Abstract
Co-culture engineering is an emerging approach for microbial biosynthesis of a variety of biochemicals. In this study, E. coli-E. coli co-cultures were developed for heterologous biosynthesis of the natural product naringenin. The co-cultures were composed of two independent E. coli strains dedicated to functional expression of different portions of the biosynthetic pathway, respectively. The co-culture biosynthesis was optimized by investigating the effect of carbon source, E. coli strain selection, timing of IPTG induction and the inoculation ratio between the co-culture strains. Compared with the mono-culture strategy, the utilization of the designed co-cultures significantly improved the naringenin production, largely due to the reduction of metabolic stress, employment of proper hosts for improving pathway enzyme activities, and flexible adjustment of the relative biosynthetic strength between the co-culture strains. The findings of this study extend the applicability of co-culture engineering in complex natural product biosynthesis.
Keywords: Co-culture engineering, Naringenin, Heterologous biosynthesis, E. coli
1. Introduction
Naringenin is a valuable natural product with identified biological activities such as anti-oxidative, anti-cancer, anti-obesity and anti-inflammatory activities [1], [2], [3]. As a member of the broad flavonoid family, naringenin possesses a characteristic 15-carbon skeleton with three ring structures. Naringenin is mainly produced as a secondary metabolite by plants, although a recent study reported that it can also be produced by prokaryote Streptomyces clavuligerus [4]. Production of naringenin through direct extraction from plants requires extensive separation and purification efforts and thus is not cost-effective from the perspective of process economy. Alternatively, employment of surrogate microbial hosts for heterologous biosynthesis can be a viable option for high-efficiency naringenin production. To this end, the biosynthetic pathway for naringenin has been intensively investigated and well characterized. As shown in Fig. 1, a simple carbon substrate is first used to produce aromatic amino acid tyrosine. Tyrosine is then converted to p-coumaric acid by tyrosine ammonia lyase (TAL), which is further utilized to make p-coumaroyl-CoA by 4-coumarate:CoA ligase (4CL). The sequential condensation of one p-coumaroyl-CoA and three malonyl-CoA leads to the formation of naringenin chalcone by chalcone synthase (CHS), which is then converted to naringenin by chalcone isomerase (CHI). The heterologous biosynthesis of naringenin has been achieved through functional reconstitution of the complex biosynthetic pathway in selected microbial hosts, such as S. cerevisiae and E. coli [5], [6], [7], [8], [9], [10], [11], [12]. Notably, these previous efforts utilized the mono-culture of an engineered microbial strain to achieve the heterologous production and relied on the traditional mono-culture engineering strategies to address the challenges of production optimization.
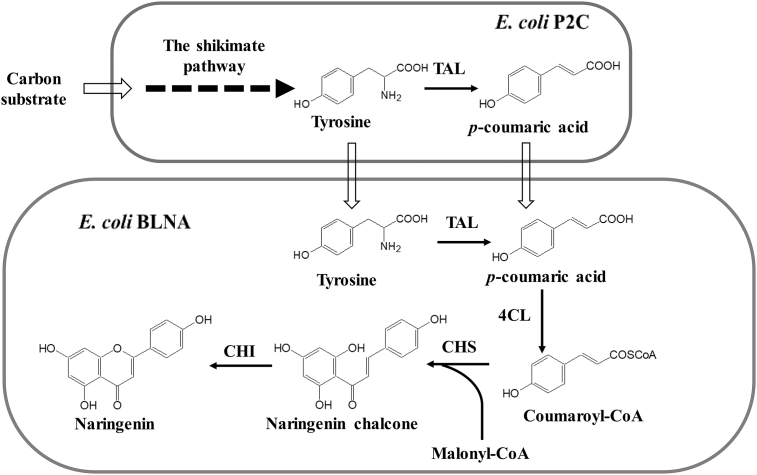
Fig. 1.
The co-culture design for biosynthesis of naringenin. The upstream strain is responsible for producing tyrosine and p-coumaric acid from simple substrates. The downstream strain is dedicated to converting tyrosine and p-coumaric acid into naringenin. Tyrosine ammonia lyase (TAL) was expressed in both upstream E. coli strain (P2C) and downstream E. coli strain (BLNA). 4-coumarate:CoA ligase (4CL), chalcone synthase (CHS) and chalcone isomerase (CHI) were expressed in the downstream BLNA strain.
On the other hand, engineering microbial co-cultures composed of multiple microbial strains has been shown to be a robust method for production of a variety of biochemicals [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23]. In particular, several studies have reported that this approach could be adapted to improve the biosynthesis of natural products with complex structures. For example, Zhou et al. employed an E. coli-S. cerevisiae co-culture system to biosynthesize oxygenated isoprenoids with significantly higher titers than the mono-culture approach [20]. Minami et al. developed E. coli-S. cerevisiae co-cultures to accommodate complex biosynthetic pathways for benzylisoquinoline alkaloids and successfully produced 7.2 mg/L magnoflorine and 8.3 mg/L scoulerine [21]. Using an E. coli-E. coli co-culture, Jones et al. achieved the 960-fold improvement of flavan-3-ols production from exogenous p-coumaric acid and caffeic acid substrates [22]. Recently, novel E. coli polycultures containing four individually engineered strains were also constructed for biosynthesis of complex natural product anthocyanins [23].
Therefore, this study aims to explore the potential of co-culture engineering for de novo naringenin heterologous biosynthesis. Specifically, the naringenin biosynthetic pathway is accommodated in two separate E. coli strains, which are individually engineered for performing the assigned biosynthesis task. This design splits the biosynthetic labor between two E. coli strains and thus reduces the metabolic burden on each strain. In addition, different E. coli strains can be screened for identification of the most suitable host to support a specific portion of the biosynthetic pathway. Importantly, through adjusting the ratio between the upstream and downstream strains, the biosynthetic strength for intermediate supply and consumption can be flexibly adjusted for bioproduction optimization. The results of this study provide a new perspective to overcome the barriers of complex natural product heterologous biosynthesis by the co-culture engineering strategies.
2. Materials and methods
2.1. Strains and cultivation medium
Strains and plasmids used in this work are listed in Table 1. All strains were cultivated at 275 rpm under 30 °C. The media used in this study are as follows. One liter of glucose medium contained 2 g of NH4Cl, 5 g of (NH4)2SO4, 3 g of KH2PO4, 7.3 g of K2HPO4, 8.4 g of 3-(N-morpholino)propanesulfonic acid, 0.5 g of NaCl, 0.24 g of MgSO4, 0.5 g of yeast extract, 40 mg of tyrosine, 40 mg of tryptophan, 10 mg of 4-HB and 5 g of glucose. For glycerol medium, 5 g of glycerol was used to replace glucose.
Table 1.
Strains and plasmids used in this study.
| Plasmids | Description | Source |
|---|---|---|
| pTrcHis2B | trc promoter, pBR322 ori, AmpR | Invitrogen |
| pCDFDuet-1 | double T7 promoters, CDF ori, SpR | Novagen |
| pOM | Vector from Evonik (derived from pUC18), contains pGAP promoter, rrnB terminator, AmpR and ColE1 origin | [7] |
| pCA1 | pTrcHis2B carrying codon optimized R.glutinis TAL | [29] |
| pCDF-trc-RgTALsyn- Pc4CLsyn | pCDFDuet-1 carrying codon-optimized R. glutinis TAL and codon-optimized P. crispus 4CL-1 with trc promoter | [7] |
| pOM-PhCHS-MsCHI |
pOM carrying P. hybrida CHS and M. sativa CHI with a single GAP (constitutive) promoter |
[7] |
| Strains |
Description |
Source |
| P2 | E. coli K12 ΔpheA ΔtyrR lacZ::PLtetO-1-tyrAfbraroGfbrtyrR::PLtetO-1-tyrAfbraroGfbr | [24] |
| P2H | P2 hisH (L82R) (DE3) | [24] |
| PMC | P2H carrying pCDF-trc-RgTALsyn- Pc4CLsyn and pOM-PhCHS-MsCHI | This study |
| P2C | P2H carrying pCA1 and pCDFDuet-1 | This study |
| BL21 (DE3) | F−ompT hsdSB (rB–, mB–) gal dcm (DE3) | Invitrogen |
| BLNA | BL21 (DE3) carrying pCDF-trc-RgTALsyn- Pc4CLsyn and pOM-PhCHS-MsCHI | This study |
| BL21 Star (DE3) | F−ompT hsdSB (rB–, mB–) gal dcm rne131 (DE3) | Life technologies |
| StarNA | BL21 Star (DE3) carrying pCDF-RgTALsyn- Pc4CLsyn and pOM-PhCHS-MsCHI | This study |
| JM109 (DE3) | F′ traD36 proA+B+lacIqΔ(lacZ)M15/Δ(lac-proAB) glnV44 e14-gyrA96 recA1 relA1 endA1 thi hsdR17 (DE3) | NEB |
| JMNA | JM109 (DE3) carrying pCDF-trc-RgTALsyn- Pc4CLsyn and pOM-PhCHS-MsCHI | This study |
| K12 (DE3) | F- lambda- ilvG- rfb-50 rph-1 (DE3) | [7] |
| K12NA | K12 (DE3) carrying pCDF-trc-RgTALsyn- Pc4CLsyn and pOM-PhCHS-MsCHI | This study |
2.2. Naringenin biosynthesis
For E. coli PMC monoculture cultivation, PMC was first grown overnight in the LB medium. The resulting culture was then centrifuged and suspended in 5 mL of glycerol or glucose medium containing 50 mg/L of streptomycin and 100 mg/L of ampicillin, followed by cultivation at 30 °C for 72 h. The initial cell density was kept the same with that of the corresponding co-culture with different inoculation ratios. 1 mM IPTG was added to the culture either at the time of inoculation or at desired points after inoculation.
For naringenin biosynthesis using E. coli-E. coli co-cultures, E. coli P2C and BLNA were first inoculated into LB medium containing 50 mg/L streptomycin and 100 mg/L ampicillin at 30 °C. Based on the need of inoculation, appropriate amounts of P2C and BLNA overnight culture were centrifuged, and the collected cell pellets of P2C and BLNA were suspended together in 5 mL medium containing 50 mg/L of streptomycin and 100 mg/L of ampicillin. The initial cell density of P2C was set to be OD600 = 0.15 and the cell density of BLNA was varied according to the desired inoculation ratio. 1 mM IPTG was added either at the time of inoculation or at designed time points after inoculation. At the end of the co-culture cultivation, the cell density was determined by measuring the absorbance at 600 nm with a UV/Visible Spectrophotometer (VWR, Radnor, PA) and the culture samples were taken for production analysis. Similar protocol was used for cultivation of P2C:K12NA, P2C:JMNA, and P2C:StarNA co-cultures.
2.3. Production analysis
For tyrosine quantification, culture samples were first centrifuged at 10,000 rpm and the supernatants were filtered through 0.45 μm PTFE membrane syringe filters (VWR, Radnor, PA). The filtered samples were analyzed by a Shimadzu HPLC system equipped with a Shimadzu Photodiode Array detector set to a wavelength of 280 nm. The analysis was performed on a Waters C18 column using 95% water and 5% acetonitrile as the mobile phase at a flow rate of 0.3 mL/min.
To quantify p-coumaric acid and naringenin, 1 mL of cell-free culture broth was extracted with 1 mL ethyl acetate. After mixing and centrifugation at 10,000 rpm, the top layer was transferred to a clean tube for evaporation to dryness, and dissolved with 1 mL ethanol. Such prepared samples containing p-coumaric acid and naringenin were analyzed on the Waters C18 column with an isocratic method of 65% water and 35% acetonitrile at a flow rate of 0.3 mL/min. The wavelength used for quantification was 290 nm for naringenin and 300 nm for p-coumaric acid, respectively.
2.4. Determination of the strain-to-strain ratio
The strain-to-strain ratio of the co-culture members was analyzed by the blue-white screening method described in a previous report [17]. Specifically, 10 μL of the co-culture sample was diluted 105–106-fold before being spread onto an LB agar plate containing IPTG and X-Gal. After 24 h of incubation, the upstream strain P2C carrying a disrupted lacZ gene generated white colonies, while the downstream strain BLNA carrying the intact lacZ gene generated blue colonies. The number of the blue and white colonies on the plates were counted for statistical analysis.
3. Results and discussion
3.1. Design of the E. coli-E. coli co-culture system for naringenin pathway accommodation
The schematic design of the E. coli-E. coli co-culture for naringenin biosynthesis is shown in Fig. 1. Specifically, the co-culture system is composed of an upstream strain for producing the pathway intermediates tyrosine and p-coumaric acid and a downstream strain for converting the intermediates to the final product naringenin. For the upstream strain, a previously engineered E. coli tyrosine overproducer P2 was modified to generate strain P2C (Table 1). P2C carried an engineered tyrosine pathway as well as the heterologous enzyme tyrosine ammonia lyase (TAL), which enabled it to perform tyrosine biosynthesis and subsequent conversion to p-coumaric acid [24], [25]. E. coli K12 (DE3), JM109 (DE3), BL21 (DE3) and BL21 Star (DE3), were individually employed as the downstream strain for functional expression of the heterologous enzymes tyrosine ammonia lyase (TAL), 4-coumarate:CoA ligase (4CL), chalcone synthase (CHS) and chalcone isomerase (CHI). Such engineered downstream strains can convert the pathway intermediates tyrosine and p-coumaric acid to the final naringenin product. Notably, TAL was expressed in both upstream and downstream strains to enhance the tyrosine-to-coumaric acid conversion, as this step has been found to be a limiting step for the naringenin biosynthesis [7]. The mass transfer between the two co-culture strains was carried out via the pathway intermediates tyrosine and p-coumaric acid, both of which can travel across E. coli cell membrane.
The co-culture design of this study allows us to reduce the metabolic stress on each strain. Instead of imposing the entire biosynthetic pathway on one single strain, the utilization of the co-culture splits the biosynthetic labor and metabolic stress between two strains, improving the cell fitness and biosynthetic capabilities of the both strains. In addition, different strains can be compared for their performance of intermediates-to-product conversion to determine the most suitable host for supporting the heterologous enzymes' activities associated with the downstream pathway. To adjust the biosynthetic strength between the upstream and downstream pathways, the corresponding strains can be inoculated into the co-culture system at designed ratios. The biosynthesis results can demonstrate the impact of the relative population size between co-culture strains on the overall bioproduction performance.
3.2. Comparison of bioproduction on glycerol and glucose
Carbon substrate selection is an important factor for microbial biosynthesis performance. Glucose is popularly used as a model carbon substrate for biosynthesis of a wide range of bioproducts. In contrast, glycerol has been reported to be a good carbon substrate for the shikimate pathway for supporting complex natural product biosynthesis [26], [27], [28]. Therefore, we first compared the naringenin bioproduction when glycerol and glucose were used as the carbon substrates. The biosynthesis was conducted in 5 mL medium containing 5 g/L carbon substrate and IPTG was supplemented at the time of inoculation. Strains P2C and BLNA (BL21 (DE3) carrying the downstream pathway) were used for construction of the co-culture, whereas strain PMC was employed as the mono-culture control (Table 1). Notably, the inoculation ratio between the two co-culture strains was changed to in-depth investigate the co-culture bioproduction on different carbon substrates.
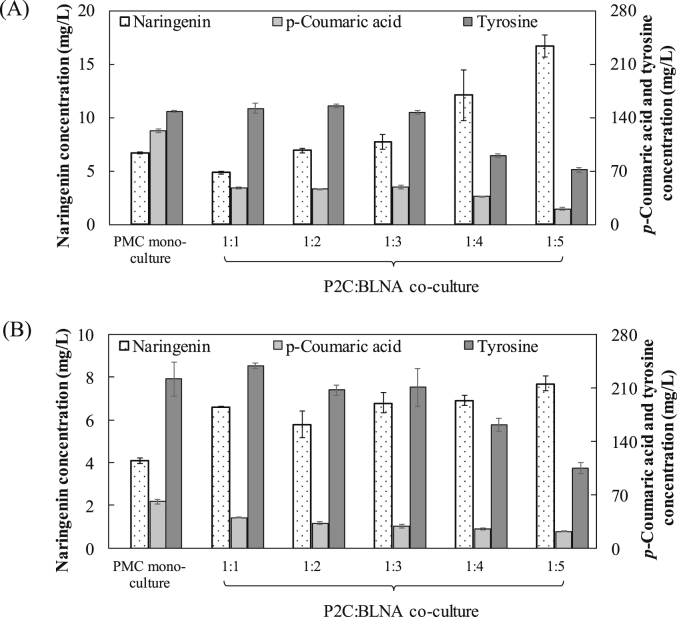
As shown in Fig. 2A, when glucose was fed as the carbon substrate, the P2C:BLNA co-culture showed varied naringenin production performance at different inoculation ratios. The highest production of 16.7 mg/L was achieved at the inoculation ratio of 1:5. It is therefore suggested that, when more downstream strain culture was inoculated (inoculation ratio changed from 1:1 to 1:5), the downstream pathway was enhanced and final naringenin product concentration was accordingly increased. In comparison, the mono-culture of the PMC strain carrying the whole pathway only produced 6.7 mg/L naringenin at initial OD 0.3 (the same with the initial OD of the co-culture inoculated at 1:1 ratio), indicating that the co-culture strategy resulted in 150% increase in the naringenin production. Also, the increase of the initial cell density in the mono-culture, which matched those of the co-culture inoculated at the rising ratio, did not significantly impact the naringenin production (Fig. S1).
Fig. 2.
Naringenin biosynthesis using (A) glucose and (B) glycerol as the carbon substrate. The bioproduction was compared with between the engineered E. coli mono-culture and E. coli-E. coli co-culture inoculated at different ratios.
The accumulation of the intermediates p-coumaric acid and tyrosine was also shown to be dependent upon the co-culture inoculation ratio. In fact, as more BLNA was inoculated, the concentrations of these pathway intermediates were reduced, indicating higher conversion to the final naringenin product by the enhanced downstream strain population. However, the concentrations of p-coumaric acid and tyrosine were still significantly higher than naringenin, which was in agreement with the previous reports [5], [6], [7], [8], [9], [10], [11]. These results demonstrate that, despite the final product biosynthesis improvement, the intermediate conversion was still limiting the overall biosynthesis performance under the co-culture design.
The naringenin production using glycerol as the carbon substrate is shown in Fig. 2B. The naringenin concentration of the P2C:BLNA co-culture rose from 6.6 mg/L to 7.7 mg/L, as the inoculation ratio was changed from 1:1 to 1:5. In comparison, the PMC mono-culture produced 4.1 mg/L naringenin at an initial OD equal to that of the 1:1 inoculated co-culture. Notably, the change of the cultivation temperature from 30 to 37 °C resulted in decreased naringenin production (Fig. S2). The naringenin biosynthesis on glycerol by the mono-culture and co-culture was all lower than the production on glucose. It was also found that, when glycerol was used, the tyrosine accumulation was more severe, whereas p-coumaric acid accumulation was lower. These findings suggest that the co-cultures grown on glucose better supported tyrosine-to-coumaric acid conversion, which contributed to higher naringenin production on glucose. Overall, our results indicate that, compared with glycerol, glucose is a better carbon substrate for the production of naringenin, as the production on glucose was improved for both the E. coli PMC mono-culture and the P2C:BLNA co-culture. Therefore, glucose was selected as the carbon substrate for the following sections of this study.
3.3. Optimization of the downstream strain selection
It has been demonstrated that microbial biosynthesis performance can be altered when different E. coli strains were used to constitute the co-culture system [18]. Therefore, four E. coli strains, including K12 (DE3), JM109 (DE3), BL21 (DE3), and BL21 Star (DE3), were individually investigated to identify the best candidate as the downstream strain for the naringenin co-culture synthesis. These strains have different metabolic characteristics. For example, K12 and JM109 strains are often used as cloning strains; BL21 (DE3) is a popular host for heterologous protein expression; BL21 Star (DE3) is an engineered BL21 (DE3) derivative strain for promoting mRNA stability. The introduction of the downstream naringenin biosynthesis pathway in these E. coli strains generated K12NA, JMNA, BLNA and StarNA, respectively. The constructed strains were co-cultivated with the upstream strain P2C to constitute four new co-cultures.
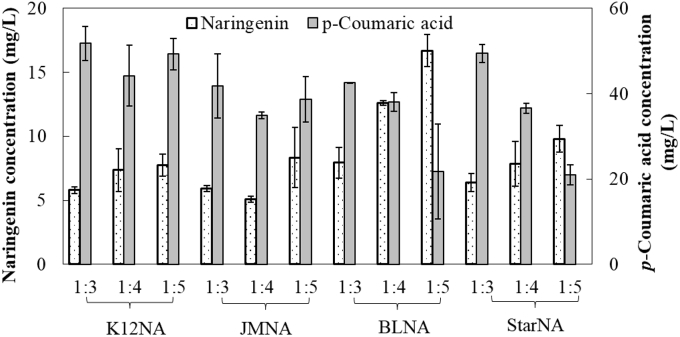
As shown in Fig. 3, the naringenin production profile did vary among these four co-cultures. The highest naringenin concentrations of 7.7 mg/L, 8.3 mg/L, 16.7 mg/L and 9.8 mg/L were achieved when K12NA, JMNA, BLNA and StarNA were employed (all at inoculation ratio of 1:5) for the co-cultures, respectively. Also, p-coumaric acid accumulation was found more severe for K12NA, JMNA and StarNA, suggesting these strains could not efficiently convert the intermediate p-coumaric acid to the final product naringenin. These findings indicate that BL21 (DE3) was the most suitable strain for accommodating the downstream portion of the biosynthetic pathway. In fact, BL21 (DE3) has unique metabolic characteristics and has been found more capable of supporting heterologous enzymes expression than K12, which is in good agreement with this study [30], [31]. On the other hand, the use of BL21 Star (DE3), a derivative of BL21 (DE3), may lead to overly strong expression of the heterologous enzymes and the potential formation of inclusion bodies, which could be the reason for reduced biosynthesis performance for StarNA. Therefore, the strain BL21 (DE3) carrying the downstream pathway enzymes, or BLNA, was selected as the downstream host as for the following studies.
Fig. 3.
Biosynthesis of naringenin by using different E. coli strains as the host for the downstream pathway. All the downstream strains, including K12NA, JMNA, StarNA and BLNA were engineered to express the heterologous TAL, 4CL, CHI and CHS enzymes.
3.4. Timing of IPTG induction
The timing of initiating IPTG induction can largely vary the protein expression profile and thus the microbial biosynthesis performance. It has been found that the naringenin bioproduction can be greatly improved through delayed IPTG induction [7]. In this section, the IPTG induction was initiated at different time points to evaluate its influence on the co-culture production of naringenin. The production was performed using the previously identified conditions, i.e. the P2C:BLNA co-culture grown on glucose under 30 °C with the inoculation ratio of 1:5.
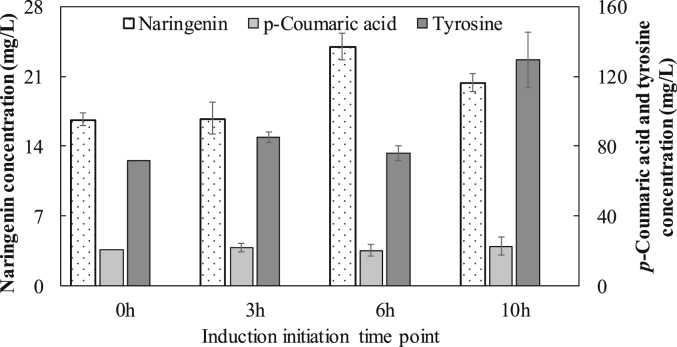
As shown in Fig. 4, our results showed that the timing of IPTG induction indeed played a significant role in the co-culture biosynthesis. As the IPTG induction was initiated at different time points ranging from 0 h to 10 h, the final naringenin concentration fluctuated accordingly. At the optimal induction initiation time of 6 h, the co-culture produced 24.0 mg/L naringenin, which was 35% higher than 17.7 mg/L naringenin produced by the mono-culture inoculated with same cell density and induced at 6 h. Compared with 0 h induction, delayed induction at 6 h improved naringenin concentration by 44%, which is consistent with the previous report [7]. Similar effect was found when glycerol was used as the carbon substrate. As shown in Fig. S3, the optimal biosynthesis on glycerol was also achieved when IPTG induction was initiated 6 h after the inoculation.
Fig. 4.
Effect of IPTG induction time points on naringenin biosynthesis. The P2C:BLNA co-culture was inoculated at the ratio of 1:5 and the IPTG was added to the culture at 0, 3, 6 and 10 h after inoculation.
The results of this section suggest that early IPTG induction imposed overly strong metabolic stress on the co-culture strains before they could thrive under the cultivation conditions, whereas late induction missed the time window for desired expression at exponential growth phase when the strains were robust for reconstituting the pathway enzymes' activities. It should be noted that, the heterologous genes' expression in both the upstream and downstream strain was induced by exogenous IPTG. As such, the induction timing impacted the growth and biosynthetic behaviors of both strains, although the extent of impact may be different. From the perspective of improving naringenin bioproduction, the optimal induction timing should be the most favorable for supporting the coordinated growth and biosynthetic performance of both co-culture strains. To this end, our results confirm that selection of a proper timing to initiate the induction plays an essential role in co-culture bioproduction optimization. Yet, such a proper induction timing is case-dependent and needs to be examined and identified for each co-culture system constructed for biosynthesis of different target products.
3.5. Co-culture bioproduction in shake flasks
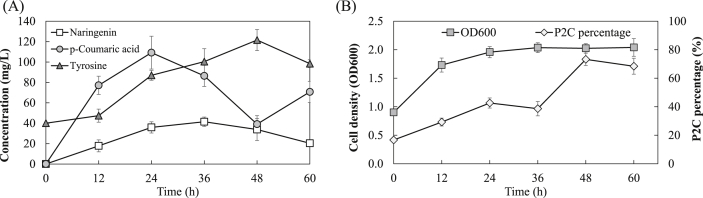
To further increase the naringenin biosynthesis, the engineered co-culture was cultivated in shake flasks containing the medium supplemented with 20 g/L glucose. The higher glucose substrate concentration and better dissolved oxygen supply under this cultivation condition were expected to improve the final product titer. The upstream and downstream strains were inoculated at the ratio of 1:5 and the IPTG induction was initiated at 6 h, as determined by the previous results. The time profiles of the intermediates and the product concentration, co-culture cell density and population composition changes in shake flask cultivation are shown in Fig. 5A and B. It was found that the concentration of naringenin first increased over the first 36 h and then declined towards the end of cultivation. The decline of naringenin concentration suggested the degradation in the culture over the extended cultivation time, although such degradation was not observed in previous studies and the potential mechanisms for this is still unknown. The highest naringenin concentration of 41.5 mg/L was achieved at 36 h, which is significantly higher than the production on 5 g/L glucose. On the other hand, the concentration of tyrosine and p-coumaric acid accumulated at 36 h were 100.4 mg/L and 86.4 mg/L, respectively, indicating that there was still high amounts of un-consumed pathway intermediates. In fact, tyrosine and p-coumaric acid accumulation was observed throughout the entire co-culture cultivation process, which suggests that the bioconversion capability of the downstream pathway is limiting the overall bioproduction performance even when the downstream strain was inoculated five times more than the upstream strain.
Fig. 5.
The co-culture bioproduction on 20 g/L glucose. (A) Time profiles of Naringenin and intermediate production. (B) Time profiles of the P2C/BLNA co-culture overall cell density (OD600) and P2C percentage.
In the meantime, it was found that the upstream strain P2C grew much faster than the downstream strain BLNA after inoculation and gradually dominated the co-culture system over the course of cultivation (Fig. 5B). In fact, the percentage of P2C increased from 16.7% (0 h) to 68.4% (60 h), indicating that the majority of the glucose was consumed to support the growth of the upstream cells. Relatively, the sub-population size of the downstream strain BLNA was constantly repressed throughout the whole bioproduction process, despite the fact that this strain took 83% of the entire co-culture population at the beginning of the cultivation. These findings indicate that BLNA was at great growth disadvantage when competing with the upstream P2C strain in the co-culture, which should be due to the substantial metabolic stress associated with the over-expression of four heterologous enzymes in BLNA. As a result, there was no sufficient tyrosine-to-naringenin conversion power to consume the accumulated tyrosine and p-coumaric acid. Such growth imbalance between the co-culture members has also been identified in previous studies [15], [17]. In fact, due to the difference in the designated biosynthetic labor and the corresponding metabolic stress, it is anticipated that the co-culture strains show different levels of growth inconsistency. To this end, manipulation of the inoculation ratio helps “pre-set” the initial strain-to-strain ratio, but the pathway balancing under this ratio can be easily disrupted due to the dynamic growth and complex interaction between the co-culture members. Nonetheless, our results indicate that, using the determined bioproduction conditions, the constructed co-culture can be grown on higher concentration glucose for naringenin production improvement. Also, imbalanced growth between the upstream and downstream strain was the main reason of the intermediate accumulation, which will be the focus of the future efforts for naringenin production improvement.
4. Conclusions
Biosynthetic pathway balancing plays an essential role for production optimization [32]. Pathway balancing, i.e., coordination of intermediate supply and consumption, in mono-cultures can be achieved by strategies such as modifying the promoter strength or changing the copy number of the plasmids carrying the pathway genes. However, these strategies are limited by the available genetic engineering tools (e.g. availability of plasmids with a wide range of different copy numbers) and often times require extensive research efforts. Co-culture engineering offers a new perspective to adjust and balance the biosynthetic strength between different pathway portions for production optimization. In fact, this can be achieved by changing the relative population size between the co-culture members without the need of additional genetic modification of either member.
This advantage is utilized in this study for improving the heterologous biosynthesis of complex natural product naringenin. The utility of two separate E. coli strains provides the unique opportunity to enhance the relatively weak downstream intermediate-to-product conversion by increasing the inoculation ratio of the downstream strain. In addition, such a design allows for the alleviation of metabolic burden on each strain as well as the employment of specialized strains for different biosynthesis tasks. In the optimized co-culture system, the highest naringenin production was achieved by the P2C:BLNA co-culture inoculated at the ratio of = 1:5, induced at 6 h after inoculation and grown on glucose under 30 °C. The production performance was found to be significantly better than that of the mono-culture. In addition, naringenin bioproduction was further improved when higher concentration of glucose was used as the starting materials. The results of this study demonstrate that the co-culture engineering indeed holds strong potential for wide application in complex natural product biosynthesis.
Acknowledgement
The authors gratefully acknowledge Professor Matthews Koffas (Rensselaer Polytechnic Institute) and Professor Gregory Stephanopoulos (Massachusetts Institute of Technology) for the generous gift of plasmids pOM-PhCHS-MsCHI, pCDF-trc-RgTALsyn- Pc4CLsyn and pCA1 in support of this study. This work is supported by startup research funds from Rutgers, The State University of New Jersey. Zhenghong Li is grateful for the Ph.D. fellowship from the China Scholarship Council.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.synbio.2017.08.003.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Hermenean A., Ardelean A., Stan M., Herman H., Mihali C.-V., Costache M. Protective effects of naringenin on carbon tetrachloride-induced acute nephrotoxicity in mouse kidney. Chem-Biol Interact. 2013;205:138–147. doi: 10.1016/j.cbi.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 2.Kumar S., Pandey A.K. Chemistry and biological activities of flavonoids: an overview. Sci World J. 2013:2013. doi: 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu C., Chen J., Zhang J., Hu X., Zhou X., Lu Z. Naringenin inhibits angiotensin ii-induced vascular smooth muscle cells proliferation and migration and decreases neointimal hyperplasia in balloon injured rat carotid arteries through suppressing oxidative stress. Biol Pharm Bull. 2013;36:1549–1555. doi: 10.1248/bpb.b13-00247. [DOI] [PubMed] [Google Scholar]
- 4.Álvarez-Álvarez R., Botas A., Albillos S.M., Rumbero A., Martín J.F., Liras P. Molecular genetics of naringenin biosynthesis, a typical plant secondary metabolite produced by Streptomyces clavuligerus. Microb Cell Fact. 2015;14:178. doi: 10.1186/s12934-015-0373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koopman F., Beekwilder J., Crimi B., van Houwelingen A., Hall R.D., Bosch D. De novo production of the flavonoid naringenin in engineered Saccharomyces Cerevisiae. Microb Cell Fact. 2012;11:155. doi: 10.1186/1475-2859-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leonard E., Lim K.-H., Saw P.-N., Koffas M.A. Engineering central metabolic pathways for high-level flavonoid production in Escherichia Coli. Appl Environ Microbiol. 2007;73:3877–3886. doi: 10.1128/AEM.00200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos C.N.S., Koffas M., Stephanopoulos G. Optimization of a heterologous pathway for the production of flavonoids from glucose. Metab Eng. 2011;13:392–400. doi: 10.1016/j.ymben.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Wu J., Du G., Chen J., Zhou J. Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a crispr interference system in Escherichia Coli. Sci Rep. 2015;5:13477. doi: 10.1038/srep13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitaker W.B., Jones J.A., Bennett R.K., Gonzalez J.E., Vernacchio V.R., Collins S.M. Engineering the biological conversion of methanol to specialty chemicals in Escherichia coli. Metab Eng. 2017;39:49–59. doi: 10.1016/j.ymben.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 10.Xu P., Ranganathan S., Fowler Z.L., Maranas C.D., Koffas M.A. Genome-scale metabolic network modeling results in minimal interventions that cooperativly force the carbon flux towards malonyl-CoA. Metab Eng. 2011;13:578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Xiu Y., Jang S., Jones J.A., Zill N.A., Linhardt R.J., Yuan Q. Naringenin-responsive riboswitch-based fluorescent biosensor module for Escherichia Coli Co-Cultures. Biotechnol Bioeng. 2017 doi: 10.1002/bit.26340. [DOI] [PubMed] [Google Scholar]
- 12.Pandey R.P., Parajuli P., Koffas M.A., Sohng J.K. Microbial production of natural and non-natural flavonoids: pathway engineering, directed evolution and systems/synthetic biology. Biotechnol Adv. 2016;34:634–662. doi: 10.1016/j.biotechadv.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 13.Minty J.J., Singer M.E., Scholz S.A., Bae C.-H., Ahn J.-H., Foster C.E. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc Natl Acad Sci. 2013;110:14592–14597. doi: 10.1073/pnas.1218447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saini M., Chen M.H., Chiang C.-J., Chao Y.-P. Potential production platform of N-Butanol in Escherichia Coli. Metab Eng. 2015;27:76–82. doi: 10.1016/j.ymben.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H., Li Z., Pereira B., Stephanopoulos G. Engineering E. Coli-E. Coli cocultures for production of muconic acid from glycerol. Microb Cell Fact. 2015;14:134. doi: 10.1186/s12934-015-0319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin H.-D., McClendon S., Vo T., Chen R.R. Escherichia Coli binary culture engineered for direct fermentation of hemicellulose to a biofuel. Appl Environ Microbiol. 2010;76:8150–8159. doi: 10.1128/AEM.00908-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H., Pereira B., Li Z., Stephanopoulos G. Engineering Escherichia Coli coculture systems for the production of biochemical products. Proc Natl Acad Sci. 2015;112:8266–8271. doi: 10.1073/pnas.1506781112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H., Stephanopoulos G. Co-culture engineering for microbial biosynthesis of 3-amino-benzoic acid in Escherichia Coli. Biotechnol J. 2016;11:981–987. doi: 10.1002/biot.201600013. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H., Wang X. Modular Co-Culture engineering, a new approach for metabolic engineering. Metab Eng. 2016;37:114–121. doi: 10.1016/j.ymben.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Zhou K., Qiao K., Edgar S., Stephanopoulos G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol. 2015;33:377–383. doi: 10.1038/nbt.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minami H., Kim J.-S., Ikezawa N., Takemura T., Katayama T., Kumagai H. Microbial production of plant benzylisoquinoline alkaloids. Proc Natl Acad Sci. 2008;105:7393–7398. doi: 10.1073/pnas.0802981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones J.A., Vernacchio V.R., Sinkoe A.L., Collins S.M., Ibrahim M.H., Lachance D.M. Experimental and computational optimization of an Escherichia Coli Co-Culture for the efficient production of flavonoids. Metab Eng. 2016;35:55–63. doi: 10.1016/j.ymben.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Jones J.A., Vernacchio V.R., Collins S.M., Shirke A.N., Xiu Y., Englaender J.A. Complete biosynthesis of anthocyanins using E. Coli polycultures. mBio. 2017;8 doi: 10.1128/mBio.00621-17. e00621-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santos C.N.S. Massachusetts Institute of Technology; 2010. Combinatorial search strategies for the metabolic engineering of microorganisms. [Google Scholar]
- 25.Santos C.N.S., Xiao W., Stephanopoulos G. Rational, combinatorial, and genomic approaches for engineering l-tyrosine production in Escherichia Coli. Proc Natl Acad Sci. 2012;109:13538–13543. doi: 10.1073/pnas.1206346109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahn J.O., Lee H.W., Saha R., Park M.S., Jung J.-K., Lee D.-Y. Exploring the effects of carbon sources on the metabolic capacity for shikimic acid production in Escherichia Coli using in silico metabolic predictions. J Microbiol Biotechnol. 2008;18:1773–1784. [PubMed] [Google Scholar]
- 27.Jiang M., Zhang H. Engineering the shikimate pathway for biosynthesis of molecules with pharmaceutical activities in E. Coli. Curr Opin Biotechnol. 2016;42:1–6. doi: 10.1016/j.copbio.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 28.Nakagawa A., Minami H., Kim J.-S., Koyanagi T., Katayama T., Sato F. A bacterial platform for fermentative production of plant alkaloids. Nat Commun. 2011;2:326. doi: 10.1038/ncomms1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H., Stephanopoulos G. Engineering E. Coli for caffeic acid biosynthesis from renewable sugars. Appl Microbiol Biotechnol. 2013;97:3333–3341. doi: 10.1007/s00253-012-4544-8. [DOI] [PubMed] [Google Scholar]
- 30.Wu J., Boghigian B., Myint M., Zhang H., Zhang S., Pfeifer B. Construction and performance of heterologous polyketide-producing K-12-and B-Derived Escherichia Coli. Lett Appl Microbiol. 2010;51:196–204. doi: 10.1111/j.1472-765X.2010.02880.x. [DOI] [PubMed] [Google Scholar]
- 31.Han M.-J. Exploring the proteomic characteristics of the Escherichia Coli B and K-12 strains in different cellular compartments. J Biosci Bioeng. 2016;122:1–9. doi: 10.1016/j.jbiosc.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 32.Jones J.A., Toparlak Ö D., Koffas M.A. Metabolic pathway balancing and its role in the production of biofuels and chemicals. Curr Opin Biotechnol. 2015;33:52–59. doi: 10.1016/j.copbio.2014.11.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.