Abstract
Metabolic engineering efforts toward rewiring metabolism of cells to produce new compounds often require the utilization of non-native enzymatic machinery that is capable of producing a broad range of chemical functionalities. Polyketides encompass one of the largest classes of chemically diverse natural products. With thousands of known polyketides, modular polyketide synthases (PKSs) share a particularly attractive biosynthetic logic for generating chemical diversity. The engineering of modular PKSs could open access to the deliberate production of both existing and novel compounds. In this review, we discuss PKS engineering efforts applied at both the protein and cellular level for the generation of a diverse range of chemical structures, and we examine future applications of PKSs in the production of medicines, fuels and other industrially relevant chemicals.
Keywords: Polyketide, Polyketide synthase, Natural products, Commodity chemical, Metabolic engineering, Synthetic biology
Abbreviations: PK, Polyketide; PKS, Polyketide synthase; FAS, Fatty acid synthases; KS, Ketosynthase; AT, Acyltransferase; ACP, Acyl carrier protein; DH, Dehydratase; KR, Ketoreductase; ER, Enoylreductase; TE, Thioesterase; LM, Loading module; CoL, CoA-Ligase; R, Reductase domain; PDB, Precursor directed biosynthesis; TKL, Triketide lactone; DE, Dimerization element; SNAC, N-acetylcysteamine; DEBS, 6-deoxyerythronolide B synthase; SARP, Streptomyces antibiotic regulatory protein; LTTR, LysR-type transcriptional regulator; PCC, Propionyl-CoA carboxylase
1. Introduction
Polyketides are one of the largest classes of natural products, possessing immense structural diversity and complex chemical architectures. Many polyketides (PKs) are among the most important secondary metabolites for their applications in medicine, agriculture, and industry. Examples include anticancer drugs (epothilone) [1], [2], antibiotics (erythromycin) [3], insecticides (spinosyn A) [4] and antifungals (amphotericin B) [5]. These particular examples of polyketides are biosynthesized by multimodular enzyme complexes known as type I modular polyketide synthases (PKSs). Working in an assembly-line fashion, multimodular PKSs assemble and tailor readily available acyl-CoAs within the host cell into large, complex, chiral molecules [6]. Each of these PKSs comprises a series of modules that can be further dissected into a series of domains responsible for the extension of the polyketide backbone through condensation and selective reductive processing of an acyl-CoA building block. The collinear architecture of these modules, apparent by inspection of the domains present and the predictive selectivity motifs harbored within, provide insights into the chemical connectivity and stereochemical configuration of the polyketide metabolite from analysis of its coding sequence.
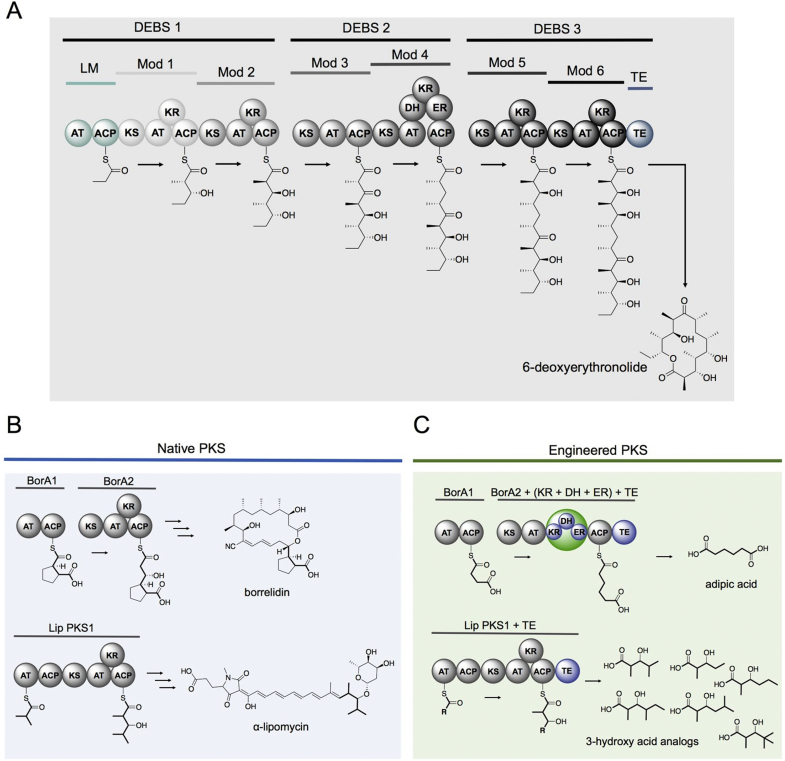
One of the best-studied PKSs is the 6-deoxyerythronolide synthase (DEBS) (Fig. 1A), which is responsible for synthesizing the macrolactone core of the antibiotic, erythromycin [7]. The catalytic domains of DEBS are expressed within modules that are each responsible for a single round of chain elongation and reductive processing. To this end, the loading acyltransferase (AT) domain loads the acyl carrier protein (ACP) with a starter unit derived from propionyl-CoA (Fig. 1A). The ketosynthase (KS) within each module catalyzes decarboxylative carbon-carbon bond formation between an acyl precursor and the ACP-bound methylmalonyl derivative. Unlike fatty acid synthases (FASs), the occurrence of reductive domains within modules varies, and PKS intermediates typically exhibit various levels of reduction. If present, the ketoreductase (KR) converts the β-ketone to an alcohol using NADPH. The dehydratase (DH) eliminates the alcohol to form an olefin, and the enoylreductase (ER) utilizes NADPH to reduce the olefin to a methylene. Finally, a thioesterase (TE) domain, located at the terminal of DEBS 3 module, catalyzes the release and cyclization to produce the macrolactone, 6-deoxyerythronolide (6-dEB). The structure and mechanism of each PKS domain is reviewed in detail elsewhere [8], [9], [10], [11], [12].
Fig. 1.
Biosynthesis of 6-deoxyerythronolide and examples of both native and engineered polyketide synthases. A) Modular biosynthesis of 6-deoxyerythronolide by the well-studied 6-deoxyerythronolide polyketide synthase. B) The carboxylic acid starter unit promiscuity by the borrelidin PKS was utilized to produce adipic acid (C) from succinic acid and malonic acid using an engineered BorA2 containing a full reductive loop (highlighted in green circle) and a thioesterase. B) The broad starter unit selectivity of the lipomycin PKS was utilized to produce 3-hydroxyacid congeners from branched acids and methylmalonyl-CoA (C). The carbon backbone for both the native and engineered polyketides are represented in bold. Grey domains represent the native pathway and blue domains represent engineered insertions.
With this collinear biosynthetic logic in mind, engineered PKSs have the potential to become an effective retrobiosynthetic platform to produce molecules that are difficult or too complex to acquire via traditional synthetic means (Fig. 1B–C). From DNA sequence, one could control chemical structure by successfully modifying and rearranging existing polyketide modules and domains [13], [14]. Moreover, rationally-designed PKSs could be introduced into a variety of engineered hosts [15], [16], [17] capable of expressing these large PKS complexes while providing the necessary precursor metabolites to biosynthesize a target chemical. In this review, we highlight PKS engineering efforts at both the protein level and the host/cellular level. We further aim to describe PKS engineering efforts within the context of metabolic engineering, and introduce the idea of successful PKS/host modifications for both traditional medicinal applications as well as the production of fuels and commodity chemicals.
2. PKS protein engineering
The ability to tailor the molecular architecture of polyketide metabolites through the inclusion of various reductive domains and/or domains with altered selectivity has long been the promise of PKSs as a retrobiosynthetic platform. In this section, we discuss the current knowledge of PKS engineering at the protein level. We have divided the PKS protein engineering section into sections based on domain type. Within the types of domains, we have selected the most engineerable targets. We will not focus on KS or ACP domain engineering in this review, as they are arguably the least targetable domains based on the chemistry and functions they perform, respectively. In addition, methyltransferase domains, which transfer an S-adenosyl-methionine-derived methyl group to the α-carbon of the β-keto intermediate, are somewhat rare and less well characterized, and thus will not be discussed here. In each section, we will first give a basic overview of the current state of knowledge regarding the specific domain(s) in question. Next, we will highlight some significant accomplishments in engineering, both via site-directed mutagenesis and/or domain swapping experiments. Because of the extensive amount of published PKS research, we cannot include all examples of PKS engineering within the scope of this review. Nevertheless, numerous representative examples are highlighted.
2.1. Loading modules
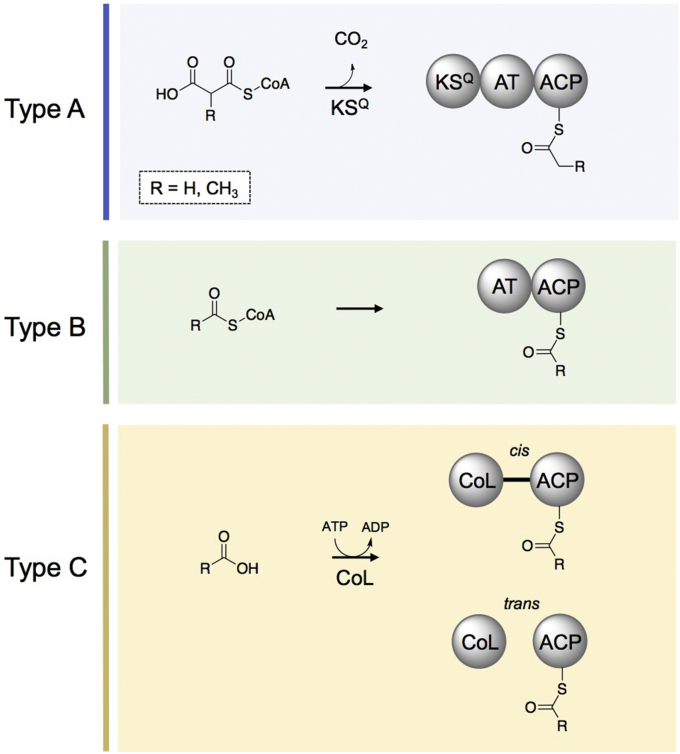
Nature has evolved several mechanisms for activating acyl substrates to initiate PK biosynthesis. To begin chain formation, modular type I PKSs employ a loading module (LM) to select the priming unit. LMs are categorized based on their domain architecture and the mechanism by which each activates substrates to begin chain formation. Although LMs are not officially characterized within the field, for simplicity within this review we will refer to each class of LMs with a representative letter (e.g. “type A” or “A-type”) so as not to confuse the reader with the type I, II, or III PKS designations used to describe the entire assembly line systems.
The most common LM organization consists of a condensation-incompetent KSQ (named for the active site C→ Q mutation), AT, and ACP domain (Type A LM) [18], [19]. In type A LMs, the KSQ decarboxylates malonyl- or methylmalonyl-CoA to yield acetyl- or propionyl starter units, respectively (Fig. 2, Fig. 3) [20]. The AT domains of these modules are strictly specific for CoA esters of dicarboxylic acids (malonyl- or methylmalonyl-CoA) [20] and share a conserved arginine residue with extender AT domains that is used to stabilize the free carboxyl moiety of the substrate [18]. Type B LMs prime polyketide biosynthesis with a much broader range of substrates (Fig. 2, Fig. 3), and their domain organization consists of an AT and ACP didomain. The AT domain selects a CoA-bound priming unit and transfers it to its cognate ACP where it is poised for transfer to the KS of the first extension module. Because type B LM ATs are not limited to utilizing β-carboxy-CoA starters, they tend to recognize a more diverse set of starter units derived from other acyl-CoAs. For example, the B-type avermectin LM natively primes with either 2-methylbutyryl-CoA or isobutyryl-CoA, and it can also accept a large number of other substrates [21]. Similarly, while the related lipomycin LM primes biosynthesis with isobutyryl-CoA in vivo, in vitro it also loads a variety of other branched fatty acyl-CoAs (Fig. 1B–C) [22]. Nevertheless, type B domain architecture does not always imply promiscuity. Another B-type LM from the borrelidin PKS is selective for dicarboxylic acid starter units both in vivo [23] and in vitro (Fig. 1B–C) [24]. The final common organization (type C) of LMs consists of a CoA-Ligase-type (CoL) domain located upstream of an ACP, which activates a carboxylic acid substrate in an ATP-dependent fashion so that it can then be loaded onto the ACP (Fig. 2, Fig. 3). While some LMs have the CoL domain in cis (e.g. rifamycin [25], rapamycin [26]), others have a separately encoded CoL domain that activates the ACP of the loading domain in trans in a mechanistically similar fashion (e.g. aureothin [27], and several mycobacterial polyketide synthases [28]). It is also common to find other accessory domains within type C LMs, such as the enoylreductase domains observed in FK506 and rapamycin biosynthesis [26], [29].
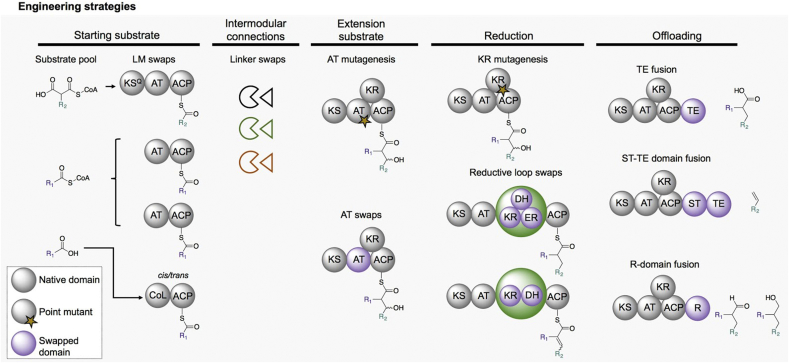
Fig. 2.
Summary of polyketide synthase engineering strategies highlighted in this review. Starter unit selectivity and incorporation is mediated by either native or non-native swapped LMs. Intermolecular linker regions allow for successful communications between domains. Chemical diversity is further increased by varying the extender building blocks. AT mutagenesis or AT swaps mediate incorporation of various extender units into a polyketide intermediate. Various degrees of reduction at the β-keto position can be accomplished by KR mutagenesis, KR swaps and/or the insertion of full or partially full reductive loops, containing DH and ER domains. Release of the polyketide intermediate is mediated by various releasing domains, which further increase chemical diversity into the final product.
Fig. 3.
Examples of loading modules. In type A LMs, the KSQ decarboxylates malonyl- or methylmalonyl-CoA to yield acetyl- or propionyl starter units, respectively. Type B LMs consist of an AT that selects a CoA-bound priming unit and transfers it to its cognate ACP. Type C LMs consists of a CoL domain located upstream of an ACP. The CoL activates a carboxylic acid substrate in an ATP-dependent fashion in order to load it onto the ACP, either in cis or in trans. It is also common to find other accessory domains within type C LMs, between the CoL and ACP domain (represented by the black line between the cis CoL and ACP).
The inherent promiscuity of certain loading modules and the wide variety of potential starter units make the initial precursor loading process an attractive protein engineering target. To date, most LM engineering efforts have focused on swapping LMs with type A or type B architectures that incorporate short fatty acyl groups into polyketides. Possibly the first LM swap successfully placed the tylosin LM (type A) into the platenolide (type A LM) biosynthetic pathway, resulting in the production of 16-methyl platenolide derived from propionyl-CoA (methylmalonyl-derived) instead of the native acetyl-CoA (malonyl-derived) starter unit [30]. In a similar experiment, Leadlay and coworkers exchanged the native 6-deoxyerythronolide LM (type B) for the avermectin LM (type B), increasing the diversity of erythromycins produced in vivo [31]. Both of these early examples demonstrated the feasibility of swapping loading modules with consistent domain architectures.
Mixed LM type swaps have also been explored. By replacing the type B LM of DEBS M1+TE with the type A LMs from the oleandomycin or tylosin pathways, production of triketide lactones (TKLs) derived from almost exclusively acetate- or propionate-derived starter units, respectively, was achieved [20]. Under the same conditions, the native type B DEBS LM architecture lends itself to broader starter unit selectivity, resulting in the production of both types of TKLs [32]. Therefore, by changing LM type, greater starter unit fidelity was achieved. In addition, an AT swap of the oleandomycin (type A LM) loading AT with the extending AT from the second rapamycin module (rapAT2, also conferring malonyl-CoA specificity) resulted in production of C13-methyl erythromycins [20]. A similar AT swap into the nystatin LM nysA (type A) resulted in the restoration of nystatin production in a nysA knockout strain of S. noursei, albeit at much lower titers than the native system. In addition, swapping in a methylmalonyl-CoA-specific AT domain resulted in no production of the nystatin analog, perhaps due to the innate specificity of the downstream nysB module or the nysA KSQ domain [33]. These swaps illustrate the potential to exercise tighter control over starter units incorporated into polyketides that normally begin with type B LMs in order to engineer systems to precisely control the metabolites produced.
LM swaps involving type C LMs could provide mechanisms for introducing even greater diversity into polyketides than type B LMs based on the range of starter units the type C LMs accept. Dihydroxycyclohexene-carboxoyl-CoA [34], benzoyl-CoA [35], and 3-amino-5-hydroxybenzyl-CoA [25] are just a few of the starter units incorporated via this type of LM. Attempts to switch the type C LM of the rimocidin PKS protein (RimA) with the type A LM from the nystatin PKS (NysA) showed that while RimA could initiate nystatin biosynthesis in the native S. noursei producer, nysA could not rescue rimocidin biosynthesis in a rimA knockout strain of S. diastaticus. In addition, RimA natively accepts both acetate and butyrate starter units, but only acetate units could be incorporated into the nystatin skeleton, suggesting that the downstream nys PKS is gatekeeping [36]. However, a swap of the non-canonical soraphen LM SorA (ACP-KS-AT-AT architecture) into DEBS1+TE produced a small amount of the expected benzoyl-CoA-derived TKL [37]. In addition, a swap of the first SorA AT domain into the DEBS LM also resulted in the production of a TKL incorporating benzoyl-CoA. These experiments illustrate that more exotic LM swaps, including type C LM swaps and domain swaps within LMs, may be tolerated by modular PKSs. However, incorporation of non-native starter units via noncognate LMs could require additional engineering of the downstream modules of the polyketide synthase, as they might not accept or act on unusual or bulky functionalities [38].
Currently, there is no flawless method for LM engineering to incorporate nonnative starter units into polyketides. The most common approach involves swapping a full LM (that selects for a desired starter) in place of the native LM. This has been met with limited success, and a more detailed understanding of where bottlenecks arise is needed before LM swaps can become a more common practice. Because LMs can occur in cis or in trans, the native protein-protein interactions that occur within each type of system should be considered. It might be best to retain the native intermodular linker for in cis LMs, and in the same way, LMs used to complement in trans may be more successful if domains are engineered with C-terminal linker domains from the native system (Fig. 2). Other possibilities for retaining native protein-protein interactions include performing AT swaps within LMs instead of swapping full modules. We hypothesize that more conservative swaps (in terms of chemical structure) will be more successful because of the gatekeeping functions of the downstream PKS modules. Interactions between the LM ACP and the first KS of the downstream extension module should also be considered, as improper protein-protein interactions will prevent chain translocation to the first extension module. However, until a more detailed and systematic study of LM swaps is published, the importance of each of these factors for each unique system is unknown. Therefore, with the current state of knowledge, each LM engineering attempt should be optimized individually and should be attempted with multiple domain boundaries in order to find the most productive system.
2.2. AT domains
The acyltransferase domain is responsible for selecting the CoA-based starter and/or extender units that form the majority of the carbon backbone of the growing polyketide. AT domains are attractive engineering targets for several reasons. First, we can easily predict the substrate specificity of most AT domains based on conserved residues. Secondly, AT domains are the primary sources of diversification at the α-carbon, a diversity that extends beyond the extent of the various oxidations states of the β-keto groups to include other heteroatoms, halogens, and unique functionalities that are otherwise unachievable via traditional polyketide chemistry [39], [40], [41], [42], [43], [44]. Finally, AT domains are frequently more promiscuous in comparison to other PKS domains [39], [45], [46], making precursor-directed mutagenesis viable. For these reasons, AT domains are the most well-studied of all PKS domains in terms of engineering via site-directed mutagenesis, domain swapping, and other host-level techniques (vide infra).
While nature uses numerous diverse extender units to form PKSs, most known ATs select for either malonyl- or methylmalonyl-CoA [47]. Well before the first KS-AT didomains structures were solved for DEBS M3 [48] and M5 [49], consensus sequences were identified within the AT domain that correspond to malonyl-(HAFH) or methylmalonyl-CoA (YASH) specificity [50]. With structural information in hand, identification of active site residues for targeted mutagenesis became much easier. Later on, more diverse extender units were identified, revealing other specificity motifs in the associated AT domains. The AT from module 3 of the epothilone cluster, for example, accepts both malonyl- and methylmalonyl-CoA and contains a “HASH” motif [51], while the allylmalonyl-CoA-specific AT from module 4 of the FK506 cluster has a “CPTH” motif at the same location [52]. Several other ATs with unique specificity-conferring residues at this position have also been discovered and their cognate extender units identified, significantly expanding the chemistries accessible via PKS-mediated biosynthesis [47], [53]. Nevertheless, simply mutating these residues to the appropriate specificity motif does not necessarily confer specificity to the desired extender unit. Mutation of the of the methylmalonyl-specific “YASH” motif to “HAFH” in DEBS AT1 [46], AT4, and AT6 [54] increased promiscuity, allowing the domains to accept and transfer both malonyl- and methylmalonyl-CoA. However, later in vitro investigations demonstrated that these modules had greatly reduced activity [55]. Mutations outside the active site can also lead to more promiscuous AT domains, although a greater amount of retention of native substrate specificity is frequently observed [54], [56], [57]. These types of mutations can even abolish the transacylation activity of the domain altogether [52]. Evidence suggests that the specificity conferred by these conserved residues could be overcome if nonnative extender unit concentrations are high enough relative to the native extender [58].
The incorporation of rare extender units can also be achieved by targeting other residues with site-directed mutagenesis (Fig. 2). For example, mutating an active site valine to an alanine in DEBS AT6 created a more promiscuous enzyme that was able to incorporate various nonnative extender units such as propargylmalonyl-, allylmalonyl-, and ethylmalonyl-CoA into the erythromycin backbone [57]. More recent work has revealed that targeted active site mutagenesis screens can be more effective at producing ATs that are more specific for nonnative extenders than the native substrate. Williams and coworkers showed that DEBS AT6 could be converted to a propargylmalonyl-CoA-specific domain by screening a library of active site mutants and identifying a Y → R mutation that changes the native specificity [44]. Combined with exogenous feeding of precursors (vide infra), AT mutagenesis screens could be used to create diverse libraries of polyketides from just one engineered PKS.
AT swapping is also commonly undertaken as a method for constructing hybrid modules capable of producing new polyketides with novel α-carbon substituents (Fig. 2). Traditionally, AT swaps were constructed via restriction enzyme-based cloning, thus relying on conserved restriction sites flanking the AT domain. One of the first attempts at a full AT swap was performed by Leadlay and coworkers in which the native AT domain from DEBS1-TE (malonyl-CoA specific) was replaced with the AT domain from module 2 of the rapamycin synthase (methylmalonyl-CoA specific) [59]. The hybrid protein was able to produce TKLs exclusively derived from methylmalonyl-CoA with little to no effect on yield, highlighting the significant potential of the technique for producing novel polyketides.
Katz and coworkers showed that, when placed in the context of a full modular PKS system, AT domain exchanges are not as efficient as the native system [40]. Swaps of malonyl-CoA-specific ATs from the pikromycin and rapamycin PKSs into DEBS modules 1 and 2, in particular swaps into the second module, resulted in significantly lower titers of the expected erythromycin derivatives [41], [42], [43]. In a similar study, a swap of the second rapamycin AT into module 6 of DEBS was capable of producing 25 mg/L of the appropriate 6-deoxyerythronolide B analogs, suggesting that the efficiency of AT-swapped modules may be affected by the position of the module within the larger context of the assembly line [60]. However, a successful swap of the ethylmalonyl-CoA-specific AT domain from niddamycin in module 4 of DEBS, suggested that other factors may be at play [61]. A functional rapamycin AT2 swap into DEBS module 4 finally illustrated that the domain boundaries for AT swaps (and any domain swap) may need to be optimized for each unique acceptor module-AT pairing [62]. Several other examples of AT domain exchanges emerged, each reporting extremely low titers due to the engineered nature of the systems at hand [63], [64]. DEBS module 4 was the last module in the model system to be successfully engineered in this manner by varying the domain boundaries used, suggesting that modules with full reductive loops may necessitate different AT swap domain boundaries because of the architectural difference due to the domains and linkers that surround the AT.
The mixed success observed with AT swaps led to the pursuit of a more fundamental understanding of the mechanisms of catalysis in native versus engineered AT systems. To this end, Khosla and coworkers systematically characterized the bottlenecks hindering successful AT swaps [43]. By analyzing the kinetics of AT acylation and KS-mediated condensation in native and engineered systems, they concluded that condensation in the hybrid modules occurred at a rate over ten times slower than the native system. In addition, limited proteolysis experiments and the poor expression of the engineered proteins suggested that the AT swapped mutants are inherently less stable and adopt a different protein conformation. AT domain swaps may minimize or interfere with the important protein-protein contacts required for condensation. Swapped domains may disturb the orientation of stabilizing residues [65] with respect to hydrogen bonding partners that help maintain the dimeric structure of the PKS or prevent the proper interaction of the ACP domain with the KS-AT linker [66]. Most recently, Keasling and coworkers performed a detailed kinetic analysis of AT-swapped modules utilizing different domain boundaries and showed that the optimized boundaries could be applied to construct functional swaps of various heterologous AT domains [67]. We hypothesize that more comprehensive biochemical and structural studies of AT domain swaps will reveal a less ambiguous set of design rules for AT replacements.
A final method for AT domain engineering involves the complementation of inactivated cis-AT PKSs with trans-acting domains. Although this review does not focus on engineering trans-AT PKS systems, we will note that the freestanding AT domains from these PKSs can successfully communicate with cis-AT PKS modules [39]. In trans-AT PKSs, the acyltransferase domain is not incorporated into the same polypeptide as the remaining catalytic domains of a typical PKS module. Instead, a freestanding trans-AT typically transfers extender units to multiple ACP partners. The utilization of trans-ATs to generate polyketide diversity in place of the activity of a native domain has been demonstrated in several contexts. Typically, an AT null (AT0) mutant is generated by converting the conserved active site serine to an alanine, destroying the active site nucleophile responsible for initial attack of the extender unit thioester. Khosla and coworkers originally showed that trans-AT domains such as DszsAT from the disorazole PKS system could be used to complement an AT0 version of DEBS1 in vitro [39]. It was later found that the DszsAT could also be used to produce fluorinated polyketides through the transfer of fluoromalonyl-CoA to several different AT-deficient modules of DEBS, both in vitro and in vivo [68]. This technique has since been further extended to a bimodular system in which DszsAT was able to communicate with multiple ACPs at once, transferring fluoromalonyl-CoA to AT0 versions of DEBS module 2 and 3 to produce a difluorinated TKL [69]. Other trans-AT domains, such as the KirCII AT, have successfully been used to install propargyl and allyl groups into polyketides, albeit so far only within the native kirromycin PKS system [45].
AT domain engineering has been extensively studied, yet similar to LMs, no consensus set of engineering rules exist. It is well established that mutation of the active site serine in a conserved “GHSxG” motif to alanine abolishes domain activity. Changing domain specificity with point mutations has also been successful, but rational engineering in this case is still unpredictable. We believe that techniques such as saturation mutagenesis of active site residues can be successful but could require large screens of many mutants. AT domain swapping is currently also a viable method for changing α-carbon substituents. Based on recent experiments, it appears that screening and optimization of various swaps has yielded a potential set of widely applicable domain swap boundaries for ATs [67]. It remains to be seen, however, if this method will be generalizable to all type I PKS systems given the potential for disrupted protein-protein interactions and gatekeeping from downstream processing. In the case of protein-protein interactions, protein stability and folding likely plays a crucial role in expressing active soluble chimeric PKSs. It remains to be seen whether domain swapping or site-directed mutagenesis will prove the more productive AT engineering method in a general. However, we anticipate that the continued improvement in DNA synthesis and sequencing technologies will facilitate more large-scale systematic experiments that result in the improved activities of engineered enzymes.
2.3. KR domains
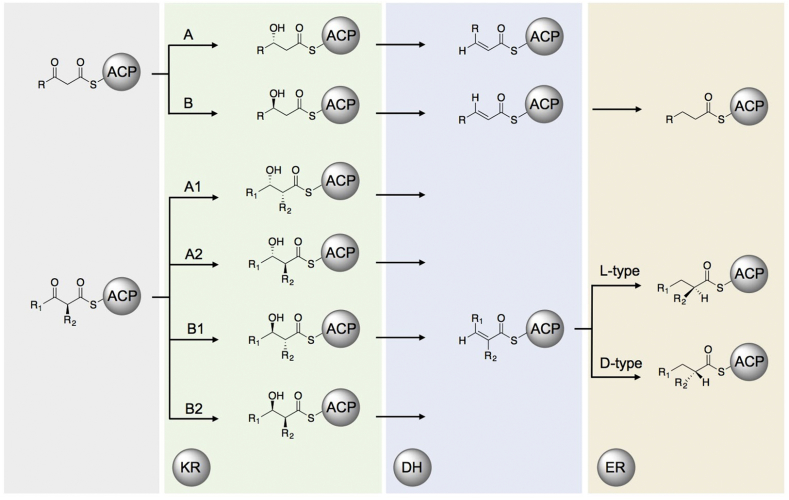
Ketoreductases perform the NADPH-mediated reduction of newly-formed β-keto groups after condensation. They are both stereospecific and stereoselective, and they are also known to epimerize α-substituents (if present) prior to reduction [70], [71]. KRs are characterized based on their stereochemical outcomes: A-type KRs generate L-configured alcohols and B-type KRs D-configured (Fig. 4). KRs are further described based on the final orientation of any α-substituents. A1 and B1 KRs produce D-configured α-substituents, and A2/B2, L-configured [8]. Finally, C-type KRs are reductase-incompetent but can retain epimerase activity [72]. Structures are available for each type of KR: A1 [73], A2 [71], B1 [74], B2 [75], and C2 [76].
Fig. 4.
Reducing domain(s) product outcomes. Potential stereochemical outcomes of each combination of β-carbon processing domains within a PKS module.
A- and B-type KRs can be distinguished not only by function, but also through the presence of certain structural elements. The presence of an “LDD” motif ∼57 residues before the catalytic tyrosine indicates a B-type KR (although frequently only the final “D” of this motif is strictly conserved) whereas it is absent in A-type KRs. Additionally, A-type KRs typically contain a tryptophan eight residues before the catalytic tyrosine. Structural insight has guided mutagenesis efforts, leading to the generation of A2-type [71] and nonstereospecific A-type KRs [73] derived from A1-type KRs. Additionally, a B2-type KR was converted to an A2-type through a single point mutant [77]. Not only can KR specificity be modified through site-directed mutagenesis, but activity can be completely abolished as well. Mutation of the catalytic tyrosine of the DEBS module 6 KR lead to production of the expected ketone, illustrating that KR inactivation is a way to generate new polyketides [78].
In addition to site-directed methods for engineering KR domains, numerous experiments have illustrated the viability of full KR domain swaps to modulate the stereochemistry and oxidation state of a given polyketide. Preliminary experiments were performed in the model DEBS system. A swap of the inactive KR domain from module 3 into module 2 resulted in the production of a TKL with a keto group in place of the alcohol generated by the native system [79]. This preliminary result showed that the surrounding domains function in the presence of a non-cognate KR partner. Complete removal of KR domains and replacement with an AT-ACP linker, on the other hand, has produced completely inactive mutants [80]. As the generation of loss-of-function mutants can easily be achieved via site-directed mutagenesis, it is the preferable method for KR inactivation because it allows for the retention of native protein-protein interactions and folding. Construction of KR-swapped PKSs with novel functions have also been reported. Successful gain-of-function KR swaps were implemented in a three module DEBS system in which KR domains from the rapamycin modules 2 or 4 were swapped into DEBS module 2, producing TKLs with non-native hydroxyl stereochemistry [81]. In a similar set of experiments, researchers from the same group showed that KR swaps could also be performed by replacing the KR in DEBS module 2 with KR5 and KR6 of DEBS [82]. Interestingly, KR6 was nonfunctional in this context. Further applying these KR swap design principles, Ashley and coworkers subsequently used KR-swapped DEBS modules to produce various derivatives of 6-deoxyerythronolide B in vivo, albeit with significant titer losses compared to the wild type system [80].
More recent efforts have focused on applying KR swaps to tailor the stereochemistry of both hydroxy groups and the adjacent α-methyl groups. Weissman and coworkers evaluated 14 total A2 and B2-type KR swaps into module 2 of DEBS1+TE using different swap sites [83]. Interestingly, there were obvious differences in activity of swaps performed using the various junctions. In addition, the A-type KR swaps were more functional than B-type swaps, suggesting that domain replacements that retain the native KR type might be more successful (DEBS module 2 contains an A1-type KR). Finally, all of the KR domains were sourced from modules with KS-AT-KR-ACP domain organization, so swaps of KR domains with neighboring reductive domains may need to be optimized separately. A subsequent study also corroborated the results from the work by Weissman and coworkers. Only A-type KR domain exchanges into a lipomycin PKS derivative (LipPKS1+TE), natively containing an A2-type KR, were active whereas all attempts to swap in B-type KRs produced PKSs incapable of reduction [84]. By comparing KR swaps of domains that contain or lack the newly-discovered KR dimerization element (DE), Keasling and coworkers also found that retaining the DE from the acceptor module almost always produced PKSs competent at performing condensation. Retaining the DE produced mutants with much higher activity than previous reports, suggesting that more systematic analyses of the structural implications of domain swaps might improve the activity of hybrid PKSs.
A few trends emerge when comparing the various strategies for altering the stereochemical outcome conferred by the KR domain. An aspect that appears to be generally consistent between studies with small molecule substrates, mutagenesis, and domain swaps is that KRs that reduce longer substrates (presumably possessing larger binding pockets) tend to be either less active on smaller substrates, or tend to generate predominantly the energetically favored A2 product. This was observed in studies by Keatinge-Clay and coworkers, wherein small N-acetylcysteamine (SNAC) mimics were treated with a panel of various type I KRs [71]. This is also consistent with the work of Keasling and coworkers, where DEBS KR6 (the KR from the terminal module) was shown to retain the least amount of selectivity among the KR+DEs swapped [84]. The notion that expanding the binding pocket affects stereochemical outcome was further corroborated by the work of Keatinge-Clay via enlarging active site through point mutations [77]. This observation has implications for choice of donor KRs to exchange in KR swapping experiments. If the donor KR does not natively reduce a substrate of similar size to the target small molecule, it is unlikely to retain stereochemical fidelity, thus impacting the processing of downstream β-carbon processing domains (Fig. 4).
2.4. DH and ER domains
Dehydratase domains are responsible for installing the majority of alkenes in polyketides. Dehydration proceeds via the syn-coplanar elimination of water and is therefore sensitive to the stereochemical configuration of the substrate [85]. Thus, DH domains are tied to the KR domains that precede them. Most DHs follow B-type KRs and catalyze trans-olefin formation (Fig. 4) [8]. However, both cis- and trans-alkenes are found in polyketide backbones. Post-PKS processing is implicated in formation of the cis-olefins in borrelidin [86] and rifamycin [87]. The cis-alkene of phoslactomycin, however, is likely installed by the DH of the first module of the synthase, Plm1 [88]. This DH succeeds an A-type KR, so syn-coplanar elimination of water from the resultant KR product would yield a cis alkene (Fig. 4). Despite having several DH crystal structures [87], [89], [90], there is no clear trend in the active site residues that govern the stereochemistry of reduction or the preference for α-substituents [91].
Although DH engineering has not been as extensively pursued as the engineering of other PKS domains, several examples of both site-directed and swap-based engineering exist. There are several known examples of naturally-inactive DH domains, such as those observed in the amphotericin [92], avermectin [93], nystatin [94], and nanchangmycin [95] clusters. Each of the inactive domains exhibits a H→ R or H→ Y mutation within the conserved active site motif “HxxxGxxxxPP”. Installing the H→ Y mutation in a DH domain within the FR-008 PKS resulted in successful inactivation of the DH and production of solely the appropriately hydroxylated products [96]. Using this same technique, a DH in the nystatin cluster could also be inactivated [97]. DH swaps have also been attempted. The DH domain from DEBS was swapped into a module of the avermectin synthase in S. avermitilis, resulting in the exclusive production of C22,23-unsaturated avermectins (although at much lower titers than the wild type) [98]. DH domain swaps were also employed in a polyketide synthase engineered to produce adipic acid, decreasing the amount of 3-hydroxyadipoyl product that had built up due to suspected non-optimal DH activity. Interestingly, the same group showed that DH domains can also be provided in trans to achieve the same effect (Fig. 1C) [99].
Even less is known about the ER domains, which reduce trans-α, β-unsaturated intermediates provided by DHs. ERs, like KR domains, exhibit epimerase activity on α-substituents as well [91]. ERs that produce L-oriented products possess a conserved tyrosine that is absent in D-type ERs [100]. A crystal structure of the ER from the second module of the spinosyn PKS (Spn2) shows this characteristic L-type tyrosine residue proximal to 4-pro-R-hydride of the bound NADP+, suggesting its role as the proton donor [101]. This structure also revealed a lysine-aspartate pair that was crucial for catalysis; the lysine is thought to act as the proton donor in D-type ERs. Finally, this structure revealed that the ER domain is inserted between the structural and catalytic domains of the KR, tying the ER to its cognate KR partner.
Early mutagenesis studies of ER domains in DEBS module 4 targeted the conserved “HAAAGGVGMA” NADPH binding motif for engineering [102]. Changing this sequence to “HAAASPVGMA,” based on the NADPH binding motif of the inactive KR domain from the same module, resulted in production of primarily Δ6,7-anhydroerythromycin C, the expected product. Moreover, mutation of the conserved L-type tyrosine in the same ER resulted in a change of stereochemistry at the α-methyl substituent, switching it from the S to R configuration [100]. Nevertheless, it should be noted that the same mutation in the rapamycin ER failed to produce the expected inversion of stereochemistry in TKL products; therefore, more studies of stereocontrol in ERs are needed before a comprehensive method for switching α-substituent orientation can be determined. Subsequent mutagenesis experiments performed by Keatinge-Clay and coworkers were unable to completely inactivate the Spn2 ER through a single mutation [101]. Instead, mutations both slowed the rate of reduction and increased the rate of the hydroxylation side reaction.
When introducing a non-native DH and ER domain into a chimeric PKS, it is important to consider the DH stereoselectivity of the hydroxylated product exercised by the upstream KR domain (Fig. 4). It might be best to pair DH domains with their cognate KR partners in order to ensure proper stereochemistry of the hydroxylated substrate. Alternatively, swaps of DH domains into modules with the same KR type could also be successful. Not only is proper substrate stereochemistry required, but substituents farther from the zone of reactivity could also come into play [99]. Another study corroborated this hypothesis, presenting structural and functional data to suggest that DHs which natively act on long-chain (C28-30) substrates are unable to dehydrate shorter chain (C4) substrates [103]. This was consistent with the structural information presented, highlighting a substrate channel that made hydrophobic interactions with the acyl chain of the long-chain substrates. Other issues involving protein-protein interactions, protein solubility and protein stability, could also arise, as different ACP partners can completely reverse the stereospecificity of DHs [87].
2.5. Reductive loops
Reductive loops, here defined as any combination of reducing domains, determine the degree of reduction of the β-carbon formed post-condensation and typically set the stereochemistry of any α-substituents (see previous sections). Typically, site-directed mutagenesis is performed on single reductive domains to achieve a desired outcome, whether it be domain inactivation or a change in activity. Schulz and coworkers, however, systematically evaluated the feasibility of “domain skipping” mutations in every reducing domain in the monensin PKS in order to evaluate the ability of downstream modules to accept less reduced substrates [104]. PKSs mutated in a way that produced less reduced intermediates were less likely to produce the desired final product. All ER inactivations reliably produced the expected products, but only two thirds of DH and one half of KR inactivations resulted in any detectable premonensin analogs. This suggests not only that downstream domains may display selectivity for their native substrates, but also that attempts to engineer more drastic changes of degree of reduction within the molecular structure may be less successful.
Full reductive loop additions into modules with fewer (or no) native reducing domains have been successfully employed in a few cases. A series of reductive loop swaps into DEBS module 2 were promising examples, demonstrating that heterologous reducing domains can be feasibly employed [105]. Excitingly, Leadlay and coworkers illustrated that the KR, DH and ER domains can not only be exchanged among PKSs, but they can also be added to modules having a partial reductive loop, natively containing a KR domain. However, the junctions of each domain swap may need to optimized on a case-by-case basis, as no single set of boundaries was optimal for every hybrid PKS. By heeding this advice, Keasling and coworkers were successfully able to engineer a PKS capable of producing a novel polyketide, adipic acid, by introducing a full reductive loop into the first module of the borrelidin PKS (Fig. 1B–C). A series of donor loops and junction sites were tested to find the most productive mutant. Although application of reductive loop swaps was promising, a significant bottleneck at the DH was observed [99].
In addition to full reductive loop swaps, other examples of reductive domain replacements have been published. A swap of the DH-KR didomain from module 11 to module 12 of the pimaricin PKS improved activity of the marginally active module 12 DH [106]. The most successful mutant retained the linkers flanking the native DH-KR, while replacing simply the DH domain proved ineffective. Additional examples include a swap of the pikromycin module 4 DH-ER didomain into the avermectin PKS [107] and replacement of the DEBS KR domains with the full reductive loop from rapamycin module 1 [80], both of which resulted in significant decreases in product yield in vivo. Since having a match between substrate size and activities of both the KRs and DHs (vide supra) have been shown to impact their levels of activity on non-native substrates (the ER has been less extensively studied), it may be important to introduce reducing loops that act on native substrates of a similar size with regard to the target donor molecule. By continuing to parse the limitations of these types of swaps, we imagine that more successful implementations of reductive domain additions and replacements will arise.
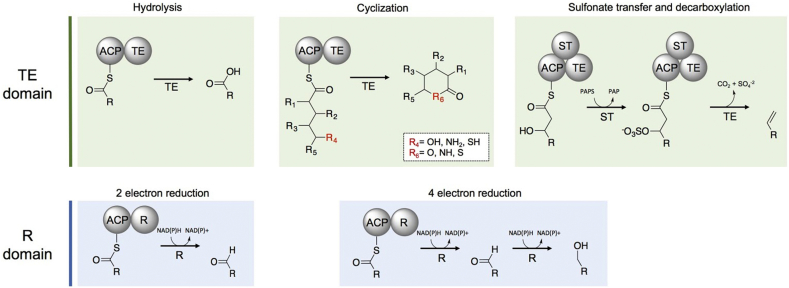
2.6. Offloading domains
To complete PKS biosynthesis, a terminal domain must release the ACP-bound polyketide intermediate in the last module. Termination domains include thioesterase (TE) domains and reductase (R) domains (Fig. 2, Fig. 5). The more common of the two release mechanisms, TEs, belong to the α/β-hydrolase superfamily of proteins and can release the polyketide as a linear acid, macrolactone, macrolactam, or macrothiolactone [9] (Fig. 2, Fig. 5). They are generally selective catalysts in the presence of native substrates, exemplified by the highly regio- [80] and stereospecific [108] cyclization performed by DEBS TE. Despite their native specificity, cyclization TEs are often promiscuous towards unnatural substrates [12]. DEBS TE, for example, cyclizes various macrolactones derived from engineered versions of DEBS that produce non-native oxidized and substituted backbones [80]. Additionally, non-native 6-, 8-, 12-, 14- and 16-membered rings [109] have been produced by either DEBS TE fusions or mutasynthesis [82], [110], [111]. Cyclizing TEs (especially the commonly used DEBS TE) are also known to hydrolyze nonnative substrates in engineered systems when cyclization is not feasible [12], [22], [99], [112], [113].
Fig. 5.
Offloading domains. Common TE-mediated release include products such as linear acids, lactones, lactams, thiolactones and olefins. The less common R-domains conduct a two-electron reduction to produce aldehyde final products or primary alcohols through a four-electron reduction.
A number of striking differences emerge when comparing TEs that natively hydrolyze their substrates and those that cyclize. The best studied of the hydrolytic TEs is from tautomycetin, which has been structurally characterized and shown to have a narrower substrate chamber than that observed in the structures of the cyclizing pikromycin and DEBS TEs [114]. The selectivity of this TE was further elucidated with chemically-synthesized SNAC intermediates, which demonstrated that its hydrolytic activity is highly stereospecific for R-β-hydroxy moieties. Further probing the hydrolytic selectivity of the tautomycetin TE, Kim and coworkers swapped the tautomycetin TE domain with the macrocyclic polyketide pikromycin TE [115]. The Pik-TE-swapped strain produced a mixture of both linear tautomycetin and a cyclized analog [115].
Although most termination TE domains are located at the C-terminus modular PKSs, the modular PKSs that encode the polyethers such as nanchangmycin [116], [117] and monensin [118] lack a C-terminally tethered (cis) thioesterase. Deng and coworkers demonstrated that the discrete TE domain NanE, also part of the α/β-hydrolase superfamily of proteins, is required for the biosynthesis of nanchangmycin. NanE catalyzes the specific hydrolysis of the polyether analog, nanchangmycin-SNAC [116], [117].
A termination mechanism that is evolutionarily related to thioesterases consists of tandem sulfotransferase (ST)-TE domains which yield terminal olefins, as found in the curacin PKS (Fig. 2, Fig. 5). The ST-TE didomains mechanistically employ the sulfotransferase domain to activate the β-hydroxy group formed by the terminal module. The activated sulfate group then undergoes a decarboxylative elimination reaction to yield the terminal olefin as the released product [90], [119], [120]. This biosynthetic logic is particularly attractive for applications of engineered polyketides to produce a variety of desirable chemicals, especially monomers for polymeric materials [121] or synthetic handles for further functionalization of natural products [122].
Another less common releasing enzyme, the R (reductive) domain, generally catalyzes the NADPH-dependent reductive release of polyketide and non-ribosomal peptide intermediates via thioester reduction (Fig. 2, Fig. 5). R domains possess a Rossmann fold [123], characteristic of nucleotide binding proteins which release acyl intermediates by either a 2- electron reduction, yielding an aldehyde final product, or by a 4-electron reduction, yielding a primary alcohol final product. Examples include the reductive release of aldehyde-containing products such as aureusimine [124] and primary alcohols in glycopeptidolipid [125] and myxalamid biosynthesis [9], [126]. Structure-guided mutagenesis of the terminal myxalamid R domain, MxaA, generated variants with increased activity towards hydrocarbon substrates that resulted in highly-reduced primary alcohols [127]. The removal of bulky residues in the C-terminal substrate binding domain facilitated reduction of non-native 10-carbon intermediates, yielding a reduced 10-carbon alcohol product [127]. This termination mechanism is of particular interest when trying to engineer polyketides that generate fuel molecules (e.g. n-butanol [128]).
2.7. Intermodular linkers
Intermodular linkers facilitate communication and chain translocation between two modules. They can take the form of C- and N-terminal docking helices that mediate contacts between modules on separate polypeptides, or they can consist of simple peptide linkers between two modules on the same polypeptide (Fig. 2) [10], [129], [130], [131]. Early attempts to improve intermodular communication between PKSs focused on covalently fusing two non-covalently-linked modules together. The expected TKLs could be produced in S. coelicolor by fusing DEBS module 1 to DEBS modules 3 or 6 by conserving the natural intermodular linker between modules 1 and 2. This technique could also facilitate communication of module 1 with rifamycin module 5 (Rif M5). Finally, by appending the C-terminal linker domain from DEBS module 2 to the end of Rif M5, the hybrid DEBS M1-Rif M5 protein could be coaxed to communicate with the downstream DEBS M3. This preliminary work laid the foundation for combinatorial biosynthesis of polyketides utilizing modules, not domains, as the fundamental unit [132].
Khosla and coworkers demonstrated that the position of modules within their native context is an important consideration for intermodular linker engineering. Swapping linker regions between module 2 and module 3 of DEBS with the module 4 and 5 linkers significantly increased the Km of the module-module interaction with little to no effect on the kcat of polyketide transfer between the modules [133]. The Khosla group also suggested that the KS domains of C-terminal modules, such as DEBS module 2 and 4, are less selective of substrates from upstream ACP partners than N-terminal modules because C-terminal KSs are already covalently tethered to the upstream ACP. The importance of ACP-KS interactions for chain transfer was again highlighted in the work of Santi and coworkers, where over 150 module-module interactions were tested for chain transfer [134]. The efficacy of 11 upstream and 14 downstream module combinations, facilitated by the C- and N-terminal linker domains from DEBS module 2 and 3, respectively, was tested for production of the expected TKL products. Although most of the individual PKSs were active on some level, less than 50% of the combined two module systems produced any product. The best combinations were between native partners, suggesting that incorrect ACP-KS interactions were a limiting factor [135].
3. Engineering at the host/cellular level
Despite all the current insights gained through PKS engineering at the amino acid, domain, and module level, progress has been slower than expected. This arises from a lack of understanding and inability to successfully express PKSs and various polyketide precursor pathways in a variety of hosts. In this section, we will highlight some significant accomplishments in polyketide precursor engineering and move towards PKS regulation, metabolic engineering and the application of synthetic biology tools for PKS host engineering. Because of the extensive amount of research in this area, this is not an exhaustive overview of host engineering for heterologous polyketide production. Nevertheless, numerous representative examples are highlighted.
3.1. Precursor directed-biosynthesis and mutasynthesis
Precursor-directed biosynthesis (PDB) has been a successful tool utilized to further understand and expand PKS substrate promiscuity by feeding analogues of their natural building blocks that are likely tolerated by the native biosynthetic PKS in the producing host. The efficiency of PDB can be enhanced by complementation with mutational biosynthesis (“mutasynthesis”), wherein the naturally occurring precursor pathways are inactivated, thus removing competition from natural precursors. In addition to precursor supplementation through feeding experiments, metabolic pathways that produce precursor analogs can be introduced heterologously to replace the deleted pathways [136].
One of the earliest and most successful examples of mutasynthesis targeted the avermectin PKS. A strain of Streptomyces avermitilis was generated wherein the enzymes required for generating the precursors 2-methylbutyryl-CoA and isobutyryl-CoA were inactivated, more specifically the branched chain fatty acid dehydrogenase complex bkd. Out of more than 800 potential precursors tested, over 40 starter unit analogs were tolerated by the avermectin PKS [21]. Via this method, a cyclohexyl-containing avermectin derivative (later named doramectin) was generated that exhibited increased antiparasitic activity against veterinary pathogens [137]. Another group later identified a shikimate-derived cyclohexyl-CoA biosynthetic pathway that, when re-introduced into S. avermitilis Δbkd, enabled the production of doramectin without cyclohexanoic acid supplementation [138], [139]. Intriguingly, while other PKSs are primed with isobutyryl or 2-methylbutyryl-CoA (such as lipomycin and tautomycin), no analogous experiments to generate other mutasynthetic analogs in Δbkd strains has been reported. While the lipomycin loading AT has been shown to be somewhat promiscuous, only six loading substrates have been characterized [22]. Thus, it is unclear if the avermectin PKS has an unusually promiscuous loading AT, or if this extreme promiscuity is a general feature of loading ATs from type B LMs that accept bulkier acyl-CoA priming substrates.
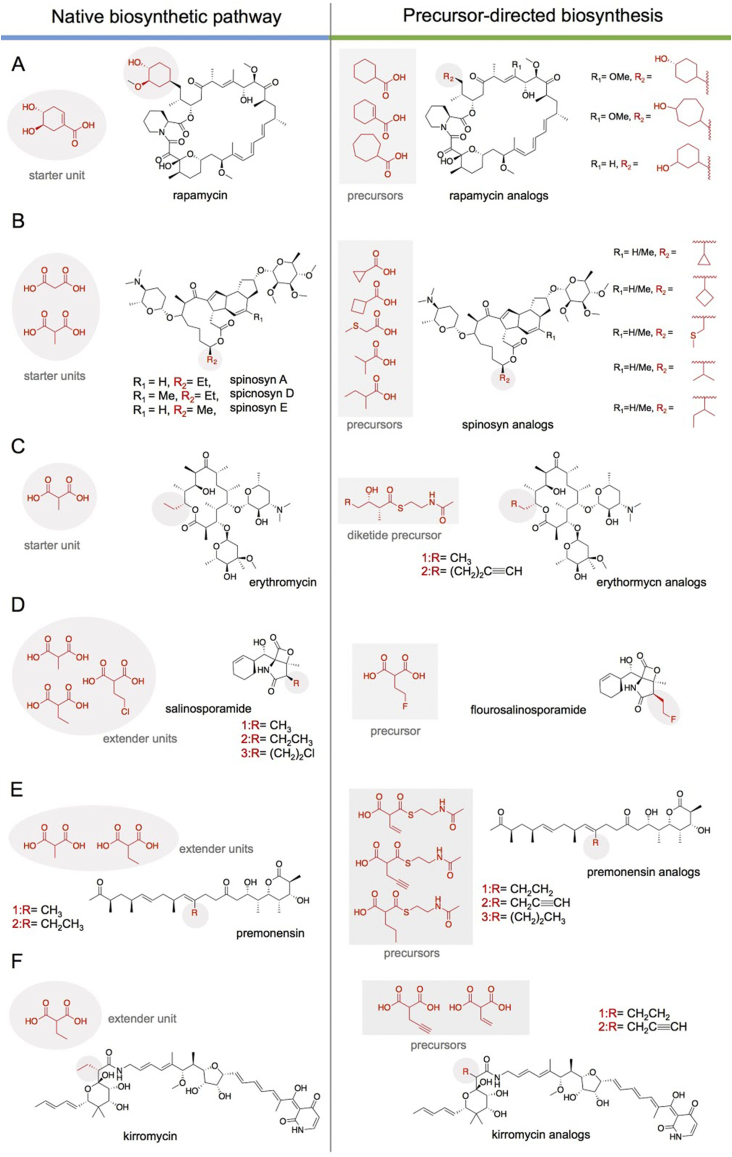
Other examples of PDB have targeted the macrolide rapamycin [140], [141]. In rapamycin biosynthesis, Leadlay and coworkers conducted feeding experiments with starter unit analogues. A series of 21 substrates consisting of monocyclic, polycyclic, branched aliphatic acids, substituted benzoic acids and heterocyclic acids, were fed to the rapamycin-producing strain of S. hygroscopicus [141]. New metabolites were observed in the presence of three monocyclic aliphatic acids, demonstrating a certain degree of starter unit promiscuity within the rapamycin LM and further suggesting some substrate tolerance within the downstream modules (Fig. 6A) [141]. In a similar approach, Martin and coworkers explored carboxylic acid starter unit tolerance in the spinosyn PKS appended with the avermectin or erythromycin LM [142]. Supplementation with a range of carboxylic acids led to the successful production of new spinosyn analogues (Fig. 6B) [142]. In a similar example, the aureothin PKS pathway demonstrated starter unit tolerance towards various aromatic priming analogs [140]. Lastly, Khosla and coworkers evolved an engineered erythromycin biosynthetic pathway in E. coli, yielding erythromycin analogs from SNAC precursors. Khosla and coworkers fed diketide SNAC precursors to E. coli cells harboring a truncated form of DEBS lacking the loading and first modules. They demonstrated the incorporation of the native substrate analog diketide SNAC and a novel diketide-SNAC containing an alkynyl moiety into the erythromycin final product (Fig. 6C). These results support the tolerance of DEBS to utilize a nonnative substrate for priming, extension and modification by the downstream DEBS PKS machinery to produce an alkynyl-containing erythromycin derivative [143], [144].
Fig. 6.
Examples of precursor-directed biosynthesis altering priming unit. A) Starter unit PDB in the rapamycin PKS. B) Starter unit tolerance in spinosyn biosynthesis. C) Extender unit PDB using a diketide SNAC precursor in the erythromycin PKS. D) Incorporation of fluoromalonyl-CoA through mutasynthesis of the salinosporamide pathway. E) Incorporation of propargyl-, propyl- and allylmalonyl-SNAC into the monensin polyketide backbone. F) Incorporation of propargyl- and allylmalonyl-CoA into the kirromycin polyketide backbone.
The selectivity and promiscuity of extending acyltransferase domains can also be targeted for PDB and/or mutasynthetic approaches to alter PKS scaffolds. While the AT domains that select for malonyl-CoA or methylmalonyl-CoA are typically quite selective for their respective extender units [41], it is common for ATs that naturally select for more uncommon and/or bulkier substrates to have more relaxed intrinsic substrate selectivity. For example, module 5 of the monensin PKS natively incorporates both methylmalonyl-CoA as well as ethylmalonyl-CoA [145], [146], [147]. In the salinosporamide PKS, while the major product is derived from SalA-AT-mediated chloroethylmalonyl-CoA incorporation (forming salinosporamide A), analogs containing ethylmalonyl-CoA (salinosporamide B) and methylmalonyl-CoA (salinosporamide D) are also generated (Fig. 6D) [148]. This inherent promiscuity can be harnessed in concert with precursor pool engineering to bias the distribution of metabolites or generate new metabolites altogether. Deletion of 5-chlorodeoxyadenosine (a precursor for chloroethylmalonyl-CoA) biosynthetic genes coupled with exogenous supplementation of 5-fluorodeoxyadenosine [149] or heterologous expression of the heterologous precursor pathways [150] resulted in the generation of a fluorinated analog, fluorosalinosporamide.
Likewise, the promiscuity of the monensin AT5 was explored to generate premonensin analogs (Fig. 6E). Schulz and coworkers determined that MonAT5 accepts propargylmalonyl, allylmalonyl, propylmalonyl, and butylmalonyl-SNAC when supplemented in the media [147]. Similar promiscuity was shown for an ethylmalonyl-CoA selective AT in the kirromycin pathway, KirCII (Fig. 6F). In vitro, KirCII accepts a range of longer malonyl derivatives including allylmalonyl-CoA, propargylmalonyl-CoA, and to a lesser extent azidoethyl-CoA [151], [152]. Rather than supplementing the culture with SNAC analogs, Williams and coworkers introduced a highly promiscuous mutant of the malonyl-CoA ligase, MatB, from Rhizobium trifolii [152], [153] into the host strain S. collinus Tü 365 and supplemented exogenous allylmalonate and propargylmalonate. The malonate derivatives were then activated by MatB and incorporated into the polyketide to generate kirromycin analogs [45]. Propargyl-containing extender units were introduced in one additional polyketide scaffold through targeted mutagenesis. A combination of structural modeling and sequence comparisons of ATs with specificity for bulkier substrates (such as ethylmalonyl-CoA) was used to expand the promiscuity of a methylmalonyl-selective AT [54], [57].
3.2. Metabolic engineering for improved precursor pools
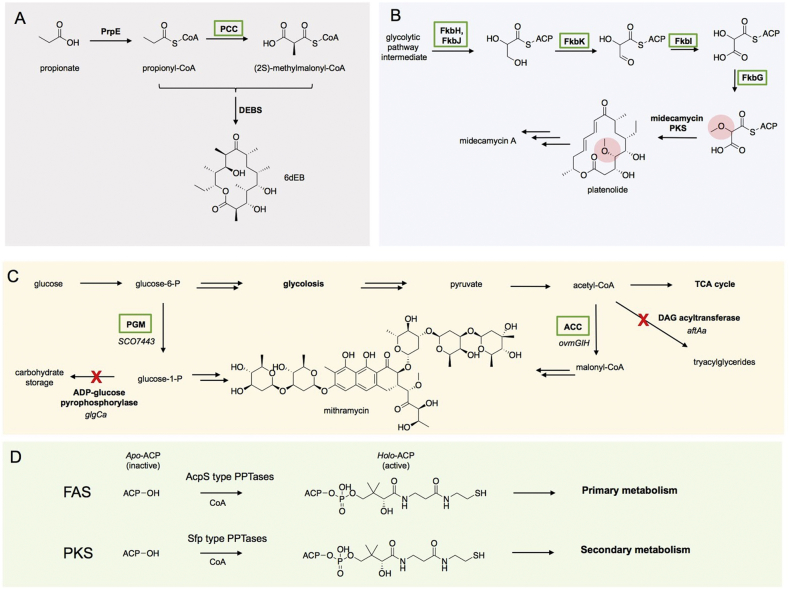
Outside the limitations of directly engineering PKSs, one approach to improve polyketide production involves increasing the availability of polyketide precursors. Pathways for the biosynthesis of polyketide metabolites often use precursors synthesized during glycolysis, the tricarboxylic acid cycle, and the pentose-phosphate pathway [154]. Metabolic engineering has successfully improved selected bottlenecks in various strains, increasing intracellular precursor pools, and thus redirecting flux towards the desired polyketide biosynthetic pathway. The deletion of the S. coelicolor phosphofructokinase gene plfkA or the S. lividans glucose-6-phosphate dehydrogenase genes zwf1 and awf2 achieved higher production of actinorhodin and undecylprodigiosin, respectively [155]. Inactivation of the glyceraldehyde-3-phosphate dehydrogenase gap1 in S. clavuligerus resulted in more clavulanic acid production [156], [157]. In contrast to deletion or knockout of genes, overexpression and knockin of genes has been utilized more often as a strategy to increase specific precursors for polyketide production. Such polyketide precursors include well-known and unique starter-/extender-units. For many type I assembly-line PKS pathways, such as DEBS, priming begins with one propionyl-CoA starter unit and extension is performed with (2S)-methylmalonyl-CoA. E. coli does not naturally produce (2S)-methylmalonyl-CoA, posing a bottleneck for expression of the erythromycin pathway in this host [158]. Hillenmeyer and coworkers have worked to overcome E. coli's (2S)-methylmalonyl-CoA limitation by introducing heterologous propionyl-CoA carboxylases (PCCs) (Fig. 7A) [159]. The propionyl-CoA carboxylase is a biotin-dependent enzyme that catalyzes the carboxylation of propionyl-CoA to form (2S)-methylmalonyl-CoA. The core PCC complex consists of two genes encoding the biotin carboxyl carrier α-subunit protein and a β-subunit containing the carboxylase activity [160], [161]. Hillenmeyer and coworkers examined the effect of heterologous expression of various PCCs on erythromycin production in E. coli by optimizing both expression of the S. coelicolor PCC complex and screening 13 homologous PCCs from diverse species [159]. The hybrid PCC complex from Myxococcus fulvus and S. coelicolor outperformed all other PCC complexes in stimulating erythromycin production.
Fig. 7.
Examples of metabolic engineering for improved precursor pools. A) Overexpression of PCC for the conversion of propionyl-CoA to (2S)-methylmalonyl-CoA allowed for the production of 6-deoxyerythronolide B in bacteria that don't natively produce methylmalonyl-CoA (such as E. coli). B) Expression of the methoxymalonyl-ACP biosynthetic pathway in a platenolide producer generated a platenolide analog containing the methoxy moiety. C) Higher titers of mithramycin were achieved by increasing the precursor supply of malonyl-CoA and glucose-1-phosphate. Green boxes denote upregulated pathways and red cross marks denote downregulation or knockout pathways. D) Post-translational modification of apo-ACPs by PPTases generated active holo-ACPs which can activate FAS biosynthetic pathways in primary metabolism or PKS pathways in secondary metabolism.
Unlike E. coli, where certain acyl-CoA extender units may be limited or nonexistent, Streptomyces already contain the common precursors to support polyketide biosynthesis. The diverse precursor pools that Streptomyces produce facilitate the heterologous expression and engineering of full pathways in these hosts. For example, in S. coelicolor and S. lividans strains heterologously expressing the DEBS PKS, Floss and coworkers constructed strains capable of producing 3 mg/L of 2-desmethyl-2-methoxy-6-deoxyerythronolide via the introduction of the methoxymalonyl-ACP biosynthetic pathway from Actinosynnema pretiosum [64]. The incorporation of methoxymalonyl-ACP was also utilized in S. fradiae, a host that natively produces three of the most common polyketide precursors: malonyl-CoA, methylmalonyl-CoA, and ethylmalonyl-CoA [162]. By introducing the biosynthetic pathway for methoxymalonyl-ACP into an S. fradiae strain heterologously expressing the midecamycin pathway, Katz and coworkers produced a methoxymalonate-platenolide analog (Fig. 7B) [162]. More recently, precursor pool engineering was also successfully employed to improve salinomycin production in S. albus. Metabolomic analysis suggested that intracellular ethylmalonyl-CoA concentrations were limiting titer, so an additional copy of the crotonyl-CoA carboxylase gene responsible for generating ethylmalonyl-CoA was integrated into the producing strain. This approach improved titers over ten fold [163].
Méndez and coworkers improved precursor metabolite pools for the production of the antitumor compound/polyketide mythramycin in S. argillaceus by increasing the precursor supply of malonyl-CoA and glucose-1-phosphate (Fig. 7C) [157]. The mythramycin PKS uses ten malonyl-CoA units for chain extension and cyclizes the extended polyketide to a tetracyclic intermediate. The tetracyclic intermediate is subsequently glycosylated by five deoxysugars derived from glucose-1-phosphate, generating mythramycin as the final product [157]. By either overexpressing the S. coelicolor phosphoglucomutase gene pgm or by inactivating the ADP-glucose pyrophosphorylase gene glgCa, they were able to increase glucose-1-phosphate production (Fig. 7C). Moreover, they increased malonyl-CoA concentrations by overexpressing the acetyl-CoA carboxylase ovmGIH gene or by inactivating the acyl-CoA diacylglycerol acyltransferase gene aftAa, the latter strategy being superior for improving mythramycin production [157].
In another example of PK precursor engineering to produce myxothiazol, Müller and coworkers identified in the S. cellulosum Soce56 genome an operon responsible for methylmalonyl-CoA production [164]. The operon, consisting of three genes, included a methylmalonyl-CoA epimerase epi, methylmalonyl-CoA mutase mcm and meaB which were sub-cloned and heterologously expressed in P. putida KT2440 containing the myxothiazol PKS/NRPS hybrid. The engineered P. putida strain containing both the myxothiazol PKS/NRPS and the three enzymatic methylmalonyl-CoA pathways produced myxothiazol [164]. Such an engineered P. putida strain provides an additional PKS expression host outside of the previously mentioned E. coli and Streptomyces species.
3.3. Phosphopantetheinyltransferase expression and regulation
Another frequent issue encountered when heterologously expressing PKSs is the need for post-translational activation of each ACP domain with a prosthetic phosphopantetheine arm. All ACP domains must be converted to the holo form by phosphopantetheinyltransferases (PPTases), which install the phosphopantetheinyl prosthetic group of ACP domains. PPTases can have a range of substrate selectivities, depending on whether they evolved to activate a carrier protein from a specific biosynthetic pathway or whether they have a pleiotropic role in the cell, activating multiple pathways [165], [166], [167]. PPTases are classified by whether they activate fatty acid biosynthetic pathways from primary metabolism (AcpS-type PPTases) or polyketide and nonribosomal peptides from secondary metabolism (Sfp-type PPTases) (Fig. 7D) [168]. In host organisms that do not natively harbor multiple polyketide and/or non-ribosomal peptide biosynthetic pathways (such as E. coli or S. cerevisiae) integration of a promiscuous Sfp-type PPTase is critical to enable the heterologous production of polyketide metabolites. The promiscuous PPTase Sfp, from the surfactin biosynthetic pathway in Bacillus subtilis, has historically been the most common choice for application in heterologous systems [169], [170]. Sfp has been integrated into E. coli to generate strains engineered specifically for polyketide production: BAP1 [171] and K207-3 [172]. Successful production of polyketide metabolites has also been accomplished in S. cerevisiae strains containing genomically-integrated Sfp [173], [174], [175]. The usefulness of many B. subtilis laboratory strains for heterologous PKS production is diminished because of a common frameshift within the sfp gene, effectively shutting down the production of multiple secondary metabolites [166], [176], [177]. Indeed, to successfully heterologously produce 6-dEB in the commonly used B. subtilis Marburg 168 strain, restoration of Sfp activity was required [177]. As an alternative to Sfp, the similarly promiscuous PPTase, Svp, from S. verticullus ATCC15003 has also been used to post-translationally modify a wide range of ACPs [178].
In most Streptomyces, heterologous polyketide production can be achieved without integration of an exogenous PPTase [179], suggesting that most Streptomyces species natively harbor promiscuous PPTases. This is unsurprising, given the wealth of secondary metabolites that are produced by Actinomycetes. However, more recently the overexpression of PPTases has been explored as a means of boosting polyketide production in various Actinomycetes. Li and coworkers demonstrated that overexpression of the endogenous PPTase SchPPT in S. chattanoogensis L10 increased the production of the polyketide natamycin by over 40% [180]. Other experiments suggest that PPTases may play a major role in the regulation of PKS expression. For example, overexpression of either Sfp or Svp in over 33 Actinomycetes activated the expression of silent biosynthetic pathways [181]. Although a basal level of PPTase expression is typically sufficient for polyketide production, these experiments suggest that overexpression of these enzymes can significantly enhance or alter the regulation and production of PKS metabolites. Thus, to achieve extremely high titers of polyketide products in Streptomyces and related Actinomycetes, manipulating the expression of both endogenous and exogenous PPTases is likely an underexplored approach.
3.4. Transcriptional regulation and refactoring of PKS genetic components
Polyketide biosynthetic gene clusters commonly contain pathway-specific regulatory elements that act as activators or repressors of various genetic elements within the cluster. In the biosynthesis of tylosin, for example, both activators and repressors have been identified and characterized [182], [183]. Activator elements often belong to the Streptomyces Antibiotic Regulatory Protein (SARP) family, a group of proteins known to enhance the production of polyketides such as daunorubicin, mithramycin and tylosin [155]. These SARPs can be effectively used as tools to increase polyketide production. Tylosin production, for example, is increased when the SARP regulators tylS and tylR are overexpressed in the native producer S. fradiae [183]. Mithramycin production in S. argillaceus can also be enhanced by overexpressing two SARPs, mtrY and mtmR, on multicopy plasmids [184], [185]. Other families of transcriptional regulators, such as the LysR-type transcriptional regulators (LTTRs), can serve as additional tools for controlling polyketide production. The role of LTTRs has been studied in the ascomycin biosynthetic cluster; in particular, Wen and coworkers have characterized the role of the LTTR fkbR1 [186]. Inactivation of fkbR1 leads to a reduction of ascomycin production, which can be restored by complementation with fkbR1. Overexpression of fkbR1, on the other hand, results in increased ascomycin production, suggesting that fkbR1 acts as a positive regulator [186]. Manipulation of negative regulators can also be used as a tool for manipulating polyketide pathways. For example, inactivation of a transcriptional repressor (tylQ) from the tylosin pathway causes production to begin at an earlier stage of S. fradiae growth [187]. On the other hand, disruption of the tylP gene that encodes a γ-butyrolactone receptor increases production [182].
DNA-based regulatory elements can be used to complement the manipulation of protein-based positive and negative regulators for the optimization of polyketide production. The introduction of heterologous promoters, for example, has been effectively used to activate multiple silent polyketide clusters in various hosts. Eliciting expression of silent polyketide pathways is often challenging due to poorly understood regulation mechanisms. In addition, these silent polyketide gene clusters often remain inactive under typical culturing conditions. With the goal of developing a generalizable method to elucidate the function of cryptic biosynthetic gene clusters, Zhao and coworkers developed a synthetic biology approach to activate silent biosynthetic gene clusters [188]. The entire silent biosynthetic gene pathway was refactored using a plug-and-play scaffold, containing a set of heterologous functional promoters and placed in a heterologous host under more traditional culturing conditions. Using this strategy, Zhao and coworkers successfully activated the previously silent spectinabilin pathway from S. orinoci [188].
More recently, Apel and coworkers used a similar plug-and-play method for the assembly of artificial genes into functional biosynthetic gene clusters [189]. As a proof of concept, the novobiocin pathway was disassembled and genetically reorganized using the artificial gene operon assembly method, a tool that consecutively assembles artificial genes into a destination vector and subsequently expresses them under the control of a de-repressed promoter in a Streptomyces host. By completely refactoring the pathway in S. coelicolor, Apel and coworkers could observe production of novobiocin precursors and novobiocin [189].
The development and improvement of omic analyses (genomics, proteomics, metabolomics and transcriptomics) further provides a useful tool to understand PK biosynthetic bottlenecks in a given host. In a recent example, Liu and coworkers utilized a targeted omics approach to identify rate-limiting steps in the heterologous production of spinosad in S. albus [190]. Both targeted metabolomics and translational analysis indicated insufficient activity of enzymes involved in two essential sugar precursors: rhamnose and forosamine. Moreover, the PKS protein SpnE and methyltranferase SpnI were not detectable by targeted proteomics analysis, suggesting other rate-limiting steps. With the aim of overexpressing the sugar biosynthetic module, the polyketide synthase SpnE, and the methyltransferase SpnI, strong constitutive promoters were replaced with native promoters. The new refactored engineered S. albus strain increased spinosad production by 1000-fold over the wildtype S. albus strain [190].
The increasing promise of the CRISPR-Cas9 system for rapid and targeted genetic manipulations [191], [192] has led to the development of protocols for using the technique in natural product-producing bacteria such as Streptomyces. Several groups laid the foundation for work with CRISPR-Cas9, first with the development of methods for performing knockouts [193], [194]. Later, Zhao and coworkers extended the usefulness of this CRISPR-Cas9 platform, using it to perform knockins to activate silent biosynthetic gene clusters in Streptomyces [195]. In a one-step strategy, multiple biosynthetic gene clusters of type I, II and III PKSs in five Streptomyces species were activated. Moreover, by introducing strategically-placed constitutive promoters, they elicited the production of a new metabolite in S. viridochromogenes: a multicyclic type II polyketide [195].
4. Host, precursor, and protein engineering synergy
Ultimately, for PKS protein engineering to be feasibly employed to produce target chemicals, it must be evaluated whether the engineered PKS activity in vivo is similar to its activity in model systems such as biochemical in vitro assays. In vitro studies afford absolute control of substrate and cofactor concentrations, which is not feasible in vivo. Thus, combinations of host and engineered PKS can result in various (and sometimes unanticipated) substrate incorporations. A study that exemplifies this phenomenon dissects the LM of DEBS. While in the native Saccharopolyspora erythrea DEBS is primed exclusively by propionyl-CoA [196], the LM of DEBS incorporates other priming units in heterologous systems [197]. Depending on the host, TKLs from DEBS1+TE can form via priming by acetyl-CoA (in S. coelicolor [198] and S. venezuelae [197]) or isobutyryl-CoA (in S. venezuelae [197]), albeit as minor products. Reynolds and coworkers interrogated the relationship between PKS architecture and starter unit incorporation in variations of DEBS1+TE expressed in S. venezuelae. It was determined that placing the LM and module 1 on separate polypeptides (with the use of heterologous docking domains) changed the distributions of acetyl, propionyl, and isobutyryl priming units incorporated in comparison to the ratios observed with both modules on the same polypeptide (i.e. the selectivity shifted from predominantly propionyl selective to isobutyryl-selective). This was rationalized through biochemical studies suggesting that the loading AT selects isobutyryl-CoA as the thermodynamic product. However, DEBS KS1 has faster kinetics for propionate compared to isobutyrate. When the loading domain is placed on a separate polypeptide from module 1, there is a resulting kinetic stall. This kinetic stall shifts the product formation towards thermodynamic control, especially when higher levels of isobutyl-CoA are present [197]. This example demonstrates that the apparent selectivity of PKS domains can be altered depending on the architecture of the PKS in concert with the precursor availability. Therefore, results of both in vitro and in vivo experiments for engineered polyketide production should be evaluated with a careful consideration of the advantages and limitations of each experimental approach.
5. Conclusions
With all the current success in PKS/host engineering, the gap between our understanding of PKS biosynthetic logic and the application of PKSs as a retrosynthetic platform is narrowing. In the past, our ability to design, build and test chimeric PKSs was limited by DNA sequencing and assembly techniques. Currently, our ability to discover and generate PKS genotypic diversity, enabled by improved DNA sequencing and synthesis technologies, allows us to rapidly build sizeable libraries of engineered PKSs [199], [200]. However, our ability to screen the resulting libraries lags. There is a need for more high-throughput approaches to test engineered PKS scaffolds. Analytical methods such as liquid chromatography coupled with mass spectrometry are improving and new techniques are being developed which allow for increased throughput with limited sample preparation [14], [201], [202], [203]. Transcription factor-based biosensors could be potentially employed to screen and select for successful engineered PKSs constructs [204]. Additionally, there are an increased number of computational and bioinformatic tools which aid the identification of candidate swaps as well as the curation and annotation of PKS pathways [205], [206], [207], [208], [209], [210]. With the increasing success of chimeric PKS scaffolds, there will also be a need for optimization of precursor flux within the target host. The metabolic engineering pathways for polyketide precursors must be integrated into hosts that express highly soluble and active engineered PKSs. By building on existing polyketide engineering and characterization strategies, harnessing current synthetic biology technologies, and utilizing advances in metabolic/host engineering, we anticipate future successes in engineering PKSs capable of producing designer polyketides for applications in medicine, fuels, and industrial products.
Acknowledgements
This work was funded by the Joint BioEnergy Institute (JBEI), which is funded by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, under Contract DE-AC02-05CH11231, by the National Science Foundation under awards MCB-1442724, NSF-GRFP DGE-1106400 and CBET-1437775, as part of the Co-Optimization of Fuels & Engines (Co-Optima) project sponsored by the U.S. Department of Energy (DOE) Office of Energy Efficiency and Renewable Energy (EERE), Bioenergy Technologies and Vehicle Technologies Offices, and by the DOE AgileBiofoundry (https://agilebiofoundry.org) supported by the U.S. Department of Energy, Energy Efficiency and Renewable Energy, Bioenergy Technologies Office, through contract DE-AC02- 05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. Additional funding was provided by the National Science Foundation Graduate Research Fellowship under Grant No. (DGE 1106400).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Julien B., Shah S., Ziermann R., Goldman R., Katz L., Khosla C. Isolation and characterization of the epothilone biosynthetic gene cluster from Sorangium cellulosum. Gene. 2000;249:153–160. doi: 10.1016/s0378-1119(00)00149-9. [DOI] [PubMed] [Google Scholar]
- 2.Bollag D.M., McQueney P.A., Zhu J., Hensens O., Koupal L., Liesch J. Epothilones, a new class of microtubule-stabilizing agents with a taxol-like mechanism of action. Cancer Res. 1995;55:2325–2333. [PubMed] [Google Scholar]
- 3.Luo G., Pieper R., Rosa A., Khosla C., Cane D.E. Erythromycin biosynthesis: exploiting the catalytic versatility of the modular polyketide synthase. Bioorg Med Chem. 1996;4:995–999. doi: 10.1016/0968-0896(96)00096-x. [DOI] [PubMed] [Google Scholar]
- 4.Waldron C., Madduri K., Crawford K., Merlo D.J., Treadway P., Broughton M.C. A cluster of genes for the biosynthesis of spinosyns, novel macrolide insect control agents produced by Saccharopolyspora spinosa. Ant Van Leeuwenhoek. 2000;78:385–390. doi: 10.1023/a:1010289901631. [DOI] [PubMed] [Google Scholar]
- 5.Khan N., Rawlings B., Caffrey P. A labile point in mutant amphotericin polyketide synthases. Biotechnol Lett. 2011;33:1121–1126. doi: 10.1007/s10529-011-0538-3. [DOI] [PubMed] [Google Scholar]
- 6.Weissman K.J., Leadlay P.F. Combinatorial biosynthesis of reduced polyketides. Nat Rev Microbiol. 2005;3:925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 7.Unable to find information for 911735, (n.d.).
- 8.Keatinge-Clay A.T. The structures of type I polyketide synthases. Nat Prod Rep. 2012;29:1050–1073. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- 9.Du L., Lou L. PKS and NRPS release mechanisms. Nat Prod Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- 10.Weissman K.J. The structural basis for docking in modular polyketide biosynthesis. Chembiochem. 2006;7:485–494. doi: 10.1002/cbic.200500435. [DOI] [PubMed] [Google Scholar]
- 11.Borgos S.E.F., Tsan P., Sletta H., Ellingsen T.E., Lancelin J.-M., Zotchev S.B. Probing the structure-function relationship of polyene macrolides: engineered biosynthesis of soluble nystatin analogues. J Med Chem. 2006;49:2431–2439. doi: 10.1021/jm050895w. [DOI] [PubMed] [Google Scholar]
- 12.Horsman M.E., Hari T.P.A., Boddy C.N. Polyketide synthase and non-ribosomal peptide synthetase thioesterase selectivity: logic gate or a victim of fate? Nat Prod Rep. 2016;33:183–202. doi: 10.1039/c4np00148f. [DOI] [PubMed] [Google Scholar]
- 13.Cummings M., Breitling R., Takano E. Steps towards the synthetic biology of polyketide biosynthesis. FEMS Microbiol Lett. 2014;351:116–125. doi: 10.1111/1574-6968.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poust S., Hagen A., Katz L., Keasling J.D. Narrowing the gap between the promise and reality of polyketide synthases as a synthetic biology platform. Curr Opin Biotechnol. 2014;30:32–39. doi: 10.1016/j.copbio.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez E., Menzella H.G., Gramajo H. Complex enzymes in microbial natural product biosynthesis, Part B: polyketides, aminocoumarins and carbohydrates. Elsevier; 2009. Chapter 15 heterologous production of polyketides in bacteria; pp. 339–365. [Google Scholar]
- 16.Yuzawa S., Kim W., Katz L., Keasling J.D. Heterologous production of polyketides by modular type I polyketide synthases in Escherichia coli. Curr Opin Biotechnol. 2012;23:727–735. doi: 10.1016/j.copbio.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 17.Stevens D.C., Hari T.P.A., Boddy C.N. The role of transcription in heterologous expression of polyketides in bacterial hosts. Nat Prod Rep. 2013;30:1391–1411. doi: 10.1039/c3np70060g. [DOI] [PubMed] [Google Scholar]
- 18.Oefner C., Schulz H., D'Arcy A., Dale G.E. Mapping the active site of Escherichia coli malonyl-CoA-acyl carrier protein transacylase (FabD) by protein crystallography. Acta Crystallogr D Biol Crystallogr. 2006;62:613–618. doi: 10.1107/S0907444906009474. [DOI] [PubMed] [Google Scholar]
- 19.Unable to find information for 675417, (n.d.).
- 20.Long P.F., Wilkinson C.J., Bisang C.P., Cortés J., Dunster N., Oliynyk M. Engineering specificity of starter unit selection by the erythromycin-producing polyketide synthase. Mol Microbiol. 2002;43:1215–1225. doi: 10.1046/j.1365-2958.2002.02815.x. [DOI] [PubMed] [Google Scholar]
- 21.Dutton C.J., Gibson S.P., Goudie A.C., Holdom K.S., Pacey M.S., Ruddock J.C. Novel avermectins produced by mutational biosynthesis. J Antibiot. 1991;44:357–365. doi: 10.7164/antibiotics.44.357. [DOI] [PubMed] [Google Scholar]
- 22.Yuzawa S., Eng C.H., Katz L., Keasling J.D. Broad substrate specificity of the loading didomain of the lipomycin polyketide synthase. Biochemistry. 2013;52:3791–3793. doi: 10.1021/bi400520t. [DOI] [PubMed] [Google Scholar]
- 23.Moss S.J., Carletti I., Olano C., Sheridan R.M., Ward M., Math V. Biosynthesis of the angiogenesis inhibitor borrelidin: directed biosynthesis of novel analogues. Chem Commun (Camb) 2006:2341–2343. doi: 10.1039/b602931k. [DOI] [PubMed] [Google Scholar]
- 24.Hagen A., Poust S., de Rond T., Yuzawa S., Katz L., Adams P.D. in vitro analysis of carboxyacyl substrate tolerance in the loading and first extension modules of borrelidin polyketide synthase. Biochemistry. 2014;53:5975–5977. doi: 10.1021/bi500951c. [DOI] [PubMed] [Google Scholar]
- 25.August P.R., Tang L., Yoon Y.J., Ning S., Müller R., Yu T.W. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 26.Schwecke T., Aparicio J.F., Molnár I., König A., Khaw L.E., Haydock S.F. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci U. S. A. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Traitcheva N., Jenke-Kodama H., He J., Dittmann E., Hertweck C. Non-colinear polyketide biosynthesis in the aureothin and neoaureothin pathways: an evolutionary perspective. Chembiochem. 2007;8:1841–1849. doi: 10.1002/cbic.200700309. [DOI] [PubMed] [Google Scholar]
- 28.Trivedi O.A., Arora P., Sridharan V., Tickoo R., Mohanty D., Gokhale R.S. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- 29.Motamedi H., Shafiee A. The biosynthetic gene cluster for the macrolactone ring of the immunosuppressant FK506. Eur J Biochem. 1998;256:528–534. doi: 10.1046/j.1432-1327.1998.2560528.x. [DOI] [PubMed] [Google Scholar]
- 30.Kuhstoss S., Huber M., Turner J.R., Paschal J.W., Rao R.N. Production of a novel polyketide through the construction of a hybrid polyketide synthase. Gene. 1996;183:231–236. doi: 10.1016/s0378-1119(96)00565-3. [DOI] [PubMed] [Google Scholar]
- 31.Marsden A.F., Wilkinson B., Cortés J., Dunster N.J., Staunton J., Leadlay P.F. Engineering broader specificity into an antibiotic-producing polyketide synthase. Science. 1998;279:199–202. doi: 10.1126/science.279.5348.199. [DOI] [PubMed] [Google Scholar]
- 32.Rowe C.J., Cortés J., Gaisser S., Staunton J., Leadlay P.F. Construction of new vectors for high-level expression in actinomycetes. Gene. 1998;216:215–223. doi: 10.1016/s0378-1119(98)00327-8. [DOI] [PubMed] [Google Scholar]
- 33.Brautaset T., Borgos S.E.F., Sletta H., Ellingsen T.E., Zotchev S.B. Site-specific mutagenesis and domain substitutions in the loading module of the nystatin polyketide synthase, and their effects on nystatin biosynthesis in Streptomyces noursei. J Biol Chem. 2003;278:14913–14919. doi: 10.1074/jbc.M212611200. [DOI] [PubMed] [Google Scholar]
- 34.Park S.R., Yoo Y.J., Ban Y.-H., Yoon Y.J. Biosynthesis of rapamycin and its regulation: past achievements and recent progress. J Antibiot. 2010;63:434–441. doi: 10.1038/ja.2010.71. [DOI] [PubMed] [Google Scholar]
- 35.Krastel P., Roggo S., Schirle M., Ross N.T., Perruccio F., Aspesi P. Nannocystin a: an elongation factor 1 inhibitor from myxobacteria with differential anti-cancer properties. Angew Chem Int Ed Engl. 2015;54:10149–10154. doi: 10.1002/anie.201505069. [DOI] [PubMed] [Google Scholar]
- 36.Heia S., Borgos S.E.F., Sletta H., Escudero L., Seco E.M., Malpartida F. Initiation of polyene macrolide biosynthesis: interplay between polyketide synthase domains and modules as revealed via domain swapping, mutagenesis, and heterologous complementation. Appl Environ Microbiol. 2011;77:6982–6990. doi: 10.1128/AEM.05781-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson C.J., Frost E.J., Staunton J., Leadlay P.F. Chain initiation on the soraphen-producing modular polyketide synthase from Sorangium cellulosum. Chem Biol. 2001;8:1197–1208. doi: 10.1016/s1074-5521(01)00087-4. [DOI] [PubMed] [Google Scholar]
- 38.Hunziker D., Yu T.-W., Hutchinson C.R., Floss H.G., Khosla C. Primer unit specificity in rifamycin biosynthesis principally resides in the later stages of the biosynthetic pathway. J Am Chem Soc. 1998;120:1092–1093. [Google Scholar]
- 39.Dunn B.J., Watts K.R., Robbins T., Cane D.E., Khosla C. Comparative analysis of the substrate specificity of trans- versus cis-acyltransferases of assembly line polyketide synthases. Biochemistry. 2014;53:3796–3806. doi: 10.1021/bi5004316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruan X., Pereda A., Stassi D.L., Zeidner D., Summers R.G., Jackson M. Acyltransferase domain substitutions in erythromycin polyketide synthase yield novel erythromycin derivatives. J Bacteriol. 1997;179:6416–6425. doi: 10.1128/jb.179.20.6416-6425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunn B.J., Khosla C. Engineering the acyltransferase substrate specificity of assembly line polyketide synthases. J R Soc Interface. 2013;10:20130297. doi: 10.1098/rsif.2013.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau J., Fu H., Cane D.E., Khosla C. Dissecting the role of acyltransferase domains of modular polyketide synthases in the choice and stereochemical fate of extender units. Biochemistry. 1999;38:1643–1651. doi: 10.1021/bi9820311. [DOI] [PubMed] [Google Scholar]
- 43.Hans M., Hornung A., Dziarnowski A., Cane D.E., Khosla C. Mechanistic analysis of acyl transferase domain exchange in polyketide synthase modules. J Am Chem Soc. 2003;125:5366–5374. doi: 10.1021/ja029539i. [DOI] [PubMed] [Google Scholar]
- 44.Koryakina I., Kasey C., McArthur J.B., Lowell A.N., Chemler J.A., Li S. Inversion of extender unit selectivity in the erythromycin polyketide synthase by acyltransferase domain engineering. ACS Chem Biol. 2017;12:114–123. doi: 10.1021/acschembio.6b00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Musiol-Kroll E.M., Zubeil F., Schafhauser T., Härtner T., Kulik A., McArthur J. Polyketide bioderivatization using the promiscuous acyltransferase kircii. ACS Synth Biol. 2017;6:421–427. doi: 10.1021/acssynbio.6b00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reeves C.D., Murli S., Ashley G.W., Piagentini M., Hutchinson C.R., McDaniel R. Alteration of the substrate specificity of a modular polyketide synthase acyltransferase domain through site-specific mutations. Biochemistry. 2001;40:15464–15470. doi: 10.1021/bi015864r. [DOI] [PubMed] [Google Scholar]
- 47.Chan Y.A., Podevels A.M., Kevany B.M., Thomas M.G. Biosynthesis of polyketide synthase extender units. Nat Prod Rep. 2009;26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang Y., Chen A.Y., Kim C.-Y., Cane D.E., Khosla C. Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase. Chem Biol. 2007;14:931–943. doi: 10.1016/j.chembiol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang Y., Kim C.-Y., Mathews I.I., Cane D.E., Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc Natl Acad Sci U. S. A. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haydock S.F., Aparicio J.F., Molnár I., Schwecke T., Khaw L.E., König A. Divergent sequence motifs correlated with the substrate specificity of (methyl)malonyl-CoA:acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett. 1995;374:246–248. doi: 10.1016/0014-5793(95)01119-y. [DOI] [PubMed] [Google Scholar]
- 51.Petković H., Sandmann A., Challis I.R., Hecht H.-J., Silakowski B., Low L. Substrate specificity of the acyl transferase domains of EpoC from the epothilone polyketide synthase. Org Biomol Chem. 2008;6:500–506. doi: 10.1039/b714804f. [DOI] [PubMed] [Google Scholar]
- 52.Jiang H., Wang Y.-Y., Guo Y.-Y., Shen J.-J., Zhang X.-S., Luo H.-D. An acyltransferase domain of FK506 polyketide synthase recognizing both an acyl carrier protein and coenzyme A as acyl donors to transfer allylmalonyl and ethylmalonyl units. FEBS J. 2015;282:2527–2539. doi: 10.1111/febs.13296. [DOI] [PubMed] [Google Scholar]
- 53.Ray L., Moore B.S. Recent advances in the biosynthesis of unusual polyketide synthase substrates. Nat Prod Rep. 2016;33:150–161. doi: 10.1039/c5np00112a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sundermann U., Bravo-Rodriguez K., Klopries S., Kushnir S., Gomez H., Sanchez-Garcia E. Enzyme-directed mutasynthesis: a combined experimental and theoretical approach to substrate recognition of a polyketide synthase. ACS Chem Biol. 2013;8:443–450. doi: 10.1021/cb300505w. [DOI] [PubMed] [Google Scholar]
- 55.Unable to find information for 911461, (n.d.).
- 56.Reeves C.D., Murli S., Ashley G.W., Piagentini M., Hutchinson C.R., McDaniel R. Alteration of the substrate specificity of a modular polyketide synthase acyltransferase domain through site-specific mutations? Biochemistry. 2001;40:15464–15470. doi: 10.1021/bi015864r. [DOI] [PubMed] [Google Scholar]
- 57.Bravo-Rodriguez K., Klopries S., Koopmans K.R.M., Sundermann U., Yahiaoui S., Arens J. Substrate flexibility of a mutated acyltransferase domain and implications for polyketide biosynthesis. Chem Biol. 2015;22:1425–1430. doi: 10.1016/j.chembiol.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 58.Koryakina I., McArthur J.B., Draelos M.M., Williams G.J. Promiscuity of a modular polyketide synthase towards natural and non-natural extender units. Org Biomol Chem. 2013;11:4449–4458. doi: 10.1039/c3ob40633d. [DOI] [PubMed] [Google Scholar]
- 59.Oliynyk M., Brown M.J., Cortés J., Staunton J., Leadlay P.F. A hybrid modular polyketide synthase obtained by domain swapping. Chem Biol. 1996;3:833–839. doi: 10.1016/s1074-5521(96)90069-1. [DOI] [PubMed] [Google Scholar]
- 60.Liu L., Thamchaipenet A., Fu H., Betlach M., Ashley G. Biosynthesis of 2-Nor-6-deoxyerythronolide B by rationally designed domain substitution. J Am Chem Soc. 1997;119:10553–10554. [Google Scholar]
- 61.Stassi D.L., Kakavas S.J., Reynolds K.A., Gunawardana G., Swanson S., Zeidner D. Ethyl-substituted erythromycin derivatives produced by directed metabolic engineering. Proc Natl Acad Sci U. S. A. 1998;95:7305–7309. doi: 10.1073/pnas.95.13.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petkovic H., Lill R.E., Sheridan R.M., Wilkinson B., McCormick E.L., McArthur H.A.I. A novel erythromycin, 6-desmethyl erythromycin D, made by substituting an acyltransferase domain of the erythromycin polyketide synthase. J Antibiot. 2003;56:543–551. doi: 10.7164/antibiotics.56.543. [DOI] [PubMed] [Google Scholar]
- 63.Revill W.P., Voda J., Reeves C.R., Chung L., Schirmer A., Ashley G. Genetically engineered analogs of ascomycin for nerve regeneration. J Pharmacol Exp Ther. 2002;302:1278–1285. doi: 10.1124/jpet.102.034264. [DOI] [PubMed] [Google Scholar]
- 64.Kato Y., Bai L., Xue Q., Revill W.P., Yu T.-W., Floss H.G. Functional expression of genes involved in the biosynthesis of the novel polyketide chain extension unit, methoxymalonyl-acyl carrier protein, and engineered biosynthesis of 2-desmethyl-2-methoxy-6-deoxyerythronolide B. J Am Chem Soc. 2002;124:5268–5269. doi: 10.1021/ja0127483. [DOI] [PubMed] [Google Scholar]
- 65.Yuzawa S., Kapur S., Cane D.E., Khosla C. Role of a conserved arginine residue in linkers between the ketosynthase and acyltransferase domains of multimodular polyketide synthases. Biochemistry. 2012;51:3708–3710. doi: 10.1021/bi300399u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kapur S., Chen A.Y., Cane D.E., Khosla C. Molecular recognition between ketosynthase and acyl carrier protein domains of the 6-deoxyerythronolide B synthase. Proc Natl Acad Sci U. S. A. 2010;107:22066–22071. doi: 10.1073/pnas.1014081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuzawa S., Deng K., Wang G., Baidoo E.E.K., Northen T.R., Adams P.D. Comprehensive in vitro analysis of acyltransferase domain exchanges in modular polyketide synthases and its application for short-chain ketone production. ACS Synth Biol. 2017;6:139–147. doi: 10.1021/acssynbio.6b00176. [DOI] [PubMed] [Google Scholar]
- 68.Walker M.C., Thuronyi B.W., Charkoudian L.K., Lowry B., Khosla C., Chang M.C.Y. Expanding the fluorine chemistry of living systems using engineered polyketide synthase pathways. Science. 2013;341:1089–1094. doi: 10.1126/science.1242345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ad O., Thuronyi B.W., Chang M.C.Y. Elucidating the mechanism of fluorinated extender unit loading for improved production of fluorine-containing polyketides. Proc Natl Acad Sci U. S. A. 2017;114:E660–E668. doi: 10.1073/pnas.1614196114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valenzano C.R., Lawson R.J., Chen A.Y., Khosla C., Cane D.E. The biochemical basis for stereochemical control in polyketide biosynthesis. J Am Chem Soc. 2009;131:18501–18511. doi: 10.1021/ja908296m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng J., Piasecki S.K., Keatinge-Clay A.T. Structural studies of an A2-type modular polyketide synthase ketoreductase reveal features controlling α-substituent stereochemistry. ACS Chem Biol. 2013;8:1964–1971. doi: 10.1021/cb400161g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie X., Garg A., Keatinge-Clay A.T., Khosla C., Cane D.E. Epimerase and reductase activities of polyketide synthase ketoreductase domains utilize the same conserved tyrosine and serine residues. Biochemistry. 2016;55:1179–1186. doi: 10.1021/acs.biochem.6b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng J., Taylor C.A., Piasecki S.K., Keatinge-Clay A.T. Structural and functional analysis of A-type ketoreductases from the amphotericin modular polyketide synthase. Structure. 2010;18:913–922. doi: 10.1016/j.str.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 74.Keatinge-Clay A.T. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 75.Keatinge-Clay A.T., Stroud R.M. The structure of a ketoreductase determines the organization of the beta-carbon processing enzymes of modular polyketide synthases. Structure. 2006;14:737–748. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 76.Zheng J., Keatinge-Clay A.T. Structural and functional analysis of C2-type ketoreductases from modular polyketide synthases. J Mol Biol. 2011;410:105–117. doi: 10.1016/j.jmb.2011.04.065. [DOI] [PubMed] [Google Scholar]
- 77.Bailey C.B., Pasman M.E., Keatinge-Clay A.T. Substrate structure-activity relationships guide rational engineering of modular polyketide synthase ketoreductases. Chem Commun (Camb) 2016;52:792–795. doi: 10.1039/c5cc07315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reid R., Piagentini M., Rodriguez E., Ashley G., Viswanathan N., Carney J. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry. 2003;42:72–79. doi: 10.1021/bi0268706. [DOI] [PubMed] [Google Scholar]
- 79.Bedford D., Jacobsen J.R., Luo G., Cane D.E., Khosla C. A functional chimeric modular polyketide synthase generated via domain replacement. Chem Biol. 1996;3:827–831. doi: 10.1016/s1074-5521(96)90068-x. [DOI] [PubMed] [Google Scholar]
- 80.McDaniel R., Thamchaipenet A., Gustafsson C., Fu H., Betlach M., Ashley G. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel “unnatural” natural products. Proc Natl Acad Sci U. S. A. 1999;96:1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kao C.M., McPherson M., McDaniel R.N., Fu H., Cane D.E., Khosla C. Alcohol stereochemistry in polyketide backbones is controlled by the β-ketoreductase domains of modular polyketide synthases. J Am Chem Soc. 1998;120:2478–2479. [Google Scholar]
- 82.McDaniel R., Kao C.M., Hwang S.J., Khosla C. Engineered intermodular and intramodular polyketide synthase fusions. Chem Biol. 1997;4:667–674. doi: 10.1016/s1074-5521(97)90222-2. [DOI] [PubMed] [Google Scholar]
- 83.Annaval T., Paris C., Leadlay P.F., Jacob C., Weissman K.J. Evaluating ketoreductase exchanges as a means of rationally altering polyketide stereochemistry. Chembiochem. 2015;16:1357–1364. doi: 10.1002/cbic.201500113. [DOI] [PubMed] [Google Scholar]
- 84.Eng C.H., Yuzawa S., Wang G., Baidoo E.E.K., Katz L., Keasling J.D. Alteration of polyketide stereochemistry from anti to syn by a ketoreductase domain exchange in a type I modular polyketide synthase subunit. Biochemistry. 2016;55:1677–1680. doi: 10.1021/acs.biochem.6b00129. [DOI] [PubMed] [Google Scholar]
- 85.Unable to find information for 912365, (n.d.).
- 86.Kandziora N., Andexer J.N., Moss S.J., Wilkinson B., Leadlay P.F., Hahn F. Uncovering the origin of Z-configured double bonds in polyketides: intermediate E-double bond formation during borrelidin biosynthesis. Chem Sci. 2014;5:3563. [Google Scholar]
- 87.Gay D., You Y.-O., Keatinge-Clay A., Cane D.E. Structure and stereospecificity of the dehydratase domain from the terminal module of the rifamycin polyketide synthase. Biochemistry. 2013;52:8916–8928. doi: 10.1021/bi400988t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alhamadsheh M.M., Palaniappan N., Daschouduri S., Reynolds K.A. Modular polyketide synthases and cis double bond formation: establishment of activated cis-3-cyclohexylpropenoic acid as the diketide intermediate in phoslactomycin biosynthesis. J Am Chem Soc. 2007;129:1910–1911. doi: 10.1021/ja068818t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Keatinge-Clay A. Crystal structure of the erythromycin polyketide synthase dehydratase. J Mol Biol. 2008;384:941–953. doi: 10.1016/j.jmb.2008.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Akey D.L., Razelun J.R., Tehranisa J., Sherman D.H., Gerwick W.H., Smith J.L. Crystal structures of dehydratase domains from the curacin polyketide biosynthetic pathway. Structure. 2010;18:94–105. doi: 10.1016/j.str.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Keatinge-Clay A.T. Stereocontrol within polyketide assembly lines. Nat Prod Rep. 2016;33:141–149. doi: 10.1039/c5np00092k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Caffrey P., Lynch S., Flood E., Finnan S., Oliynyk M. Amphotericin biosynthesis in Streptomyces nodosus: deductions from analysis of polyketide synthase and late genes. Chem Biol. 2001;8:713–723. doi: 10.1016/s1074-5521(01)00046-1. [DOI] [PubMed] [Google Scholar]
- 93.Ikeda H., Nonomiya T., Usami M., Ohta T., Omura S. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc Natl Acad Sci U. S. A. 1999;96:9509–9514. doi: 10.1073/pnas.96.17.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brautaset T., Sekurova O.N., Sletta H., Ellingsen T.E., StrŁm A.R., Valla S. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem Biol. 2000;7:395–403. doi: 10.1016/s1074-5521(00)00120-4. [DOI] [PubMed] [Google Scholar]
- 95.Sun Y., Zhou X., Dong H., Tu G., Wang M., Wang B. A complete gene cluster from Streptomyces nanchangensis NS3226 encoding biosynthesis of the polyether ionophore nanchangmycin. Chem Biol. 2003;10:431–441. doi: 10.1016/s1074-5521(03)00092-9. [DOI] [PubMed] [Google Scholar]
- 96.Zhou Y., Li J., Zhu J., Chen S., Bai L., Zhou X. Incomplete beta-ketone processing as a mechanism for polyene structural variation in the FR-008/candicidin complex. Chem Biol. 2008;15:629–638. doi: 10.1016/j.chembiol.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 97.Brautaset T., Sletta H., Nedal A., Borgos S.E.F., Degnes K.F., Bakke I. Improved antifungal polyene macrolides via engineering of the nystatin biosynthetic genes in Streptomyces noursei. Chem Biol. 2008;15:1198–1206. doi: 10.1016/j.chembiol.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 98.Yong J.-H., Byeon W.-H. Alternative production of avermectin components in Streptomyces avermitilis by gene replacement. J Microbiol. 2005;43:277–284. [PubMed] [Google Scholar]
- 99.Hagen A., Poust S., de Rond T., Fortman J.L., Katz L., Petzold C.J. Engineering a polyketide synthase for in vitro production of adipic acid. ACS Synth Biol. 2016;5:21–27. doi: 10.1021/acssynbio.5b00153. [DOI] [PubMed] [Google Scholar]
- 100.Kwan D.H., Sun Y., Schulz F., Hong H., Popovic B., Sim-Stark J.C.C. Prediction and manipulation of the stereochemistry of enoylreduction in modular polyketide synthases. Chem Biol. 2008;15:1231–1240. doi: 10.1016/j.chembiol.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 101.Zheng J., Gay D.C., Demeler B., White M.A., Keatinge-Clay A.T. Divergence of multimodular polyketide synthases revealed by a didomain structure. Nat Chem Biol. 2012;8:615–621. doi: 10.1038/nchembio.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Unable to find information for 911445, (n.d.).
- 103.Faille A., Gavalda S., Slama N., Lherbet C., Maveyraud L., Guillet V. Insights into substrate modification by dehydratases from type I polyketide synthases. J Mol Biol. 2017;429:1554–1569. doi: 10.1016/j.jmb.2017.03.026. [DOI] [PubMed] [Google Scholar]
- 104.Kushnir S., Sundermann U., Yahiaoui S., Brockmeyer A., Janning P., Schulz F. Minimally invasive mutagenesis gives rise to a biosynthetic polyketide library. Angew Chem Int Ed Engl. 2012;51:10664–10669. doi: 10.1002/anie.201202438. [DOI] [PubMed] [Google Scholar]
- 105.Kellenberger L., Galloway I.S., Sauter G., Böhm G., Hanefeld U., Cortés J. A polylinker approach to reductive loop swaps in modular polyketide synthases. Chembiochem. 2008;9:2740–2749. doi: 10.1002/cbic.200800332. [DOI] [PubMed] [Google Scholar]
- 106.Qi Z., Zhou Y., Kang Q., Jiang C., Zheng J., Bai L. Directed accumulation of less toxic pimaricin derivatives by improving the efficiency of a polyketide synthase dehydratase domain. Appl Microbiol Biotechnol. 2017;101:2427–2436. doi: 10.1007/s00253-016-8074-7. [DOI] [PubMed] [Google Scholar]
- 107.Zhang X., Chen Z., Li M., Wen Y., Song Y., Li J. Construction of ivermectin producer by domain swaps of avermectin polyketide synthase in Streptomyces avermitilis. Appl Microbiol Biotechnol. 2006;72:986–994. doi: 10.1007/s00253-006-0361-2. [DOI] [PubMed] [Google Scholar]
- 108.Pinto A., Wang M., Horsman M., Boddy C.N. 6-Deoxyerythronolide B synthase thioesterase-catalyzed macrocyclization is highly stereoselective. Org Lett. 2012;14:2278–2281. doi: 10.1021/ol300707j. [DOI] [PubMed] [Google Scholar]
- 109.Unable to find information for 911689, (n.d.).
- 110.Kao C.M., Luo G., Katz L., Cane D.E., Khosla C. Engineered biosynthesis of structurally diverse tetraketides by a trimodular polyketide synthase. J Am Chem Soc. 1996;118:9184–9185. [Google Scholar]
- 111.Kao C.M., McPherson M., McDaniel R.N., Fu H., Cane D.E., Khosla C. Gain of function mutagenesis of the erythromycin polyketide synthase. 2. Engineered biosynthesis of an eight-membered ring tetraketide lactone. J Am Chem Soc. 1997;119:11339–11340. [Google Scholar]
- 112.Sharma K.K., Boddy C.N. The thioesterase domain from the pimaricin and erythromycin biosynthetic pathways can catalyze hydrolysis of simple thioester substrates. Bioorg Med Chem Lett. 2007;17:3034–3037. doi: 10.1016/j.bmcl.2007.03.060. [DOI] [PubMed] [Google Scholar]
- 113.Harper A.D., Bailey C.B., Edwards A.D., Detelich J.F., Keatinge-Clay A.T. Preparative, in vitro biocatalysis of triketide lactone chiral building blocks. Chembiochem. 2012;13:2200–2203. doi: 10.1002/cbic.201200378. [DOI] [PubMed] [Google Scholar]
- 114.Akey D.L., Kittendorf J.D., Giraldes J.W., Fecik R.A., Sherman D.H., Smith J.L. Structural basis for macrolactonization by the pikromycin thioesterase. Nat Chem Biol. 2006;2:537–542. doi: 10.1038/nchembio824. [DOI] [PubMed] [Google Scholar]
- 115.Tripathi A., Choi S.-S., Sherman D.H., Kim E.-S. Thioesterase domain swapping of a linear polyketide tautomycetin with a macrocyclic polyketide pikromycin in Streptomyces sp. CK4412. J Ind Microbiol Biotechnol. 2016;43:1189–1193. doi: 10.1007/s10295-016-1790-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu T., You D., Valenzano C., Sun Y., Li J., Yu Q. Identification of NanE as the thioesterase for polyether chain release in nanchangmycin biosynthesis. Chem Biol. 2006;13:945–955. doi: 10.1016/j.chembiol.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 117.Liu T., Lin X., Zhou X., Deng Z., Cane D.E. Mechanism of thioesterase-catalyzed chain release in the biosynthesis of the polyether antibiotic nanchangmycin. Chem Biol. 2008;15:449–458. doi: 10.1016/j.chembiol.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Harvey B.M., Hong H., Jones M.A., Hughes-Thomas Z.A., Goss R.M., Heathcote M.L. Evidence that a novel thioesterase is responsible for polyketide chain release during biosynthesis of the polyether ionophore monensin. Chembiochem. 2006;7:1435–1442. doi: 10.1002/cbic.200500474. [DOI] [PubMed] [Google Scholar]
- 119.Gehret J.J., Gu L., Gerwick W.H., Wipf P., Sherman D.H., Smith J.L. Terminal alkene formation by the thioesterase of curacin A biosynthesis: structure of a decarboxylating thioesterase. J Biol Chem. 2011;286:14445–14454. doi: 10.1074/jbc.M110.214635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gu L., Wang B., Kulkarni A., Gehret J.J., Lloyd K.R., Gerwick L. Polyketide decarboxylative chain termination preceded by o-sulfonation in curacin a biosynthesis. J Am Chem Soc. 2009;131:16033–16035. doi: 10.1021/ja9071578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dong Y., Matson J.B., Edgar K.J. Olefin cross-metathesis in polymer and polysaccharide chemistry: a review. Biomacromolecules. 2017;18:1661–1676. doi: 10.1021/acs.biomac.7b00364. [DOI] [PubMed] [Google Scholar]
- 122.Ciardiello J.J., Galloway W.R.J.D., O'Connor C.J., Sore H.F., Stokes J.E., Wu Y. An expedient strategy for the diversity-oriented synthesis of macrocyclic compounds with natural product-like characteristics. Tetrahedron. 2016;72:3567–3578. [Google Scholar]
- 123.Kavanagh K.L., Jörnvall H., Persson B., Oppermann U. Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci. 2008;65:3895–3906. doi: 10.1007/s00018-008-8588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wyatt M.A., Mok M.C.Y., Junop M., Magarvey N.A. Heterologous expression and structural characterisation of a pyrazinone natural product assembly line. Chembiochem. 2012;13:2408–2415. doi: 10.1002/cbic.201200340. [DOI] [PubMed] [Google Scholar]
- 125.Chhabra A., Haque A.S., Pal R.K., Goyal A., Rai R., Joshi S. Nonprocessive [2 + 2]e- off-loading reductase domains from mycobacterial nonribosomal peptide synthetases. Proc Natl Acad Sci U. S. A. 2012;109:5681–5686. doi: 10.1073/pnas.1118680109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Silakowski B., Nordsiek G., Kunze B., Blöcker H., Müller R. Novel features in a combined polyketide synthase/non-ribosomal peptide synthetase: the myxalamid biosynthetic gene cluster of the myxobacterium Stigmatella aurantiaca Sga1511This article is dedicated to Prof. Dr. E. Leistner on the occasion of his 60th birthday. Chem Biol. 2001;8:59–69. doi: 10.1016/s1074-5521(00)00056-9. [DOI] [PubMed] [Google Scholar]
- 127.Barajas J.F., Phelan R.M., Schaub A.J., Kliewer J.T., Kelly P.J., Jackson D.R. Comprehensive structural and biochemical analysis of the terminal myxalamid reductase domain for the engineered production of primary alcohols. Chem Biol. 2015;22:1018–1029. doi: 10.1016/j.chembiol.2015.06.022. [DOI] [PubMed] [Google Scholar]
- 128.Jin C., Yao M., Liu H., Lee C.F., Ji J. Progress in the production and application of n-butanol as a biofuel. Renew Sustain Energy Rev. 2011;15:4080–4106. [Google Scholar]
- 129.Dorival J., Annaval T., Risser F., Collin S., Roblin P., Jacob C. Characterization of intersubunit communication in the virginiamycin trans-acyl transferase polyketide synthase. J Am Chem Soc. 2016;138:4155–4167. doi: 10.1021/jacs.5b13372. [DOI] [PubMed] [Google Scholar]
- 130.Whicher J.R., Smaga S.S., Hansen D.A., Brown W.C., Gerwick W.H., Sherman D.H. Cyanobacterial polyketide synthase docking domains: a tool for engineering natural product biosynthesis. Chem Biol. 2013;20:1340–1351. doi: 10.1016/j.chembiol.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Buchholz T.J., Geders T.W., Bartley F.E., Reynolds K.A., Smith J.L., Sherman D.H. Structural basis for binding specificity between subclasses of modular polyketide synthase docking domains. ACS Chem Biol. 2009;4:41–52. doi: 10.1021/cb8002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gokhale R.S., Tsuji S.Y., Cane D.E., Khosla C. Dissecting and exploiting intermodular communication in polyketide synthases. Science. 1999;284:482–485. doi: 10.1126/science.284.5413.482. [DOI] [PubMed] [Google Scholar]
- 133.Wu N., Cane D.E., Khosla C. Quantitative analysis of the relative contributions of donor acyl carrier proteins, acceptor ketosynthases, and linker regions to intermodular transfer of intermediates in hybrid polyketide synthases. Biochemistry. 2002;41:5056–5066. doi: 10.1021/bi012086u. [DOI] [PubMed] [Google Scholar]
- 134.Menzella H.G., Reid R., Carney J.R., Chandran S.S., Reisinger S.J., Patel K.G. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat Biotechnol. 2005;23:1171–1176. doi: 10.1038/nbt1128. [DOI] [PubMed] [Google Scholar]
- 135.Klaus M., Ostrowski M.P., Austerjost J., Robbins T., Lowry B., Cane D.E. Protein-protein interactions, not substrate recognition, dominate the turnover of chimeric assembly line polyketide synthases. J Biol Chem. 2016;291:16404–16415. doi: 10.1074/jbc.M116.730531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kennedy J. Mutasynthesis, chemobiosynthesis, and back to semi-synthesis: combining synthetic chemistry and biosynthetic engineering for diversifying natural products. Nat Prod Rep. 2008;25:25–34. doi: 10.1039/b707678a. [DOI] [PubMed] [Google Scholar]
- 137.Mehlhorn H., Jones H.L., Weatherley A.J., Schumacher B. Doramectin, a new avermectin highly efficacious against gastrointestinal nematodes and lungworms of cattle and pigs: two studies carried out under field conditions in Germany. Parasitol Res. 1993;79:603–607. doi: 10.1007/BF00932246. [DOI] [PubMed] [Google Scholar]
- 138.Cropp T.A., Wilson D.J., Reynolds K.A. Identification of a cyclohexylcarbonyl CoA biosynthetic gene cluster and application in the production of doramectin. Nat Biotechnol. 2000;18:980–983. doi: 10.1038/79479. [DOI] [PubMed] [Google Scholar]
- 139.Zhao X., Wang Y., Wang S., Chen Z., Wen Y., Song Y. Construction of a doramectin producer mutant from an avermectin-overproducing industrial strain of Streptomyces avermitilis. Can J Microbiol. 2009;55:1355–1363. doi: 10.1139/W09-098. [DOI] [PubMed] [Google Scholar]
- 140.Werneburg M., Busch B., He J., Richter M.E.A., Xiang L., Moore B.S. Exploiting enzymatic promiscuity to engineer a focused library of highly selective antifungal and antiproliferative aureothin analogues. J Am Chem Soc. 2010;132:10407–10413. doi: 10.1021/ja102751h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lowden P.A.S., Böhm G.A., Metcalfe S., Staunton J., Leadlay P.F. New rapamycin derivatives by precursor-directed biosynthesis. Chembiochem. 2004;5:535–538. doi: 10.1002/cbic.200300758. [DOI] [PubMed] [Google Scholar]
- 142.Sheehan L.S., Lill R.E., Wilkinson B., Sheridan R.M., Vousden W.A., Kaja A.L. Engineering of the spinosyn PKS: directing starter unit incorporation. J Nat Prod. 2006;69:1702–1710. doi: 10.1021/np0602517. [DOI] [PubMed] [Google Scholar]
- 143.Lee H.Y., Harvey C.J.B., Cane D.E., Khosla C. Improved precursor-directed biosynthesis in E. coli via directed evolution. J Antibiot. 2011;64:59–64. doi: 10.1038/ja.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kibwage I.O., Janssen G., Busson R., Hoogmartens J., Vanderhaeghe H., Verbist L. Identification of novel erythromycin derivatives in mother liquor concentrates of Streptomyces erythraeus. J Antibiot. 1987;40:1–6. doi: 10.7164/antibiotics.40.1. [DOI] [PubMed] [Google Scholar]
- 145.Oliynyk M., Stark C.B.W., Bhatt A., Jones M.A., Hughes-Thomas Z.A., Wilkinson C. Analysis of the biosynthetic gene cluster for the polyether antibiotic monensin in Streptomyces cinnamonensis and evidence for the role of monB and monC genes in oxidative cyclization. Mol Microbiol. 2003;49:1179–1190. doi: 10.1046/j.1365-2958.2003.03571.x. [DOI] [PubMed] [Google Scholar]
- 146.Bhatt A., Stark C.B.W., Harvey B.M., Gallimore A.R., Demydchuk Y.A., Spencer J.B. Accumulation of an E, E,E-triene by the monensin-producing polyketide synthase when oxidative cyclization is blocked. Angew Chem Int Ed Engl. 2005;44:7075–7078. doi: 10.1002/anie.200501757. [DOI] [PubMed] [Google Scholar]
- 147.Bravo-Rodriguez K., Ismail-Ali A.F., Klopries S., Kushnir S., Ismail S., Fansa E.K. Predicted incorporation of non-native substrates by a polyketide synthase yields bioactive natural product derivatives. Chembiochem. 2014;15:1991–1997. doi: 10.1002/cbic.201402206. [DOI] [PubMed] [Google Scholar]
- 148.Eustáquio A.S., McGlinchey R.P., Liu Y., Hazzard C., Beer L.L., Florova G. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc Natl Acad Sci U. S. A. 2009;106:12295–12300. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Eustáquio A.S., Moore B.S. Mutasynthesis of fluorosalinosporamide, a potent and reversible inhibitor of the proteasome. Angew Chem Int Ed Engl. 2008;47:3936–3938. doi: 10.1002/anie.200800177. [DOI] [PubMed] [Google Scholar]
- 150.Eustáquio A.S., O'Hagan D., Moore B.S. Engineering fluorometabolite production: fluorinase expression in Salinispora tropica Yields Fluorosalinosporamide. J Nat Prod. 2010;73:378–382. doi: 10.1021/np900719u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ye Z., Musiol E.M., Weber T., Williams G.J. Reprogramming acyl carrier protein interactions of an Acyl-CoA promiscuous trans-acyltransferase. Chem Biol. 2014;21:636–646. doi: 10.1016/j.chembiol.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Koryakina I., McArthur J., Randall S., Draelos M.M., Musiol E.M., Muddiman D.C. Poly specific trans-acyltransferase machinery revealed via engineered acyl-CoA synthetases. ACS Chem Biol. 2013;8:200–208. doi: 10.1021/cb3003489. [DOI] [PubMed] [Google Scholar]
- 153.Koryakina I., Williams G.J. Mutant malonyl-CoA synthetases with altered specificity for polyketide synthase extender unit generation. Chembiochem. 2011;12:2289–2293. doi: 10.1002/cbic.201100383. [DOI] [PubMed] [Google Scholar]
- 154.Rokem J.S., Lantz A.E., Nielsen J. Systems biology of antibiotic production by microorganisms. Nat Prod Rep. 2007;24:1262–1287. doi: 10.1039/b617765b. [DOI] [PubMed] [Google Scholar]
- 155.Olano C., Lombó F., Méndez C., Salas J.A. Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng. 2008;10:281–292. doi: 10.1016/j.ymben.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 156.Li R., Townsend C.A. Rational strain improvement for enhanced clavulanic acid production by genetic engineering of the glycolytic pathway in Streptomyces clavuligerus. Metab Eng. 2006;8:240–252. doi: 10.1016/j.ymben.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 157.Zabala D., Braña A.F., Flórez A.B., Salas J.A., Méndez C. Engineering precursor metabolite pools for increasing production of antitumor mithramycins in Streptomyces argillaceus. Metab Eng. 2013;20:187–197. doi: 10.1016/j.ymben.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 158.Zhang H., Boghigian B.A., Pfeifer B.A. Investigating the role of native propionyl-CoA and methylmalonyl-CoA metabolism on heterologous polyketide production in Escherichia coli. Biotechnol Bioeng. 2010;105:567–573. doi: 10.1002/bit.22560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vandova G.A., O'Brien R.V., Lowry B., Robbins T.F., Fischer C.R., Davis R.W. Heterologous expression of diverse propionyl-CoA carboxylases affects polyketide production in Escherichia coli. J Antibiot. 2017 doi: 10.1038/ja.2017.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Arabolaza A., Shillito M.E., Lin T.-W., Diacovich L., Melgar M., Pham H. Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor. Biochemistry. 2010;49:7367–7376. doi: 10.1021/bi1005305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ray L., Valentic T.R., Miyazawa T., Withall D.M., Song L., Milligan J.C. A crotonyl-CoA reductase-carboxylase independent pathway for assembly of unusual alkylmalonyl-CoA polyketide synthase extender units. Nat Commun. 2016;7:13609. doi: 10.1038/ncomms13609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rodriguez E., Ward S., Fu H., Revill W.P., McDaniel R., Katz L. Engineered biosynthesis of 16-membered macrolides that require methoxymalonyl-ACP precursors in Streptomyces fradiae. Appl Microbiol Biotechnol. 2004;66:85–91. doi: 10.1007/s00253-004-1658-7. [DOI] [PubMed] [Google Scholar]
- 163.Lu C., Zhang X., Jiang M., Bai L. Enhanced salinomycin production by adjusting the supply of polyketide extender units in Streptomyces albus. Metab Eng. 2016;35:129–137. doi: 10.1016/j.ymben.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 164.Gross F., Ring M.W., Perlova O., Fu J., Schneider S., Gerth K. Metabolic engineering of Pseudomonas putida for methylmalonyl-CoA biosynthesis to enable complex heterologous secondary metabolite formation. Chem Biol. 2006;13:1253–1264. doi: 10.1016/j.chembiol.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 165.Bunet R., Riclea R., Laureti L., Hôtel L., Paris C., Girardet J.-M. A single Sfp-type phosphopantetheinyl transferase plays a major role in the biosynthesis of PKS and NRPS derived metabolites in Streptomyces ambofaciens ATCC23877. PLoS ONE. 2014;9:e87607. doi: 10.1371/journal.pone.0087607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen X.-H., Vater J., Piel J., Franke P., Scholz R., Schneider K. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J Bacteriol. 2006;188:4024–4036. doi: 10.1128/JB.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Volokhan O., Sletta H., Sekurova O.N., Ellingsen T.E., Zotchev S.B. An unexpected role for the putative 4'-phosphopantetheinyl transferase-encoding gene nysF in the regulation of nystatin biosynthesis in Streptomyces noursei ATCC 11455. FEMS Microbiol Lett. 2005;249:57–64. doi: 10.1016/j.femsle.2005.05.052. [DOI] [PubMed] [Google Scholar]
- 168.Beld J., Sonnenschein E.C., Vickery C.R., Noel J.P., Burkart M.D. The phosphopantetheinyl transferases: catalysis of a post-translational modification crucial for life. Nat Prod Rep. 2014;31:61–108. doi: 10.1039/c3np70054b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Quadri L.E., Weinreb P.H., Lei M., Nakano M.M., Zuber P., Walsh C.T. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 170.Peypoux F., Bonmatin J.M., Wallach J. Recent trends in the biochemistry of surfactin. Appl Microbiol Biotechnol. 1999;51:553–563. doi: 10.1007/s002530051432. [DOI] [PubMed] [Google Scholar]
- 171.Pfeifer B.A., Admiraal S.J., Gramajo H., Cane D.E., Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 172.Murli S., Kennedy J., Dayem L.C., Carney J.R., Kealey J.T. Metabolic engineering of Escherichia coli for improved 6-deoxyerythronolide B production. J Ind Microbiol Biotechnol. 2003;30:500–509. doi: 10.1007/s10295-003-0073-x. [DOI] [PubMed] [Google Scholar]
- 173.Mutka S.C., Bondi S.M., Carney J.R., Da Silva N.A., Kealey J.T. Metabolic pathway engineering for complex polyketide biosynthesis in Saccharomyces cerevisiae. FEMS Yeast Res. 2006;6:40–47. doi: 10.1111/j.1567-1356.2005.00001.x. [DOI] [PubMed] [Google Scholar]
- 174.Kealey J.T., Liu L., Santi D.V., Betlach M.C., Barr P.J. Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc Natl Acad Sci U. S. A. 1998;95:505–509. doi: 10.1073/pnas.95.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mootz H.D., Schörgendorfer K., Marahiel M.A. Functional characterization of 4'-phosphopantetheinyl transferase genes of bacterial and fungal origin by complementation of Saccharomyces cerevisiae lys5. FEMS Microbiol Lett. 2002;213:51–57. doi: 10.1111/j.1574-6968.2002.tb11285.x. [DOI] [PubMed] [Google Scholar]
- 176.Zobel S., Kumpfmüller J., Süssmuth R.D., Schweder T. Bacillus subtilis as heterologous host for the secretory production of the non-ribosomal cyclodepsipeptide enniatin. Appl Microbiol Biotechnol. 2015;99:681–691. doi: 10.1007/s00253-014-6199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kumpfmüller J., Methling K., Fang L., Pfeifer B.A., Lalk M., Schweder T. Production of the polyketide 6-deoxyerythronolide B in the heterologous host Bacillus subtilis. Appl Microbiol Biotechnol. 2016;100:1209–1220. doi: 10.1007/s00253-015-6990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sánchez C., Du L., Edwards D.J., Toney M.D., Shen B. Cloning and characterization of a phosphopantetheinyl transferase from Streptomyces verticillus ATCC15003, the producer of the hybrid peptide-polyketide antitumor drug bleomycin. Chem Biol. 2001;8:725–738. doi: 10.1016/s1074-5521(01)00047-3. [DOI] [PubMed] [Google Scholar]
- 179.Baltz R.H. Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol. 2010;37:759–772. doi: 10.1007/s10295-010-0730-9. [DOI] [PubMed] [Google Scholar]
- 180.Jiang H., Wang Y.-Y., Ran X.-X., Fan W.-M., Jiang X.-H., Guan W.-J. Improvement of natamycin production by engineering of phosphopantetheinyl transferases in Streptomyces chattanoogensis L10. Appl Environ Microbiol. 2013;79:3346–3354. doi: 10.1128/AEM.00099-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang B., Tian W., Wang S., Yan X., Jia X., Pierens G.K. Activation of natural products biosynthetic pathways via a protein modification level regulation. ACS Chem Biol. 2017;12:1732–1736. doi: 10.1021/acschembio.7b00225. [DOI] [PubMed] [Google Scholar]
- 182.Stratigopoulos G., Gandecha A.R., Cundliffe E. Regulation of tylosin production and morphological differentiation in Streptomyces fradiae by TylP, a deduced gamma-butyrolactone receptor. Mol Microbiol. 2002;45:735–744. doi: 10.1046/j.1365-2958.2002.03044.x. [DOI] [PubMed] [Google Scholar]
- 183.Stratigopoulos G., Bate N., Cundliffe E. Positive control of tylosin biosynthesis: pivotal role of TylR. Mol Microbiol. 2004;54:1326–1334. doi: 10.1111/j.1365-2958.2004.04347.x. [DOI] [PubMed] [Google Scholar]
- 184.Garcia-Bernardo J., Braña A.F., Méndez C., Salas J.A. Insertional inactivation of mtrX and mtrY genes from the mithramycin gene cluster affects production and growth of the producer organism Streptomyces argillaceus. FEMS Microbiol Lett. 2000;186:61–65. doi: 10.1111/j.1574-6968.2000.tb09082.x. [DOI] [PubMed] [Google Scholar]
- 185.Lombó F., Braña A.F., Méndez C., Salas J.A. The mithramycin gene cluster of Streptomyces argillaceus contains a positive regulatory gene and two repeated DNA sequences that are located at both ends of the cluster. J Bacteriol. 1999;181:642–647. doi: 10.1128/jb.181.2.642-647.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Song K., Wei L., Liu J., Wang J., Qi H., Wen J. Engineering of the LysR family transcriptional regulator FkbR1 and its target gene to improve ascomycin production. Appl Microbiol Biotechnol. 2017;101:4581–4592. doi: 10.1007/s00253-017-8242-4. [DOI] [PubMed] [Google Scholar]
- 187.Stratigopoulos G., Cundliffe E. Expression analysis of the tylosin-biosynthetic gene cluster: pivotal regulatory role of the tylQ product. Chem Biol. 2002;9:71–78. doi: 10.1016/s1074-5521(01)00095-3. [DOI] [PubMed] [Google Scholar]
- 188.Shao Z., Rao G., Li C., Abil Z., Luo Y., Zhao H. Refactoring the silent spectinabilin gene cluster using a plug-and-play scaffold. ACS Synth Biol. 2013;2:662–669. doi: 10.1021/sb400058n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Basitta P., Westrich L., Rösch M., Kulik A., Gust B., Apel A.K. AGOS: a plug-and-play method for the assembly of artificial gene operons into functional biosynthetic gene clusters. ACS Synth Biol. 2017;6:817–825. doi: 10.1021/acssynbio.6b00319. [DOI] [PubMed] [Google Scholar]
- 190.Tan G.-Y., Deng K., Liu X., Tao H., Chang Y., Chen J. Heterologous biosynthesis of spinosad: an omics-guided large polyketide synthase gene cluster reconstitution in Streptomyces. ACS Synth Biol. 2017;6:995–1005. doi: 10.1021/acssynbio.6b00330. [DOI] [PubMed] [Google Scholar]
- 191.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 192.Zhang M.M., Wang Y., Ang E.L., Zhao H. Engineering microbial hosts for production of bacterial natural products. Nat Prod Rep. 2016;33:963–987. doi: 10.1039/c6np00017g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Huang H., Zheng G., Jiang W., Hu H., Lu Y. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin (Shanghai) 2015;47:231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 194.Cobb R.E., Wang Y., Zhao H. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth Biol. 2015;4:723–728. doi: 10.1021/sb500351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang M.M., Wong F.T., Wang Y., Luo S., Lim Y.H., Heng E. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat Chem Biol. 2017 doi: 10.1038/nchembio.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Khosla C., Tang Y., Chen A.Y., Schnarr N.A., Cane D.E. Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu Rev Biochem. 2007;76:195–221. doi: 10.1146/annurev.biochem.76.053105.093515. [DOI] [PubMed] [Google Scholar]
- 197.Yan J., Hazzard C., Bonnett S.A., Reynolds K.A. Functional modular dissection of DEBS1-TE changes triketide lactone ratios and provides insight into Acyl group loading, hydrolysis, and ACP transfer. Biochemistry. 2012;51:9333–9341. doi: 10.1021/bi300830q. [DOI] [PubMed] [Google Scholar]
- 198.Kao C.M., Katz L., Khosla C. Engineered biosynthesis of a complete macrolactone in a heterologous host. Science. 1994;265:509–512. doi: 10.1126/science.8036492. [DOI] [PubMed] [Google Scholar]
- 199.Hertweck C. Decoding and reprogramming complex polyketide assembly lines: prospects for synthetic biology. Trends Biochem Sci. 2015;40:189–199. doi: 10.1016/j.tibs.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 200.Licona-Cassani C., Cruz-Morales P., Manteca A., Barona-Gomez F., Nielsen L.K., Marcellin E. Systems biology approaches to understand natural products biosynthesis. Front Bioeng Biotechnol. 2015;3:199. doi: 10.3389/fbioe.2015.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pérez-Victoria I., Martín J., Reyes F. Combined LC/UV/MS and NMR strategies for the dereplication of marine natural products. Planta Med. 2016;82:857–871. doi: 10.1055/s-0042-101763. [DOI] [PubMed] [Google Scholar]
- 202.Covington B.C., McLean J.A., Bachmann B.O. Comparative mass spectrometry-based metabolomics strategies for the investigation of microbial secondary metabolites. Nat Prod Rep. 2017;34:6–24. doi: 10.1039/c6np00048g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hoffmann T., Krug D., Hüttel S., Müller R. Improving natural products identification through targeted LC-MS/MS in an untargeted secondary metabolomics workflow. Anal Chem. 2014;86:10780–10788. doi: 10.1021/ac502805w. [DOI] [PubMed] [Google Scholar]
- 204.Rogers J.K., Guzman C.D., Taylor N.D., Raman S., Anderson K., Church G.M. Synthetic biosensors for precise gene control and real-time monitoring of metabolites. Nucleic Acids Res. 2015;43:7648–7660. doi: 10.1093/nar/gkv616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Tremblay N., Hill P., Conway K.R., Boddy C.N. The use of clustermine360 for the analysis of polyketide and nonribosomal peptide biosynthetic pathways. Methods Mol Biol. 2016;1401:233–252. doi: 10.1007/978-1-4939-3375-4_15. [DOI] [PubMed] [Google Scholar]
- 206.Khater S., Gupta M., Agrawal P., Sain N., Prava J., Gupta P. SBSPKSv2: structure-based sequence analysis of polyketide synthases and non-ribosomal peptide synthetases. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ichikawa N., Sasagawa M., Yamamoto M., Komaki H., Yoshida Y., Yamazaki S. DoBISCUIT: a database of secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2013;41:D408–D414. doi: 10.1093/nar/gks1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Blin K., Wolf T., Chevrette M.G., Lu X., Schwalen C.J., Kautsar S.A. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Conway K.R., Boddy C.N. ClusterMine360: a database of microbial PKS/NRPS biosynthesis. Nucleic Acids Res. 2013;41:D402–D407. doi: 10.1093/nar/gks993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K. Minimum information about a biosynthetic gene cluster. Nat Chem Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]