Abstract
Microbial production of chemicals and proteins from biomass-derived and waste sugar streams is a rapidly growing area of research and development. While the model yeast Saccharomyces cerevisiae is an excellent host for the conversion of glucose to ethanol, production of other chemicals from alternative substrates often requires extensive strain engineering. To avoid complex and intensive engineering of S. cerevisiae, other yeasts are often selected as hosts for bioprocessing based on their natural capacity to produce a desired product: for example, the efficient production and secretion of proteins, lipids, and primary metabolites that have value as commodity chemicals. Even when using yeasts with beneficial native phenotypes, metabolic engineering to increase yield, titer, and production rate is essential. The non-conventional yeasts Kluyveromyces lactis, K. marxianus, Scheffersomyces stipitis, Yarrowia lipolytica, Hansenula polymorpha and Pichia pastoris have been developed as eukaryotic hosts because of their desirable phenotypes, including thermotolerance, assimilation of diverse carbon sources, and high protein secretion. However, advanced metabolic engineering in these yeasts has been limited. This review outlines the challenges of using non-conventional yeasts for strain and pathway engineering, and discusses the developed solutions to these problems and the resulting applications in industrial biotechnology.
Keywords: CRISPR-Cas9, Kluyveromyces lactis, Kluyveromyces marxianus, Scheffersomyces stipitis, Yarrowia lipolytica, Hansenula polymorpha, Pichia pastoris
Abbreviations: HR, homologous recombination; NHEJ, nonhomologous end-joining; DSB, double strand break; CRISPR, Clustered regularly interspaced short palindromic repeats; TALEN, transcription activator-like effector nucleases; sgRNA, short (or single) guide RNA; PAM, protospacer adjacent motif
1. Introduction
The microbial production of fuels and chemicals from biomass and other renewable carbon sources is an attractive alternative to petroleum-derived products. One of the largest scale example of this is ethanol production by the yeast Saccharomyces cerevisiae— in 2015, over 25 billion gallons were produced worldwide from starch, waste sugar streams, and biomass-derived sugars (www.afdc.energy.gov/data/10331). S. cerevisiae is the organism of choice because of its high rate of production and tolerance to ethanol titers upwards of 120 g L−1 [1], [2]. These phenotypes, among others, have led to the widespread study of S. cerevisiae and its development as a model eukaryotic host for chemical biosynthesis. A valuable approach to metabolic engineering is identifying organisms with desirable phenotypes and developing new synthetic biology tools to enhance these phenotypes. Bioethanol production in S. cerevisiae is a good example of this, and illustrates the potential of identifying other hosts and phenotypes to synthesize bioproducts other than ethanol. A number of examples of this strategy already exist in industry, where non-conventional yeasts with unique and advantageous phenotypes are used to produce proteins, lipids, and commodity chemicals. Metabolic engineering in these yeasts is, however, more challenging in comparison with S. cerevisiae, because less is known about their metabolism and genomics, and advanced genetic engineering tools are limited.
In this review, we focus on six non-conventional yeasts (Table 1): Kluyveromyces lactis, K. marxianus, Scheffersomyces (Pichia) stipitis, Yarrowia lipolytica, Hansenula polymorpha, and Pichia pastoris. In contrast to S. cerevisiae, these yeasts are Crabtree negative and favor respiration over fermentation; phenotypes that are particularly useful for protein production as well as the biosynthesis of chemicals other than ethanol [3]. K. lactis is discussed here because of its capacity to metabolize inexpensive substrates such as waste whey and because of its use as a host for heterologous protein production in the food, feed, and pharmaceutical industries [4]. The Kluyveromyces species K. marxianus is also industrially relevant because of its wide substrate spectrum, fast growth characteristics, and thermotolerance to ∼50 °C [5], [6]. Native strains of K. marxianus are also known to synthesize ethyl acetate at rates above 2 g L−1 h−1 in aerated bioreactors [7], [8]. S. stipitis is capable of fermenting xylose at high rates compared to other yeasts and has been widely studied for ethanol production from biomass-derived sugars [9], [10]. Y. lipolytica is a well-studied oleaginous yeast and has attracted interest due to its ability to synthesize and accumulate high levels of intracellular lipids [11], [12], [13]. The methylotrophic yeast H. polymorpha has been studied as a model system for peroxisome function as well as for its methanol and nitrate assimilation pathways [14], [15]. Significant efforts have gone into heterologous protein production in H. polymorpha due to its efficient secretion pathways, effective glycosylation machinery, and tightly controlled expression systems [16]. H. polymorpha is also thermotolerant to temperatures comparable to K. marxianus and can assimilates various substrates, thus making it a potential alternative host for ethanol production [17]. The methylotrophic yeast P. pastoris has similar protein secretion and glycosylation capabilities to H. polymorpha and has been widely used for heterologous protein production [18]. Its capacity to grow to extremely high cell densities and high capacity for membrane protein expression also provide inherent advantages over other yeast hosts [19], [20].
Table 1.
Overview of non-conventional yeast species, their industrially-relevant phenotypes, common uses in biotechnology, and comparison with S. cerevisiae.
| Yeast | Beneficial Phenotype | Products | Reference |
|---|---|---|---|
| K. lactis | High protein secretion Growth on lactose |
Proteins for food and feed industry Pharmaceutical enzymes |
[4] |
| K. marxianus | Thermotolerance Fast growth characteristics High ethyl acetate production Growth on a range of sugars |
Ethanol and volatile acetate esters | [5] |
| S. stipitis | High ethanol production from xylose | Ethanol fermentation from biomass derived carbohydrates | [21] |
| Y. lipolytica | Efficient production of lipids Growth on glycerol and alkanes |
Lipids and oleochemicals | [12] |
| H. polymorpha | Thermotolerance Tightly regulated expression system Beneficial glycosylation for therapeutics |
Heterologous protein High temperature ethanol fermentation |
[17], [18] |
| P. pastoris | Tightly regulated expression system High cell density on minimal media Beneficial glycosylation for therapeutics Efficient production of membrane proteins |
Pharmaceuticals and industrial enzymes | [18] |
| S. cerevisiae | High ethanol production High HR capacity Well known genomics and physiology Advanced synthetic biology tools |
Ethanol in fermented beverages and as biofuel Commodity and specialty chemicals Pharmaceuticals |
[2], [22] |
Despite these many advantages, metabolic engineering of non-conventional yeasts is limited by a lack of sophisticated genome editing tools and an incomplete understanding of their genetics, metabolism, and cellular physiology. In this review, we discuss the challenges and solutions that have arisen in engineering non-conventional yeasts for metabolic engineering and synthetic biology applications. We begin our review with a discussion of the challenges to genetic engineering, followed by a discussion of strategies for improving genome and pathway engineering. Finally, we discuss representative examples of metabolic engineering in each of the selected yeasts. While the presented examples are not exhaustive, they are exemplative of current and past research efforts that exploit the yeasts' advantageous phenotypes. Reviews that provide comprehensive discussions on engineering each of the non-conventional yeasts described here are available elsewhere [4], [12], [17], [21], [23], [24].
2. Genetic engineering challenges in non-conventional yeasts
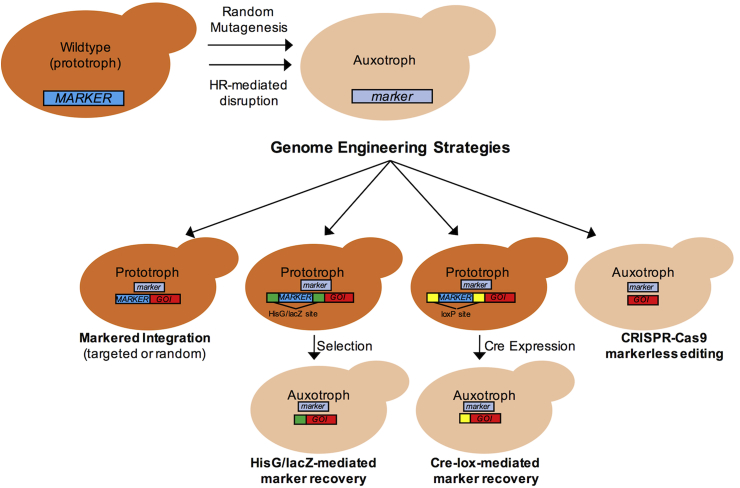
A basic requirement for metabolic engineering is the ability to express a gene (native or heterologous) from an expression cassette. Most strategies for heterologous gene expression in yeasts utilize auxotrophic markers to provide selective pressure for the maintenance of heterologous DNA containing the expression cassette (Fig. 1). Gene expression of native or heterologous genes is most often accomplished through episomal vectors or by integration of the gene(s)-of-interest into the host genome. In S. cerevisiae, transformation and expression from replicating plasmids is widely used due to the availability of stable and high copy number vectors [25], [26]. In non-conventional yeasts, options for stable plasmids are more limited. Plasmids are initially generated by combining centromeric regions of the organism’s genome and autonomous replicating sequences with a selectable auxotrophic marker [27]. While functional plasmids are available for most non-conventional yeasts, they tend to be low copy number and show variable expression across cells in a single population, an effect that is due to imperfect partitioning of plasmids upon cell division [28], [29].
Fig. 1.
Schematic diagram of the generation and utilization of auxotrophic markers for engineering yeast. Random mutagenesis of host DNA or homologous recombination of a cassette that inactivates an essential gene for nutrient synthesis can be used to produce stable auxotrophic strains. The presence of an auxotrophy allows more advanced genome editing and pathway engineering tools to be applied in the yeast species of interest. Shown here are 1) targeted and random integration using a selectable marker (bottom, left), 2) HisG/lacZ-mediated marker recovery (bottom, middle), 3) Cre-lox-mediated marker recovery (bottom, middle), and 4) Markerless editing by CRISPR-Cas9 (bottom, right).
The preferred strategy for heterologous gene expression in industrial strains is integration into the host's genome. Genomic integration leads to more homogenous expression levels across the population, increases the stability of the expression cassette over extended culture times, and eliminates the need for constant selection of a genetic marker [30]. Transformation with a linear DNA fragment containing an expression cassette and a selectable marker results in genomic integration in one of two ways: heterologous DNA is either incorporated into the genome at a random locus (often called illegitimate recombination [31]), or the cassette is targeted to a specific site in the genome by homology to the site of interest (Fig. 1). Both types of integrations are performed by native DNA repair pathways. Random integration proceeds through nonhomologous end-joining (NHEJ), while targeted integration occurs by homologous recombination (HR) [32]. Integration via HR is often preferred, because it enables control over the integration loci, avoids disrupting essential genes, and allows for integration into a site with a consistent expression profile [33]. Integration via HR can also be used to knockout native genes.
In S. cerevisiae, HR is the dominant DNA repair pathway. The high capacity for HR makes genome engineering relatively efficient and has facilitated the development of a wide range of in vivo DNA assembly tools [34], [35]. This is not the case in most other yeasts, where NHEJ is the favored DNA repair pathway and genome engineering by HR is inefficient. As a result, engineering of non-conventional yeasts is frequently accomplished by random integration. The random integration of the transformed expression cassette can lead to unwanted disruptions of open reading frames or other genomic elements. In addition, expression levels of heterologous cassettes have been shown to be highly dependent on the integration site, and so random integration can result in variable expression across transformants [33], [36].
An additional challenge to engineering multi-gene pathways is the limited number of viable selectable markers. To overcome this experimental challenge, researchers have developed several techniques for marker recovery (Fig. 1). The most commonly used systems are Cre-loxP, hisG, and lacZ [37], [38]. In these cases, the selectable marker (e.g., an antibiotic resistance gene or auxotrophic marker) is surrounded by hisG, lacZ, or loxP sequences. After genome integration of an expression or knockout cassette, the marker is excised by spontaneous HR or Cre recombinase activity. While the hisG and lacZ systems are effective, the Cre-loxP method is more common because hisG and lacZ systems require a counter-selection such as growth on media supplemented with 5-fluoroorotic acid (5-FOA) for URA3 excision [39], [40], [41], [42], [43]. While Cre-loxP systems are available for use in the non-conventional yeasts discussed in this review, this marker recovery technique does not solve the challenge of random, unknown integration sites that is problematic with illegitimate recombination.
3. Enhancing HR in non-conventional yeasts
A widely used strategy to enhance HR in non-conventional yeasts is disruption of genes essential for the NHEJ pathway, such as KU70 or KU80. K. lactis provides an early example of this strategy, where disruption of KU80 produced a strain capable of integrating heterologous DNA via HR at a rate of 97% [44]. Similarly, disruption of KU80 in S. stipitis resulted in an increase in the rate of HR-mediated integration of transformed linear donors [45]. In Y. lipolytica, disruption of KU70 or KU80 produced significant increases in HR rates, and allowed HR to occur with homology regions down to 50 bp [46], [47]. In H. polymorpha, KU80 knockout produced an increase in alcohol oxidase gene knockout rates (AOX2-8) from an average of 19% in the wildtype background to 76% in the KU80 deficient strain [48]. In P. pastoris, knockout of KU70 enabled HR rates as high as 90% [49]. A similar result was found in K. marxianus, where KU70 knockout increased HR rates to as high as 95% [50]. KU80 disruption in K. marxianus was similarly effective, with HR rates increasing to upwards of 70% [51].
A second strategy that has had success in increasing HR is cell cycle synchronization. Natively, the activity of the HR DNA repair pathway is dependent on cell cycle [52]. When a single copy of chromosomal DNA is present, as in G1 phase, NHEJ is favored. Genes required for HR tend to only be expressed during phases of the cell cycle when multiple copies of chromosomes are available, i.e., S phase and G2 phase. Cell cycle synchronization has been widely used for fundamental biochemistry studies, and a recent work took advantage of this strategy to stall cells in S phase with the intent of increasing HR [53]. By adding hydroxyurea to cultures undergoing exponential growth, the authors demonstrated that S phase stalling resulted in enhanced HR in Y. lipolytica, K. lactis, and P. pastoris.
An alternative strategy to achieve efficient HR is the introduction of a genomic double strand break (DSB) using a programmable endonuclease in the presence of a homologous repair template [54]. Due to the deleterious effects of DSBs on cell viability, native repair pathways attempt to repair the cut. If a repair template with adequate homology to the region flanking the break is present, the host may use the template as a donor for HR. This strategy has the added benefit of not requiring a selectable marker on the integrated DNA fragment. Several programmable tools exist for targeted DSB, including dimeric meganucleases, zinc finger nucleases, transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated 9 (CRISPR-Cas9) [54]. The first three of these have primarily been developed and applied in S. cerevisiae, although TALENs were recently used in Y. lipolytica [55], [56]. CRISPR-Cas9, however, has been widely applied in non-conventional yeasts as described in the following section.
4. CRISPR-Cas9 genome editing and transcriptional control
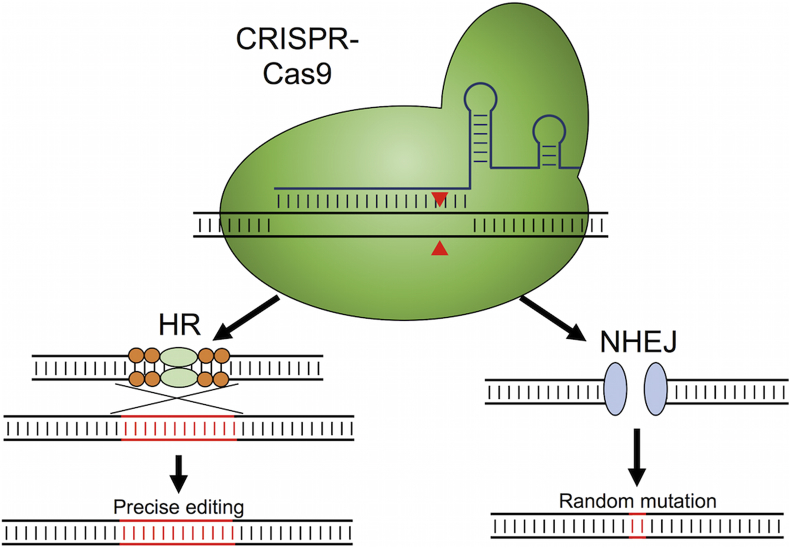
In recent years, the application of CRISPR-Cas9 technology has revolutionized genome editing. Specifically, the type II CRISPR-Cas9 system from Streptococcus pyogenes has been widely adopted to enable targeted DSB generation in a wide number of organisms [57], [58]. Functional expression of CRISPR-Cas9 in yeast has two main requirements. First, a codon-optimized Cas9 expression cassette is generated, with a nuclear localization tag fused to its C-terminus. A nuclear localization tag is needed because S. pyogenes is a bacterium, and so unmodified Cas9 would localize to the cytosol in yeast. The second component of CRISPR-Cas9 systems (as commonly applied for genome editing) is a short (or single) guide RNA (sgRNA) [59]. The sgRNA has two main roles. The first 20 bp at the 5′ end are known as the spacer and are responsible for genome targeting through complementation to the desired locus. The sgRNA also contains a structural region encoded downstream of the spacer that facilitates the interaction of the sgRNA and Cas9. Upon formation of the CRISPR-Cas9 ribonucleoprotein, the complex unwinds double stranded DNA and begins scanning for a sequence complementary to the spacer region of the sgRNA. When a complementary sequence is found, and if there is an appropriate protospacer adjacent motif (PAM; for S. pyogenes a genomic “NGG” found immediately 3′ of the targeted sequence), the nuclease domains of Cas9 cleave both strands of the DNA [57], [59]. The introduction of this DSB must then be repaired to avoid host cell death (Fig. 2). Repair of the DSB by NHEJ commonly results in indel mutations and gene inactivation. Providing a homology repair template induces repair of the break by HR and allows for a desired sequence to be inserted at the cut site.
Fig. 2.
CRISPR-Cas9-mediated genome editing. The Cas9-sgRNA complex scans DNA until finding a complementary sequence. Upon binding, the endonuclease domains cleave both DNA strands 3 bases upstream of the PAM sequence. The double strand break is then repaired either by homologous recombination (HR) if an appropriate homology donor is present, or by nonhomologous end-joining (NHEJ). Repair via HR allows for precise genome editing at the target site, while NHEJ introduces short insertions or deletions.
To date, CRISPR-Cas9 systems have been developed to allow gene disruptions and/or markerless integrations in all 6 of the non-conventional yeasts discussed in this review (Table 2). While most systems use a similar strategy for Cas9 expression and nuclear localization (commonly an SV40 C-terminal tag), a variety of strategies for sgRNA expression have been developed. In S. cerevisiae, the native SNR52 RNA polymerase III promoter is often used, as it allows for proper 5′ and 3′ maturation of the expressed sgRNA [57]. A similar sgRNA strategy was used in K. lactis and the resulting CRISPR-Cas9 system was demonstrated by simultaneously introducing three DSBs for multiplexed HR-mediated gene integration, successfully engineering a six gene pathway in a single transformation [35]. A native SNR52 promoter was also used to enable functional CRISPR-Cas9 genome editing in S. stipitis, where gene disruption rates upwards of 80% were achieved [27]. Two different CRISPR-Cas9 systems have been used in Y. lipolytica: the first relies on synthetic RNA polymerase III promoters for sgRNA expression, while the second used an RNA polymerase II promoter with ribozymes flanking the sgRNA to ensure proper 5′ and 3′ maturation [60], [61]. Both systems have been shown to achieve efficient gene disruption and gene integration rates. An analogous synthetic RNA polymerase III strategy was recently used for sgRNA expression to adapt the CRISPR-Cas9 system for use in K. marxianus, where gene disruption rates of 66% have been reported [62]. Successful adaptation of CRISPR-Cas9 to P. pastoris required the use of an RNA polymerase II promoter and ribozymes flanking the sgRNA, achieving efficiencies up to 100% for disruptions [63]. In H. polymorpha, a CRISPR-Cas9 system was developed by using tRNALeu as a promoter to drive sgRNA expression [64]. This system uses the endogenous tRNA processing system for proper sgRNA maturation and disruption efficiencies of up to 71% were achieved.
Table 2.
CRISPR-Cas9 systems for genome editing in non-conventional yeasts.
| Yeast | Cas9 expression | sgRNA expression | Gene disruption rate | HR rate | Reference |
|---|---|---|---|---|---|
| K. lactis | ScFBA1 promoter Genome integrated |
SNR52 promoter | N/A | 2% (3 integrations simultaneously) in NHEJ deficient strain | [35] |
| K. marxianus | ScTEF1 promoter codon-optimized |
RPR1′-tRNAGly | 66% | N/A | [62] |
| S. stipitis | eno1 promoter codon-optimized |
SNR52 promoter | 80% | N/A | [27] |
| Y. lipolytica | TEFintron promoter codon-optimized |
TEFintron promoter, flanked by hammerhead and hepatitis delta virus ribozymes | 85% | 11% in wildtype up to 100% in NHEJ deficient strain |
[60] |
| Y. lipolytica | UAS1B8-TEF promoter codon-optimized |
SCR1′-tRNAGly promoter | 92% | 64% in wildtype up to 100% in NHEJ deficient strain |
[61] |
| H. polymorpha | DH3 promoter human codon-optimized |
tRNALeu | 71% | 47% (marker integration with selection) | [64] |
| P. pastoris | HTX1 promoter human codon-optimized |
HTX1 promoter, flanked by hammerhead and hepatitis delta virus | 100% | 20% | [63] |
To further extend the yeast CRISPR-Cas9 toolbox, Cas9 can be mutated to deactivate its endonuclease activity while retaining its DNA targeting and binding capacity (dCas9). Targeting dCas9 to the promoter region of a gene can sterically block the RNA polymerase machinery from assembling, thus suppressing transcription; a technology referred to as CRISPR interference (CRISPRi) [65]. Fusion of a transcriptional repressor to dCas9 can result in a more effective CRISPRi system. In yeasts, the Mxi1 protein domain has proven most effective to date [66], [67]. In the context of non-conventional yeasts, CRISPRi has so far only been demonstrated in Y. lipolytica [68]. In this case, the synthetic RNA polymerase III system of sgRNA expression and Mxi1 fusion to dCas9 reduced target gene expression to as low as 10% of native expression levels. Finally, the fusion of a transcriptional activator to Cas9 has enabled CRISPR activation (CRISPRa), where native genes can be overexpressed by targeting CRISPRa to a gene's promoter [69]. To date, however, CRISPRa has not been demonstrated in the yeasts discussed in this review. Targeted transcriptional control represents a novel experimental ability in non-conventional yeasts, where well-characterized promoters and tightly tunable inducible promoters are less common relative to model organisms.
5. Bioprocessing and metabolic engineering with non-conventional yeasts
Despite the challenges of engineering non-conventional yeasts in comparison to the model host S. cerevisiae, a variety of successful bioprocesses have been developed. Here we present selected metabolic engineering examples that exploit desirable phenotypes expressed by K. lactis, K. marxianus, S. stipitis, Y. lipolytica, H. polymorpha, and P. pastoris and discuss the genetic engineering tools used to create new strains of these yeasts. Exemplative products produced from these hosts are presented in Table 3.
Table 3.
Expemplative list of non-conventional yeast products.
| Yeast | Products | Reference |
|---|---|---|
| K. lactis | Proteins | |
| Native β-galactosidase | ||
| Chymosin | ||
| Brazzein | Reviewed in Ref. [4] | |
| Human serum albumin (HSA) | ||
| Human interleukin 1-β | ||
| Interferon-α f | ||
| Chemicals | ||
| Glycolic acid | Reviewed in Ref. [3] | |
| Lactic acid | ||
| K. marxianus | Proteins | |
| Native inulinase | ||
| Native β-galactosidases | Reviewed in Ref. [24] | |
| Native pectinases | ||
| Chemicals | ||
| Ethanol from dairy waste or lignocellulosic feeds | Reviewed in Ref. [5] | |
| 2-Phenylethanol/2-phenyl ethyl acetate | [70] | |
| Ethyl acetate | [7] | |
| Y. lipolytica | Proteins | |
| Lipases | ||
| Proteases | ||
| α-amylases | Reviewed in Ref. [71] | |
| β-mannases | ||
| Chemicals | ||
| Lipids | Reviewed in Refs. [12] and [3] | |
| α-Ketoglutaric acid (KGA) | Reviewed in Ref. [71] | |
| Lycopene | [36] | |
| Omega-3 eicosapentaenoic acid (EPA) | [72] | |
| Citric acid | Reviewed in Ref. [3] | |
| S. stipitis | Chemicals | |
| Ethanol from lignocellulosic feeds | Reviewed in Ref. [21] | |
| Fumaric acid | [73] | |
| Lactic acid | [74] | |
| Xylitol | [75] | |
| H. polymorpha | Proteins | |
| Hepatitis B surface antigen (HBsAg) | ||
| Insulin | ||
| IFNα-2a | Reviewed in Ref. [76] | |
| Hexose oxidase | ||
| Phytase | ||
| Chemicals | ||
| Ethanol from various carbon sources | Reviewed in Ref. [17] | |
| P. pastoris | Proteins | |
| Ecallantide | ||
| Ocriplasmin | ||
| Phytase | Reviewed in Refs. [23] and [77] | |
| Trypsin | ||
| Phospholipase C | ||
| Chemicals | ||
| (+)-nootkatone | [78] | |
| violacein | [79] | |
| β-carotene | [79] |
5.1. Kluyveromyces lactis
Over the past three decades, considerable efforts have been put towards developing K. lactis as a yeast host for heterologous protein expression. To date, over 100 proteins have been produced, with more than 20% of these demonstrations occurring in the past five years [4]. Examples, such as β-galactosidase and the endopeptidase chymosin, sold by DSM under the trade names of Maxilact and Maxiren (DSM), respectively, have been produced at industrial-scale [4], [80]. The economic success of these processes is, in part, due to the ability of K. lactis to secrete high titers of protein and the ability metabolize inexpensive carbon sources such as waste whey streams produced in the dairy industry.
Strain development for K. lactis bioprocesses is most often achieved through an established and commercially available gene integration technology, pKLac2. The plasmid contains an acetamidase selection marker that allows for growth on acetamide as sole nitrogen source and facilitates multiple integrations into the genome of K. lactis. A mutant variant of the strongly inducible Lac4 promoter eliminates recognition of the promoter by E. coli and thus enables cloning of constructs toxic to E. coli. Efficient protein secretion is achieved using a K. lactis α-mating factor secretion domain [81]. One example of the successful use of the pKLac2 system was the production of cardosin B chymosin, a coagulant essential for cheese production, from galactose media [82]. Another example is the sweetener brazzein, which was produced from galactose with protein titers reaching 104 mg L−1 [83].
In comparison to protein synthesis, the use of K. lactis as a host for chemical biosynthesis has been limited. New CRISPR-Cas9 genome editing systems are, however, enabling multiplexed engineering and driving the field forward. For example, a recent work engineered a synthetic muconic acid pathway by simultaneous integrating six heterologous genes into three different K. lactis loci by HR. While triple integration efficiency was low at 2.1%, the desired strain was constructed in a time-efficient manner and produced ∼0.9 g L−1 muconic acid [35].
5.2. Kluyveromyces marxianus
One of the reasons that K. marxianus has attracted interest is its high capacity to produce the volatile short chain ester ethyl acetate [6]. Wild type strains of K. marxianus have been shown to produce ethyl acetate at yields of 0.265 g g−1 glucose (51.4% of maximum) and pilot-scale plants with productivity upwards of 2 g L−1 h−1 using waste whey as a feed stock have been demonstrated [7], [8]. In addition to ethyl acetate, K. marxianus is able to produce fusel alcohols and their corresponding acetate esters. This capacity has been harnessed for 2-phenylethanol production from phenylalanine at industrial scale [5], [70]. Biosynthesis of 2-phenylethanol from glucose and the synthesis of phenylethyl acetate from phenylalanine feeds have also been demonstrated [70], [84].
K. marxianus has also been considered as a host for bioethanol production from lignocellulosic biomass hydrolysates and crude waste whey streams [85], [86]. Commercial production plants have been built or are under consideration in the United States, Ireland, and New Zealand, and rely on production from dairy waste streams [5]. Ethanol fermentation has also been engineered using metabolic engineering and cofactor balancing approaches [87], [88]. In one study, ethanol production from xylose was enhanced by 1) overexpressing heterologous xylose reductase (XYL1) and xylitol dehydrogenase (XYL2), and 2) increasing the capacity of the pentose phosphate pathway and flux towards ethanol through the overexpression of native xylulokinase (XYL3), l-ribulose-5-phosphate 4-epimerase (RPE1), ribose-5-phosphate isomerase (RKI1), transketolase (TKL1), transaldolase (TAL1) genes as well as pyruvate decarboxylase (PDC1) and alcohol dehydrogenase (ADH2) [87]. The heterologous XYL1 and XYL2 genes were selected due to their preference of NADP(H) over NAD(H), thus helping to rectify an imbalance in co-factors when grown on xylose. Further improvement of fermentation efficiencies was achieved by eliminating glycerol production through the disruption of glycerol-3-phosphate dehydrogenase (GPD1). The resulting strain contained disruptions to three native genes and overexpression of two heterologous and seven native genes, and was able to produce ethanol from xylose at rates of 2.49 g L−1 h−1. In this case, strain engineering was achieved by markered gene disruption via HR and sequential random integration facilitated by a URA3 marker. Prior to each gene integration the marker was inactivated by HR with a truncated URA3 cassette and selection on 5-FOA containing media [87], [89].
K. marxianus' high capacity for NHEJ can, for many applications, limit genome editing. However, some researchers have exploited this capacity for multiplexed gene integration. For example, a five-gene pathway for the production of hexanoic acid was integrated in a single transformation by selection on uracil dropout media [90]. In this case, each integrated gene was accompanied by a URA3 selectable marker, resulting in a 50% success rate for full pathway integration. Random integration still proved to be problematic as hexanoic acid production varied widely between successful transformants, likely due to gene integration at critical genomic loci.
5.3. Scheffersomyces stipitis
A primary advantage of S. stipitis over other yeasts is its ability to ferment xylose at high rates [9], [10]. This phenotype has been exploited for ethanol production from biomass-derived and pure xylose streams. S. stipitis has also been engineered for higher ethanol tolerance as well as growth inhibiters present in biomass hydrolysates [91]. Due to a lack of efficient genome editing tools, engineering of S. stipitis has been limited to random mutagenesis through UV radiation, adaptive evolution, protoplast fusion, and genome shuffling [92], [93], [94], [95]. Single gene deletions have also been achieved through genomic integration using a selectable genetic marker. For example, a HR method was used to create a HXK1 deficient strain lacking glucose repression and a XYL2 deficient strain that produces xylitol from xylose [75]. In another example, S. stipitis was engineered to efficiently produce lactic acid through random integration of a heterologous LDH gene, with engineered strains producing up to 58 g L−1 lactate from 100 g L−1 xylose [74].
More complex pathway engineering has also been achieved. In one case, the deletion of two genes coupled with the overexpression of four heterologous genes led to the production of 4.67 g L−1 fumaric acid from 20 g L−1 xylose [73]. The plasmid-based pathway was comprised of a fumaric acid biosynthesis steps from Rhizopus oryzae and a fumaric acid transporter from Schizosaccharomyces pombe. Disruption of reaction steps competing with fumaric acid production, such as fumarase genes FUM1 and FUM2, was achieved via HR with a URA3 selectable marker and marker recovery by Cre-loxP. A critical lesson from this work was the need for codon optimization of the heterologous genes, as S. stipitis has an unusual usage of CTG, which it uses to code for serine instead of leucine, as in other yeasts [42].
5.4. Yarrowia lipolytica
The oleaginous nature of Y. lipolytica has made it the focus of considerable efforts to convert a range of carbon sources into neutral lipids and lipid-derived compounds [11], [96], [97]. These efforts have been extensively reviewed elsewhere (see Refs. [12], [13] and references therein). Y. lipolytica has also been used for heterologous protein production, but we focus here on its capacity for lipid biosynthesis [71]. In one recent work, high levels of triacylglycerides were engineered [98]. By engineering the conversion of glycolytic NADH to lipid precursors, specifically NADPH and acetyl-CoA, lipid production was increased by ∼25% to 0.27 g g−1 glucose while reducing oxygen requirements of the strain. These improvements, along with a resulting high rate of lipid production (1.2 g L−1h−1), help to move this process closer to industrial feasibility.
In another example, researchers from DuPont used Y. lipolytica as a host for the biosynthesis of the nutritional supplement omega-3 eicosapentaenoic acid (EPA) [72]. The resulting strain gave rise to two commercial products, Newharvest™ EPA oil, a supplement for human consumption, and Verlasso®, a salmon feed with the high EPA biomass. Random integration of 30 copies of nine homologous and heterogeneous genes along with the disruption of β-oxidation resulted in an industrial production strain capable of producing EPA at 15% of dry cell weight and 57% of the total fatty acid content by weight. The aforementioned project relied on genome editing by random integration, thus necessitating marker recovery at each integration step. The recent adaptation of CRISPR-Cas9 for use in Y. lipolytica has alleviated this challenge by enabling site specific, markerless integration [36].
5.5. Hansenula polymorpha
The methylotrophic yeast H. polymorpha (previously Pichia angusta or Ogataea polymorpha) was first studied as model organism for peroxisome function as well as nitrate assimilation [14], [15], [76], [99]. The availability of a strong inducible expression system coupled with effective protein secretion and glycosylation has also made H. polymorpha a successful host for protein production. While S. cerevisiae is able to N-glycosylate proteins, it tends to hyperglycosylate with alpha-1,3-linked mannose residues, which triggers immunogenicity in humans [100]. H. polymorpha's glycosylation machinery does not produce alpha-1,3-linked residues and is less prone to hyperglycosylation [76]. Moreover, significant efforts have been put towards optimization of human-like glycosylation [101]. Industrially produced biopharmaceutical examples include, but are not limited to, insulin under the trade name AgB, IFNα-2a sold as Wosulin, and proteins for hepatitis B vaccine HepaVax Gene [16], [18], [102]. With respect to the methylotrophic nature of H. polymorpha, the compartmentalized methanol assimilation pathway has been exploited for the overexpression of peroxisome-dependent pathways. For example, penicillin production is localized, in part, to the peroxisomes. Growth on methanol promotes peroxisome proliferation, efficient heterologous protein expression, and consequently penicillin production [103].
H. polymorpha is also a good candidate for chemical biosynthesis. Thermotolerance, broad substrate utilization, and resistance to a variety of growth inhibitors match well with lignocellulosic as well as crude substrate streams [17]. Ethanol biosynthesis has been engineered with glycerol, cellulose hydrolysate, and starch-derived sugars as process inputs [104], [105], [106], [107], [108]. Most commonly, pathway engineering has been achieved by random integration or integration into the telomeric regions of the H. polymorpha genome by HR. While H. polymorpha easily accepts integration of heterologous genes, high NHEJ capacity makes it hard to disrupt genes. In a recent study, disruption of a CAT8, a transcriptional activator that is involved in gluconeogenesis, respiration, the glyoxylic cycle and ethanol catabolism, increased ethanol yields to 12.5 g/L at 45 °C. This work produced disruption efficiencies of 2.5% and below using a homology donor with a resistance marker [109]. The recent development of a CRISPR-Cas9 in H. polymorpha has enabled gene disruption rates of up to 71%, significantly facilitating future metabolic engineering approaches [64].
5.6. Pichia pastoris
Similar to H. polymorpha, early interest in P. pastoris was driven by its ability to grow on methanol as sole carbon source, with research and development focusing on the production of single-cell protein [18]. Its use in bioprocessing is also similar to H. polymorpha, as P. pastoris is a common yeast host for protein production for the pharmaceutical, and feed and food industries [18], [77]. Processes benefit from effective protein secretion as well as strong constitutive and inducible promoters engineered from the methanol assimilation pathway [18], [110]. Glycosylation pathways have been extensively engineered, thus facilitating mammalian protein production [111], [112], [113]. Bioprocesses also benefit from P. pastoris' ability to grow to high cell density and efficiently produce membrane proteins [18], [19], [20]. These characteristics have been exploited for the industrial production of several proteins including ecallantide (trade name Kalbitor® produced by Dyax), a recombinant protein inhibitor of the plasma protease kallikrein, and ocriplasmin (trade name Jetrea® produced by ThromboGenics), a truncated recombinant form of human plasmin [22], [23].
Due to low expression from plasmids, heterologous genes are usually integrated into the genome [18]. These traditional techniques have also been used for metabolic pathway engineering. For example, a biosynthetic pathway for the terpenoid (+)-nootkatone was engineered in P. pastoris in a KU70 deficient strain [78]. The pathway required the integration of four heterologous and one homologous overexpression cassettes, which was achieved by targeted integration. Central to the success of this pathway was expression of two membrane-associated cytochrome P450 enzymes. The resulting strain produced upwards of 200 mg L−1 of (+)-nootkatone in a high cell density fermentation. In a different example, P. pastoris was used to construct a nine gene polycistronic pathway using a 2A sequence that causes a ribosomal skip that terminates translation at the final proline codon of its C-terminally located conserved sequence ‘‘NPGP’. This allows for production of multiple proteins from a single mRNA [79]. The system was used to produce the pigments violacein and β-carotene. This study served as a proof-of-concept for stable and balanced multi-enzyme pathway expression using a single promoter, and will facilitate future metabolic engineering approaches in P. pastoris.
6. Perspectives
Non-conventional yeasts have been extensively used for a range of biotechnological applications. So far, wild type strains and straightforward pathway engineering that leverages advantageous phenotypes native to the host have been the focus. With the increasing availability of next generation sequencing, genome editing tools, and the development of system wide –omics studies, more advanced understanding of the unique metabolisms and physiologies of non-conventional yeast has become attainable. Future engineering efforts will need to leverage these emerging systems and synthetic biology tools to address a critical lack of fundamental biochemical information, maximize the desired phenotypes, and increase productivity to reach industrially relevant production yields of new products.
Non-conventional yeast engineering will also be advanced by the application of genome-wide engineering tools. Tools such as yeast oligo-mediated genome engineering (YOGE, a recombineering strategy) and the yeast deletion collection in S. cerevisiae demonstrate the power that functional genomics studies can have in yeast [116], [117]. While neither YOGE nor a full deletion collection are feasible in each non-conventional yeast of interest, an alternative strategy seems poised to fill this niche. Genome-wide CRISPR-Cas9 loss of function screens will allow researchers to perform analogous functional genomics studies by transforming pooled plasmids to introduce an indel into each gene in the genome separately [118], [119]. Already widely used and validated in mammalian studies, the application of genome-wide CRISPR-Cas9 screens will greatly advance engineering in non-conventional yeasts, and will allow for further enhancement of desirable phenotypes.
While this review mainly focuses on genome and pathway engineering, other methods and techniques, such as genome-scale modeling and metabolic flux balance analysis, have been used to guide strain engineering. For example, such models and analyses were used to optimize lipid production in Y. lipolytica and assess the biotechnological potential of P. pastoris and S. stipitis [114], [115]. Culture condition optimization has also been prominently featured in process development with non-conventional yeast. For example, low iron content media and in situ product removal strategies have led to the high rate production of ethyl acetate and 2-phenylethanol in wild type strains of K. marxianus [7], [70].
Most often, the limits of metabolic engineering and synthetic biology have been pushed using common lab strains of S. cerevisiae and E. coli. At the same time, many industrial biotechnology efforts have relied on wild type strains and traditional mutagenesis methods to create viable bioprocesses from non-conventional yeasts. As new systems and synthetic biology methods and tools are adapted for use in non-conventional yeasts, we expect that new bioprocesses that exploit desired phenotypes in non-conventional yeasts will be developed and that these yeasts will become new model strain on their own merits.
Acknowledgements
This work was supported by NSF CBET-1510697 and -1403264.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Qiu Z., Jiang R. Improving Saccharomyces cerevisiae ethanol production and tolerance via RNA polymerase II subunit Rpb7. Biotechnol Biofuels. 2017;10:125. doi: 10.1186/s13068-017-0806-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nielsen J., Larsson C., van Maris A., Pronk J. Metabolic engineering of yeast for production of fuels and chemicals. Curr Opin Biotechnol. 2013;24(3):398–404. doi: 10.1016/j.copbio.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 3.Wagner J.M., Alper H.S. Synthetic biology and molecular genetics in non-conventional yeasts: current tools and future advances. Fungal Genet Biol. 2016;89:126–136. doi: 10.1016/j.fgb.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Spohner S.C., Schaum V., Quitmann H., Czermak P. Kluyveromyces lactis: an emerging tool in biotechnology. J Biotechnol. 2016;222:104–116. doi: 10.1016/j.jbiotec.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 5.Varela J.A., Gethins L., Stanton C., Ross P., Morrissey J.P. Applications of Kluyveromyces marxianus in biotechnology. In: Satyanarayana T., Kunze G., editors. Yeast diversity in human welfare. Springer; Singapore: 2017. pp. 439–453. [Google Scholar]
- 6.Lobs A.K., Lin J.L., Cook M., Wheeldon I. High throughput, colorimetric screening of microbial ester biosynthesis reveals high ethyl acetate production from Kluyveromyces marxianus on C5, C6, and C12 carbon sources. Biotechnol J. 2016;11(10):1274–1281. doi: 10.1002/biot.201600060. [DOI] [PubMed] [Google Scholar]
- 7.Loser C., Urit T., Stukert A., Bley T. Formation of ethyl acetate from whey by Kluyveromyces marxianus on a pilot scale. J Biotechnol. 2013;163(1):17–23. doi: 10.1016/j.jbiotec.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Loser C., Urit T., Keil P., Bley T. Studies on the mechanism of synthesis of ethyl acetate in Kluyveromyces marxianus DSM 5422. Appl Microbiol Biotechnol. 2014;99(3):1131–1144. doi: 10.1007/s00253-014-6098-4. [DOI] [PubMed] [Google Scholar]
- 9.Agbogbo F.K., Coward-Kelly G. Cellulosic ethanol production using the naturally occurring xylose-fermenting yeast, Pichia stipitis. Biotechnol Lett. 2008;30(9):1515–1524. doi: 10.1007/s10529-008-9728-z. [DOI] [PubMed] [Google Scholar]
- 10.Dupreez J.C., Bosch M., Prior B.A. The fermentation of hexose and pentose sugars by Candida-Shehatae and Pichia-stipitis. Appl Microbiol Biotechnol. 1986;23(3–4):228–233. [Google Scholar]
- 11.Blazeck J., Hill A., Liu L.Q., Knight R., Miller J., Pan A. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat Commun. 2014:5. doi: 10.1038/ncomms4131. [DOI] [PubMed] [Google Scholar]
- 12.Hussain M.S., Rodriguez G.M., Gao D.F., Spagnuolo M., Gambill L., Blenner M. Recent advances in bioengineering of the oleaginous yeast Yarrowia lipolytica. Aims Bioeng. 2016;3(4):493–514. [Google Scholar]
- 13.Beopoulos A., Cescut J., Haddouche R., Uribelarrea J.L., Molina-Jouve C., Nicaud J.M. Yarrowia lipolytica as a model for bio-oil production. Prog Lipid Res. 2009;48(6):375–387. doi: 10.1016/j.plipres.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 14.van der Klei I.J., Yurimoto H., Sakai Y., Veenhuis M. The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Bba-Mol Cell Res. 2006;1763(12):1453–1462. doi: 10.1016/j.bbamcr.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Siverio J.M. Assimilation of nitrate by yeasts. Fems Microbiol Rev. 2002;26(3):277–284. doi: 10.1111/j.1574-6976.2002.tb00615.x. [DOI] [PubMed] [Google Scholar]
- 16.Gellissen G. Heterologous protein production in methylotrophic yeasts. Appl Microbiol Biotechnol. 2000;54(6):741–750. doi: 10.1007/s002530000464. [DOI] [PubMed] [Google Scholar]
- 17.Dmytruk K., Kurylenko O., Ruchala J., Ishchuk O., Sibirny A. Yeast diversity in human welfare. Springer; 2017. Development of the thermotolerant methylotrophic yeast Hansenula polymorpha as efficient ethanol producer; pp. 257–282. [Google Scholar]
- 18.Gellissen G., Kunze G., Gaillardin C., Cregg J.M., Berardi E., Veenhuis M. New yeast expression platforms based on methylotrophic Hansenula polymorpha and Pichia pastoris and on dimorphic Arxula adeninivorans and Yarrowia lipolytica - a comparison. Fems Yeast Res. 2005;5(11):1079–1096. doi: 10.1016/j.femsyr.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Jahic M., Veide A., Charoenrat T., Teeri T., Enfors S.O. Process technology for production and recovery of heterologous proteins with Pichia pastoris. Biotechnol Prog. 2006;22(6):1465–1473. doi: 10.1021/bp060171t. [DOI] [PubMed] [Google Scholar]
- 20.Byme B. Pichia pastoris as an expression host for membrane protein structural biology. Curr Opin Struct Biol. 2015;32:9–17. doi: 10.1016/j.sbi.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Dmytruk K.V., Kurylenko O.O., Ruchala J., Abbas C.A., Sibirny A.A. Biotechnology of yeasts and filamentous fungi. Springer; 2017. Genetic improvement of conventional and nonconventional yeasts for the production of first-and second-generation ethanol; pp. 1–38. [Google Scholar]
- 22.Walsh G. Biopharmaceutical benchmarks 2014. Nat Biotechnol. 2014;32(10):992–1000. doi: 10.1038/nbt.3040. [DOI] [PubMed] [Google Scholar]
- 23.Ahmad M., Hirz M., Pichler H., Schwab H. Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol. 2014;98(12):5301–5317. doi: 10.1007/s00253-014-5732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lane M.M., Morrissey J.P. Kluyveromyces marxianus: a yeast emerging from its sister's shadow. Fungal Biol Rev. 2010;24(1):17–26. [Google Scholar]
- 25.Mead D.J., Gardner D.C.J., Oliver S.G. The yeast 2-Mu plasmid - strategies for the survival of a selfish DNA. Mol Mol Gen Genet. 1986;205(3):417–421. doi: 10.1007/BF00338076. [DOI] [PubMed] [Google Scholar]
- 26.Lee M.E., DeLoache W.C., Cervantes B., Dueber J.E. A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth Biol. 2015;4(9):975–986. doi: 10.1021/sb500366v. [DOI] [PubMed] [Google Scholar]
- 27.Cao M., Gao M., Lopez-Garcia C.L., Wu Y., Seetharam A.S., Severin A.J. Centromeric DNA facilitates nonconventional yeast genetic engineering. ACS Synth Biol. 2017;6(8):1545–1553. doi: 10.1021/acssynbio.7b00046. [DOI] [PubMed] [Google Scholar]
- 28.Jensen N.B., Strucko T., Kildegaard K.R., David F., Maury J., Mortensen U.H. EasyClone: method for iterative chromosomal integration of multiple genes in Saccharomyces cerevisiae. Fems Yeast Res. 2014;14(2):238–248. doi: 10.1111/1567-1364.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vernis L., Poljak L., Chasles M., Uchida K., Casaregola S., Kas E. Only centromeres can supply the partition system required for ARS function in the yeast Yarrowia lipolytica. J Mol Biol. 2001;305(2):203–217. doi: 10.1006/jmbi.2000.4300. [DOI] [PubMed] [Google Scholar]
- 30.Da Silva N.A., Srikrishnan S. Introduction and expression of genes for metabolic engineering applications in Saccharomyces cerevisiae. Fems Yeast Res. 2012;12(2):197–214. doi: 10.1111/j.1567-1364.2011.00769.x. [DOI] [PubMed] [Google Scholar]
- 31.Kegel A., Martinez P., Carter S.D., Astrom S.U. Genome wide distribution of illegitimate recombination events in Kluyveromyces lactis. Nucleic Acids Res. 2006;34(5):1633–1645. doi: 10.1093/nar/gkl064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flagfeldt D.B., Siewers V., Huang L., Nielsen J. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast. 2009;26(10):545–551. doi: 10.1002/yea.1705. [DOI] [PubMed] [Google Scholar]
- 34.Shao Z., Zhao H., Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37(2):e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horwitz A.A., Walter J.M., Schubert M.G., Kung S.H., Hawkins K., Platt D.M. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst. 2015;1(1):88–96. doi: 10.1016/j.cels.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Schwartz C., Shabbir-Hussain M., Frogue K., Blenner M., Wheeldon I. Standardized markerless gene integration for pathway engineering in Yarrowia lipolytica. ACS Synth Biol. 2017;6(3):402–409. doi: 10.1021/acssynbio.6b00285. [DOI] [PubMed] [Google Scholar]
- 37.Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7(6):2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alani E., Cao L., Kleckner N. A method for gene disruption that allows repeated use of Ura3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116(4):541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheon S.A., Choo J., Ubiyvovk V.M., Park J.N., Kim M.W., Oh D.B. New selectable host-marker systems for multiple genetic manipulations based on TRP1, MET2 and ADE2 in the methylotrophic yeast Hansenula polymorpha. Yeast. 2009;26(9):507–521. doi: 10.1002/yea.1701. [DOI] [PubMed] [Google Scholar]
- 40.Pecota D.C., Rajgarhia V., Da Silva N.A. Sequential gene integration for the engineering of Kluyveromyces marxianus. J Biotechnol. 2007;127(3):408–416. doi: 10.1016/j.jbiotec.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 41.Pan R.Q., Zhang J., Shen W.L., Tao Z.Q., Li S.P., Yan X. Sequential deletion of Pichia pastoris genes by a self-excisable cassette. Fems Yeast Res. 2011;11(3):292–298. doi: 10.1111/j.1567-1364.2011.00716.x. [DOI] [PubMed] [Google Scholar]
- 42.Laplaza J.M., Torres B.R., Jin Y.S., Jeffries T.W. Sh ble and Cre adapted for functional genomics and metabolic engineering of Pichia stipitis. Enzyme Microb Tech. 2006;38(6):741–747. [Google Scholar]
- 43.Steensma H.Y., Ter Linde J.J.M. Plasmids with the Cre-recombinase and the dominant nat marker, suitable for use in prototrophic strains of Saccharomyces cerevisiae and Kluyveromyces lactis. Yeast. 2001;18(5):469–472. doi: 10.1002/yea.696. [DOI] [PubMed] [Google Scholar]
- 44.Kooistra R., Hooykaas P.J., Steensma H.Y. Efficient gene targeting in Kluyveromyces lactis. Yeast. 2004;21(9):781–792. doi: 10.1002/yea.1131. [DOI] [PubMed] [Google Scholar]
- 45.Maassen N., Freese S., Schruff B., Passoth V., Klinner U. Nonhomologous end joining and homologous recombination DNA repair pathways in integration mutagenesis in the xylose-fermenting yeast Pichia stipitis. Fems Yeast Res. 2008;8(5):735–743. doi: 10.1111/j.1567-1364.2008.00383.x. [DOI] [PubMed] [Google Scholar]
- 46.Verbeke J., Beopoulos A., Nicaud J.M. Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnol Lett. 2013;35(4):571–576. doi: 10.1007/s10529-012-1107-0. [DOI] [PubMed] [Google Scholar]
- 47.Kretzschmar A., Otto C., Holz M., Werner S., Hubner L., Barth G. Increased homologous integration frequency in Yarrowia lipolytica strains defective in non-homologous end-joining. Curr Genet. 2013;59(1–2):63–72. doi: 10.1007/s00294-013-0389-7. [DOI] [PubMed] [Google Scholar]
- 48.Saraya R., Krikken A.M., Kiel J.A., Baerends R.J., Veenhuis M., van der Klei I.J. Novel genetic tools for Hansenula polymorpha. Fems Yeast Res. 2012;12(3):271–278. doi: 10.1111/j.1567-1364.2011.00772.x. [DOI] [PubMed] [Google Scholar]
- 49.Naatsaari L., Mistlberger B., Ruth C., Hajek T., Hartner F.S., Glieder A. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS One. 2012;7(6):e39720. doi: 10.1371/journal.pone.0039720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abdel-Banat B.M., Nonklang S., Hoshida H., Akada R. Random and targeted gene integrations through the control of non-homologous end joining in the yeast Kluyveromyces marxianus. Yeast. 2010;27(1):29–39. doi: 10.1002/yea.1729. [DOI] [PubMed] [Google Scholar]
- 51.Choo J.H., Han C., Kim J.Y., Kang H.A. Deletion of a KU80 homolog enhances homologous recombination in the thermotolerant yeast Kluyveromyces marxianus. Biotechnol Lett. 2014;36(10):2059–2067. doi: 10.1007/s10529-014-1576-4. [DOI] [PubMed] [Google Scholar]
- 52.Chapman J.R., Taylor M.R.G., Boulton S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47(4):497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 53.Tsakraklides V., Brevnova E., Stephanopoulos G., Shaw A.J. Improved gene targeting through cell cycle synchronization. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0133434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Z., Liang Y., Ang E.L., Zhao H. A new era of genome integration-simply cut and paste! ACS Synth Biol. 2017;6(4):601–609. doi: 10.1021/acssynbio.6b00331. [DOI] [PubMed] [Google Scholar]
- 55.David F., Siewers V. Advances in yeast genome engineering. Fems Yeast Res. 2015;15(1):1–14. doi: 10.1111/1567-1364.12200. [DOI] [PubMed] [Google Scholar]
- 56.Rigouin C., Gueroult M., Croux C., Dubois G., Borsenberger V., Barbe S. Production of medium chain fatty acids by Yarrowia lipolytica: combining molecular design and TALEN to engineer the fatty acid synthase. ACS Synth Biol. 2017 doi: 10.1021/acssynbio.7b00034. Article ASAP. [DOI] [PubMed] [Google Scholar]
- 57.DiCarlo J.E., Norville J.E., Mali P., Rios X., Aach J., Church G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41(7):4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stovicek V., Holkenbrink C., Borodina I. CRISPR/Cas system for yeast genome engineering: advances and applications. Fems Yeast Res. 2017;17(5) doi: 10.1093/femsyr/fox030. fox030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao S., Tong Y., Wen Z., Zhu L., Ge M., Chen D. Multiplex gene editing of the Yarrowia lipolytica genome using the CRISPR-Cas9 system. J Ind Microbiol. 2016;43(8):1085–1093. doi: 10.1007/s10295-016-1789-8. [DOI] [PubMed] [Google Scholar]
- 61.Schwartz C.M., Hussain M.S., Blenner M., Wheeldon I. Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR-Cas9-mediated genome editing in Yarrowia lipolytica. ACS Synth Biol. 2016;5(4):356–359. doi: 10.1021/acssynbio.5b00162. [DOI] [PubMed] [Google Scholar]
- 62.Lobs A.K., Engel R., Schwartz C., Flores A., Wheeldon I. CRISPR-Cas9-enabled genetic disruptions for understanding ethanol and ethyl acetate biosynthesis in Kluyveromyces marxianus. Biotechnol Biofuels. 2017;10:164. doi: 10.1186/s13068-017-0854-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weninger A., Hatzl A.M., Schmid C., Vogl T., Glieder A. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J Biotechnol. 2016;235:139–149. doi: 10.1016/j.jbiotec.2016.03.027. [DOI] [PubMed] [Google Scholar]
- 64.Numamoto M., Maekawa H., Kaneko Y. Efficient genome editing by CRISPR/Cas9 with a tRNA-sgRNA fusion in the methylotrophic yeast Ogataea polymorpha. J Biosci Bioeng. 2017 doi: 10.1016/j.jbiosc.2017.06.001. (in press) [DOI] [PubMed] [Google Scholar]
- 65.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith J.D., Suresh S., Schlecht U., Wu M., Wagih O., Peltz G. St Onge RP: quantitative CRISPR interference screens in yeast identify chemical-genetic interactions and new rules for guide RNA design. Genome Biol. 2016;17:45. doi: 10.1186/s13059-016-0900-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwartz C., Frogue K., Ramesh A., Misa J., Wheeldon I. CRISPRi repression of nonhomologous end-joining for enhanced genome engineering via homologous recombination in Yarrowia lipolytica. Biotechnol Bioeng. 2017 doi: 10.1002/bit.26404. Accepted Author Manuscript. [DOI] [PubMed] [Google Scholar]
- 69.Gilbert L.A., Larson M.H., Morsut L., Liu Z.R., Brar G.A., Torres S.E. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Etschmann M.M.W., Schrader J. An aqueous-organic two-phase bioprocess for efficient production of the natural aroma chemicals 2-phenylethanol and 2-phenylethylacetate with yeast. Appl Microbiol Biotechnol. 2006;71(4):440–443. doi: 10.1007/s00253-005-0281-6. [DOI] [PubMed] [Google Scholar]
- 71.Madzak C. Yarrowia lipolytica: recent achievements in heterologous protein expression and pathway engineering. Appl Microbiol Biotechnol. 2015;99(11):4559–4577. doi: 10.1007/s00253-015-6624-z. [DOI] [PubMed] [Google Scholar]
- 72.Xue Z., Sharpe P.L., Hong S.P., Yadav N.S., Xie D., Short D.R. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat Biotechnol. 2013;31(8):734–740. doi: 10.1038/nbt.2622. [DOI] [PubMed] [Google Scholar]
- 73.Wei L., Liu J., Qi H.S., Wen J.P. Engineering Scheffersomyces stipitis for fumaric acid production from xylose. Bioresour Technol. 2015;187:246–254. doi: 10.1016/j.biortech.2015.03.122. [DOI] [PubMed] [Google Scholar]
- 74.Ilmen M., Koivuranta K., Ruohonen L., Suominen P., Penttila M. Efficient production of L-lactic acid from xylose by Pichia stipitis. Appl Environ Microbiol. 2007;73(1):117–123. doi: 10.1128/AEM.01311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dashtban M., Wen X., Bajwa P.K., Ho C.Y., Lee H. Deletion of hxk1 gene results in derepression of xylose utilization in Scheffersomyces stipitis. J Ind Microbiol. 2015;42(6):889–896. doi: 10.1007/s10295-015-1614-9. [DOI] [PubMed] [Google Scholar]
- 76.Kunze G., Kang H.A., Gellissen G. Yeast biotechnology: diversity and applications. Springer; 2009. Hansenula polymorpha (Pichia angusta): biology and applications; pp. 47–64. [Google Scholar]
- 77.Spohner S.C., Muller H., Quitmann H., Czermak P. Expression of enzymes for the usage in food and feed industry with Pichia pastoris. J Biotechnol. 2015;202:118–134. doi: 10.1016/j.jbiotec.2015.01.027. [DOI] [PubMed] [Google Scholar]
- 78.Wriessnegger T., Augustin P., Engleder M., Leitner E., Muller M., Kaluzna I. Production of the sesquiterpenoid (+)-nootkatone by metabolic engineering of Pichia pastoris. Metab Eng. 2014;24:18–29. doi: 10.1016/j.ymben.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 79.Geier M., Fauland P., Vogl T., Glieder A. Compact multi-enzyme pathways in P. pastoris. Chem Commun. 2015;51(9):1643–1646. doi: 10.1039/c4cc08502g. [DOI] [PubMed] [Google Scholar]
- 80.Vandenberg J.A., Vanderlaken K.J., Vanooyen A.J.J., Renniers T.C.H.M., Rietveld K., Schaap A. Kluyveromyces as a host for heterologous gene-expression - expression and secretion of prochymosin. Bio-Technol. 1990;8(2):135–139. doi: 10.1038/nbt0290-135. [DOI] [PubMed] [Google Scholar]
- 81.Colussi P.A., Taron C.H. Kluyveromyces lactis LAC4 promoter variants that lack function in bacteria but retain full function in K. lactis. Appl Environ Microbiol. 2005;71(11):7092–7098. doi: 10.1128/AEM.71.11.7092-7098.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Almeida C.M., Gomes D., Faro C., Simoes I. Engineering a cardosin B-derived rennet for sheep and goat cheese manufacture. Appl Microbiol Biotechnol. 2015;99(1):269–281. doi: 10.1007/s00253-014-5902-5. [DOI] [PubMed] [Google Scholar]
- 83.Jo H.J., Noh J.S., Kong K.H. Efficient secretory expression of the sweet-tasting protein brazzein in the yeast Kluyveromyces lactis. Protein Expres Purif. 2013;90(2):84–89. doi: 10.1016/j.pep.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 84.Kim T.Y., Lee S.W., Oh M.K. Biosynthesis of 2-phenylethanol from glucose with genetically engineered Kluyveromyces marxianus. Enzyme Microb Technol. 2014;61–62:44–47. doi: 10.1016/j.enzmictec.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 85.Ballesteros M., Oliva J.M., Negro M.J., Manzanares P., Ballesteros I. Ethanol from lignocellulosic materials by a simultaneous saccharification and fermentation process (SFS) with Kluyveromyces marxianus CECT 10875. Process Biochem. 2004;39(12):1843–1848. [Google Scholar]
- 86.Zafar S., Owais M. Ethanol production from crude whey by Kluyveromyces marxianus. Biochem Eng J. 2006;27(3):295–298. [Google Scholar]
- 87.Zhang J., Zhang B., Wang D.M., Gao X.L., Sun L.H., Hong J. Rapid ethanol production at elevated temperatures by engineered thermotolerant Kluyveromyces marxianus via the NADP(H)-preferring xylose reductase-xylitol dehydrogenase pathway. Metab Eng. 2015;31:140–152. doi: 10.1016/j.ymben.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 88.Yanase S., Hasunuma T., Yamada R., Tanaka T., Ogino C., Fukuda H. Direct ethanol production from cellulosic materials at high temperature using the thermotolerant yeast Kluyveromyces marxianus displaying cellulolytic enzymes. Appl Microbiol Biotechnol. 2010;88(1):381–388. doi: 10.1007/s00253-010-2784-z. [DOI] [PubMed] [Google Scholar]
- 89.Wang R.L., Li L.L., Zhang B., Gao X.L., Wang D.M., Hong J. Improved xylose fermentation of Kluyveromyces marxianus at elevated temperature through construction of a xylose isomerase pathway. J Ind Microbiol. 2013;40(8):841–854. doi: 10.1007/s10295-013-1282-6. [DOI] [PubMed] [Google Scholar]
- 90.Cheon Y., Kim J.S., Park J.B., Heo P., Lim J.H., Jung G.Y. A biosynthetic pathway for hexanoic acid production in Kluyveromyces marxianus. J Biotechnol. 2014;182:30–36. doi: 10.1016/j.jbiotec.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 91.Slininger P.J., Shea-Andersh M.A., Thompson S.R., Dien B.S., Kurtzman C.P., Balan V. Evolved strains of Scheffersomyces stipitis achieving high ethanol productivity on acid- and base-pretreated biomass hydrolyzate at high solids loading. Biotechnol Biofuels. 2015;8:60. doi: 10.1186/s13068-015-0239-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shi J., Zhang M., Zhang L., Wang P., Jiang L., Deng H. Xylose-fermenting Pichia stipitis by genome shuffling for improved ethanol production. Microb Biotechnol. 2014;7(2):90–99. doi: 10.1111/1751-7915.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pereira S.R., Sanchez I.N.V., Frazao C.J., Serafim L.S., Gorwa-Grauslund M.F., Xavier A.M. Adaptation of Scheffersomyces stipitis to hardwood spent sulfite liquor by evolutionary engineering. Biotechnol Biofuels. 2015;8:50. doi: 10.1186/s13068-015-0234-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang W., Geng A.L. Improved ethanol production by a xylose-fermenting recombinant yeast strain constructed through a modified genome shuffling method. Biotechnol Biofuels. 2012:5. doi: 10.1186/1754-6834-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hughes S.R., Gibbons W.R., Bang S.S., Pinkelman R., Bischoff K.M., Slininger P.J. Random UV-C mutagenesis of Scheffersomyces (formerly Pichia) stipitis NRRL Y-7124 to improve anaerobic growth on lignocellulosic sugars. J Ind Microbiol. 2012;39(1):163–173. doi: 10.1007/s10295-011-1012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rodriguez G.M., Hussain M.S., Gambill L., Gao D.F., Yaguchi A., Blenner M. Engineering xylose utilization in Yarrowia lipolytica by understanding its cryptic xylose pathway. Biotechnol Biofuels. 2016;9 doi: 10.1186/s13068-016-0562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu J., Liu N., Qiao K., Vogg S., Stephanopoulos G. Application of metabolic controls for the maximization of lipid production in semicontinuous fermentation. Proc Natl Acad Sci U. S. A. 2017;114(27):E5308–E5316. doi: 10.1073/pnas.1703321114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qiao K.J., Wasylenko T.M., Zhou K., Xu P., Stephanopoulos G. Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat Biotechnol. 2017;35(2):173–177. doi: 10.1038/nbt.3763. [DOI] [PubMed] [Google Scholar]
- 99.Gellissen G., Veenhuis M. The methylotrophic yeast Hansenula polymorpha: its use in fundamental research and as a cell factory. Yeast. 2001;18(3) doi: 10.1002/1097-0061(200102)18:3<::AID-YEA695>3.0.CO;2-9. i-iii. [DOI] [PubMed] [Google Scholar]
- 100.Ballou C.E. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 1990;185:440–470. doi: 10.1016/0076-6879(90)85038-p. [DOI] [PubMed] [Google Scholar]
- 101.Cheon S.A., Kim H., Oh D.B., Kwon O., Kang H.A. Remodeling of the glycosylation pathway in the methylotrophic yeast Hansenula polymorpha to produce human hybrid-type N-glycans. J Microbiol. 2012;50(2):341–348. doi: 10.1007/s12275-012-2097-2. [DOI] [PubMed] [Google Scholar]
- 102.Muller F., II, Tieke A., Waschk D., Muhle C., Muller I.F., Seigelchifer M. Production of IFN alpha-2a in Hansenula polymorpha. Process Biochem. 2002;38(1):15–25. [Google Scholar]
- 103.Gidijala L., Kiel J.A., Douma R.D., Seifar R.M., van Gulik W.M., Bovenberg R.A. An engineered yeast efficiently secreting penicillin. PLoS One. 2009;4(12):e8317. doi: 10.1371/journal.pone.0008317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kurylenko O.O., Ruchala J., Hryniv O.B., Abbas C.A., Dmytruk K.V., Sibirny A.A. Metabolic engineering and classical selection of the methylotrophic thermotolerant yeast Hansenula polymorpha for improvement of high-temperature xylose alcoholic fermentation. Microb Cell Fact. 2014;13 doi: 10.1186/s12934-014-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kata I., Semkiv M.V., Ruchala J., Dmytruk K.V., Sibirny A.A. Overexpression of the genes PDC1 and ADH1 activates glycerol conversion to ethanol in the thermotolerant yeast Ogataea (Hansenula) polymorpha. Yeast. 2016;33(8):471–478. doi: 10.1002/yea.3175. [DOI] [PubMed] [Google Scholar]
- 106.Voronovsky A.Y., Rohulya O.V., Abbas C.A., Sibirny A.A. Development of strains of the thermotolerant yeast Hansenula polymorpha capable of alcoholic fermentation of starch and xylan. Metab Eng. 2009;11(4–5):234–242. doi: 10.1016/j.ymben.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 107.Ryabova O.B., Chmil O.M., Sibirny A.A. Xylose and cellobiose fermentation to ethanol by the thermotolerant methylotrophic yeast Hansenula polymorpha. Fems Yeast Res. 2003;4(2):157–164. doi: 10.1016/S1567-1356(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 108.Hong W.K., Kim C.H., Heo S.Y., Luo L., Oh B.R., Seo J.W. Enhanced production of ethanol from glycerol by engineered Hansenula polymorpha expressing pyruvate decarboxylase and aldehyde dehydrogenase genes from Zymomonas mobilis. Biotechnol Lett. 2010;32(8):1077–1082. doi: 10.1007/s10529-010-0259-z. [DOI] [PubMed] [Google Scholar]
- 109.Ruchala J., Kurylenko O.O., Soontorngun N., Dmytruk K.V., Sibirny A.A. Transcriptional activator Cat8 is involved in regulation of xylose alcoholic fermentation in the thermotolerant yeast Ogataea (Hansenula) polymorpha. Microb Cell Facts. 2017;16 doi: 10.1186/s12934-017-0652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Macauley-Patrick S., Fazenda M.L., McNeil B., Harvey L.M. Heterologous protein production using the Pichia pastoris expression system. Yeast. 2005;22(4):249–270. doi: 10.1002/yea.1208. [DOI] [PubMed] [Google Scholar]
- 111.Jacobs P.P., Geysens S., Vervecken W., Contreras R., Callewaert N. Engineering complex-type N-glycosylation in Pichia pastoris using GlycoSwitch technology. Nat Protoc. 2009;4(1):58–70. doi: 10.1038/nprot.2008.213. [DOI] [PubMed] [Google Scholar]
- 112.Hamilton S.R., Gerngross T.U. Glycosylation engineering in yeast: the advent of fully humanized yeast. Curr Opin Biotechnol. 2007;18(5):387–392. doi: 10.1016/j.copbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 113.Hamilton S.R., Davidson R.C., Sethuraman N., Nett J.H., Jiang Y.W., Rios S. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science. 2006;313(5792):1441–1443. doi: 10.1126/science.1130256. [DOI] [PubMed] [Google Scholar]
- 114.Kavscek M., Bhutada G., Madl T., Natter K. Optimization of lipid production with a genome-scale model of Yarrowia lipolytica. BMC Syst Biol. 2015;9 doi: 10.1186/s12918-015-0217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Caspeta L., Shoaie S., Agren R., Nookaew I., Nielsen J. Genome-scale metabolic reconstructions of Pichia stipitis and Pichia pastoris and in silico evaluation of their potentials. BMC Syst Biol. 2012;6 doi: 10.1186/1752-0509-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.DiCarlo J.E., Conley A.J., Penttila M., Jantti J., Wang H.H., Church G.M. Yeast oligo-mediated genome engineering (YOGE) ACS Synth Biol. 2013;2(12):741–749. doi: 10.1021/sb400117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Giaever G., Nislow C. The yeast deletion collection: a decade of functional genomics. Genetics. 2014;197(2):451–465. doi: 10.1534/genetics.114.161620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]