Abstract
Organisms’ reactions to adverse events result in the generation of immune effectors, which, in the case of insects, may be produced from the direct activation of pathways such as Toll, Jak-STAT, Imd, or RNAi or may be derived from the crosstalk of different intracellular pathways. One such pathway, the unfolded protein response (UPR), has the primary objective of restoring homeostasis in the endoplasmic reticulum. In addition, the UPR participates in signaling crosstalk with the immune pathways, generating protection against pathogenic organisms. Dengue virus is a plus-strand RNA virus belonging to the Flavivirus genus that uses the ER as a replication site; during the infection, there are indicators of the activation of the UPR, which in turn, induces the synthesis of internal membranes and preferential translation of viral proteins enhancing the replication. One of the dengue virus proteins, the NS4B can block the pathway of α/β interferon in mammals. However, what happen in insects is interesting because the lack of the main antiviral pathway, the interferon and the role of the NS4B protein in the UPR-immunity relationship can be better understood. Thus, in this study, we demonstrated that the DENV2/16681 NS4B protein is capable of modulating the immune effectors that result from the activation of the UPR in insect cells.
Keywords: UPR, ER stress, Innate immune, NS4B, Dengue
Introduction
The endoplasmic reticulum (ER) is the cell compartment that sense the synthesis and folding of local polypeptides or those intended to continue along the secretory pathway. Post-translational modifications such as the formation of disulfide bonds and N-glycosylation occur in the ER environment and contribute to final protein conformation (Bulleid 2012; Chavan and Lennarz 2006). There are several situations that may generate ER stress and that have implications for protein folding. The cell mechanism that allows the adaptation of the cell, under both physiological and pathological stress conditions, is called the UPR (Unfolded Protein Response) (Schroder and Kaufman 2005; Walter and Ron 2011). The UPR is a pathway of transcriptional regulation that was discovered in Saccharomyces cerevisiae, where the activation of the response is mediated only by one protein: Ire1p (Cox and Walter 1996; Mori et al. 1996). Ire1p has kinase and endoribonuclease activities, and its primary target is splicing of the mRNA encoding the basic leucine zipper (bZIP) transcription factor HAC1 mRNA (XBP1 in higher eukaryotes), that binds to a DNA sequence motif termed the UPR element (UPRE), which in turn upregulates many of the genes associated with the secretory pathway (Hetz et al. 2011). By contrast, UPR signaling in metazoans is more complex; i.e., ER homeostasis is recovered through three sensor transmembrane proteins: IRE1 (inositol requiring kinase 1), ATF6 (activating transcription factor 6) and PERK (double-stranded RNA-activated protein kinase PKR-like endoplasmic reticulum kinase) (Bernales et al. 2006; Bukau et al. 2006). Each of these folding sensors may be regulated, directly or indirectly, by the ER molecular chaperone BiP/GRP78. These folding sensors in the ER initiate signaling pathways that communicate with the nucleus (Harding et al. 1999), where they improve the expression of genes that help increase ER folding capacity (Kozutsumi et al. 1988), modulate protein translation, and they boost the degradation of misfolded ER proteins through ER-associated degradation (ERAD) (Smith et al. 2011; Travers et al. 2000; Yoshida et al. 2003). Finally, when these measures are unsuccessful in resolving ER stress, cell death may be induced (Rutkowski et al. 2006).
Although the UPR genes appear to be conserved between organisms, it is not known whether the functions are equivalent between them. The mammalian specific protein functions may not be represented in invertebrates; in which it seems that the general functions of the UPR are conserved allowing the adaptation of the cells to stress. For example, transcriptional regulation, which is a key aspect of the UPR in higher eukaryotes, is not widely employed by protozoans, which appear to lack IRE1 and XBP1. Nonetheless, ER stress in Trypanosoma brucei does result in gene expression changes similar to those seen during the UPR in other eukaryotes, including the upregulation of many genes involved in protein secretion. Although the proteins that mediate this response are not known, the upregulation of several targets is accomplished through mRNA stabilization. Interestingly, a large fraction of RNAs downregulated by stress in T. brucei encode proteins destined for the ER, reminiscent of the RIDD pathway in higher eukaryotes (Goldshmidt et al. 2010; Gosline et al. 2011). By contrast, in Caenorhabditis elegans and Drosophila melanogaster, only a few genes depend on the ATF6 pathway for the induction of the response to ER stress, ER chaperones and ERAD components are fully induced in the atf-6 knockdown worm, suggesting that the IRE1 pathway is responsible for the induction of most canonical UPR target genes, including ER chaperones and ERAD components. These findings imply that the major regulator of ER quality control proteins switched from IRE1 to ATF6 during evolution from fly to mammals and that the mechanisms that relieve stress within the ER are preserved between species.
Recent findings have stimulated interest in the relationship between the UPR and immune response. Those studies have shown that the activation of IRE1/XBP1 arm promotes NF-κB, JNK and IRF3 activation, generating the transcription of immune effectors such as cytokines and reactive oxygen species that generate a proinflammatory phenotype protecting the cells against invading organisms (Kim et al. 2015; Urano et al. 2000). This mechanism is notably essential in organisms in which the defense against infectious agents relies exclusively on innate immunity. For instance, a study carried out in C. elegans showed that IRE1 plays a central protective role in a challenge against Pseudomona aeruginosa, protecting against “excessive” activation of innate immunity (Richardson et al. 2010). Aedes aegypti is another example that launches an UPR-dependent defense mechanism in the presence of Cry11Aa toxin to promote cell survival (Bedoya-Perez et al. 2013).
On the other hand, when some pathogens interfere with ER function as part of their replication cycle, for example, in mammals, an infection caused by Dengue virus (DENV) there are indicators of UPR activation. This activation seems to be modulated by the virus to generate membrane expansion due to the activation of the IRE1/XBP1 axis (Pena and Harris 2011; Umareddy et al. 2007; Yu et al. 2006) what in turn regulates the viral load by generating structures called convoluted membranes that increase the concentration of viral and host components to enhance the replication and assembly of virions, and hide these antigenic proteins from the innate immune system (Netherton et al. 2007; Novoa et al. 2005; Salonen et al. 2005; Welsch et al. 2009). There is another widely described viral mechanism that improves replication, is the capacity to block the α/β interferon pathway, the main antiviral response in chordates. DENV-NS4B (744 pb, 248 aa, 25 kDa) is a hydrophobic protein that is associated with the ER lumen. It plays a crucial role as a scaffolding protein for membrane expansion and the formation of replication complexes, blocking the α/β interferon pathway by inhibiting STAT1 phosphorylation, dimerization, and translocation into the nucleus (Munoz-Jordan et al. 2003, 2005). Additionally, recent studies have described a role for DENV-NS4B, which, during infection, associates with mitochondrial proteins and induces mitochondrial elongation in contact with the convoluted membranes by inactivation of the mitochondrial fission factor dynamin-related protein-1 (DRP1). Therefore, this restructuring inhibited the activation of the immune response resulting from the mitochondrial antiviral-signaling protein (MAVS)-dependent interaction of ER-mitochondrion to promote infection (Chatel-Chaix et al. 2016).
With these examples, we can postulate the protein DENV-NS4B as one of the proteins with higher pro-viral characteristics because it is involved in mechanisms that make viral replication more efficient and blocks several of the immune signaling pathways in mammals. Therefore, it would be interesting to know what occurs in insects and their own immune signaling pathways concerning this protein. In the case of insect vectors of this virus, we know that there is a complementary activation of more than one defense pathway in response to DENV infection (Sim and Dimopoulos 2010). For example, when the Toll pathway becomes activated, an increase in Rel1 (homolog of NF-κB) and different antimicrobial peptides are observed, resulting in a visible reduction in viral replication (Xi et al. 2008). The antiviral role of the Jak-STAT pathway was demonstrated by the RNAi suppression of Jak or DOME, two components of the pathway, which results in an increase in viral replication (and the contrary effect when suppressing its negative regulator) (Souza-Neto et al. 2009). It has also been shown that infection with the 4 serotypes of the DENV in D. melanogaster cells produces siRNA targeting the DENV genome and knockdown of any of the components of the RNAi pathway causes an increase in viral replication (Mukherjee and Hanley 2010).
These data suggest that insect vectors represent an interesting model for study as a highly conserved system that can be used to understand the adaptive stress of cells to persistent infection and immune effector regulation. Importantly, because insects lack the α/β interferon pathway, the UPR-immunity relationship can be interpreted more easily. Here, we describe how the DENV2-NS4B protein, which is stably expressed in an insect cell line, can modulate UPR activation-dependent immune effectors as an adaptive mechanism to ER stress.
Methods
Cell culture, virus and treatments
The techniques and procedures used in this work comply with the approval of the Internal Review Board both ethically (research with no risk, CI1005-1057) and regarding biosafety (CI1005-996, May 31, 2011), which, in turn, comply with international standards. The Drosophila melanogaster CME W1 cI.8+ cell line was purchased from the Drosophila Genomics Resource Center (DGRC). CME W1 cI.8 cells and the clones H (DENV-NS4B protein high expression), L (DENV-NS4B protein low expression) and G (only GFP expression) were cultured at 27 °C in M3 medium (Sigma) supplemented with 2% heat inactivated fetal bovine serum (FBS) (Invitrogen), 5 μg/ml insulin and 2.5% fly extract that was prepared according to the protocol given on the DGRC website (Cherbas 2016). Briefly, for the fly extract we collected adult flies, put them in the freezer for at least 45 min, weighed them, and then transferred them to a glass homogenizer for homogenization under cold conditions. Next, we spun the homogenate at 1500×g at 4 °C for 15 min, incubated the supernatant at 60 °C for 5 min, spun the supernatant at 1500×g at 4 °C for 90 min, filter-sterilized the extract through a 0.22-μm filter and stored it at 20 °C. ER Stress was induced with tunicamycin (TM; Calbiochem Cat# 654380). PMA and STF083010 were purchased from Sigma-Aldrich (St. Louis, MO). DENV-2 (New Guinea C strain) was kindly donated by InDRE México.
Plasmid construction
Standard molecular biology techniques were used for cloning. The vector pCAGGS containing DENV-2 16,681 strain NS4B cDNA (a kind gift of Adolfo García-Sastre, School of Medicine at Mount Sinai, New York, USA) served as a template for polymerase chain reaction amplification (PCR). For the expression of NS4B/GFP fusion protein, DNA fragments encoding the aminoacids of the respective DENV-NS4B protein were generated by PCR with the sense primer 5-GAGGTACCCAAAATGAACGAGATGGGTTTCCTAGA-3 and antisense primer 5-GAGAATTCCCTTCTTGTGTTGGTTGTGTT-3, followed by insertion into the insect expression vector Ac5-STABLE1-blast (a kind gift of James Sutherland, CIC-bioGUNE, Spain PMID: 22355594) into the KpnI and Eco RI sites in frame with the GFP sequence. The DENV2-NS4B/GFP plasmid was expressed and purified following the protocol of the plasmid purification Maxi Kit (Qiagen). All constructs were confirmed by restriction analysis, and nucleotide sequences were analyzed in the Unidad de Síntesis y Secuenciación by Institute of Biotechnology, UNAM.
Infection and cell viability assays
Infection of clones with DENV-2 (New Guinea C strain) was carried out at the MOI of 0.1 in 2% FBS/M3 medium at 27 °C for 5 days. The cisplatin assay is commonly used to determinate cell viability as an inductor of apoptosis. After incubation of the cells with a range of concentrations of cisplatin, cell viability control was used to obtain an IC50 value of 50 μM. Cells were plated in 24-well plates and were treated with cisplatin or a combination of TM and cisplatin at the indicated drug dosage. At the indicated time, the medium was removed, and fresh medium was added to each well. Thereafter, the cells were incubated at 27 °C for 8 h. The supernatant was removed, and the Scepter ® automated cell counter was used.
Transfection and FACS
Drosophila melanogaster CME W1 cI.8+ cells seeded in 6-well plates were simultaneously transfected with either the empty vector (only GFP) or the DENV2-NS4B/GFP plasmid using Attractene (Qiagen) according to the manufacturer’s protocol. To obtain a clonal cell population expressing the plasmid, transfected cells were subject to increasing dilutions of the parent cell culture, and then the cells were incubated in complete media containing 30 μg/ml blasticidin (Calbiochem). To quantify the expression levels of intracellular NS4B protein, fluorescence-activated cell sorting was utilized. Cells were sorted using a FACSAria II Cell Sorter (BD Biosciences) in the Unidad de Citometría at the Biomedicas Institute, UNAM, and data were analyzed using FlowJo software (Tree Star Inc.).
Western blotting
For western blots, cell lysates from cI.8+ cells and clones were prepared in lysis buffer containing halt protease inhibitor cocktail (ROCHE) and phosphoSafe (Millipore). After centrifugation at 16,000×g for 15 min at 4 °C, the protein concentration was determined by the BCA protein assay kit (Pierce). Next, the supernatant was boiled with Laemmli loading buffer (250 nM Tris-HCl, pH 6.8, 50% Glycerol, 0.5 M β-Mercaptoethanol, 10% SDS and 0.1% bromo phenol blue) for 10 min, and then 30 μg was loaded onto a 4–12% polyacrylamide gel. The proteins were transferred onto nitrocellulose membranes and after blocking with 3% non-fat milk in TBS-Tween, the membrane was incubated with primary antibody 1:1000 rabbit polyclonal anti-DENV-NS4B (Genetex GTX124250), 1:1000 for mouse monoclonal eIF2α (Cell Signaling), 1:1000 rabbit monoclonal phosphorylated eIF2α at S51 (Cell Signaling), and 1:2000 for anti-β actin (Santa Cruz) overnight at 4 °C. The membrane was then washed, incubated similarly with secondary antibody (goat anti-rabbit or anti-mouse conjugated (Jackson Immuno Research) to HRP 1:10,000 for 1 h at room temperature and probed with SuperSignal™ West Femto Maximum Sensitivity Substrate (Pierce/Thermo). Antibody-protein complexes were detected using the C-Digit Western blotting detection system (Li-Cor). Quantification of the signals was performed with a computer running Image Studio Digits Li-Cor Inc. software.
Real-time RT-PCR and XBP1 mRNA splicing
Total RNA was extracted from clone cells using TRIzol (Invitrogen) according to the manufacturer’s guidelines. After quantitation, first-strand cDNA was synthesized from 1 μg of RNA using Oligo-dT and the RevertAid Minus first-strand cDNA synthesis kit (Thermo). Real-time qPCR was performed using gene-specific primers with the Verso SYBR Green 1 step qRT-PCR Rox mix (Thermo) and ViiA 7 (Applied Biosystems). For relative quantification, the expression of each target gene was measured with respect to the actin in each sample, and the concentration was calculated from a standard curve. Melting curves were used to verify the specificities of products. All of the samples were analyzed in triplicate, and experiments were repeated at least twice. ΔΔCt was calculated using the equation described in the manufacturer’s protocol. PCR primer pairs were as follows: Ero1:L- TGCTCCAAATACCTGCTGTC- R-CGCTTGTGGTTTCTGGACTA-, EDEM L- ACCATCCTTCTGGAATTTGC- R- TTAGCTTCCACAGTGCATCC-, Cactus L- TGCCTTGACAGAGAAGTTCG- R- TCGTAGTTGCGAATCTCCAG-, iNOS- L- TACAGAGGTCTGCACCAAGC- R- CAGCGGAACTTCCAGTATCA-, Rel1 L- GAAAGTAGCGATGCTGGTCA- R-TGTTGTCCATTTCGGTGTCT-, Def L- TGGATCCAATTCCAGAGGAT- R- CACTTGGAGAGTAGGTCGCA, BiP/GRP78 L: CGCATCG AAATTGAATCCTT- R- TTCAGGGTGGAACGGAATAG. The graphs were plotted as the fold increases over the control value. All of the results were expressed as the relative expression of the target genes with respect to actin using the G, L and H group without stimulus as a reference. Activation of IRE1 was determined by measuring the splicing of its substrate, the mRNA encoding the XBP1 transcription factor. Total RNA was harvested with TRIzol reagent (Invitrogen) immediately after the completion of stress treatments. Typically, 1 μg of RNA was used as the template in every 25 μl reaction mixture to make first-strand cDNA using the RevertAid Minus first-strand cDNA synthesis kit (Thermo) according to the manufacturer’s recommendations, and 1 μl of synthesized first-strand cDNA was used as the template in each 25-μl PCR reaction mixture. To amplify D. melanogaster XBP1splicing/unsplicing, the PCR mixture was heated at 94 °C for 5 min, and then it was amplified for 30 cycles of 94 °C for 30 s; 58 °C for 30 s, and 72 °C for 30s with 5 min in the final cycle, using 5 CTGGAACAGCAAGTGGTAGA-3 and 5-CTGGGTCCTTCTGGGTAGAC-3 primers with Taq DNA Polymerase (PROMEGA). Next, 10 μl of PCR products was subjected to electrophoresis on a 3% agarose gel, followed by quantification by densitometry.
NO (nitrite/nitrate) and antimicrobial peptide concentration in the supernatant
The nitric oxide (nitrite/nitrate) concentration in the supernatant of stimulated cells was determined using the Griess reaction [18]. Briefly, 50 μl of the supernatant of stimulated clone cells seeded in 12-well plates was mixed with 50 μl of 1% sulfanilamide and 50 μl of 0.1% naphthyl-ethylene-diamine (Sigma). After incubation for 10 min at RT, the absorbance was recorded at 540 nm in a plate reader. NO was quantified using the NaNO2 (1–100 μM) standard reference curve for each assay. Bacteria were cultured in Luria-Bertani (LB) broth at 37 °C overnight with shaking at 225 rpm. Bacterial cultures were then centrifuged at 16,100×g for 5 min. The ATP level in bacterial culture was determined using the BacTiter-Glo™ Microbial Cell Viability Assay Reagent (Promega, Madison, WI) subjected to a luciferase-based assay, and the ATP level was determined by measuring luminescence levels and comparing them to an ATP standard curve. 100 μl of culture supernatant mixed with 100 μl of the clone cell-treated supernatant was incubated for 3 h at RT. Thereafter, an equal volume of BacTiter-Glo™ Microbial Cell Viability Assay Reagent was added to the mixture in a 96-well opaque plate, followed by incubation at room temperature for 5 min. After incubation, luminescence was read in a GloRunner plate reader (Turner BioSystem). ATP (A2383, Sigma Aldrich, St. Louis, MO), standard curve using 10-fold dilutions of ATP standard solutions prepared in H2O was included in each experiment. LB, H2O or buffer was included in all assays as negative controls.
Statistics
We performed an analysis of variance (ANOVA) test to determine differences between groups, followed by a post hoc test (with Bonferroni correction). Differences were considered to be significant if P < 0.05. Statistical analysis was performed using SPSS for OS X Yosemite, version 20 X 0 (SPSS Inc., Chicago, IL, USA).
Results
UPR activation in cells that express DENV-NS4B protein
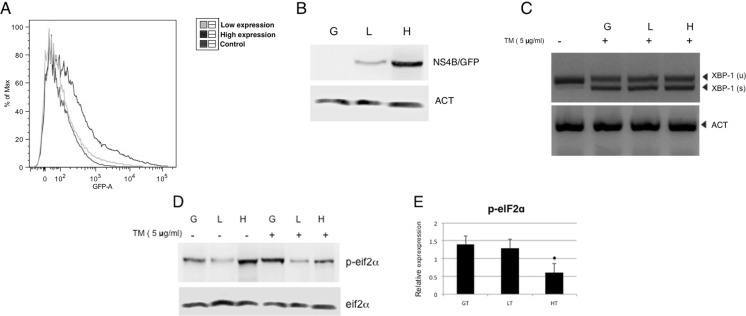
To perform our experiments, we used a Drosophila CME W1 cI.8+ cell line purchased from the Drosophila Genomics Resource Center (DGRC) at the Indiana University/ Bloomington Campus, USA. We used the Drosophila melanogaster cell line because it has been previously demonstrated that C6/36 cell line (Aedes albopictus) exhibit a weak, and possibly incomplete, RNAi response, which may contribute to their ability to support high levels of DENV replication (Brackney et al. 2010). The Aag2 cell line (Aedes aegypti) in contrast is infected by an insect Flavivirus called CFAV (cell fusion agent virus), so the interpretation under both infections would be confusing. Finally, Sessions et al. validated the use of the Drosophila screen to identify dipteran dengue-host interactions (Sessions et al. 2009). Thus, the cI.8 cell line was stably transfected with a plasmid containing DENV2/16681 NS4B protein tagged with GFP. We subsequently subjected the transfected cells to blasticidin, the antibiotic of resistance. Then, fluorescence-activated cell sorting of cells with high (H) and low (L) expression of the protein was compared with untransfected controls (Fig. 1a) or those transfected with the same plasmid bearing GFP only. The level of protein expression in the sorted clones was validated by Western blotting (Fig. 1b). The percentage of cells expressing the transgene was 70–80% in clone L but was 30–40% in clone H. This percentage in both clones were sufficient to observe a dose response relationship as shown in the following experiments. We then measured UPR activation with tunicamycin (TM, 5 μg/mL for 4 h), a drug that promotes protein misfolding through the inhibition of N-glycosylation, and which stimulated XBP1 splicing (from activation of IRE1 endoribonuclease) (Fig. 1c). Moreover, the activation of PERK due to the stress generated by the drug was measured by the formation of phospho eIF2α compared with the total of eIF2α (Fig. 1d, e). In the control cells (G) stimulated with TM, eIF2α phosphorylation was increased, whereas phospho-eIF2α was decreased significantly in H clones. These results demonstrate that clones L and H express the DENV-NS4B protein and are sensitive to UPR activation, and the increase in NS4B protein expression decreased PERK activation.
Fig. 1.
Activation of UPR in cells expressing DENV-NS4B protein. a CME W1 cI.8+ of Drosophila cells were transfected with the original plasmid with only GFP (G clone) and the plasmid expressing NS4B-GFP (L and H clones). Thereafter, they were sorted, and b the level of expression of NS4B was measured by Western immunoblotting (WB) where actin (ACT) was used as a control. c Clones G, L, and H were treated with TM (5 μg/ml) for 4 h, and RT-PCR was carried out to measure XBP1 splicing as an indicator of UPR activation. d Clones G, L, and H were treated with TM (5 μg/ml) for 4 h. Subsequently, WB was carried out to determine the total expression of p-eIF2α and eIF2α. e Relative expression of p-eIF2α compared with that of the total. The data shown are representative of three independent experiments for each clone. The asterisks shown are p < 0.05 compared with the G clone treated with TM
DENV-NS4B protein expression modulates UPR transcriptional activation
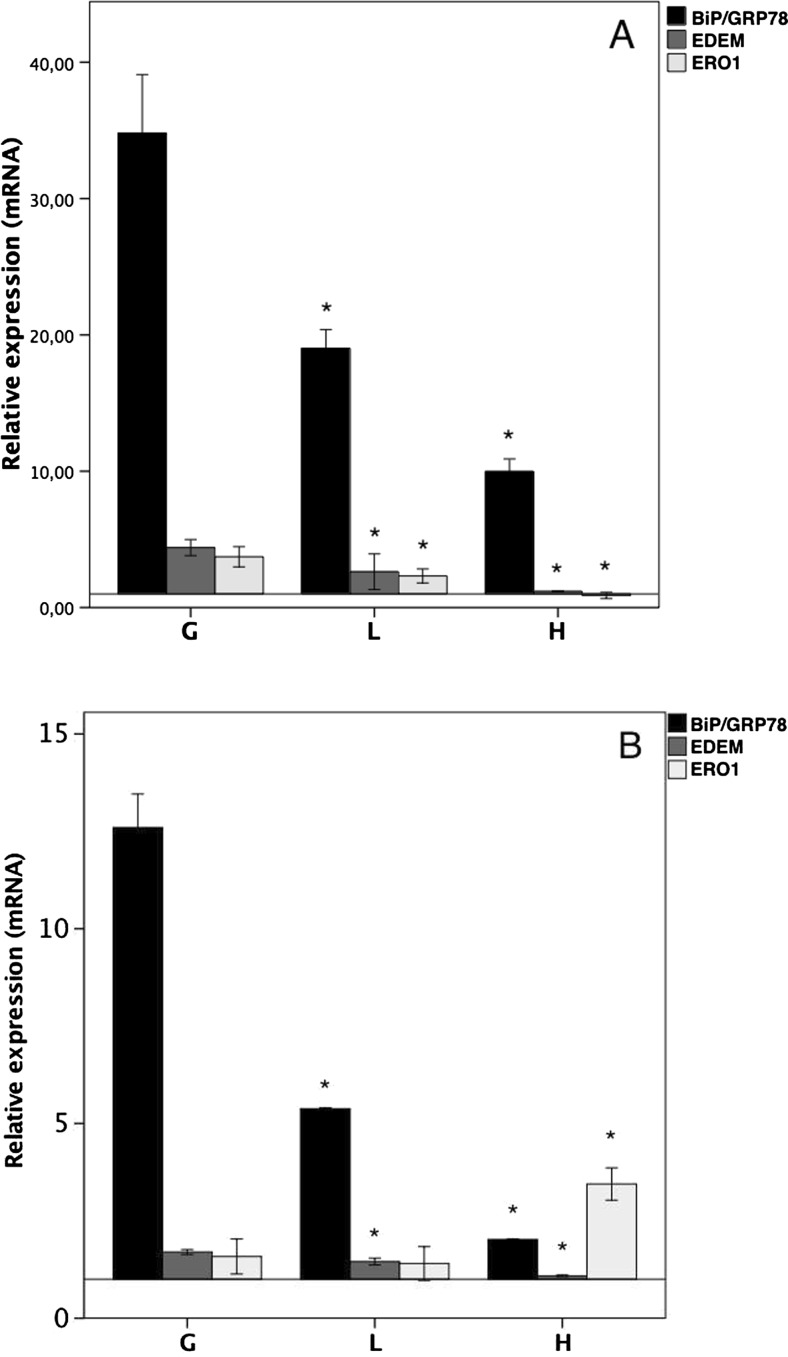
To confirm whether DENV-NS4B protein expression induces changes in the UPR at the transcriptomic level, we treated clones G, L, and H with TM over an 8- or 24-h time course. Through qRT-PCR (Fig. 2), we measured the levels of mRNAs encoding proteins that are activated to relieve ER stress: BiP/GRP78, an ER chaperone, which controls the structural maturation of nascent glycoproteins, EDEM1 (an ER degradation-enhancing α-mannosidase like protein) and Ero1 (ER oxidoreductin-1α). After 8 h of stimulation in clone G, BiP/GRP78 expression was increased 35-fold, EDEM was increased 4.5-fold, and Ero1 was increased 3.5-fold compared with the control clones. However, in the NS4B-expressing clones L and H, there was a concentration-dependent effect because the transcription of these markers was decreased significantly as the DENV-NS4B protein expression was increased. In contrast, at 24 h f stimulation, we found a decrease in the expression of these effectors regarding the measurements at 8 h and the same pattern of decrease in the expression of BiP/GRP78 and EDEM with respect to clone G, which did not express the DENV-NS4B protein, although Ero1 expression was actually increased in clone H. The transcriptome following UPR activation is highly regulated. The transcription of BiP/GRP78 may result from the activation of both ATF6 and XBP1 arms, whereas the activation of Ero1 is almost exclusively a result of PERK arm activation. However, in the early hours, the decrease in PERK activation in clones expressing NS4B appears to be reversed after 24 h. These results show that NS4B protein expression suppresses the induction of BiP/GRP78 and EDEM but not Ero1 after 24 h of TM treatment.
Fig. 2.
Expression of DENV-NS4B protein modulates the activation of UPR transcriptionally. Clones G, L, and H were treated with TM (5 μg/ml) for 8 h (a) and 24 h (b) the levels of BiP/GRP78, EDEM and Ero1 expression were measured through qRT-PCR. All of the results are expressed as the relative expression of the target genes with respect to actin, using the G, L, and H groups without stimulus as references. Analysis of variance (ANOVA) test with post hoc Bonferroni correction test: *P < 0.05
Activation of UPR results in the transcription of immune effectors modulated by the expression of DENV-NS4B protein
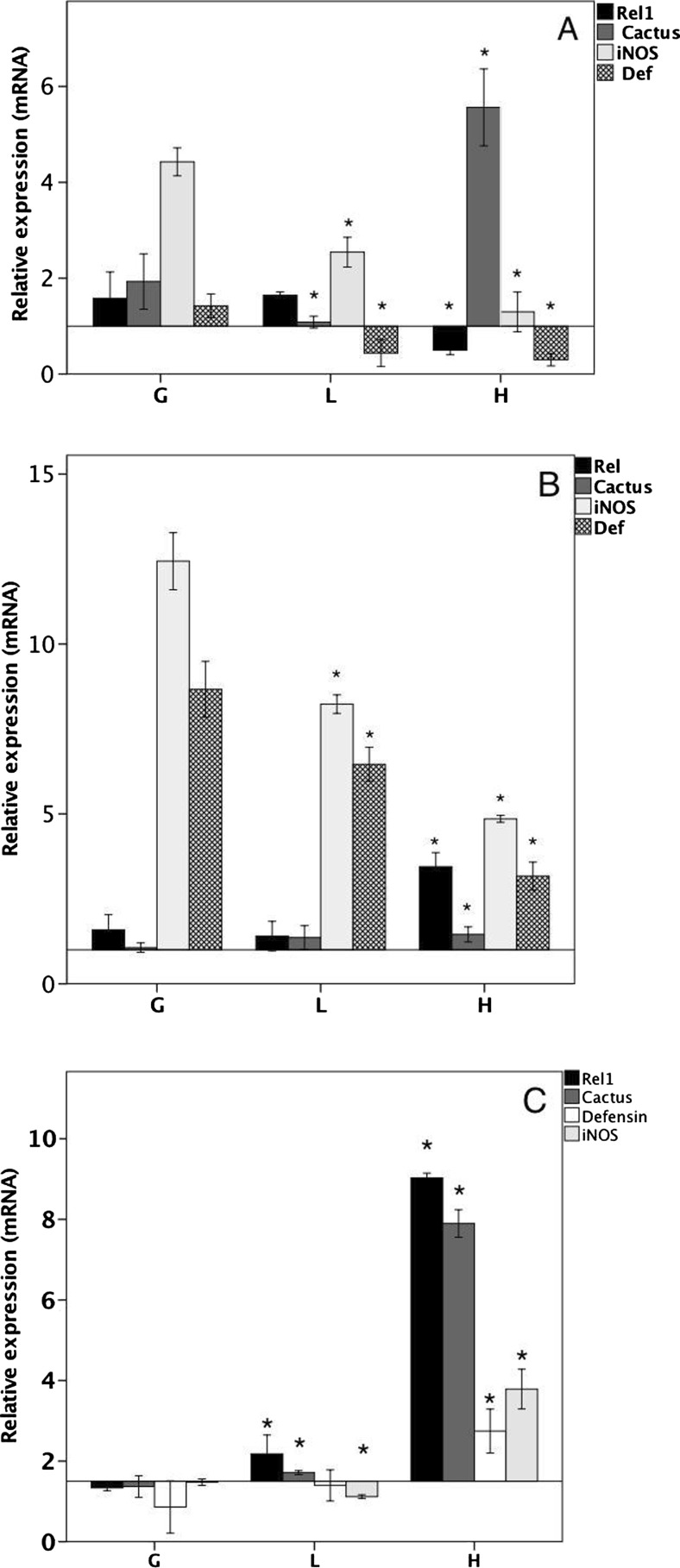
Several previous studies have shown that the activation of IRE1 arm activates NF-κB transcription factor, resulting in the downstream production of proinflammatory cytokines (Urano et al. 2000). To determine the effect of DENV2-NS4B protein over UPR-dependent immune effectors at the mRNA transcript level, we treated clones with TM for 8 and 24 h and measured Rel1 (a homolog of the transcription factor NF-κB in insects), cactus (a homolog of the negative regulator of NF-κB, Iκk), iNOS (an enzyme capable of catalyzing nitric oxide) and defensin (a family of peptides with antimicrobial activity). As shown in Fig. 3a, after 8 h of stimulation, the expression of Rel1 was unchanged, but that of cactus was significantly increased in clone H. Interestingly, the iNOS levels were increased 3.5 times in the control clones, but both the iNOS and defensin mRNA levels were significantly decreased in cells with high DENV-NS4B protein expression. Assuming that the downstream activation of immune effectors required a longer time, we conducted a 24-h analysis. After 24 h of stimulation with TM (Fig. 3b), the expression of Rel1 remained unchanged, cactus tended to increase, and defensin and iNOS had decreased transcription levels in the cells expressing the DENV-NS4B protein. To determine the specificity of the transcription of these genes, we stimulated the clones with PMA (phorbol myristate acetate) to activate the immune response and found differential induction with respect to clones stimulated with TM (Fig. 3c) but also, and more importantly, PMA regulates the RNAi pathway by inhibiting the formation of diverse microRNA effector complexes (Wu et al. 2013). Thus, it was important to discard cross-linking with this pathway. Altogether, these results demonstrate that the expression of UPR-dependent immune effectors is downregulated in cells that stably express the DENV-NS4B protein.
Fig. 3.
The expression of DENV-NS4B protein modulates the expression of UPR-dependent immune effectors. Clones G, L, and H were treated with TM (5 μg/ml) for 8 h (a) and 24 h (b), and the levels of Rel1, Cactus, iNOS and defensin expression were measured by qRT-PCR. c The clones were treated with PMA for 8 h to determine the specificity of the activation pathway. The data are presented as the means ± SD of three independent determinations. All of the results are expressed as the relative expression of the target genes with respect to actin, using the G, L, and H groups without stimulus as references. Analysis of variance (ANOVA) test with post hoc Bonferroni correction test: *P < 0.05
DENV-NS4B protein decreases antimicrobial activity and nitric oxide (NO) of UPR activation-dependent production
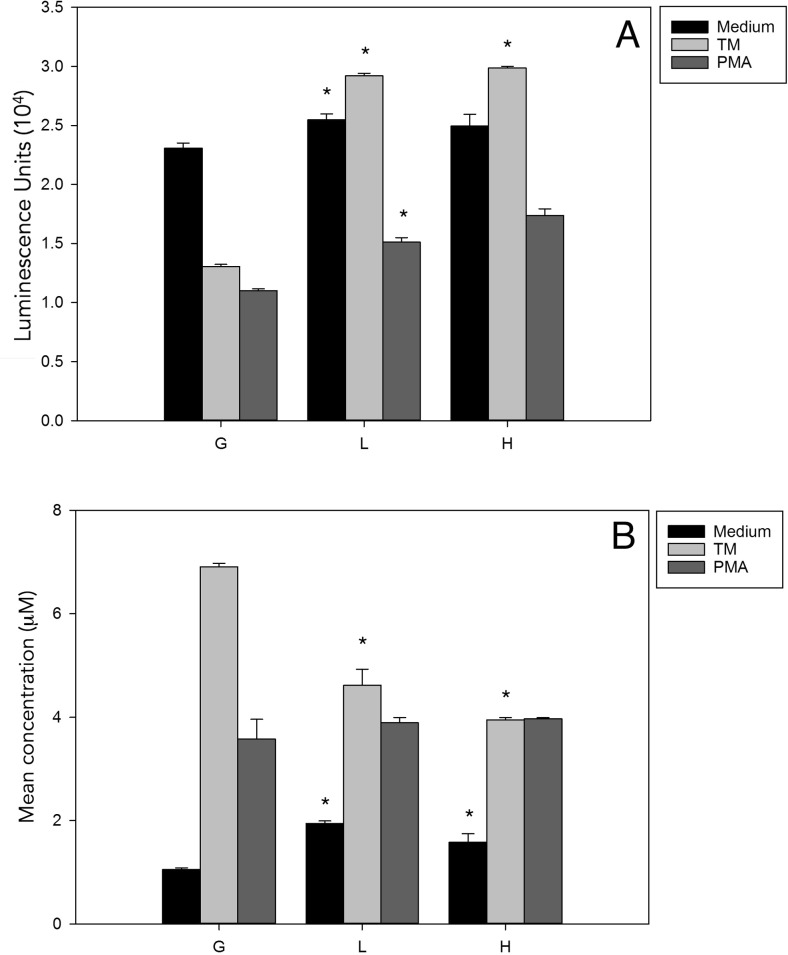
By demonstrating that UPR increases defensin mRNA in G clones and decreases clones expressing DENV-NS4B protein, we looked for antimicrobial activity in the supernatant of the clones stimulated with TM for 24 h. For this, we used a luminescence assay to measure the viability of Gram-negative bacteria (E. coli ATCC 25922) after incubation with the supernatant of the stimulated clones. As shown in Fig. 4a, in clone G stimulated with TM, there was a significant reduction in the viability of bacteria, whereas clones L and H yielded an increase in bacterial viability. The results are consistent with the possibility that by limiting the induction of defensin mRNA in cells expressing DENV-NS4B protein, functional antimicrobial activity is inhibited. To monitor iNOS as another immune effector, we measured the nitrite and nitrate concentrations in the cell culture supernatant (Fig. 4b). In the G clones, stimulation with TM for 24 h resulted in a significant increase in nitrite/nitrate, but the stable expression of DENV-NS4B protein decreased the concentration of these metabolites. The clones stimulated with phorbol-12-myristate-13-acetate (PMA, at a concentration of 5 ng/mL) showed different behavior from those stimulated with TM. These results demonstrate that the expression of DENV-NS4B protein inhibits the UPR-mediated production of both antimicrobial peptides and nitrite/nitrate as indicators of activation of NO.
Fig. 4.
The stable expression of DENV-NS4B protein decreases antimicrobial peptide production and UPR activation-dependent nitric oxide. a Clones G, L, and H were treated with TM for 24 h. To determine the antimicrobial activity in their supernatant, we used the Bac-Titer Glo assay, which determines bacteria viability based on the quantification of luminescence. b Clones G, L, and H were treated with TM for 24 h to determine NO production in the supernatant using the Griess reaction assay. Analysis of variance (ANOVA) test with post hoc Bonferroni correction test: *P < 0.05
DENV-NS4B protein effect on DENV infection and insect cell survival under ER stress
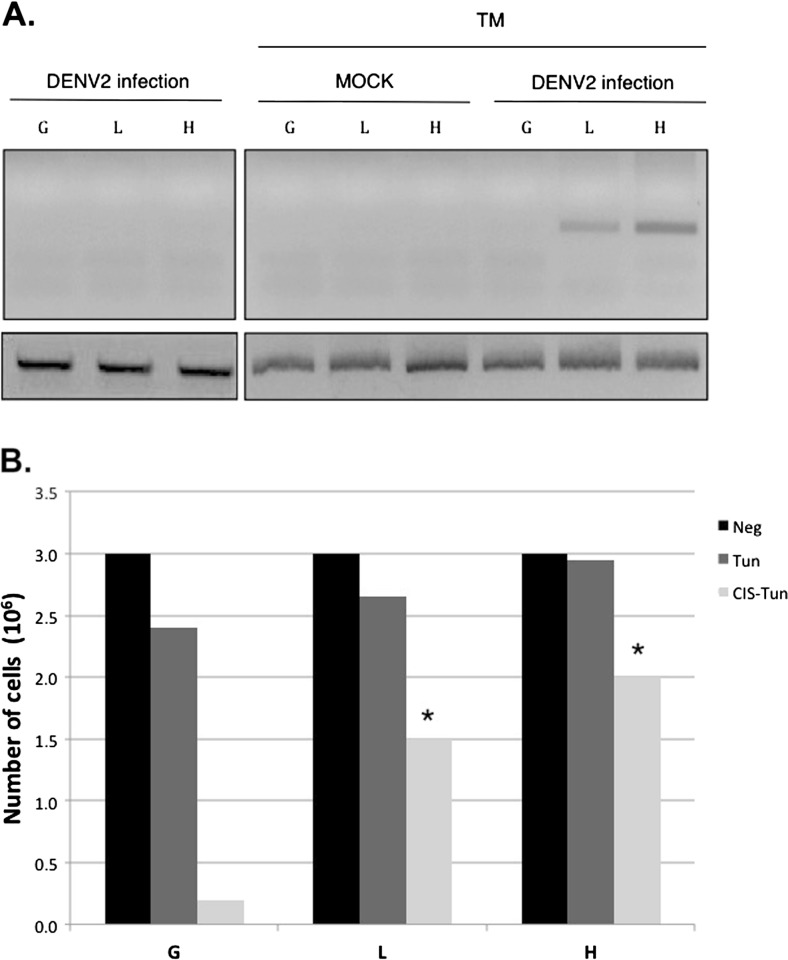
To evaluate susceptibility or resistance to DENV infection, we used the different cell clones described above. First, we tried to infect the clones with DENV2 virus without any treatment, but the cells were not infected. Later, we treated the cells with TM for 8 h, after which we expose the clones to infection with the DENV2. Remarkably, as shown in Fig. 5a, when we expose the clones to infection by DENV-2, only the clones expressing the DENV-NS4B protein and stimulated with TM became infected. One possible explanation for switching from the non-susceptible to susceptible phenotype is that clones expressing DENV-NS4B protein avoid lysis such that viral replication can increase to levels that are detectable by RT-PCR. To test this notion, we designed an experiment in which we simulated the stress effect caused by the infection with DENV, exposing cells to TM known to have induced UPR following exposure to an apoptosis inducer such as cisplatin, at a concentration that would induce approximately 50% apoptosis. Therefore, as shown in Fig. 5b, exposure to TM alone induced a reduction in the viability of approximately 17% in G cells, whereas in H cells, no change in viability was detected. When exposed to both TM and cisplatin, the protective effect is even clearer: whereas G cells decreased to almost 96% viability, L cells decreased to only ~50% and H cells to ~30%. These experiments demonstrate that by decreasing immune effectors, NS4B protein enables viral replication, and clones that express NS4B protein are partially protected from UPR-mediated cell death.
Fig. 5.
Effect of the expression of DENV-NS4B protein in dengue virus infection in insect cells undergoing ER stress. a Clones undergoing ER stress through incubation with TM (5 μg/ml) for 12 h were infected with DENV2 for 7 days. Subsequently, they were analyzed through standardized nested RT-PCR for Flavivirus. b To demonstrate the protection given to the lysis in the clones by NS4B, the clones were subjected to incubation with TM. Subsequently, they were treated with cisplatin, and cellular viability after 3 h was measured. Analysis of variance (ANOVA) test with post hoc Bonferroni correction test: *P < 0.05. Data representative of three independent experiments, compared with the G clone treated with TM and cisplatin
XBP1 splicing is the main regulator of the expression of immune effectors in insect cells
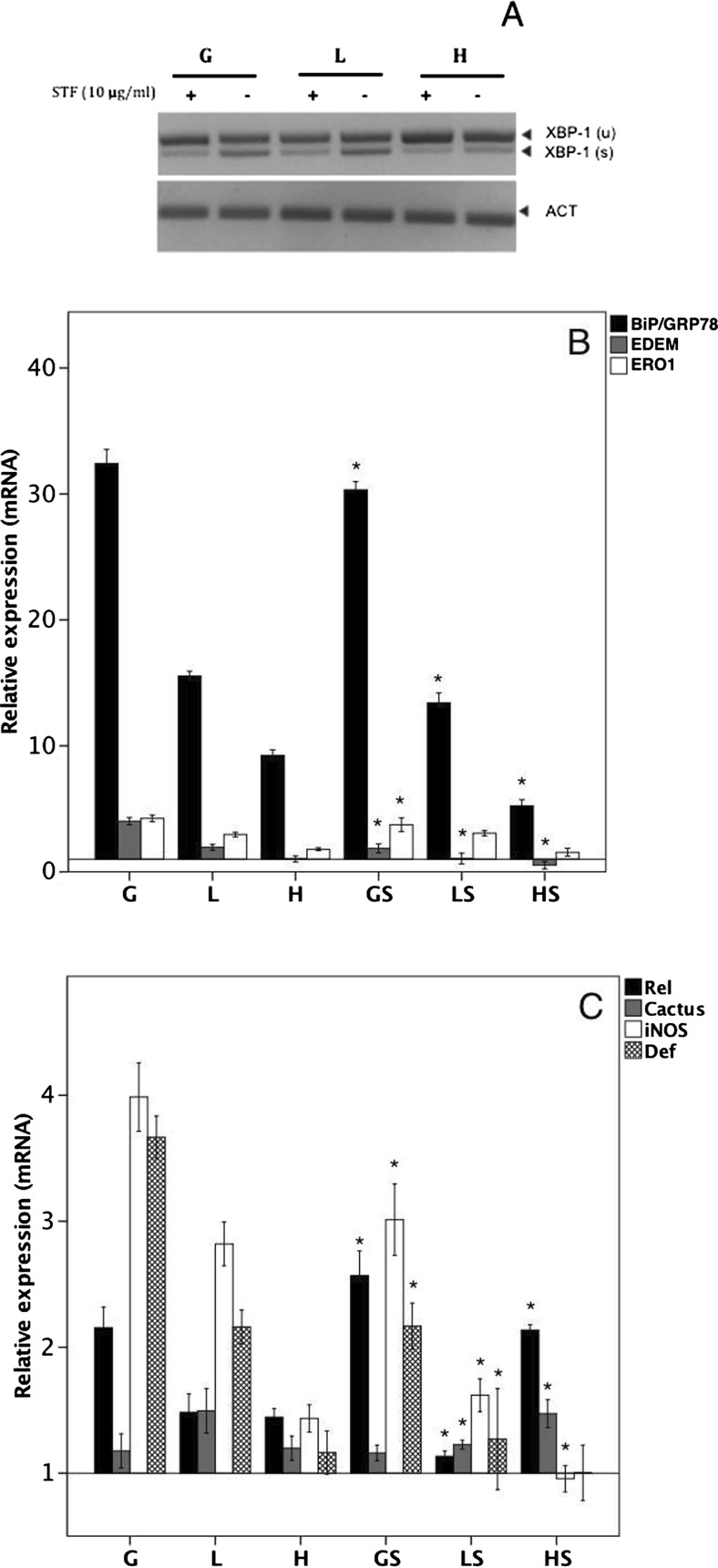
Recent evidence has demonstrated that the IRE1/XBP1 signaling is the site where UPR metabolic and innate responses intersect. To test this, we used STF080310, an inhibitor of the endoribonuclease activity of IRE1, thereby inhibiting XBP1 splicing (Fig. 6a). In cells treated with TM + STF-080310 for 8 h, there was a significant reduction in BiP/GRP78 and EDEM mRNA (Fig. 6b), with no change in Ero1. In contrast, as shown in Fig. 6c, we found differences in the mRNA levels of defensin and iNOS in cells stimulated with TM + STF-080310 compared with TM alone. These results suggest that IRE1 underlies the generation and expression of immune effectors when the UPR is activated.
Fig. 6.
IRE1 RNAse activity is the main regulator of the expression of immune factors. Clones G, L, and H were incubated with TM (5 μg/ml) in the absence or presence of STF083010 (GS, LS, and HS), an inhibitor of IRE1 endoribonuclease activity, at a concentration of 30 μM (a). The expression levels of UPR molecules in b and immune effectors in c were determined quantitatively by qRT-PCR with actin as the standard. The data are presented as the means ±SD of three independent determinations. All of the results are expressed as the relative expression of the target genes with respect to actin (ACT), using the G, L, and H groups without stimulus as references
Post-transcriptional silencing of DENV-NS4B protein recovers the phenotype of UPR-dependent immune effectors
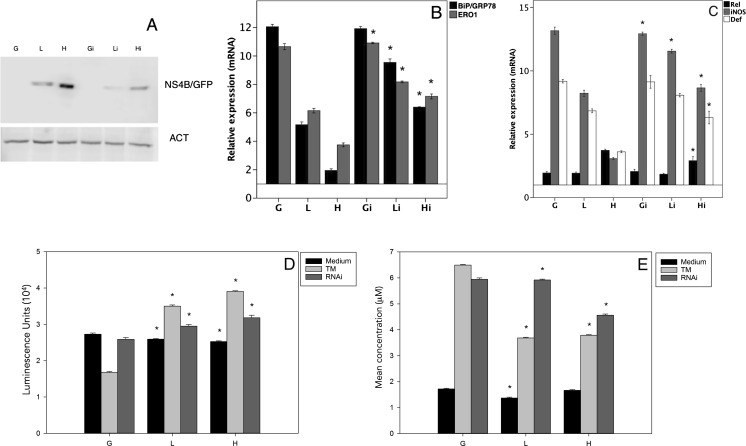
To confirm that the expression of DENV-NS4B protein can regulate the expression of immune effectors, we found that siRNA-mediated knockdown of NS4B protein restored the original phenotype of control cells. At 72 h after siRNA transfection, DENV-NS4B protein was significantly reduced (Fig. 7a), and stimulation with TM significantly enhanced the expression of BiP/GRP78 and Ero1 (Fig. 7b) and of the immune effectors iNOS and defensin (Fig. 7c). Later, we measured the nitric oxide and antimicrobial peptide levels in the supernatant of knockdown clones to observe the phenotype recuperation (Fig. 7d, e). These results strongly suggest that the magnitude of the UPR-stimulated immune effector response is negatively regulated by the DENV-NS4B protein, thereby limiting UPR-mediated cell death and enhancing viral replication.
Fig. 7.
Post-transcriptional silencing of DENV-NS4B protein recovers the phenotype of UPR-dependent immune effectors. The clones were transfected with NS4B protein RNAi (Gi, Li, and Hi) we measure the expression by WB (a) and then were incubated with TM for 24 h, followed by measurement of BiP/GRP78 and Ero1 mRNA in b and measurement of the immune effector levels in c. We used the supernatant of the cells with RNAi to measure antimicrobial activity in d and nitrite and nitrate production in e. The data are presented as the means ±SD of three independent determinations. All of the results are expressed as the relative expression of the target genes with respect to actin (ACT), using the G, L, and H groups without stimulus as references
Discussion
In this report, we contribute evidence supporting the role of DENV-NS4B protein as a modulator of the UPR-mediated immune response in insect cells. Organisms that lack the IFN-I pathway must activate the immune response to a viral infection via several pathways, such as the Toll, IMD, Jak-STAT and RNAi pathways, as demonstrated in arthropods (Mukherjee and Hanley 2010; Sim and Dimopoulos 2010; Souza-Neto et al. 2009; Xi et al. 2008); nevertheless, recently, some other non-canonical pathways have been described. In the case of viruses that use the RE as a replication site, the UPR is activated. In insects specifically, this pathway could be one of these protective pathways and is the main focus of the current study.
The UPR helps maintain proteostasis, in part, by downstream signaling emanating from the phosphorylation of the eIF2α translational initiation factor and by the direct transcriptional activation of chaperone genes such as EDEM witch recognize defective proteins for degradation and Ero1 witch maintain redox homeostasis and proper ER folding activity. When the UPR is activated by Flavivirus infection, two outcomes have been described, on one hand as a proviral mechanisms; the induction of the synthesis of internal membranes (Lei et al. 2013; Xie et al. 2013) and the preferential translation of viral proteins as mechanisms that make replication efficient and by the other hand the antiviral mechanisms that activate the immune response and cell death if the cell does not adapt to what is generating the damage. In this report, we examined the consequences of UPR activation in insect cells and stable transformants of these cells expressing DENV2-NS4B protein. Similar to DENV infection, Chikungunya virus (CHIKV) induces PERK activation but delays eIF2α phosphorylation. The expression of CHIKV NSP4, which is the RNA-dependent-RNA polymerase, contributed to the suppression of eIF2α phosphorylation, thus ensuring the translation of viral proteins (Rathore et al. 2013). In our case the expression of NS4B protein appears to be inversely proportional to the phosphorylation of eIF2α.
Furthermore, the expression of NS4B protein can regulate EDEM and Ero1 genes. Consequently, we may infer that the IRE1 arm of the UPR is modulated, with attenuation to the response of the PERK arm. In the context of infection, this effect appears beneficial for viral replication because it inhibits protein degradation induced by ER stress, and it makes the cells more resistant to death, which results in enhanced viral replication. Ambrose et al. demonstrated the viral manipulation of UPR signaling with West Nile infection in mammalian cells and suggested that UPR signaling can have a limiting effect on WNV replication but only in the early stages of replication, when the level of viral components is not sufficient to exert significant effects on downstream effectors (Ambrose and Mackenzie 2011).
Through different mechanisms, each of the three arms of the UPR may contribute to the activation of NF-κB/Rel1, which regulates the expression of cytokines such as IL-1B, TNFα and IL-6, C-reactive protein, and serum amyloid P (or DOME and VAGO in arthropods) (Hasnain et al. 2012; Smith et al. 2008; Urano et al. 2000; Zhang et al. 2006). Here, by measuring mRNA encoding immune effectors, we verified an UPR-dependent immune response. However, at 8 h of TM treatment, there was a detectable change in the expression of the NF-κB negative regulator (cactus); at 24 h, more important changes were apparent. In addition to the increase in the homolog of NF-κB Rel1, the activated UPR triggered an increase in the transcription of iNOS and defensin. However, these responses were blunted in cells expressing the DENV NS4B protein until NF-κB protein began to increase (possibly as a feedback defense mechanism).
We also found that UPR regulation in NS4B-expressing cells is not limited to transcriptomic differences; indeed, we also found a decreased level of reactive nitrites and nitrates and decreased antimicrobial activity. These findings established that the DENV-NS4b could modulate the cross talk of UPR signaling pathways with the immune response. Similarly, the inhibition of endoribonuclease activity through the pharmacological inhibitor STF083010 demonstrated a significant reduction in the expression of Ero1, NF-κB, iNOS and defensin but did not significantly alter BiP/GRP78 and EDEM mRNA levels because the latter response may come from a different arm of the UPR.
Previous studies have demonstrated that DENV-NS4B protein at the steady state is located in the ER, with a small fraction in the secretory pathway in eukaryotic cells and another fraction possibly at the junctions between the ER and mitochondria that can interfere with the formation of convoluted membranes. Another role described is in the RNAi pathway, in which DENV-NS4B protein acts as a viral RNAi suppressor by directly inhibiting the dicing process (Kakumani et al. 2013). The role of IRE1 in the activation of the UPR-immune axis leads us to speculate that IRE1 may interact with NS4B protein and, depending on its state of oligomerization, as in the RIDD mechanism (Aragon et al. 2009; Korennykh et al. 2009), enhance pro-viral responses and modulate immune signaling without the activation of PERK, which would lead to cell apoptosis. Thus, rather than have the IRE1-PERK working in opposition, multiple lines of evidence have suggested that a continuum of graded activation states (dependent on the strength of upstream stress) is available to either IRE1 or PERK, both of which, under sustained activation, undergo an adaptation promoting homeostasis against promoting cell death.
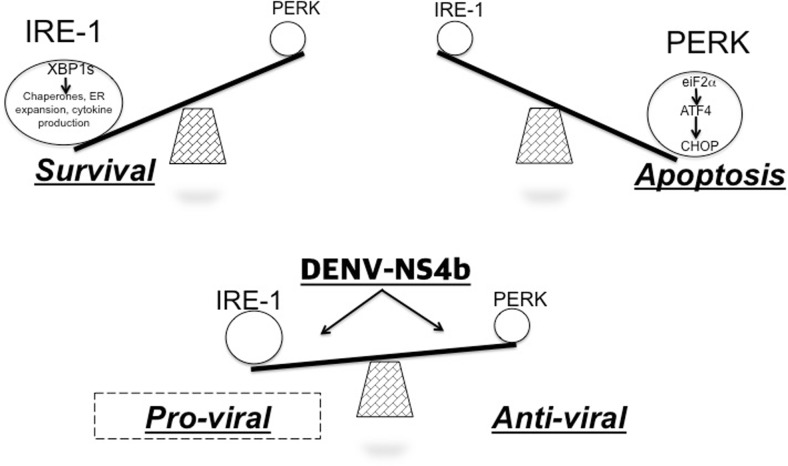
Considering Fig. 8, we suggest, based on the data found in this study, that infection in these cells only occurs when the UPR is activated and the NS4B protein is expressed. DENV-NS4B protein must modulate the IRE1 axis of the UPR, which, in Drosophila cells, is responsible for the induction of most canonical UPR target genes (Hollien and Weissman 2006). This modulation could be dependent on the manner in which IRE1 is activated by sensing the “amount of stress” generated by virus replication. Indeed, the balance between UPR/immune activity in the vector and viral circumvention mechanisms might be a determinant in the maintenance of persistent virus infection. In conclusion for insects that lack the elemental antiviral response of α/β interferon, our current findings provide further support that DENV-NS4B protein play an important role inhibiting the activation-dependent immune response of UPR, as in other several antiviral-signaling pathways already described, leading to enhance virus replication.
Fig. 8.
DENV-NS4B protein regulates the expression of UPR activation-dependent effectors through a possible interaction with IRE1. NS4B is one of the proteins with higher pro-viral characteristics because it is involved in mechanisms that make viral replication more efficient and blocks several of the immune signaling pathways in mammals. We hypothesized that NS4B could be regulating the two outputs of the UPR to benefit the replication of the virus. Thus, on the one hand, it allows the translation of viral proteins and blockade of cell death by apoptosis by decreasing the PERK pathway. On the other hand, it decreases the activation of the effectors dependent on XBP1 splicing
This study was performed on D. melonogaster cells, which are not the natural vector of the virus. However, Sessions et al. validated the use of the Drosophila screen to identify the dipteran dengue-host interaction because of the complex spatio-temporal dynamics of DENV infection in the mosquito, since the mosquitoes are genetically polymorphic and there is inherent variability of blood meal infections (Sessions et al. 2009). Further research is required to uncover the interactions that enable the DENV-NS4B protein to modulate these responses and carry out KO of the IRE1 and PERK genes in mosquito vectors of the virus to confirm the results found in this model. It is also important to develop more studies to describe the role of the UPR as an adaptive/survival response in cells facing viral challenge leading to a persistent infection.
Acknowledgements
This article was part of the doctoral thesis of KJ Sepulveda-Salinas, who is a doctoral student from the Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM). We thank to the Consejo Nacional de Ciencia y Tecnología (CONACyT) for the PhD Fellowship (344684). This work was funded by a Basic Science grant from SEP-CONACYT CB0129497 to JRC. We are grateful to Cecilia Noecker and Dr. Peter Arvan for his assistance and critical review of the manuscript.
Author contributions
KJSS and JRC conceived and design the experiments, KJSS performed the experiments, KJSS and JRC analyzed the data and wrote the paper. All authors reviewed the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The Internal Ethics Committee (CI1005-1057) approved this protocol on June 1, 2011. No informant consent was necessary.
References
- Ambrose RL, Mackenzie JM. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J Virol. 2011;85:2723–2732. doi: 10.1128/JVI.02050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoya-Perez LP, Cancino-Rodezno A, Flores-Escobar B, Soberon M, Bravo A. Role of UPR pathway in defense response of Aedes aegypti against Cry11Aa toxin from Bacillus thuringiensis. Int J Mol Sci. 2013;14:8467–8478. doi: 10.3390/ijms14048467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- Brackney DE, et al. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis. 2010;4:e856. doi: 10.1371/journal.pntd.0000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Bulleid NJ (2012) Disulfide bond formation in the mammalian endoplasmic reticulum. Cold Spring Harb Perspect Biol 4(11). doi:10.1101/cshperspect.a013219 [DOI] [PMC free article] [PubMed]
- Chatel-Chaix L, et al. Dengue virus perturbs mitochondrial morphodynamics to dampen innate immune responses. Cell Host Microbe. 2016;20:342–356. doi: 10.1016/j.chom.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavan M, Lennarz W. The molecular basis of coupling of translocation and N-glycosylation. Trends Biochem Sci. 2006;31:17–20. doi: 10.1016/j.tibs.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Cherbas L (2016) Additions to tissue culture medium. https://dgrc.bio.indiana.edu/include/file/additions_to_medium.pdf. Accessed Protocol
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/S0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Goldshmidt H, Matas D, Kabi A, Carmi S, Hope R, Michaeli S. Persistent ER stress induces the spliced leader RNA silencing pathway (SLS), leading to programmed cell death in Trypanosoma brucei. PLoS Pathog. 2010;6:e1000731. doi: 10.1371/journal.ppat.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosline SJ, Nascimento M, McCall LI, Zilberstein D, Thomas DY, Matlashewski G, Hallett M. Intracellular eukaryotic parasites have a distinct unfolded protein response. PLoS One. 2011;6:e19118. doi: 10.1371/journal.pone.0019118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Hasnain SZ, Lourie R, Das I, Chen AC, MA MG. The interplay between endoplasmic reticulum stress and inflammation. Immunol Cell Biol. 2012;90:260–270. doi: 10.1038/icb.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol Rev. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Kakumani PK, et al. Role of RNA interference (RNAi) in dengue virus replication and identification of NS4B as an RNAi suppressor. J Virol. 2013;87:8870–8883. doi: 10.1128/JVI.02774-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, et al. Endoplasmic reticulum stress-induced IRE1alpha activation mediates cross-talk of GSK-3beta and XBP-1 to regulate inflammatory cytokine production. J Immunol. 2015;194:4498–4506. doi: 10.4049/jimmunol.1401399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh AV, et al. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- Lei S, et al. ROCK is involved in vimentin phosphorylation and rearrangement induced by dengue virus. Cell Biochem Biophys. 2013;67:1333–1342. doi: 10.1007/s12013-013-9665-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Hanley KA. RNA interference modulates replication of dengue virus in Drosophila melanogaster cells. BMC Microbiol. 2010;10:127. doi: 10.1186/1471-2180-10-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A. 2003;100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol. 2005;79:8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netherton C, Moffat K, Brooks E, Wileman T. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv Virus Res. 2007;70:101–182. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell. 2005;97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena J, Harris E. Dengue virus modulates the unfolded protein response in a time-dependent manner. J Biol Chem. 2011;286:14226–14236. doi: 10.1074/jbc.M111.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore AP, Ng ML, Vasudevan SG. Differential unfolded protein response during Chikungunya and Sindbis virus infection: CHIKV nsP4 suppresses eIF2alpha phosphorylation. Virol J. 2013;10:36. doi: 10.1186/1743-422X-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CE, Kooistra T, Kim DH. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 2010;463:1092–1095. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, et al. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A, Ahola T, Kaariainen L. Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. ER stress and the unfolded protein response. Mutat Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- Sessions OM, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Dimopoulos G. Dengue virus inhibits immune responses in Aedes aegypti cells. PLoS One. 2010;5:e10678. doi: 10.1371/journal.pone.0010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Turner MJ, DeLay ML, Klenk EI, Sowders DP, Colbert RA. Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-beta induction via X-box binding protein 1. Eur J Immunol. 2008;38:1194–1203. doi: 10.1002/eji.200737882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc Natl Acad Sci U S A. 2009;106:17841–17846. doi: 10.1073/pnas.0905006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Umareddy I, Pluquet O, Wang QY, Vasudevan SG, Chevet E, Gu F. Dengue virus serotype infection specifies the activation of the unfolded protein response. Virol J. 2007;4:91. doi: 10.1186/1743-422X-4-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Welsch S, et al. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PH, Isaji M, Carthew RW. Functionally diverse microRNA effector complexes are regulated by extracellular signaling. Mol Cell. 2013;52:113–123. doi: 10.1016/j.molcel.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Gayen S, Kang C, Yuan Z, Shi PY. Membrane topology and function of dengue virus NS2A protein. J Virol. 2013;87:4609–4622. doi: 10.1128/JVI.02424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4:265–271. doi: 10.1016/S1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Yu CY, Hsu YW, Liao CL, Lin YL. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol. 2006;80:11868–11880. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]