Abstract
Derived from the bacterial adaptive immune system, CRISPR technology has revolutionized conventional genetic engineering methods and unprecedentedly facilitated strain engineering. In this review, we outline the fundamental CRISPR tools that have been employed for strain optimization. These tools include CRISPR editing, CRISPR interference, CRISPR activation and protein imaging. To further characterize the CRISPR technology, we present current applications of these tools in microbial systems, including model- and non-model industrial microorganisms. Specially, we point out the major challenges of the CRISPR tools when utilized for multiplex genome editing and sophisticated expression regulation. To address these challenges, we came up with strategies that place emphasis on the amelioration of DNA repair efficiency through CRISPR-Cas9-assisted recombineering. Lastly, multiple promising research directions were proposed, mainly focusing on CRISPR-based construction of microbial ecosystems toward high production of desired chemicals.
Keywords: CRISPR-Cas9, CRISPR interference, CRISPR activation, DNA repair, Homologous recombination
1. Introduction
One central goal of microbial metabolic engineering and synthetic biology is to overproduce intended metabolites including green chemicals and fuels. To this end, genome editing and expression control are performed by recruiting a series of genetic tools, including RecA-, Red-based homologous recombination, zinc-finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN) [1], transposon mutagenesis [2], RNA interference (RNAi) [3] and antisense RNA approaches [4]. Implementation of these approaches is laborious and time-consuming compared to the prevailing CRISPR-Cas9 technology. The emergence of CRISPR-Cas9 tools has augmented our ability to sculpt the genome and program the gene expression of model industrial microorganisms, such as Saccharomyces cerevisiae [5]and Escherichia coli [6]. However, the growing interest in other microorganisms, whereby genome editing options remain limited, necessitates further research and development of such engineering tools. In this review, we portray a picture of CRISPR-Cas9-based tools for genome editing and transcription regulation of both model and non-model microbes. Additionally we propose strategies for further improvement and expansion of these tools toward overproduction of desired chemicals.
2. Mechanisms of CRISPR-Cas9 tools
CRISPR (clustered regularly interspaced short palindromic repeats) technology was derived from the immune systems in bacteria and archaea such as Streptococcus pyogenes [7] and Staphylococcus epidermidis [8]. For naturally occurring CRISPR systems, invasive DNA is integrated into a tandem array at the CRISPR locus. This locus can be transcribed and processed into a CRISPR RNA (crRNA). In type II CRISPR system, the crRNA constitutes a complex with transactivating CRISPR RNA (tracrRNA) and CRISPR-associated protein Cas9. The crRNA directs Cas9-RNA complex to its target DNA, and Cas9 causes a double-strand break (DSB). Unlike type II CRISPR system that needs only Cas9 as endonuclease, the type I and type III CRISPR systems employ a large complex of Cas proteins for crRNA-guided targeting [9]. This attribute facilitates genetic engineering applications. The S. pyogenes tracrRNA and crRNA can be replaced by a single RNA molecule designated guide RNA (gRNA). To ensure recognition by Cas9-RNA complex, a protospacer adjacent motif (PAM) immediately downstream of the gRNA target sequence in the genome is required. The PAM sequence varies depending on Cas9 protein, and in S. pyogenes, the PAM is NGG.
The S. pyogenes type II prokaryotic CRISPR-Cas adaptive immune system has been demonstrated to execute RNA-guided site-specific DNA cleavage [9]. In the CRISPR-Cas9 system, Cas9 is an RNA-guided double-strand DNA nuclease, and a single guide RNA (sgRNA) directs Cas9 to trigger specific DSBs. Mutations H840A and D10A in the HNH and RuvC domains in dCas9, respectively, abolish cleavage (dCas9) but retain DNA-binding activity. The dCas9 in conjunction with sgRNA has been developed as gene interference (CRISPRi) and activation (CRISPRa) tools [10], [11], [12], [13], [14] (Fig. 1). To date, CRISPR-Cas tools have been widely applied to edit or regulate genes in both bacteria such as Lactobacillus reuteri [15], E. coli [16], [17], [18], Streptococcus pneumoniae [17], Bacillus subtilis [19] and Streptomyces [20], and fungi such as S. cerevisiae [5], [21], Yarrowia lipolytica [22], Talaromyces atroroseus [23] and Aspergillus niger [24], [25].
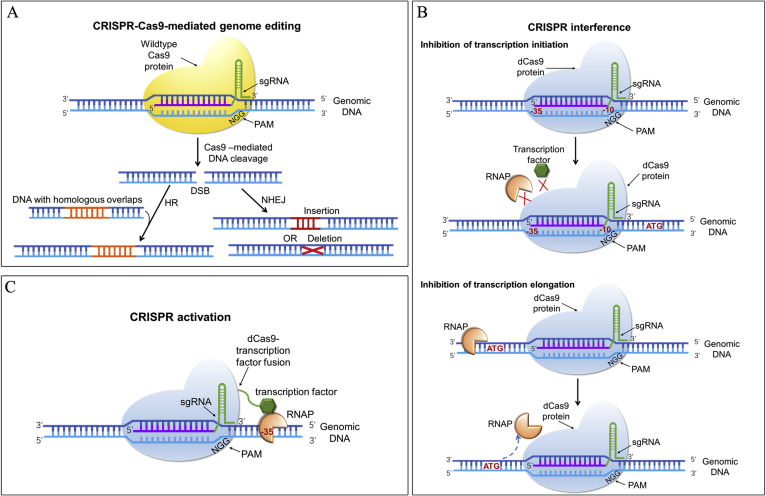
Fig. 1.
CRISPR tools for genome editing, CRISPR interference and CRISPR activation. (A) CRISPR-Cas9-mediated genome editing. Cas9 protein complexes with sgRNA and binds the target site of genomic DNA, creating a double-strand break (DSB) at 3 or 4 nucleotides upstream of the PAM sequence. DSB is repaired by Non-homologous end joining (NHEJ) or Homologous recombination (HR). In NHEJ, random insertions and deletions are introduced into the genome. In HR, precise mutations are integrated into the target genomic location by providing a donor sequence that has homology arms with the DSB site. (B) CRISPR interference. There are two ways to silence gene expression. The dCas9-sgRNA complex targets the promoter or enhancer sequence to block the RNAP and/or transcription factor, the transcription initiation is inhibited. The dCas9-sgRNA complex targets the gene sequence or its 5′ UTR sequence to prevent the transcription elongation. (C) CRISPR activation. Catalytically inactive Cas9 (dCas9) is fused with a transcription factor, which is targeted to upstream of the target gene and delivers the transcription factor to the promoter, which facilitates the combination of RNA polymerase (RNAP) and transcription factor for enhancing the transcriptional efficiency.
2.1. CRISPR-Cas9-mediated genome editing
One main application of CRISPR-Cas9 tools is DNA cleavage (Fig. 1A). Cas9 is directed by a sgRNA to induce precise DNA cleavage at endogenous genomic loci. Cas9 can also be mutated into a nicking enzyme to participate in homology-directed repair with minimal mutagenic activity [26]. Specially, multiple guide sequences can be aggregated as a single CRISPR array to enable simultaneous editing of multiple chromosomal sites [5]. Presumably, this powerful tool accelerates strain evolution. For example, the mevalonate titer in S. cerevisiae was elevated by 41-fold compared to the wild-type strain by simultaneously targeting five chromosomal sites [21], thereby demonstrating the enormous potential of CRISPR-Cas9 technology in strain engineering.
2.2. CRISPR-dCas9-mediated expression control
In addition to DNA cleavage, CRISPR-Cas9 tools exhibit functions of transcription control, including CRISPRi [10] (Fig. 1B) and CRISPRa (Fig. 1C) [11]. The dCas9-sgRNA complex represses gene expression when dCas9 binds a promoter or an open reading frame (ORF) [10], [11]. However, this complex activates gene expression when dCas9 is fused with the omega subunit of RNA polymerase [11].
2.2.1. CRISPRi
The dCas9 lacks endonuclease activity, however, when coexpressed with a guide RNA, the resulting dCas9-gRNA complex can specifically retard RNA polymerase binding, transcriptional elongation, or transcription factor binding (Fig. 1B). It has been proven that CRISPRi efficiently repressed the expression of targeted genes in E. coli [10], Synechococcus elongates PCC 7942 [13], and Actinomycetes [14]. Compared with CRISPR, CRISPRi is independent of DNA repair mechanism.
2.2.2. CRISPRa
As aforementioned, transcription is upregulated when dCas9 is fused with the omega subunit of RNA polymerase and downregulated when dCas9 binds to a promoter or an ORF [10], [11] (Fig. 1C). This finding is theoretically consistent with the experimental conclusion of the Voigt group from Massachusetts Institute of Technology (MIT), where they recognized that RNA polymerase is actually a controllable “resource allocator” that governs carbon flux allocation [27]. In accordance with the above findings, years ago the Stephanopoulos group from MIT proposed an approach named gTME (global transcription machinery engineering) based on the binding of a library of RNA polymerase sigma factors to numerous promoters in the genome [28]. All the above studies collectively highlight that RNA polymerase is a key regulatory node, which severely affects global transcription and resource distribution. We therefore may conclude that linking the CRISPR-Cas9 system to RNA polymerase may lead to an occurrence of novel phenotypes.
3. Applications of CRISPR-Cas9 tools
3.1. Knockout of competing pathways
As mentioned above, one fundamental goal of strain engineering is to increase the production of desired chemicals. In order to divert metabolic flux to desired pathways, competing pathways should be eliminated or attenuated. Deletion of competing pathways conventionally relies on RecA- or Red-dependent recombineering methods. The knocking down of pathways is completed by RNAi for eukaryotic cells [29] or antisense RNA approach for both eukaryotes and prokaryotes [4]. CRISPR-Cas9 technology outperforms RecA- and Red-based recombineering methods in knocking out pathways mainly due to simplicity: one easily engineered gRNA is employed to target a gene sequence, and Cas9 is already expressed in the cells.
3.2. Knockdown of competing pathways
For mechanisms fundamental to knocking down genes, CRISPRi is completely different from RNAi. RNAi acts on mRNA via a double-strand RNA; in contrast, CRISPRi acts on DNA by blocking transcription initiation or elongation depending on whether a promoter or open reading frame is targeted [11] (Fig. 1B). Moreover, CRISPRi can be easily applied in microbes lacking the non-homologous end joining (NHEJ) pathway or for which no efficient homologous recombination (HR) approach is available. Lastly, CRISPRi can be an important tool for functional identification of key genes [9].
3.3. CRISPR-Cas9-mediated integration of signaling pathways
Biosynthesis of desired metabolites relies on multiple factors, including substrate provision, availability of energy and cofactors, as well as signal transduction. Of the factors affecting fermentation, signal transduction remains elusive. Rewiring of signaling pathways represents a frontier in metabolic engineering and synthetic biology [30]. Integration of the signaling pathways was inspired by a recent research done concerning CRISPR-Cas9-based ‘signal conductors’ that govern the transcription of endogenous genes in response to external or internal stimuli [31]. Therefore, we anticipate that CRISPR-based reprogramming of cellular signaling pathways is a promising alternative to strain improvement.
3.4. Enhancing cell tolerance to metabolites stress
Accumulation of desired metabolites leads to byproducts formation, metabolites inhibition, and slow cell growth. Strategies for mitigating these outcomes include blocking competing pathways, in situ recovery of metabolites, and adjustment of the pH value of fermentation broth to alleviate cell tolerance. It has been shown that a panel of regulators confer cell tolerance [32], [33], [34], [35], and metabolic engineering of these regulators significantly improve cell tolerance [34], [35]. From the viewpoint of genetics, cell tolerance is ascribed to quantitative trait loci (QTLs); thus, multiplex CRISPR editing or gene regulation of QTLs may ameliorate cell tolerance. For example, based on high-resolution QTL mapping, a total of 17 QTLs that differentiate hydrolysate tolerance between an industrially related and a laboratory strain were identified. Guided by this “genetic blueprint”, multikilobase loci were replaced using a dual-guide Cas9-based method, and a strain with superior hydrolysate tolerance than reference strain was engineered [36].
3.5. Dynamic control of fermentation process and cell fate
Considering that metabolites buildup generally impedes cell growth, dynamic control of fermentation process is often required. Moving forward, the fermentation process is divided into two phases: biomass accumulation and metabolite formation. Briefly, the CRISPR-Cas9 system is linked to regulatory genes or sensors, and the engineered strain responds to metabolite stress at a defined time point. As of today, programmable temperature-, light- and chemical-inducible Cas9 have been engineered [37], [38], [39], and these studies provide solid basis for dynamic control of the fermentation process.
Research has shown that cell size affects metabolite production. Cell size depends on a myriad factors including nutrient conditions and cell division [40]. The Chen group from Tsinghua University reported that E. coli was enlarged by overexpression of the sulA gene, resulting in the formation of filamentary E. coli that manifested larger internal space for polyhydroxyalkanoates accumulation compared with rod shape E. coli. Consequently, more poly(3-hydroxybutyrate) was produced compared with its reference strain [41]. The Smolke group from Stanford University announced that yeast cell fate can be switched by handling different sets of regulators [42]. Furthermore, the binding of CRISPR/dCas9 to any position within origin or replication blocks the initiation of replication, and altering the temperature from 37 to 42 °C releases the CRISPR/dCas9 replication inhibition. This method of controlling the bacterial cell cycle is useful for automatous control of DNA-replication and cell size [43]. Overall, the above studies pave the way for automatous control of cell fate.
3.6. CRISPR/dCas9-mediated protein imaging
Direct visualization of genomic loci facilitates deep understanding of the spatial organization of microbial genome and gene expression. Fluorescence in situ hybridization (FISH) is a conventional method for labeling genetic loci especially repetitive sequences. However, FISH involves denaturing dsDNA and hybridizing fluorescent nucleic acid probes, and thus is time-consuming and cost-ineffective. The sgRNA/dCas9 complex is an ideal probe to target genetic loci. In general, dCas9 is fused with green or yellow fluorescence protein, resulting in multicolor CRISPR/dCas9 complex that can bind the genetic loci matching sgRNA [44]. In addition, an array of sgRNA library along with dCas9 allows for labeling of multiple loci. To date, this method has been developed in diverse microbes such as E. coli [45] and Staphylococcus aureus [46]. Combination of CRISPR imaging and DNA sequential FISH allows for multiplexed dynamic imaging of genomic Loci [47]. Given CRISPR imaging technique is effective to identify repetitive sequences such as isoenzymes, transposon, and RNAs which substantially affect biosynthesis of desired metabolites, we envision this technique will contribute a lot to strain engineering.
4. Challenges of CRISPR-Cas9 applications
So far, the applications of CRISPR-Cas9 tools still face limitations. These setbacks include the lack of methods for generation and delivery of a sgRNA array, low efficiency of DNA repair in non-model microorganisms, limited strategies to couple with other genetic tools, and off-target effects.
4.1. Difficulty in generation and delivery of sgRNA array
Since metabolite production relies heavily on coordinated expression of multiple genes, a gRNA array is required to simultaneously direct Cas9/dCas9 to defined sites for editing, activating or repressing genes. So far, the multiplex editing efficiency of the CRISPR-Cas9 system remains to be constrained by the gRNA-expressing device [48]. To address this, Xie et al. developed a general strategy and platform for precise processing and efficient production of numerous gRNAs in vivo from a synthetic polycistronic gene via the endogenous tRNA-processing system [49]. This strategy is shown to significantly enhance CRISPR/Cas9 multiplex editing efficiency and has broad applications for small RNA expression and genome engineering. Apart from the generation of the sgRNA library, the delivery of CRISPR-Cas9 to intended genetic loci is also vital for efficient genome engineering. To address this concern, Xu et al. harnessed the piggyback transposon as a method to deliver a gRNA library for in vivo screening [50]. Given the availability of transposon systems [2], we believe that they could be feasible vehicles for the navigation of gRNA array in cells.
4.2. Low efficiency of DNA repair system
In eukaryotes, double-strand DNA breaks (DSBs) are repaired by NHEJ mechanism in an error-prone manner, leading to insertion/deletion mutations that frequently manifest frame shifts and gene disruption. In contrast to eukaryotic genomes that all contain NHEJ-like systems, only a percentage of prokaryotic genomes harbor NHEJ machinery (Ku/LigD) [51]. In fact, bacteria mainly rely on HR to repair DNA breaks [52]. Several studies have shown that coupling recombineering with Cas9 counterselection facilitates genome editing. These topics involve four aspects: (1) single-strand DNA recombineering (SSDR); (2) double-strand DNA recombineering (DSDR); (3) non-recombineering-based homologous recombination (NrHR); and (4) NHEJ.
-
(1)
Single-Strand DNA Recombineering (SSDR)
The first SSDR-CRISPR-Cas9-based genome editing tool was reported in 2013 [17]. E. coli was transformed with the vector pCas9 for heterologous expression of SpyCas9 and the tracrRNA molecule in order to edit its genome. E. coli was simultaneously transformed with a linear ssDNA oligonucleotide for SSDR and with the pCRISPR vector. The pCRISPR vector harbored a CRISPR array with one spacer that targeted the gene of interest, which lead to a CRISPR-Cas9-based counterselection against the wild-type genes. This tandem arrangement of an SSDR system followed by CRISPR-Cas9-based counterselection constitutes an efficient genome editing tool [17]. A similar tool was developed for Lactobacillus reuteri [15]. This tool was based on a SpyCas9 and tracrRNA expressing vector and the strain L. reuteri 6475, which expresses the phage-derived ssDNA-binding protein RecT required for SSDR. The SpyCas9 and tracrRNA expressing strain L. reuteri 6475 were simultaneously transformed with a CRISPR array expressing vector and ssDNA recombineering fragments. When no homologous template was supplied, transformation with targeting Cas9 constructs failed to generate transformants, indicating the lethality of using Cas9 [15]. Moreover, scientists realized that coupling CRISPR-Cas9 system with lambda Red recombineering allows not only highly efficient recombination of short single-strand oligonucleotides but also replacement of long DNA fragment [53].
-
(2)
Double-Strand DNA Recombineering (DSDR)
A DSDR-CRISPR-Cas9-based counter selection editing system was also developed for E. coli [18]. This system relies on a λ-Red and SpyCas9 expressing vector, and a gRNA expressing plasmid. When dsDNA fragments were used as editing templates, the single gene deletion efficiency was comparable to the efficiency previously reported [17]; however, the gene insertion efficiency was low and proportional to the length of the homologous regions when using the HR template fragments. In the same study, a variant of this tool was developed, based on a SpyCas9 and λ-Red expressing plasmid, and a sgRNA expressing plasmid containing a HR template [18]. The editing tool was used for the simultaneous insertion of two genes or the deletion of three genes in E. coli and the deletion of two genes from the chromosome of Tatumella citrea [18]. Even though this tool was not based on a classic DSDR system, as the provided editing templates were plasmid-borne and not linear dsDNA fragments, it is interesting that the efficiency of this tool dropped in the absence of a functional λ-Red mechanism.
-
(3)
Non-recombineering-based Homologous Recombination (NrHR)
Since NHEJ-dependent repair of DNA double-strand breaks (DSBs) gives rise to unwanted insertions or deletions, DNA-nicking enzyme (nickase) was employed to generate single-strand breaks (SSBs) instead of DSBs, which can be repaired by error-free HR rather than mutagenic NHEJ. For example, since Cas9-induced DSBs are lethal to Clostridium cellulolyticum due to weak expression of NHEJ components, Cas9 nickase, an enzyme causing single-stranded breaks in duplex DNA, was applied to achieve single-nick-triggered HR that allows one-step editing at intended genomic loci [54]. In addition, an NrHR-CRISPR-Cas9 based counterselection editing tool was developed to target insertion sequences native to E. coli genome, leading to large chromosomal deletions [55]. This approach addresses the concern of CRISPR-Cas9-induced lethal DSBs, and may work efficiently in the genome editing of prokaryotes that lack well-developed recombineering systems.
-
(4)
Non-Homologous End Joining (NHEJ)
Tong et al. developed a CRISPR-Cas9-NHEJ tool in Actinomycetales. When no templates for homology-directed repair (HDR) were present, the site-specific DNA DSBs introduced by Cas9 were repaired through the error-prone NHEJ pathway. The resulting deletions were of variable sizes surrounding around the targeted sequence (Table 1). However, if templates for HDR were supplied at the same time, precise deletions of the targeted gene were achieved [56]. Clearly, in all cases, highly efficient DNA repair system is critical for CRISPR-assisted recombineering.
Table 1.
CRISPR-assisted recombineering systems.
| CRISPR tools | Host cells | Vectors | Repair mechanisms | Ref. |
|---|---|---|---|---|
| Single-strand DNA recombineering (SSDR) | E. coli; Lactobacillus reuteri | pCas9 plasmid carrying tracrRNA, Cas9 and a resistance cassette, and a pCRISPR kanamycin-resistant plasmid carrying CRISPR spacers. | Homologous recombination | [15], [17] |
| Double-strand DNA recombineering (DSDR) | E. coli; | λ-Red and SpyCas9 expressing vector and gRNA expressing plasmid | Homologous recombination | [18] |
| Non-recombineering-based Homologous Recombination (NrHR) | Clostridium cellulolyticum | Vector pCas9 expressing Cas9 nickase (D10A), and vector pGRNA expression guide RNA | Nickase, single-nick-triggered homologous recombination | [54], [55] |
| Non-Homologous End Joining (NHEJ) | Actinomycetales | pCRISPRCas9 for gene deletion or replacement, and pCRISPR-dCas9 for gene expression control; and vector pCRISPR-sgRNA carrying sgRNA scaffold | Non-homologous end joining | [56] |
4.3. Arrangements of CRISPR-Cas system in chromosome or plasmid
Vector-dependent overexpression of key enzymes is a common strategy for directing carbon flux to desired pathways. However, this strategy shows its drawbacks by hindering cell growth and overconsuming cellular resources. Ideally, Cas9 or dCas9 is inserted into chromosome and subjected to induction expression. By contrast, sgRNAs should be user-defined since its expression device is vector-borne or located in the chromosome depending on certain requirements. For instance, a CRISPR-Cas9 toolkit was developed for the engineering of Bacillus subtilis by chromosomal expression of Cas9 and chromosomal transcription of gRNAs using a gRNA transcription cassette and counterselectable gRNA delivery vectors. This design avoids the need for multicopy plasmids which can be unstable and impede cell viability [19].
4.4. Coupling CRISPR-Cas9 with other genetic tools
CRISPR tools are versatile but will not work well for all genetic engineering purposes. For instance, the generation of Cas9 relies on translation machinery and therefore consumes more resources when compared to RNAi where no translation process is involved. Clearly, RNAi remains a cost-effective strategy for transcription control. Additionally, while the Cas9-based gene editing system relies on repair of DNA breaks in prokaryotic cells, both antisense RNA and CRISPRi are independent of any DNA repair system. More interestingly, Lee et al. demonstrated that the gene target repressed by the CRISPR system can be undone by expressing an antisense RNA that sequesters a small guide RNA [57]. Hence, the combination of CRISPR tools with RNAi or antisense RNA approach could be a clever solution to multidimensional control of gene expression.
4.5. Off-target effects
CRISPR/Cas9 can trigger off-target mutations and chromosomal rearrangements, which may alter microbial phenotypes. For rational-design strain engineering, off-target effects are not desirable. Given that off-target effects are ascribed mainly to sgRNA, unique target sequences should be different from any other sites in the genome by at least three nucleotides in 20-nucleotide sequences. The Adli group from University of Virginia developed a web-based tool named CHOP-IT to predict off-targets and to improve guide-RNA design [58]. The sgRNAs can also be designed by harnessing online software CRISPRdirect (http://crispr.dbcls.jp/doc/) [59]. In principle, sgRNA sequence should match transcription initiation site especially that in non-template strand [10]. In addition, sgRNA sequence can be used as a query to search against genome to avoid targeting homologous sequence. So far, off-target effects remain the major limitation for CRISPR applications. Thus, in-depth exploration of the underlying mechanisms are required.
5. Concluding remarks and prospective
The exploitation of CRISPR-Cas9 tools marks a breakthrough for strain engineering due to their versatility and ease of targeting the RNA-guided Cas9 to any user-defined DNA sequence. Derived from the bacterial adaptive immune system, CRISPR-Cas9 has been repurposed for many functions, including CRISPRi (Fig. 1B), CRISPRa (Fig. 1C) [10], [11] and protein imaging [60]. Presumably, Cas9 cleavage, CRISPRa, CRISPRi, or a combination thereof could generate diverse phenotypes beyond imagination. Likewise, novel applications of CRISPR-Cas9 tools may emerge owing to ongoing interdisciplinary studies. Among numerous applications reported to date, multiplex pathway engineering may largely facilitate strain engineering. Towards genome editing and global regulation, a gRNA array directs Cas9/dCas9 to target multiple chromosomal sites, and the targeted sites can be visualized due to labeled Cas9/dCas9 [61]. More surprisingly, the CRISPR-Cas-driven gene flow may occur across various microbial communities [62]. This CRISPR-driven ecological engineering is of great attractiveness for all walks of biologists. Evidently, CRISPR-Cas9 tools allow genetic engineering at all scales. As the pace of CRISPR study accelerates and grows in complexity, we envision that microbial genomes could be completely reprogrammed and partial mutants may overproduce desired metabolites. In conclusion, the CRISPR technology has significantly contributed to the disciplines adjacent to metabolic engineering, including synthetic biology, systems biology, genetics and ecology, which in turn have advanced strain engineering as well as bioproduction of chemicals.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (No. 21276014, 21476011), National High Technology Research and Development Program (863 Program) (No. 2015AA021003), Fundamental Research Funds for the Central Universities (YS1407) and 111 project (B13005). We also acknowledge the support from the College of Engineering, The University of Georgia, Athens.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Liu P., Wang W., Wei D. Use of transcription activator-like effector for efficient gene modification and transcription in the filamentous fungus Trichoderma reesei. J Ind Microbiol Biotechnol. 2017 doi: 10.1007/s10295-017-1963-7. [DOI] [PubMed] [Google Scholar]
- 2.Sharpe P.L., Dicosimo D., Bosak M.D., Knoke K., Tao L., Cheng Q. Use of transposon promoter-probe vectors in the metabolic engineering of the obligate methanotroph Methylomonas sp. strain 16a for enhanced C40 carotenoid synthesis. Appl Environ Microb. 2007;73(6):1721–1728. doi: 10.1128/AEM.01332-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee J.J., Crook N., Sun J., Alper H.S. Improvement of lactic acid production in Saccharomyces cerevisiae by a deletion of ssb1. J Ind Microbiol Biotechnol. 2016;43(1):87–96. doi: 10.1007/s10295-015-1713-7. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y., Lin Y., Li L., Linhardt R.J., Yan Y. Regulating malonyl-CoA metabolism via synthetic antisense RNAs for enhanced biosynthesis of natural products. Metab Eng. 2015;29:217–226. doi: 10.1016/j.ymben.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Mans R., Van Rossum H.M., Wijsman M., Backx A., Kuijpers N.G., Van Den Broek M. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015;15(2):fov004. doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zerbini F., Zanella I., Fraccascia D., Konig E., Irene C., Frattini L.F. Large scale validation of an efficient CRISPR/Cas-based multi gene editing protocol in Escherichia coli. Microb Cell Fact. 2017;16(1):68–95. doi: 10.1186/s12934-017-0681-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haft D.H., Selengut J., Mongodin E.F., Nelson K.E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1(6):e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322(5909):1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doudna J.A., Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 10.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41(15):7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiani S., Chavez A., Tuttle M., Hall R.N., Chari R., Ter-Ovanesyan D. Cas9 gRNA engineering for genome editing, activation and repression. Nat Methods. 2015;12(11):1051–1054. doi: 10.1038/nmeth.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C.H., Shen C.R., Li H., Sung L.Y., Wu M.Y., Hu Y.C. CRISPR interference (CRISPRi) for gene regulation and succinate production in cyanobacterium S. elongatus PCC 7942. Microb Cell Fact. 2016;15(1):196–206. doi: 10.1186/s12934-016-0595-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong Y., Charusanti P., Zhang L., Weber T., Lee S.Y. CRISPR-Cas9 based engineering of Actinomycetal genomes. ACS Synth Biol. 2015;4(9):1020–1029. doi: 10.1021/acssynbio.5b00038. [DOI] [PubMed] [Google Scholar]
- 15.Oh J.H., Van Pijkeren J.P. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014:gku623. doi: 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H., Cheng Q.X., Liu A.M., Zhao G.P., Wang J. A novel and efficient method for bacteria genome editing employing both CRISPR/Cas9 and an antibiotic resistance cassette. Front Microbiol. 2017;8:812–822. doi: 10.3389/fmicb.2017.00812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y., Chen B., Duan C., Sun B., Yang J., Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microb. 2015;81(7):2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westbrook A.W., Moo-Young M., Chou C.P. Development of a CRISPR-Cas9 tool kit for comprehensive engineering of Bacillus subtilis. Appl Environ Microb. 2016;82(16):4876–4895. doi: 10.1128/AEM.01159-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang M.M., Wong F.T., Wang Y., Luo S., Lim Y.H., Heng E. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat Chem Biol. 2017 doi: 10.1038/nchembio.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakociunas T., Bonde I., Herrgard M., Harrison S.J., Kristensen M., Pedersen L.E. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng. 2015;28:213–222. doi: 10.1016/j.ymben.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz C.M., Hussain M.S., Blenner M., Wheeldon I. Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR–Cas9-mediated genome editing in Yarrowia lipolytica. ACS Synth Biol. 2016;5(4):356–359. doi: 10.1021/acssynbio.5b00162. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen M.L., Isbrandt T., Rasmussen K.B., Thrane U., Hoof J.B., Larsen T.O. Genes linked to production of secondary metabolites in Talaromyces atroroseus revealed using CRISPR-Cas9. PloS One. 2017;12(1) doi: 10.1371/journal.pone.0169712. e0169712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarkari P., Marx H., Blumhoff M.L., Mattanovich D., Sauer M., Steiger M.G. An efficient tool for metabolic pathway construction and gene integration for Aspergillus niger. Bioresour Technol. 2017 doi: 10.1016/j.biortech.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Kuivanen J., Wang Y.J., Richard P. Engineering Aspergillus niger for galactaric acid production: elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9. Microb Cell Fact. 2016;15(1):210. doi: 10.1186/s12934-016-0613-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segall-Shapiro T.H., Meyer A.J., Ellington A.D., Sontag E.D., Voigt C.A. A ‘resource allocator’for transcription based on a highly fragmented T7 RNA polymerase. Mol Syst Biol. 2014;10(7):742. doi: 10.15252/msb.20145299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alper H., Stephanopoulos G. Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab Eng. 2007;9(3):258–267. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Williams T.C., Espinosa M.I., Nielsen L.K., Vickers C.E. Dynamic regulation of gene expression using sucrose responsive promoters and RNA interference in Saccharomyces cerevisiae. Microb Cell Fact. 2015;14(1):43. doi: 10.1186/s12934-015-0223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peisajovich S.G., Garbarino J.E., Wei P., Lim W.A. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science. 2010;328(5976):368–372. doi: 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Zhan Y., Chen Z., He A., Li J., Wu H. Directing cellular information flow via CRISPR signal conductors. Nat Methods. 2016;13(11):938–944. doi: 10.1038/nmeth.3994. [DOI] [PubMed] [Google Scholar]
- 32.Cheville A., Arnold K., Buchrieser C., Cheng C., Kaspar C. rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157: H7. Appl Environ Microb. 1996;62(5):1822–1824. doi: 10.1128/aem.62.5.1822-1824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alper H., Moxley J., Nevoigt E., Fink G.R., Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314(5805):1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 34.Sugimoto S., Higashi C., Matsumoto S., Sonomoto K. Improvement of multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 under conditions of thermal stress by heterologous expression of Escherichia coli dnaK. Appl Environ Microb. 2010;76(13):4277–4285. doi: 10.1128/AEM.02878-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y., Stabryla L., Wei N. Improved acetic acid resistance in Saccharomyces cerevisiae by overexpression of the WHI2 gene identified through inverse metabolic engineering. Appl Environ Microb. 2016;82(7):2156–2166. doi: 10.1128/AEM.03718-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurer M.J., Sutardja L., Pinel D., Bauer S., Muehlbauer A.L., Ames T.D. QTL-guided metabolic engineering of a complex trait. bioRxiv. 2016:079764. doi: 10.1021/acssynbio.6b00264. [DOI] [PubMed] [Google Scholar]
- 37.Richter F., Fonfara I., Bouazza B., Schumacher C.H., Bratovič M., Charpentier E. Engineering of temperature-and light-switchable Cas9 variants. Nucleic Acids Res. 2016;44(20):10003–10014. doi: 10.1093/nar/gkw930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu K.I., Ramli M.N.B., Woo C.W.A., Wang Y., Zhao T., Zhang X. A chemical-inducible CRISPR-Cas9 system for rapid control of genome editing. Nat Chem Biol. 2016;12(11):980–987. doi: 10.1038/nchembio.2179. [DOI] [PubMed] [Google Scholar]
- 39.Maji B., Moore C.L., Zetsche B., Volz S.E., Zhang F., Shoulders M.D. Multidimensional chemical control of CRISPR-Cas9. Nat Chem Biol. 2017;13(1):9–11. doi: 10.1038/nchembio.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weart R.B., Lee A.H., Chien A.C., Haeusser D.P., Hill N.S., Levin P.A. A metabolic sensor governing cell size in bacteria. Cell. 2007;130(2):335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Wu H., Jiang X., Chen G.Q. Engineering Escherichia coli for enhanced production of poly (3-hydroxybutyrate-co-4-hydroxybutyrate) in larger cellular space. Metab Eng. 2014;25:183–193. doi: 10.1016/j.ymben.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 42.Galloway K.E., Franco E., Smolke C.D. Dynamically reshaping signaling networks to program cell fate via genetic controllers. Science. 2013;341(6152):1235005. doi: 10.1126/science.1235005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiktor J., Lesterlin C., Sherratt D.J., Dekker C. CRISPR-mediated control of the bacterial initiation of replication. Nucleic Acids Res. 2016:gkw214. doi: 10.1093/nar/gkw214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng W., Shi X., Tjian R., Lionnet T., Singer R.H. CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc Natl Acad Sci U. S. A. 2015;112(38):11870–11875. doi: 10.1073/pnas.1515692112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lane A.B., Strzelecka M., Ettinger A., Grenfell A.W., Wittmann T., Heald R. Enzymatically generated CRISPR libraries for genome labeling and screening. Dev Cell. 2015;34(3):373–378. doi: 10.1016/j.devcel.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen B., Hu J., Almeida R., Liu H., Balakrishnan S., Covill-Cooke C. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. 2016:gkv1533. doi: 10.1093/nar/gkv1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takei Y., Shah S., Harvey S., Qi L.S., Cai L. Multiplexed dynamic imaging of genomic loci by combined CRISPR imaging and DNA sequential FISH. Biophys J. 2017;112(9):1773–1776. doi: 10.1016/j.bpj.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nowak C.M., Lawson S., Zerez M., Bleris L. Guide RNA engineering for versatile Cas9 functionality. Nucleic Acids Res. 2016;44(20):9555–9564. doi: 10.1093/nar/gkw908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie K., Minkenberg B., Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S. A. 2015;112(11):3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu C., Qi X., Du X., Zou H., Gao F., Feng T. piggyBac mediates efficient in vivo CRISPR library screening for tumorigenesis in mice. Proc Natl Acad Sci U S. A. 2017;114(4):722–727. doi: 10.1073/pnas.1615735114. 201615735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bowater R., Doherty A.J. Making ends meet: repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet. 2006;2(2):e8. doi: 10.1371/journal.pgen.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Datta S., Costantino N., Zhou X. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc Natl Acad Sci U S. A. 2008;105(5):1626–1631. doi: 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pyne M.E., Moo-Young M., Chung D.A., Chou C.P. Coupling the CRISPR/Cas9 system with lambda Red recombineering enables simplified chromosomal gene replacement in Escherichia coli. Appl Environ Microb. 2015;81(15):5103–5114. doi: 10.1128/AEM.01248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu T., Li Y., Shi Z., Hemme C.L., Li Y., Zhu Y. Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase. Appl Environ Microb. 2015;81(13):4423–4431. doi: 10.1128/AEM.00873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Standage-Beier K., Zhang Q., Wang X. Targeted large-scale deletion of bacterial genomes using CRISPR-nickases. ACS Synth Biol. 2015;4(11):1217–1225. doi: 10.1021/acssynbio.5b00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tong Y., Charusanti P., Zhang L., Weber T., Lee S.Y. CRISPR-Cas9 based engineering of Actinomycetal genomes. ACS Synth Biol. 2015;4(9):1020–1029. doi: 10.1021/acssynbio.5b00038. [DOI] [PubMed] [Google Scholar]
- 57.Lee Y.J., Hoynes-O'connor A., Leong M.C., Moon T.S. Programmable control of bacterial gene expression with the combined CRISPR and antisense RNA system. Nucleic Acids Res. 2016:gkw056. doi: 10.1093/nar/gkw056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh R., Kuscu C., Quinlan A., Qi Y., Adli M. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res. 2015;43(18):e118. doi: 10.1093/nar/gkv575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naito Y., Hino K., Bono H., Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen B., Hu J., Almeida R., Liu H., Balakrishnan S., Covill-Cooke C. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. 2016;44(8):e75. doi: 10.1093/nar/gkv1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deng W., Shi X., Tjian R., Lionnet T., Singer R.H.C.A.S.F.I.S.H. CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc Natl Acad Sci U S. A. 2015;112(38):11870–11875. doi: 10.1073/pnas.1515692112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dicarlo J.E., Chavez A., Dietz S.L., Esvelt K.M., Church G.M. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat Biotechnol. 2015;33(12):1250–1255. doi: 10.1038/nbt.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]