, Kim OJ, Kim SY, Oh SH, Oh D, Kim OJ, Shin BS, Kim NK. Association of the miR-146a, miR-149, miR-196a2, and miR-499 polymorphisms with ischemic stroke and silent brain infarction risk. Arterioscler Thromb Vasc Biol. 2013;33:420–430. doi: 10.1161/ATVBAHA.112.300251. [DOI] [PubMed] [Google Scholar]
- 45.Zhu R, Liu X, He Z, Li Q. miR-146a and miR-196a2 polymorphisms in patients with ischemic stroke in the northern Chinese Han population. Neurochem Res. 2014;39:1709–1716. doi: 10.1007/s11064-014-1364-5. [DOI] [PubMed] [Google Scholar]
- 46.Qu JY, Xi J, Zhang YH, Zhang CN, Song L, Song Y, Hui RT, Chen JZ. Association of the MicroRNA-146a SNP rs2910164 with Ischemic Stroke Incidence and Prognosis in a Chinese Population. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17050660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang S, Zhou S, Zhang Y, Lv Z, Li S, Xie C, Ke Y, Deng P, Geng Y, Zhang Q, Chu X, Yi Z, Zhang Y, et al. Association of the genetic polymorphisms in pre-microRNAs with risk of ischemic stroke in a Chinese population. PLoS One. 2015;10:e117007. doi: 10.1371/journal.pone.0117007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen QY, Liu N, Ma J, Fang Y, Cao Y, Li H, Liu YC. Effect of a pre-microRNA-149 (miR-149) genetic variation on the risk of ischemic stroke in a Chinese Han population. Genet Mol Res. 2015;14:2582–2589. doi: 10.4238/2015.March.30.17. [DOI] [PubMed] [Google Scholar]