Abstract
Gastric cancer is one of the most common cancers and has the highest mortality rate worldwide. It is worthwhile to explore the mechanism of gastric cancer progression. An increasing number of studies have found that non-coding RNAs including miRNA and lncRNA play important roles in gastric cancer progression. This review summarized the role of ectopic miRNA in gastric cancer proliferation, growth, migration, invasion and apoptosis. Meantime, aberrantly expressed miRNA also received a great deal of attention as potential biomarker for gastric cancer diagnosis and therapy. Over the last decade, lncRNA was considered to regulate gastric cancer progression at the transcript and post-transcript level. At the transcript level, lncRNA induced gastric cancer progression by changing chromatin modification and mRNA stabilization to regulate mRNA and miRNA expression. Furthermore, lncRNA regulated gastric cancer progression by completely combining with miRNA to produce ceRNA or promote protein stabilization at the post-transcript level. Greater attention of miRNA and lncRNA in gastric cancer can provide new insight of mechanism of cancer development and may be acted as a new anticancer target.
Keywords: gastric cancer, miRNA, lncRNA
INTRODUCTION
Gastric cancer is the fourth most common cancer and the third leading cause of cancer mortality worldwide [1]. In China, the most recent statistics indicate that gastric cancer is second most common cancer in terms of incidence (679.1 per 100,000) and mortality (498.0 per 100,000) [2]. In U.S., it was estimated that 26,370 new cases of gastric cancer were diagnosed in 2016 with 10,370 estimated deaths [3]. In recent years, the 5-year mortality was significantly reduced for the early gastric cancer given the development of enterscopy and surgical technique. However, for advanced gastric cancer, the 5-year mortality remains 30% to 50% [4]. Advanced gastric cancer mortality is associated with peritoneal dissemination, hematogenous spread and lymph node metastasis. Thus, it was worthwhile to explore the mechanism of gastric cancer progression such as proliferation, growth, migration, invasion and apoptosis. Numerous studies have demonstrated that non-coding RNAs play important roles in gastric cancer progression [5–7]. In this study, we will review the role of non-coding RNA family in gastric cancer progression.
Non-coding RNA is an RNA molecule that cannot code protein. Based on the length, non-coding RNA are divided into two major classes. Large RNAs (greater than 50 nt) includes long non-coding RNA (lncRNA), small nucleolar RNA, circular RNA (ceRNA), tRNA and rRNA [8]. Small RNA (less than 50 nt) includes microRNA (miRNA), siRNA and piRNA [9]. Currently, numerous studies have found that miRNA and lncRNA play important roles in gastric cancer progression.
miRNA involved in gastric cancer progression
miRNA is a highly conserved non-coding sequences that is generally 18 to 24 nucleotides in length [10–12]. First, miRNA is transcribed by RNA polymerase II and RNA polymerase III from introns, exons, intro-exon junctions or their own genes as pri-miRNA. After Drosha and DGCR8 processing in nucleus, pri-miRNA is transcribed as pre-miRNA and subsequently mature miRNA. Mature miRNA combines with other associated proteins to form the active RNA-induced silencing complex (RISC). RISC combines with the 5′UTR, ORF or 3′UTR of a target gene mRNA to suppress its translation or induce its degradation [13–16]. One miRNA can combined with several different mRNAs at several different points. A target gene mRNA can also combine with several miRNAs [12, 17].
miRNAs in gastric cancer progression
To date, greater than 2500 miRNAs have been identified (miRbase database) and their ectopic expression is associated with tumor proliferation, invasion, metastasis, tumor growth and apoptosis. Both onco-miRNA and suppressing miRNAs play important roles in gastric cancer progression. Changes in miRNA expression and its role in gastric cancer are summarized in Table 1. For example, a previous study found that the onco-miRNA miR-130 was over-expressed in gastric cancer. The over-expressed miR-130 promoted gastric cancer proliferation and metastasis by combining with the 3′UTR of TGF-β and inhibiting its expression [18]. Our laboratory found that miR-1266, miR-1207-5p and miR-1182 exhibited relatively low expression in gastric cancer and promote cancer cell proliferation and metastasis. Further research found that miR-1266 and miR-1207-5p combine with the 3′UTR of hTERT to inhibit its expression and cancer cell proliferation and metastasis [19]. However, miR-1182 cannot combine with of the 3′UTR of hTERT. After a literature review and additional experiments, we found that miR-1182 inhibit hTERT expression by targeting its open reading frame (ORF) regions [20]. This result indicates that miRNA always combine with its target gene 3′UTR. However, the miRNA may occasionally combine with the ORF region. For tumor growth, a previous study found that over-expressed miR-24 can promote gastric cancer growth and suppress apoptosis by inhibiting BCL2L11 expression [21]. Zhang et al. found that Sirt7 can significantly inhibit miR-34a expression in gastric cancer and ultimately promote tumor growth and inhibit apoptosis [22]. Previous studies found that miRNA clusters (miR-15b, miR-16, miR-34, miR-181b, miR-181c, and miR-497) exhibit relatively low expression in gastric cancer and promote the expression BCL-2 which inhibits apoptosis [23]. A cluster of onco-miRNAs (miR-25, miR-93, miR-106b, and miR-130) is overexpressed in gastric cancer and inhibits gastric cancer cell apoptosis by suppress Bim expression. Since Bim is considered as a pro-apoptosis protein [24].
Table 1. The ectopic expression of miRNA in gastric cancer.
| miRNA | Relative expression | Target mRNA | Cell biology | Clinicopathologic feature | Reference |
|---|---|---|---|---|---|
| miR-17-5p/20a | Up | TP53INP1 | Cell proliferation and apoptosis | Tumor size | Wang et al. [25] |
| miR-100 | Down | ZBTB7A | tumor growth, invasion and metastasis | lymphatic metastasis | Shi et al. [26] |
| miR-125b | Up | PPP1CA | cellular proliferation, migration, and invasion | tumor size and depth of invasion, lymph nodes, distant metastasis, TNM stage and poor prognosis. | Wu et al. [27] |
| miR-133b | Down | PTBP1 | Tumor growth | No association with the clinicopathological future | Sugiyama et al. [28] |
| miR-145 | down | Ets1 | migration, invasion, and angiogenesis | tumor invasion and metastasis | Zheng et al. [29] |
| miR-148a | Down | ROCK1 | Migration and invasion | TNM stage and lymph node-metastasis | Zheng et al. [30] |
| miR-196a/-196b | Up | radixin | cell migration and invasion | TNM stage, lymph node metastasis, poor prognosis | Tsai et al. [31] |
| miR-199a-5p | Up | klotho | cell migration and invasion | Tumor size, TNM stage and lymph node metastasis | He et al. [32] |
| miR-302 | Down | IL-8 | Migration and invasion | metastasis and prognosis | Chen et al. [33] |
| miR-506 | Down | ETS1 | Angiogenesis and EMT | Poor prognosis | Li et al. [34] |
| miR-940 | Up | ZNF24 | Migration and invasion | poor prognosis | Liu et al. [35] |
| miR-1182 | Down | hTERT | cell proliferation, migration and invasion | Tumor size, differentiation, metastasis, TNM Stage and poor prognosis | Zhang et al. [20] |
| miR-1207-5p/miR-1266 | Down | hTERT | cell proliferation, migration and invasion | Tumor size, differentiation, metastasis, TNM Stage and poor prognosis | Chen et al. [19] |
| miR-29a/c | Down | VEGF | vascular cell growth, metastasis, and tube formation | - | Zhang et al. [36] |
| miR-29b/c | Down | DNMT3A | Migration and invasion | Invasion | Cui et al. [37] |
miRNA in gastric cancer diagnosis
Tumor diagnosis is an important process that may influence the risk of progression, recurrence and death. However, early detection methods or screening methods must be explored for various types of cancers. Thus, multiple studies are focused on exploring the biomarkers for cancer detection and progression. Numerous miRNAs are aberrantly expressed in the plasma and serum of gastric cancer patients [6, 38–41]. For example, miR-223, miR-233, miR-378, miR-421, miR-451, miR-486-5p and miR-199-3p are over-expressed in sera of gastric cancer patient [42–45]. Wang and colleagues found that miR-233 was over-expressed in gastric cancer patient serum at a normalized cutoff of 0.21. miR-233 yields a receiver operating characteristic (ROC) area under the curve (AUC) of 0.85, a sensitivity of 81% and specificity of 78% [42]. The level of miR-233 expression in patient serum was positively associated with tumor differentiation grade, TNM stage, tumor size and metastasis status [42]. Wu and colleagues found that miR-421 was overexpressed in 90 cases of gastric cancer patient sera compared with 90 controls with an AUC, sensitivity and specificity of 0.821, 95.5% and 89.1%, respectively. These values were increased compared with CA125 and CEA for gastric cancer detection. The high expression of miR-421 in mononuclear cells acts as a biomarker for gastric cancer circulating tumor cells, which may be used for early diagnosis for gastric metastasis [43]. Furthermore, in vivo and in vitro experiments demonstrated that the onco-miR-421 promotes tumor proliferation, invasion and metastasis [46]. Jiang et al. also found that miR-421 was overexpressed in gastric cancer tissues but had no significant association with the clinic-pathological feature. [47].
In contrast, let-7a, miR-375, miR-20a-5p and miR-320 expression was relatively reduced in gastric cancer patient serum [48, 49]. A previous study demonstrated that let-7a exhibited relatively low expression in plasma of gastric cancer patient compared with healthy controls, whereas the expression of miR-17-5p, miR-106a, miR-106b and miR-21 was significantly elevated in gastric cancer plasma [50]. Further analysis found that the miR-106a/let-7a ratio in 69 gastric cancer patients and 30 controls revealed a maximum AUC of 0.879, with a sensitivity of 85.5 and specificity of 80.0% [51]. Tang and colleagues also found that let-7a was suppressed in gastric cancer tissue. In vivo and in vitro experiments showed that let-7a overexpression significantly suppressed PMK2 expression, which inhibited tumor proliferation, migration and invasion [52]. Previous studies demonstrated that miR-375 was suppressed in gastric cancer. Overexpression of miR-375 suppresses gastric cancer progression by targeting p53, JAK2, ERBB2 and STAT3 [53, 54]. Zhang and colleagues found that miR-375 expression was significantly suppressed in both distal gastric adenocarcinoma tissues and patient serum compared with healthy controls. At a normalized cutoff of 0.218, the AUC of serum miR-375 was 0.835 with a sensitivity of 85% and specificity of 80% [48]. These studies indicate miRNA severs as an interesting diagnostic biomarker. However, large-scale clinical research is needed to demonstrate that miRNA can serve as a diagnostic biomarker for gastric cancer.
miRNA in gastric cancer therapy
Recent studies have found that miRNA-based cancer therapy may be a promising and challenging path. On one hand, miRNA-based drugs that overexpress the suppressed-miRNA or inhibit the onco-miRNA can inhibited tumor progression by suppressing relative signal pathway [40, 55]. For example, miR-34 suppresses miRNA in a few tumors including breast cancer, liver cancer, lung cancer and gastric cancer [56–58]. Recently, a clinical trial of MRX34 (Mirna Therapeutics, TX, USA) constructed a miR-34 mimetics to restore miR-34 expression in cancer cells. The agent was used to treat liver cancer and liver metastasis of other cancers in phase I clinical trial protocol [40, 59]. Meantime, in a pre-clinical study of non small cell lung cancer, MRX34 treatment significantly reduced the expression of the checkpoint signal PD-L1 and increase the infiltrating CD8+ cells in tumor tissues [60]. However, the MRX34 clinical trial was stopped in 2016 since multiple serious immune-related side effects were observed in patients. In detail, one patient receiving MRX34 experienced a prolonged partial response and lasted 48 weeks. Meantime, six others presented with stable disease of the total 47 patients. So the safety of MRX34 still needs for further research [61]. For gastric cancer, Ji and colleagues found that restoration of miR-34 significantly inhibits gastric cancer tumorspheres by promoting expression of tumor suppressing mutant p53 [62]. The miR-34 family targeted the 3′UTR of Yin Yang1 to inhibit its expression, reduced tumor growth and metastasis [63].
On the other hand, miRNA also played critical roles in drug resistance. An increasing number of studies have demonstrated that miRNA can significantly influence drug transporters, drug-metabolizing enzymes, transcription factor and nuclear receptors. Yang and colleagues found that miR-21 was over expressed in the cisplatin resistant cell line SGC7901. Knock-down of miR-21 expression can significantly increase the anti-proliferative effects and apoptosis induced by cisplatin [64]. Xia et al. found that miR-15/miR-16 family was significantly decreased in multidrug resistance (MDR) of gastric cancer cells. Over expression of miR-15 dramatically reduces the expression of BCL2, which reverses the MDR in gastric cancer cells by modulating cell apoptosis [65]. However, some problems should be considered, as one miRNA can target multiple genes and signaling, the off-target effect is unexpected. Thus based on miRNA therapy system should be explored with better specificity [36].
In total, we discussed the role of miRNA in gastric cancer progression. Both onco-miRNA and suppressing miRNA play critical roles in gastric cancer proliferation, growth, metastasis and apoptosis. The overexpression of miR-421 in the serum of gastric cancer patients may act as an early diagnostic marker. Meantime, overexpression of miR-34 significantly inhibited cancer progression in a few cancers and miR-34 may be used for cancer therapy in the future.
The role of lncRNA in gastric cancer
In contrast with miRNA, lncRNA is an RNA transcript longer than 200 nt with no capacity for coding protein. Based on their location and orientation, lncRNAs are classified as intronic lncRNAs, intergenic lncRNAs, pseudogenes, sense or antisense transcripts, and retrotransposons [66, 67]. Functionally, lncRNAs regulate gene expression at any level, including chromatin modification, transcription, and post-transcriptional processing. A large number of studies have found that lncRNAs played important roles in numerous diseases and disease processing, including cardiovascular and cancer progression [68–71]. Recently, studies have found that the ectopic lncRNA expression, including that of H19, TUSC7, MEG3 and MALAT1 significantly regulate gastric cancer cell proliferation, cell cycle, apoptosis, invasion, migration, metastasis, and tumorigenicity [7, 51, 72–74]. In this section, we will summarize the roles of lncRNA in gastric cancer progression.
lncRNA regulates gene expression at the transcriptional level
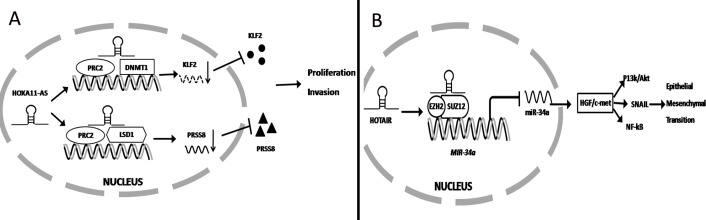
Previous studies have demonstrated that 38% of lncRNA cooperate with at least one of multiple histone-modifying complexes, inducing DNA methylation and chromatin modification that ultimately leads to silence the expression of target mRNA [75, 76]. For example, Sun and colleagues discovered that only lncRNA HOXA11-AS was specifically over-expressed in gastric cancer but not in other cancers using the sequencing data from the TCGA project and microarray profile from GEO. High expression of lncRNA HOXA11-AS in gastric cancer patients was associated with poor prognosis. Knockdown of HOXA11-AS expression significantly reduces gastric cancer cell proliferation and promotes apoptosis. FISH and subcellular fractionation experiments demonstrated that HOXA11-AS was more prevalent in the nucleus. Another study found that HOXA11-AS acts as a scaffold to reduce KLF2 and PRSS8 expression at the transcriptional level, given that it can directly bind to the RNA binding proteins PRC2, LSD1 and DNMT1. Finally lncRNA-mediated assembly of PRC2 and LSD1 coordinates targeting of PRC2 and LSD1 for coupled H3K27 methylation and H3K4 demethylation [77]. (Figure 1A) In contrast, Xu et al. found that low expression lncRNA FENDRR in gastric cancer patients was associated with poor survival. Over-expression of FENDRR can significantly reduce gastric cancer metastasis [78]. FENDRR combines with PRC2 to form FENDRR/PRC complex, which inhibits MMP2/9 expression [78]. Sun et al. also found that lncRNA GClnc1 was significantly over-expressed in gastric cancer. Mechanistically, GClnc1 binds to WDR5 and KAT2A histone acetyltransferase, acts as a modular scaffold of WDR5 and KAT2A complexes, coordinates their localization and consequently alters gastric cancer proliferation, migration and invasion [79].
Figure 1. lncRNA regulates gene expression at the transcriptional level.
(A) HOXA11-AS binds to several RNA binding proteins (PRC2, LSD1 and DNMT1) and function as a scaffold to inhibit PRSS8 and KLF2 at the transcriptional level to promote gastric cancer proliferation and invasion. (B) HOTAIR binding to PRC2 significantly inhibited miR-34a expression at the transcript level ultimately activating HGF/c-met signaling to promote EMT.
Recently, it was found that lncRNA epigenetically silences miRNA expression at the transcript level to promote gastric cancer progression. Previous studies have found that over-expression of lncRNA HOTAIR contributed to gastric cancer development and predicted a poor prognosis. Liu and colleagues further demonstrated that HOTAIR combined with EZH2 and SUZ12 to form a complex that directly binds to the miR-34a promoter to silence its expression via H3K27me3 modification. Finally, low expression of miR-34a significantly promotes HGF/c-met activation, which induces SNAIL, PI3K/Akt and NF-kB signaling to promote cancer development [80]. (Figure 1B) Zhang et al. reported that over-expression of lncRNA ANRIL was significantly associated with gastric cancer progression and can serve as an independent predictor for patient survival. Mechanistically, E2F1 directly binds to the ANRIL promoter to induce its expression. Then high expression of ANRIL combined with PRC2 significantly silences miR-99a/miR-449a expression at the transcriptional level, which ultimately increase mTOR, CDK6, and E2F1 expression. miR-99a/miR-449a binds to the 3′UTR of mTOR, CDK6, and E2F1 to inhibit their expression. In total, high expression of E2F1 induced ANRIL expression. Then the ANRIL/PRC2 complex inhibited miR-99a/miR-449a expression and promoted mTOR, CDK6 and E2F1 expression. Conversely, E2F1 promoted ANRIL expression which formed a positive feedback loop to promote gastric cancer cell proliferation [81].
lncRNAs also regulate target gene expression by directly interacting with its mRNA. For example, Xu et al. found that high expression of nuclear factor SP1 which is significantly expressed in gastric cancer, binds to the promoter of lncRNA TINCR to promote its expression. Silencing the expression of TINCR significantly reduces cancer cell proliferation, tumorigenicity and apoptosis. Mechanistic analyses indicated that TINCR was mostly present in the cytoplasm. RNA IP and pull-down assay showed that TINCR combined with STAU1 acts as a staufen-mediated mRNA decay (SMD) factor. Furthermore RNA immunoprecipitation (RIP) and RNA pull down assay showed that TINCR/STAU1 complexes directly interact with KLF2 mRNA, decrease KLF2 mRNA stability, and inhibit its protein expression. Subsequently, reduced KLF2 expression decreases CDKN2B/P15 and CDKN1A/P21 transcripts which finally promote cancer cell proliferation, migration, invasion and tumorigenicity [82]. In contrast, Yang and colleagues found that lncRNA GHET1 was significantly over-expressed in gastric cancer patients compared with healthy controls. Over-expression of GHET1 in MKN45 and AGS of gastric cancer cells significantly promotes cell proliferation in vitro. In vivo experiments showed that over-expression of GHET1 in MKN45 cells significantly promotes growth of xenograft tumors in nude mice. Mechanistically, RIP and RNA pull-down assay first showed that GHET1 can directly combine with insulin growth factor2 binding protein 1 (IGF2BP1). RIP assay and qRT-PCR results showed that IGF2BP1 combined with the mRNA of c-myc. Together, these results showed that lncRNA GHET1/IGF2BP1 complex combines with c-myc mRNA to increase its stability and promote protein expression. Finally, high expression of c-myc promoted gastric cancer cell proliferation [72].
lncRNA regulates gene expression at the post-transcriptional level
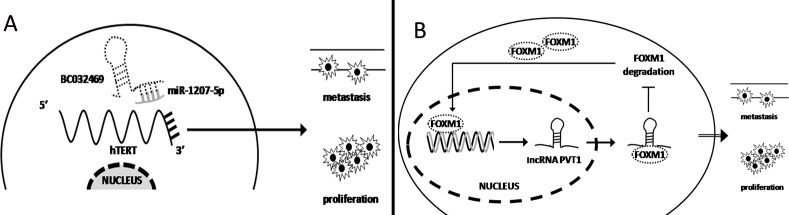
As previously described, miRNA regulates target gene expression at the post-transcriptional level. Thus, high expression of lncRNA can competitively combine with miRNA, acting as a sponge to induce miRNA disability and promote cancer progression. Our laboratory previously demonstrated low miR-1207-5p expression in gastric cancer. Over-expression of miR-1207-5p significantly inhibits cancer cell proliferation and metastasis by combining with the 3′UTR of hTERT to inhibit its expression. Recently, the further research found that lncRNA BC032469 expression was positively associated with hTERT expression and significantly promotes gastric cancer cell proliferation and metastasis. Mechanistically, RIP and Northern blot demonstrated that BC032469 directly binds to miR-1207-5p. Thus, BC032469 functions as a ceRNA to impair miR-1207-5p-dependent hTERT down-regulation, suggesting that it may a poor prognostic marker for gastric cancer [83]. (Figure 2A) Furthermore, Liu et al. found that over-expression of lncRNA HOTAIR in gastric cancer competes with miR-331-3p expression, which functions as a ceRNA to promote HER expression and promote gastric cancer progression [84].
Figure 2. lncRNA regulates gene expression at the transcriptional level.
(A) lncRNA BC032469 functions as a ceRNA to sponge for miR-1207-5p, promoting hTERT expression to induce gastric cancer cell proliferation and metastasis. (B) FOXM1 induced lncRNA PVT1 expression at the transcriptional level. Then PVT1 binds to FOXM1 protein to increase its stabilization, forming a positive feedback loop and promoting gastric cancer cell proliferation and metastasis.
lncRNA also regulate protein stabilization at the post-transcriptional level to promote gastric cancer progression. Xu and colleagues demonstrated that the promoter of lncRNA PVT1 contains binding sites for the transcriptional factor FOXM1. High FOXM1 expression in gastric cancer significantly induces PVT1 expression. Furthermore, high expression of PVT1 did not influence FOXM1 mRNA expression but can reversely bind to FOXM1 protein to increase its stabilization and inhibit 26s proteasome-mediated degradation. Thus, high expression of FOXM1 and PVT1 form a positive feedback loop that promotes gastric cancer proliferation and metastasis [85]. (Figure 2B).
In summary, lncRNA significantly regulates gene expression both in the nucleus and cytoplasm. In the nucleus, lncRNAs bind the polycomb group protein (PcG) complex, induce histone trimethylation and regulate relative gene mRNA expression at the transcriptional level. Meantime, lncRNAs directly bind with the promoter to regulate gene expression. In the cytoplasm, lncRNA also directly bind to mRNA to influence its stability and expression at the transcriptional level. Meantime, cytoplasmic lncRNA can regulate gene expression at the post-transcriptional level. As previously described, lncRNAs act as a sponge with miRNA and induce a ‘ceRNA’ to regulate gene expression. On the othe hand, lncRNA also influence protein stability and inhibit expression at the post-transcriptional level. Finally, we summarized the ectopic expression of lncRNA and its target gene and role in gastric cancer progression in Table 2.
Table 2. The ectopic expression of lncRNA in gastric cancer.
| lncRNA | Relative Expression | Molecular Mechanism | Cell biology | Clinicopathologic feature | Reference |
|---|---|---|---|---|---|
| LINC00673 | up | functioning as a scaffold for LSD1 and EZH2 and repressing KLF2 and LATS2 expression | Proliferation, tumor growth, invasion, metastasis and apoptosis | Tumor size, TNM Stage, lymphatic metastasis and poor prognosis | Huang et al. [86] |
| SNHG5 | down | Sponge with miR-32 to regulate KLF4 expression | Proliferation and migration | negatively associated with miR-32 | Zhao et al. [87] |
| LincRNAFEZF1-AS1 | up | functioning as a scaffold for LSD1 and repressing p21 by inducing H3K4me2 demethylation | Proliferation, tumor growth and apoptosis | Tumor size, TNM Stage and poor prognosis | Liu et al. [88] |
| PVT1 | up | PVT1 directly bound with FOXM1 protein to increase its stability, FOXM1 reversely bound to PVT1 promoter to promote its expression | Proliferation and metastasis | Tumor size, distant metastasis and poor prognosis | Xu et al. [89] |
| lncRNA-GHET1 | up | GHET1 combined with IGF2BP1 protien up-regulates c-Myc by increasing c-Myc mRNA stability | proliferation | Tumor size, invasion and poor prognosis | Yang et al. [72] |
| TINCR | up | TINCR binds to STAU1 down-regulate KLF2 expression by influence its mRNA stability | Proliferation and apoptosis | TNM stage and lymphatic metastasis | Xu et al. [82] |
| ANRIL | up | ANRIL binds with PRC2 decreased miR-99a/miR-449a expression by interact with its promoter | proliferation | Tumor size, TNM stage and poor prognosis | Zhang et al. [81] |
| LincHOTAIR | up | HOTAIR silenced miR34a expression by recruiting PRC2 and finally activated HGF/c-Met /Snail pathway to promote gastric cancer EMT | Invasion and metastasis | Invasion depth, lymphatic metastasis and poor prognosis | Liu et al. [80] |
| GAPLINC | up | GAPLINC sponge with miR211-3p to regulate CD44 associated downstream signal | Proliferation and Invasion | Tumor size, lymphatic metastasis, TNM stage and poor prognosis | Hu et al. [90] |
| FENDRR | down | decreased FENDRR expression induces FN1 expression and activates MMPs family | Migration and Invasion | Invasion depth, TNM Stage, lymphatic metastasis and poor prognosis | Xu et al. [78] |
| H19 | up | H19 directly binds with ISM1 to regulate its expression | proliferation, migration, invasion and metastasis | TNM Stage, lymphatic metastasis and poor prognosis | Li et al. [91] |
| FER1L4 | down | FER1L4 liberated miR-106a-5p, downregulated PTEN expression | Proliferation and cell cycle | Tumor size, lymphatic metastasis, TNM stage and invasion depth | Xia et al. [92] Liu et al. [93] |
| GAS5 | down | lncRNA GAS5 reduces the YBX1 protein and subsequently decreases YBX1-transactivated p21 expression | Cell cycle | - | Liu et al. [94] |
| nc886 | down | nc886 knockdown activation of oncogenic FOS, NF-κB, and MYC as well as other pathways | proliferation | Poor prognosis | Lee et al. [95] |
| HOXA11-AS | up | HOXA11-AS functions as a scaffold for EZH2 and LSD1, HOXA11-AS acts as a ceRNA for miR-1297 | Proliferation, cell growth, migration, invasion, and apoptosis | Tumor size, lymphatic metastasis, TNM stage and poor prognosis | Sun et al. [77] |
| GClnc1 | up | GClnc1 bound WDR5 and KAT2A histone acetyltransferase, acted as a modular scaffold of WDR5 and KAT2A complexes, specified the histone modification pattern | proliferation, migration and invasion | Tumor size, Vascular metastasis, TNM stage and poor prognosis | Sun et al. [79] |
| BC032469 | up | BC032469 bind to miR-1207-5p and effectively functioned as a sponge to modulate the derepression of hTERT | proliferation | Tumor size, poor proliferation and poor prognosis | Lu et al. [83] |
CONCLUSIONS
In this study, we discussed the role of miRNA and lncRNA in gastric cancer progression. Most miRNA bind to the 3′UTR of the target gene to regulate its expression and new research found that miRNAs also bind to the ORF region of target genes to influence expression. This information may broaden our understanding of whether miRNA can directly bind to the 5′UTR of target gene. Most studies have focused on the role of miRNA in cancer progression and the target gene of miRNA in the downstream. But few studies reported that which gene can induced the miRNA ectopic expression, so this part need to further research.
We summarized that lncRNA significantly binds to miRNA, mRNAs and proteins in gastric cancer, regulating gene expression at both the transcriptional and post-transcriptional levels. However, research on lncRNA in normal gastric cells is relatively limited. In the future, more research will focus on the role of lncRNAs in gastritis, atypical hyperplasia and early carcinoma. We hope that diagnosis and treatment based on lncRNAs will be realized in the future.
Footnotes
CONFLICTS OF INTEREST
None.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Hamashima C, Shabana M, Okada K, Okamoto M, Osaki Y. Mortality reduction from gastric cancer by endoscopic and radiographic screening. Cancer Sci. 2015;106:1744–1749. doi: 10.1111/cas.12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 6.Xuan Y, Yang H, Zhao L, Lau WB, Lau B, Ren N, Hu Y, Yi T, Zhao X, Zhou S, Wei Y. MicroRNAs in colorectal cancer: small molecules with big functions. Cancer Lett. 2015;360:89–105. doi: 10.1016/j.canlet.2014.11.051. [DOI] [PubMed] [Google Scholar]
- 7.Deng K, Wang H, Guo X, Xia J. The cross talk between long, non-coding RNAs and microRNAs in gastric cancer. Acta Biochim Biophys Sin (Shanghai) 2016;48:111–116. doi: 10.1093/abbs/gmv120. [DOI] [PubMed] [Google Scholar]
- 8.Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K, Vandesompele J, Mestdagh P. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246–251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 12.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 16.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 17.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Duan J, Zhang H, Qu Y, Deng T, Huang D, Liu R, Zhang L, Bai M, Zhou L, Ying G, Ba Y. Onco-miR-130 promotes cell proliferation and migration by targeting TGFbetaR2 in gastric cancer. Oncotarget. 2016;7:44522–44533. doi: 10.18632/oncotarget.9936. https://doi.org/10.18632/oncotarget.9936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Lu MH, Zhang D, Hao NB, Fan YH, Wu YY, Wang SM, Xie R, Fang DC, Zhang H, Hu CJ, Yang SM. miR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis. 2014;5:e1034. doi: 10.1038/cddis.2013.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang D, Xiao YF, Zhang JW, Xie R, Hu CJ, Tang B, Wang SM, Wu YY, Hao NB, Yang SM. miR-1182 attenuates gastric cancer proliferation and metastasis by targeting the open reading frame of hTERT. Cancer Lett. 2015;360:151–159. doi: 10.1016/j.canlet.2015.01.044. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Duan J, Qu Y, Deng T, Liu R, Zhang L, Bai M, Li J, Ning T, Ge S, Wang X, Wang Z, Fan Q, et al. Onco-miR-24 regulates cell growth and apoptosis by targeting BCL2L11 in gastric cancer. Protein & cell. 2016;7:141–151. doi: 10.1007/s13238-015-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S, Chen P, Huang Z, Hu X, Chen M, Hu S, Hu Y, Cai T. Sirt7 promotes gastric cancer growth and inhibits apoptosis by epigenetically inhibiting miR-34a. Sci Rep. 2015;5:9787. doi: 10.1038/srep09787. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Nishizawa T, Suzuki H. The role of microRNA in gastric malignancy. Int J Mol Sci. 2013;14:9487–9496. doi: 10.3390/ijms14059487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, Agarwal R, Paun BC, Jin Z, Olaru AV, Selaru FM, Hamilton JP, Yang J, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689–1700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang M, Gu H, Qian H, Zhu W, Zhao C, Zhang X, Tao Y, Zhang L, Xu W. miR-17-5p/20a are important markers for gastric cancer and murine double minute 2 participates in their functional regulation. Eur J Cancer. 2013;49:2010–2021. doi: 10.1016/j.ejca.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 26.Shi DB, Wang YW, Xing AY, Gao JW, Zhang H, Guo XY, Gao P. C/EBPalpha-induced miR-100 expression suppresses tumor metastasis and growth by targeting ZBTB7A in gastric cancer. Cancer Lett. 2015;369:376–385. doi: 10.1016/j.canlet.2015.08.029. [DOI] [PubMed] [Google Scholar]
- 27.Wu JG, Wang JJ, Jiang X, Lan JP, He XJ, Wang HJ, Ma YY, Xia YJ, Ru GQ, Ma J, Zhao ZS, Zhou R. MiR-125b promotes cell migration and invasion by targeting PPP1CA-Rb signal pathways in gastric cancer, resulting in a poor prognosis. Gastric Cancer. 2015;18:729–739. doi: 10.1007/s10120-014-0421-8. [DOI] [PubMed] [Google Scholar]
- 28.Sugiyama T, Taniguchi K, Matsuhashi N, Tajirika T, Futamura M, Takai T, Akao Y, Yoshida K. MiR-133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle-splicer polypyrimidine tract-binding protein 1. Cancer Sci. 2016;107:1767–1775. doi: 10.1111/cas.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng L, Pu J, Qi T, Qi M, Li D, Xiang X, Huang K, Tong Q. miRNA-145 targets v-ets erythroblastosis virus E26 oncogene homolog 1 to suppress the invasion, metastasis, and angiogenesis of gastric cancer cells. Mol Cancer Res. 2013;11:182–193. doi: 10.1158/1541-7786.MCR-12-0534. [DOI] [PubMed] [Google Scholar]
- 30.Zheng B, Liang L, Wang C, Huang S, Cao X, Zha R, Liu L, Jia D, Tian Q, Wu J, Ye Y, Wang Q, Long Z, et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574–7583. doi: 10.1158/1078-0432.CCR-11-1714. [DOI] [PubMed] [Google Scholar]
- 31.Tsai MM, Wang CS, Tsai CY, Chen CY, Chi HC, Tseng YH, Chung PJ, Lin YH, Chung IH, Chen CY, Lin KH. MicroRNA-196a/-196b promote cell metastasis via negative regulation of radixin in human gastric cancer. Cancer Lett. 2014;351:222–231. doi: 10.1016/j.canlet.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 32.He XJ, Ma YY, Yu S, Jiang XT, Lu YD, Tao L, Wang HP, Hu ZM, Tao HQ. Up-regulated miR-199a-5p in gastric cancer functions as an oncogene and targets klotho. BMC cancer. 2014;14:218. doi: 10.1186/1471-2407-14-218. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Chen L, Min L, Wang X, Zhao J, Chen H, Qin J, Chen W, Shen Z, Tang Z, Gan Q, Ruan Y, Sun Y, Qin X, et al. Loss of RACK1 Promotes Metastasis of Gastric Cancer by Inducing a miR-302c/IL8 Signaling Loop. Cancer Res. 2015;75:3832–3841. doi: 10.1158/0008-5472.CAN-14-3690. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Liu Z, Dong S, Zhang J, Tan J, Wang Y, Ge C, Li R, Xue Y, Li M, Wang W, Xiang X, Yang J, et al. miR-506 Inhibits Epithelial-to-Mesenchymal Transition and Angiogenesis in Gastric Cancer. Am J Pathol. 2015;185:2412–2420. doi: 10.1016/j.ajpath.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Ge X, Zhang Z, Zhang X, Chang J, Wu Z, Tang W, Gan L, Sun M, Li J. MicroRNA-940 promotes tumor cell invasion and metastasis by downregulating ZNF24 in gastric cancer. Oncotarget. 2015;6:25418–25428. doi: 10.18632/oncotarget.4456. https://doi.org/10.18632/oncotarget.4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai MM, Wang CS, Tsai CY, Huang HW, Chi HC, Lin YH, Lu PH, Lin KH. Potential Diagnostic, Prognostic and Therapeutic Targets of MicroRNAs in Human Gastric Cancer. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17060945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui H, Wang L, Gong P, Zhao C, Zhang S, Zhang K, Zhou R, Zhao Z, Fan H. Deregulation between miR-29b/c and DNMT3A is associated with epigenetic silencing of the CDH1 gene, affecting cell migration and invasion in gastric cancer. PloS one. 2015;10:e0123926. doi: 10.1371/journal.pone.0123926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu HS, Xiao HS. MicroRNAs as potential biomarkers for gastric cancer. World J Gastroenterol. 2014;20:12007–12017. doi: 10.3748/wjg.v20.i34.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishiguro H, Kimura M, Takeyama H. Role of microRNAs in gastric cancer. World J Gastroenterol. 2014;20:5694–5699. doi: 10.3748/wjg.v20.i19.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pichler M, Calin GA. MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br J Cancer. 2015;113:569–573. doi: 10.1038/bjc.2015.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y, Lin J, Kong D, Huang M, Xu C, Kim TK, Etheridge A, Luo Y, Ding Y, Wang K. Current State of Circulating MicroRNAs as Cancer Biomarkers. Clin Chem. 2015;61:1138–1155. doi: 10.1373/clinchem.2015.241190. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Wang L, Wu Z, Sun R, Jin H, Ma J, Liu L, Ling R, Yi J, Wang L, Bian J, Chen J, Li N, et al. Three dysregulated microRNAs in serum as novel biomarkers for gastric cancer screening. Med Oncol. 2014;31:298. doi: 10.1007/s12032-014-0298-8. [DOI] [PubMed] [Google Scholar]
- 43.Wu J, Li G, Yao Y, Wang Z, Sun W, Wang J. MicroRNA-421 is a new potential diagnosis biomarker with higher sensitivity and specificity than carcinoembryonic antigen and cancer antigen 125 in gastric cancer. Biomarkers. 2015;20:58–63. doi: 10.3109/1354750X.2014.992812. [DOI] [PubMed] [Google Scholar]
- 44.Liu H, Zhu L, Liu B, Yang L, Meng X, Zhang W, Ma Y, Xiao H. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 2012;316:196–203. doi: 10.1016/j.canlet.2011.10.034. [DOI] [PubMed] [Google Scholar]
- 45.Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo X, Mao XH, Zou QM, Yu PW, Zuo QF, Li N, Tang B, Liu KY, et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PloS one. 2012;7:e41629. doi: 10.1371/journal.pone.0041629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou H, Xiao B, Zhou F, Deng H, Zhang X, Lou Y, Gong Z, Du C, Guo J. MiR-421 is a functional marker of circulating tumor cells in gastric cancer patients. Biomarkers. 2012;17:104–110. doi: 10.3109/1354750X.2011.614961. [DOI] [PubMed] [Google Scholar]
- 47.Jiang Z, Guo J, Xiao B, Miao Y, Huang R, Li D, Zhang Y. Increased expression of miR-421 in human gastric carcinoma and its clinical association. J Gastroenterol. 2010;45:17–23. doi: 10.1007/s00535-009-0135-6. [DOI] [PubMed] [Google Scholar]
- 48.Zhang WH, Gui JH, Wang CZ, Chang Q, Xu SP, Cai CH, Li YN, Tian YP, Yan L, Wu B. The identification of miR-375 as a potential biomarker in distal gastric adenocarcinoma. Oncol Res. 2012;20:139–147. doi: 10.3727/096504012x13522227232156. [DOI] [PubMed] [Google Scholar]
- 49.Xu Q, Dong QG, Sun LP, He CY, Yuan Y. Expression of serum miR-20a-5p, let-7a, and miR-320a and their correlations with pepsinogen in atrophic gastritis and gastric cancer: a case-control study. BMC Clin Pathol. 2013;13:11. doi: 10.1186/1472-6890-13-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang YK, Yu JC. Circulating microRNAs and long non-coding RNAs in gastric cancer diagnosis: An update and review. World J Gastroenterol. 2015;21:9863–9886. doi: 10.3748/wjg.v21.i34.9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang R, Yang C, Ma X, Wang Y, Luo D, Huang C, Xu Z, Liu P, Yang L. MiR-let-7a inhibits cell proliferation, migration, and invasion by down-regulating PKM2 in gastric cancer. Oncotarget. 2016;7:5972–5984. doi: 10.18632/oncotarget.6821. https://doi.org/10.18632/oncotarget.6821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miao L, Liu K, Xie M, Xing Y, Xi T. miR-375 inhibits Helicobacter pylori-induced gastric carcinogenesis by blocking JAK2-STAT3 signaling. Cancer Immunol Immunother. 2014;63:699–711. doi: 10.1007/s00262-014-1550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Xing R, Zhang X, Dong W, Zhang J, Yan Z, Li W, Cui J, Lu Y. miR-375 targets the p53 gene to regulate cellular response to ionizing radiation and etoposide in gastric cancer cells. DNA repair. 2013;12:741–750. doi: 10.1016/j.dnarep.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 55.Riquelme I, Letelier P, Riffo-Campos AL, Brebi P, Roa JC. Emerging Role of miRNAs in the Drug Resistance of Gastric Cancer. Int J Mol Sci. 2016;17:424. doi: 10.3390/ijms17030424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang R, Ma J, Wu Q, Xia J, Miele L, Sarkar FH, Wang Z. Functional role of miR-34 family in human cancer. Curr Drug Targets. 2013;14:1185–1191. doi: 10.2174/13894501113149990191. [DOI] [PubMed] [Google Scholar]
- 57.Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, Zarone MR, Gulla A, Tagliaferri P, Tassone P, Caraglia M. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang DG, Zheng JN, Pei DS. P53/microRNA-34-induced metabolic regulation: new opportunities in anticancer therapy. Mol Cancer. 2014;13:115. doi: 10.1186/1476-4598-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farooqi AA, Fayyaz S, Shatynska-Mytsyk I, Javed Z, Jabeen S, Yaylim I, Gasparri ML, Panici PB. Is miR-34a a Well-equipped Swordsman to Conquer Temple of Molecular Oncology? Chem Biol Drug Des. 2016;87:321–334. doi: 10.1111/cbdd.12634. [DOI] [PubMed] [Google Scholar]
- 60.Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, Araujo L, Carbone DP, Shilo K, Giri DK, Kelnar K, Martin D, Komaki R, et al. PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst. 2016:108. doi: 10.1093/jnci/djv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35:180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang AM, Huang TT, Hsu KW, Huang KH, Fang WL, Yang MH, Lo SS, Chi CW, Lin JJ, Yeh TS. Yin Yang 1 is a target of microRNA-34 family and contributes to gastric carcinogenesis. Oncotarget. 2014;5:5002–5016. doi: 10.18632/oncotarget.2073. https://doi.org/10.18632/oncotarget.2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang SM, Huang C, Li XF, Yu MZ, He Y, Li J. miR-21 confers cisplatin resistance in gastric cancer cells by regulating PTEN. Toxicology. 2013;306:162–168. doi: 10.1016/j.tox.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 65.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 66.Esposti DD, Hernandez-Vargas H, Voegele C, Fernandez-Jimenez N, Forey N, Bancel B, Le Calvez-Kelm F, McKay J, Merle P, Herceg Z. Identification of novel long non-coding RNAs deregulated in hepatocellular carcinoma using RNA-sequencing. Oncotarget. 2016;7:31862–31877. doi: 10.18632/oncotarget.7364. https://doi.org/10.18632/oncotarget.7364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 68.Yu F, Zheng J, Mao Y, Dong P, Lu Z, Li G, Guo C, Liu Z, Fan X. Long Non-coding RNA Growth Arrest-specific Transcript 5 (GAS5) Inhibits Liver Fibrogenesis through a Mechanism of Competing Endogenous RNA. J Biol Chem. 2015;290:28286–28298. doi: 10.1074/jbc.M115.683813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Shao N, Ding X, Tan B, Song Q, Wang N, Jia Y, Ling H, Cheng Y. Crosstalk between transforming growth factor-beta signaling pathway and long non-coding RNAs in cancer. Cancer Lett. 2016;370:296–301. doi: 10.1016/j.canlet.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 70.Archer K, Broskova Z, Bayoumi AS, Teoh JP, Davila A, Tang Y, Su H, Kim IM. Long Non-Coding RNAs as Master Regulators in Cardiovascular Diseases. Int J Mol Sci. 2015;16:23651–23667. doi: 10.3390/ijms161023651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Y, Zhang R, Ying K. Long noncoding RNAs: novel links in respiratory diseases (review) Mol Med Rep. 2015;11:4025–4031. doi: 10.3892/mmr.2015.3290. [DOI] [PubMed] [Google Scholar]
- 72.Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y, Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802–813. doi: 10.1111/febs.12625. [DOI] [PubMed] [Google Scholar]
- 73.Xie SS, Jin J, Xu X, Zhuo W, Zhou TH. Emerging roles of non-coding RNAs in gastric cancer: Pathogenesis and clinical implications. World J Gastroenterol. 2016;22:1213–1223. doi: 10.3748/wjg.v22.i3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang ZG, Gao L, Guo XB, Shi YL. Roles of long non-coding RNAs in gastric cancer metastasis. World J Gastroenterol. 2015;21:5220–5230. doi: 10.3748/wjg.v21.i17.5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xie X, Tang B, Xiao YF, Xie R, Li BS, Dong H, Zhou JY, Yang SM. Long non-coding RNAs in colorectal cancer. Oncotarget. 2016;7:5226–5239. doi: 10.18632/oncotarget.6446. https://doi.org/10.18632/oncotarget.6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7:8601–8612. doi: 10.18632/oncotarget.6926. https://doi.org/10.18632/oncotarget.6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, Xie M, Xu L, De W, Wang Z, Wang J. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 78.Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, Chen WM, Han L, Zhang EB, Kong R, De W, Shu YQ. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. doi: 10.1186/s13045-014-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, Tang JY, Bao YJ, Hu Y, Lin Y, Sun D, Chen YX, Hong J, et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016;6:784–801. doi: 10.1158/2159-8290.CD-15-0921. [DOI] [PubMed] [Google Scholar]
- 80.Liu YW, Sun M, Xia R, Zhang EB, Liu XH, Zhang ZH, Xu TP, De W, Liu BR, Wang ZX. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802. doi: 10.1038/cddis.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Chen J. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi: 10.18632/oncotarget.1902. https://doi.org/10.18632/oncotarget.1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen WM, Huang MD, Shu YQ. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648–5661. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- 83.Lu MH, Tang B, Zeng S, Hu CJ, Xie R, Wu YY, Wang SM, He FT, Yang SM. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524–3534. doi: 10.1038/onc.2015.413. [DOI] [PubMed] [Google Scholar]
- 84.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, De W, Wang KM, Wang ZX. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Molecular cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kong R, Zhang EB, Yin DD, You LH, Xu TP, Chen WM, Xia R, Wan L, Sun M, Wang ZX, De W, Zhang ZH. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015;14:82. doi: 10.1186/s12943-015-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F, Wang Z, Sun M. Long Noncoding RNA LINC00673 Is Activated by SP1 and Exerts Oncogenic Properties by Interacting with LSD1 and EZH2 in Gastric Cancer. Mol Ther. 2017;25:1014–1026. doi: 10.1016/j.ymthe.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y, Shan B, Zheng D, Shi J. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017;31:893–903. doi: 10.1096/fj.201600994R. [DOI] [PubMed] [Google Scholar]
- 88.Liu YW, Xia R, Lu K, Xie M, Yang F, Sun M, De W, Wang C, Ji G. LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol Cancer. 2017;16:39. doi: 10.1186/s12943-017-0588-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu MD, Wang Y, Weng W, Wei P, Qi P, Zhang Q, Tan C, Ni SJ, Dong L, Yang Y, Lin W, Xu Q, Huang D, et al. A Positive Feedback Loop of lncRNA-PVT1 and FOXM1 Facilitates Gastric Cancer Growth and Invasion. Clin Cancer Res. 2017;23:2071–2080. doi: 10.1158/1078-0432.CCR-16-0742. [DOI] [PubMed] [Google Scholar]
- 90.Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, Chen H, Hong J, Zou W, Chen Y, Xu J, Fang JY. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890–6902. doi: 10.1158/0008-5472.CAN-14-0686. [DOI] [PubMed] [Google Scholar]
- 91.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. https://doi.org/10.18632/oncotarget.1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xia T, Chen S, Jiang Z, Shao Y, Jiang X, Li P, Xiao B, Guo J. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep. 2015;5:13445. doi: 10.1038/srep13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu Z, Shao Y, Tan L, Shi H, Chen S, Guo J. Clinical significance of the low expression of FER1L4 in gastric cancer patients. Tumor Biol. 2014;35:9613–9617. doi: 10.1007/s13277-014-2259-4. [DOI] [PubMed] [Google Scholar]
- 94.Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang G, Zhang Y. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep. 2015;5:10159. doi: 10.1038/srep10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee KS, Park JL, Lee K, Richardson LE, Johnson BH, Lee HS, Lee JS, Kim SB, Kwon OH, Song KS, Kim YS, Ashktorab H, Smoot DT, et al. nc886, a non-coding RNA of anti-proliferative role, is suppressed by CpG DNA methylation in human gastric cancer. Oncotarget. 2014;5:3944–3955. doi: 10.18632/oncotarget.2047. https://doi.org/10.18632/oncotarget.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]