Abstract
Crustaceans are intensively farmed in aquaculture facilities where they are vulnerable to parasites, bacteria, or viruses, often severely compromising the rearing success. The ubiquitin-proteasome system (UPS) is crucial for the maintenance of cellular integrity. Analogous to higher vertebrates, the UPS of crustaceans may also play an important role in stress resistance and pathogen defense. We studied the general properties of the proteasome system in the hemocytes of the whiteleg shrimp, Penaeus vannamei, and the European brown shrimp Crangon crangon. The 20S proteasome was the predominant proteasome population in the hemocytes of both species. The specific activities of the trypsin-like (Try-like), chymotrypsin-like (Chy-like), and caspase-like (Cas-like) enzymes of the shrimp proteasome differed between species. P. vannamei exhibited a higher ratio of Try-like to Chy-like activities and Cas-like to Chy-like activities than C. crangon. Notably, the Chy-like activity of P. vannamei showed substrate or product inhibition at concentrations of more than 25 mmol L−1. The K M values ranged from 0.072 mmol L−1 for the Try-like activity of P. vannamei to 0.309 mmol L−1 for the Cas-like activity of C. crangon. Inhibition of the proteasome of P. vannamei by proteasome inhibitors was stronger than in C. crangon. The pH profiles were similar in both species. The Try-like, Chy-like, and Cas-like sites showed the highest activities between pH 7.5 and 8.5. The proteasomes of both species were sensitive against repeated freezing and thawing losing ~80–90% of activity. This study forms the basis for future investigations on the shrimp response against infectious diseases, and the role of the UPS therein.
Keywords: Proteasome, 20S, Hemocytes, Crustacea, Crangon crangon, Penaeus vannamei
Introduction
The ATP/ubiquitin-dependent proteasome pathway is a highly complex and tightly regulated process of intracellular protein degradation (Ciechanover 2005). Proteins designated for degradation are precisely recognized, ubiquitinated by E1 to E3 ligase cascades, and directed to the proteasome (Hershko and Ciechanover 1998; Glickmann and Ciechanover 2002). Protein degradation via the ubiquitin-proteasome system (UPS) has various essential cellular functions for auto-surveillance as it is involved in cell cycle control (Ghislain et al. 1993), apoptosis (Drexler 1997; Cui et al. 1997), gene expression (Orian et al. 1995; Muratani and Tansey 2003), or stress resistance (Grune 2000). In humans, impaired UPS are associated with various disorders such as Huntington’s disease (Hipp et al. 2012) or Parkinson’s disease (Ebrahimi-Fakhari et al. 2012).
The proteasome, a large multimeric protein complex with a barrel-like structure, is predominantly located in the cytoplasm, but also occurs in the nucleus. It comprises a catalytic core (20S proteasome) to which a maximum of two regulatory protein complexes may be bound forming the 26S proteasome (Baumeister et al. 1998). The core complex comprises 28 subunits, which are organized in four stacked rings. In eukaryotes, the two outer rings consist of seven α-subunits, while the two inner rings are composed of seven β-subunits. The two inner β-rings form a central chamber carrying six threonine proteolytic active sites (Coux et al. 1996). Each of the two catalytic sites, arranged inside the 20S core, possesses the cleavage preference after which they are named: chymotrypsin-like (Chy-like), trypsin-like (Try-like), and caspase-like (Cas-like) activities. Controlled degradation is achieved in the core chamber by allosteric interactions of the catalytic sites (Kisselev et al. 1999). In vivo, the 20S and 26S proteasome populations exist. The 20S can be complexed to form the 26S proteasome degrading only ubiquitinated, and primarily short-living proteins. However, the 20S proteasome can be active on its own, degrading mainly oxidatively damaged proteins in an ubiquitin-independent manner (Grune 2000; Ciechanover 2005). Moreover, as a result of post-translational modification, single tissues/organs exhibit variable proteasome subtypes differing in their catalytic properties (Dahlmann et al. 2000).
Crustaceans belong to the most important marine taxa regarding both ecology and economy. They are abundant in the marine realm and occupy ecological key positions in coastal, neritic, and oceanic habitats. Additionally, some species have gained extraordinary relevance as a protein-rich human food source, provided either by fisheries or by aquaculture (FAO 2012). Species intensively farmed in aquaculture facilities are particularly vulnerable to diseases caused by parasites, bacteria, and viruses (Sánchez-Paz 2010).
Within the class of crustaceans, the UPS has been investigated only in few species. For example, proteasome-mediated degradation was reported to be involved in the molting process of adult and juvenile lobsters (Mykles 1999; Götze and Saborowski 2011) or in the process of learning and memory of crabs (Merlo and Romano 2007). Furthermore, it has been found that shrimp, infected with the white spot syndrome virus (WSSV), may elicit an immune response directed toward the UPS to prevent the deleterious damage caused by this virus (Jarrousse et al. 1999). The hemocytes of the circulatory system of shrimp are considered the primary target for some viruses such as the WSSV (Jiravanichpaisal et al. 2006; Feng et al. 2008). It has been recently described that an open reading frame (ORF; wsv249) located on the WSSV genome encodes a protein containing a predicted RING-H2 domain (Li et al. 2009). RING finger domains are the largest class of E3 ubiquitin ligases involved in several key processes such as cell cycle control, apoptosis, and viral replication control. Additionally, another WSSV protein (WSSV222) functions as a RING-dependent E3 ligase, and it inhibits apoptosis by ubiquitin-mediated degradation of the shrimp tumor suppressor-like protein (TSL) (He et al. 2006). Furthermore, changes in the expression of a proteasome subunit of the hemocytes of P. vannamei during an experimental infection with the Taura syndrome virus (TSV) were observed (Chongsatja et al. 2007). Even though the ubiquitination machinery has been at least partially identified in decapod crustaceans, proteasome functionality itself has not been characterized yet.
Therefore, the present study is aimed at identifying and characterizing the proteasome activities in the hemocytes of the two shrimp species, Penaeus vannamei (Dendrobranchiata), from the Mexican Pacific Ocean, and Crangon crangon (Pleocyemata, Caridea) from the European North Sea. These two species belong to the same order (Decapoda) but different taxonomic subgroups. They were chosen as model organisms to identify proteasome activities in hemocytes, to investigate catalytic properties, and to examine whether taxon-related differences in the proteasome characteristics may be present. This study forms the basis for future investigations on proteasome-mediated infection processes of shrimp by viruses.
Materials and methods
Origin of animals and sampling of hemocytes
Adult specimens of P. vannamei (n = 300) (11.7 ± 1.4 cm total body length) were obtained from a commercial farm located in Hermosillo (Sonora, Mexico). The animals were acclimated in laboratory indoor facilities for 15 days in 3000-L plastic tanks containing 1000 L of purified, aerated, and UV-treated seawater at 28 °C and 34 psu. The organisms were fed ad libitum twice daily with the commercial feed Camaronina 35® (Purina). Food residues and solid excreta were removed regularly.
Due to the prevalence of the white spot syndrome virus (WSSV), and the decapod Penstyldensovirus (PstDV-1) in the area, specimens were tested against these pathogens as follows: after acclimation, one volume (400 μL) of hemolymph samples was extracted individually from the base of the fifth pereiopod of each shrimp with a 1-mL syringe containing 400 μL of pre-cooled (4 °C) shrimp anticoagulant solution (450 mmol L−1 NaCl, 10 mmol L−1 KCl, 10 mmol L−1 Na2-EDTA, 10 mmol L−1 HEPES, pH 7.3) (Vargas-Albores et al. 1993). Hemolymph was centrifuged at 400×g for 5 min, and the resulting hemocyte pellet was resuspended in 150 μL of lysis buffer (100 mmol L−1 NaCl, 50 mmol L−1 Tris, 10 mmol L−1 EDTA, and 1% SDS) and homogenized over ice with a Teflon disposable pestle. Genomic DNA was isolated by using glassmilk (GeneClean Spin kit, MP Biomedicals) according to the manufacturer’s instructions. Samples were individually tested by PCR against WSSV by using the IQ2000 WSSV PCR kit and against PstDV-1 following a previously described procedure (Tang et al. 2007). Animals diagnosed as free of WSSV and PstDV-1 (n = 34) were then transferred into a different 3000-L tank containing approximately 1000 L of seawater under the same conditions as described above.
Subsequently, hemolymph was obtained from healthy adult specimens of P. vannamei by using 1-mL sterile syringes with 25-gauge needles filled with 0.4 mL of an anticoagulant solution (300 mmol L−1 NaCl, 10 mmol L−1 KCl, 10 mmol L−1EDTA, 10 mmol L−1 HEPES, pH 7.3) (Vargas-Albores et al. 1993). Syringes were carefully injected into the ventral blood sinus of shrimps. Up to 0.4 mL of hemolymph was obtained from each shrimp. Three hemolymph pools were taken from five individuals each (n = 3). Each pool was gently mixed and transferred into reaction tubes. Hemocytes were pelleted by centrifugation at 600×g for 10 min (4 °C), and supernatants were carefully decanted. The cell pellets were shock-frozen in liquid nitrogen and preserved at −80 °C until further analysis.
North Sea shrimp, C. crangon, were sampled with a bottom trawl in the estuary of the Weser River (Germany, 53° 50′ N, 08° 15′ E). Individuals of more than 5 cm total length were selected from the catch and transferred to the AWI laboratories in Bremerhaven. Specimens of C. crangon were not tested against WSSV or PstDV-1 as definitive cases of outbreaks have never been reported in Germany (Stentiford and Lightner 2011). Hemocytes were obtained as described for P. vannamei. Three pools of hemolymph from five individuals each were taken. Samples were centrifuged, and the isolated hemocytes were shock-frozen in liquid nitrogen and stored at −80 °C until further analysis (n = 3).
Fluorescence imaging of hemocytes
Freshly withdrawn hemolymph of C. crangon was transferred into a reaction tube and incubated for 30 min at room temperature (RT) with the substrates for either the Try-like, Chy-like, or Cas-like site of the proteasome. The substrates were dissolved in DMSO and added to the hemolymph at final concentrations of 1.5 mmol L−1 Boc-Leu-Arg-Arg-AMC for the Try-like activity, 0.3 mmol L−1 Suc-Leu-Leu-Val-Tyr-AMC for the Chy-like activity, and 2.0 mmol L−1 Ac-Gly-Pro-Leu-Asp-AMC for the Cas-like activity. In an additional assay, the hemolymph was pre-incubated for 5 min in 0.1 mmol L−1 epoxomicin before the substrates were added. The hemolymph containing the hemocytes was carefully pipetted into an Utermöhl cell chamber and observed and photographed under a laser scanning microscope (Leitz, True Confocal Scanner, PCS-SP5 II). The excitation wavelength was 360 nm (Excitation Beam Splitter FW-RT 30/70), and the emission bandwidth was 427–482 nm. The same settings were used for all samples. The objective was a ×63 HCX Pl APO CS 63.0 × 1.40 Oil UV, and the image processing software was Leica Application Suite.
Native gel electrophoresis (native-PAGE)
Shock-frozen hemocytes from C. crangon and P. vannamei were homogenized in 50 μL buffer (20 mmol L−1 Tris·HCl, 1 mmol L−1 EDTA, 5 mmol L−1 MgCl2, pH 7.5) with a micropestle. The homogenate was centrifuged for 15 min at 13,000×g, and 4 °C. Ten microliters of the supernatants was mixed with 10 μL of native sample buffer (50 mmol L−1 Tris·HCl pH 6.8, 50% glycerol, 2% bromophenol blue). Electrophoresis was carried out in a recirculating water cooler system with a Hoefer Mighty Small SE-260 chamber using NativePAGE™ 3–12% Bis-Tris gels (Invitrogen, BN1001BOX) and a running buffer (50 mmol L−1 Tris·HCl, 50 mmol L−1 Tricine, pH 6.8). Electrophoresis was run at 40 V and maximum 300 mA per gel. After separation, the gels were washed briefly with CAPS buffer (50 mmol L−1 CAPS, 0.5 mmol L−1 DTT, pH 10.5) and incubated for 15 min at 37 °C in the dark in CAPS buffer containing 0.3 mmol L−1 substrate for the proteasomal Try-like activity (Boc-Leu-Arg-Arg-AMC). The fluorescent activity bands were photographed under UV illumination in a gel documentation system (BioRad, ChemiDoc, PDQuest). Purified 26S and 20S proteasomes from human erythrocytes were used as positive control.
Preparation of enzyme extracts
Hemocytes of C. crangon were homogenized with a micropestle in 250 μL of buffer solution (50 mmol L−1 Tris·HCl, pH 7.5). Then, the samples were centrifuged for 15 min at 13,000 g and 4 °C. The supernatants were aliquoted and stored at −80 °C until further analysis. Hemocytes of P. vannamei were homogenized in 500 μL of the same buffer as described for C. crangon. Due to the low stability of the proteasome, the extracts were prepared and immediately used to determine the enzyme activities. The samples were not frozen and thawed again.
Activity measurement
The activities of the three catalytic sites of the proteasome were assayed with common fluorogenic substrates (Table 1). All assays were carried out in a NanoDrop device (Thermo Scientific, ND3300) as described previously (Götze and Saborowski 2011). In brief, the total reaction mixture of 25 μL contained 17.5 μL buffer (50 mmol L−1 Tris·HCl pH 7.5 at 30 °C), 5 μL enzyme extract yielding 10 μg protein on average, 1.25 μL of substrate solution, and either 1.25 μL dimethyl sulfoxide (DMSO) or 1.25 μL of the inhibitor epoxomicin. Substrates were dissolved in DMSO and prepared as 20-fold concentrated stock solutions freshly before use. The standard substrate concentrations in the assays were 0.75 mmol L−1 for Try-like, 0.15 mmol L−1 for Chy-like, and 1 mmol L−1 for the Cas-like activity. All samples were assayed in triplicate. The reaction mixtures were incubated in microtubes for 1 h at 30 °C. Thereafter, 2.5 μL was taken and pipetted onto the optical pathway of the NanoDrop device. The released fluorescence of the product AMC was measured at 365 nm (ex) and 437 nm (em). Blanks containing buffer, substrate, and DMSO but no enzyme were run in parallel to determine the autolysis rate of the substrates. Fluorescence was quantified via a standard series of different concentrations of the product 7-amino-4-methylcoumarin (AMC, Fluka 08440) ranging from 0 to 10 μmol L−1.
Table 1.
Substrates used for the investigation of proteasome subunit function
| Enzyme | Substrate | Source | Cat. no. |
|---|---|---|---|
| Trypsin-like | Boc-Leu-Arg-Arg-AMC | Enzo Life Sciences | BML-BW8515 |
| Chymotrypsin-like | Suc-Leu-Leu-Val-Tyr-AMC | Enzo Life Sciences | P-802 |
| Caspase-like | Ac-Gly-Pro-Leu-Asp-AMC | Enzo Life Sciences | BML-AW9560 |
Soluble protein content of each sample was determined with the Coomassie-dye reaction (Bradford 1976). Micro-plate assays were carried out with a commercial dye reagent (Biorad 500-0006). Bovine serum albumin (BSA, BioRad, 76290A) was used as standard.
Stability
The stability of the proteasome was investigated by measuring the Try-like and the Chy-like activities of fresh (F), shock-frozen (SF), and repeatedly frozen and thawed samples (F1, SF1, F2, SF2). Immediately after sampling, each of three hemolymph pools of each species was divided into two subsamples. The hemocytes of both subsamples were pelleted by centrifugation as described above. Thereafter, one pellet was directly homogenized in 500 μL buffer (50 mmol L−1 Tris·HCl, pH 7.5) as described above, while the other pellet was first shock-frozen in liquid nitrogen and thereafter homogenized in buffer. Activities of both extracts were measured following the standard procedure. The remaining extracts of both subsamples were frozen at −80 °C and then thawed, and measured again. The activities were determined over two freezing and thawing cycles.
Inhibition assays
The degrees of inhibition were investigated for all three catalytic sites of the proteasome with four commonly used proteasome inhibitors. Epoxomicin (PeptaNova, 4381), gliotoxin (Applichem, APPA7665), lactacystin (Cayman chemical company, Cas 133343-34-7), and MG132 (Sigma, C2211) were dissolved in DMSO, aliquoted, and stored at −20 °C. The final concentrations of inhibitors in the assays were epoxomicin 50 mmol L−1 (Götze and Saborowski 2011), lactacystin 1 mmol L−1 (Dick et al. 1996), gliotoxin 100 mmol L−1 (Kroll et al. 1999), and MG132 25 mmol L−1 (Luker et al. 2003). Enzymatic assays were performed as described above (n = 3).
As this study evaluates the efficacy of several proteasome inhibitors, it is worth briefly describing some of their chemical properties. Epoxomicin covalently binds to the LMP7, X, MECL1, and Z catalytic subunits of the proteasome with concomitant modification of the amino-terminal catalytic Thr residue of the 20S proteasome (Groll et al. 2000; Meng et al. 1999). Gliotoxin, a noncompetitive inhibitor of the chymotrypsin-like activity of the 20S proteasome, acts by reversible covalent modification involving mixed disulfide bonds at or near the active site of the chymotrypsin-like activity (Kroll et al. 1999). The natural product lactacystin targets the 20S proteasome by irreversibly inhibiting both the chymotrypsin-like and trypsin-like activities of the 20S proteasome, by covalent modification of the amino-terminal Thr of the catalytic β-subunit (Fenteany et al. 1995). Finally, the peptide aldehyde MG132 interacts tightly with the Thr residue in the binding pocket of the β5 catalytic subunit of the 20S proteasome by forming several stable H-bonds and maintaining a stable antiparallel β-sheet structure (Huang and Chen 2009; Zhang et al. 2009).
pH profiles and kinetics
Determination of the optimum pH of the catalytic sites was assayed at different pH according to the standard assay conditions. The pH range was chosen within the physiologically relevant range from pH 6.5 to 8.5.
The reaction velocities were measured under standard conditions as described above using different substrate concentrations. The Try-like and Chy-like activities were measured at substrate concentrations from 0 to 1.5 mmol L−1. The Cas-like activity was measured from 0 to 0.75 mmol L−1. Triplicate measurements were carried out for each substrate concentration. Additionally, the degree of inhibition by epoxomicin (standard concentration in the assays 50 mmol L−1) was measured at all substrate concentrations.
Statistics
Data sets were analyzed for normal distribution and homogeneity of variances. Percent data were transformed. Comparison between two groups was performed using Student’s t test. Comparison between more than two groups was performed with one-way ANOVA and Tukey post hoc test. The significance level was set at p < 0.05.
The results are presented as means and standard deviations. Statistical differences were indicated in graphs by letters or asterisks. All statistical analyses were carried out with the computer software SigmaStat (ver. 12) or GraphPad Prism 5 (ver. 5.4).
Results
Fluorescence imaging
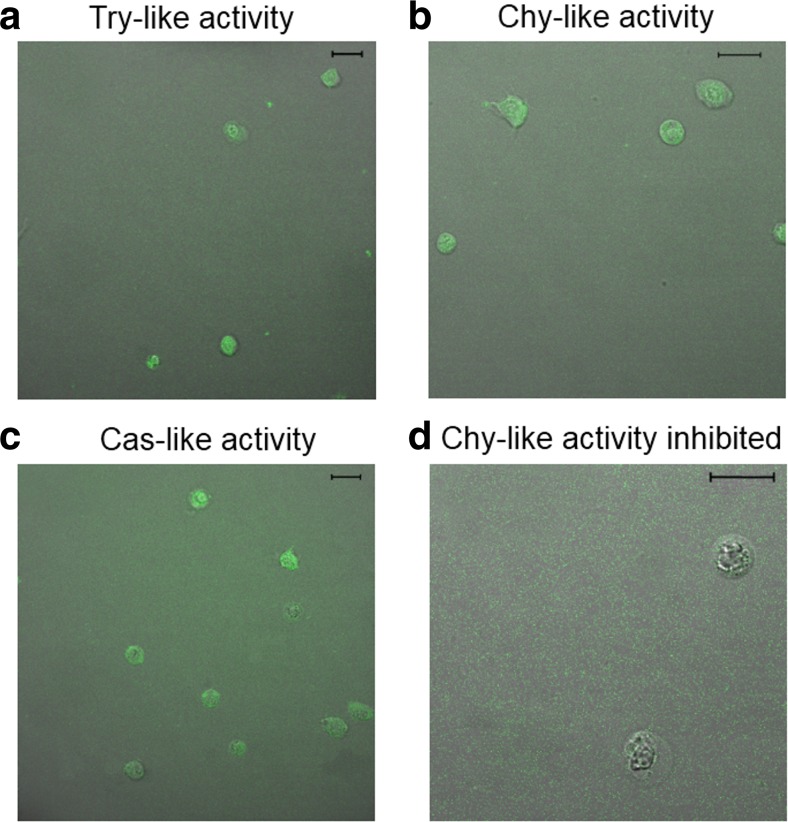
The presence of integer proteasome populations after sampling was investigated in C. crangon by fluorescence imaging after incubation of intact hemocytes with the respective fluorogenic substrate. The substrate penetrated into the hemocytes and was degraded by the proteasome into the fluorescent product (Fig. 1a–c). Simultaneous incubation of the cells with the substrate for the Chy-like activity and the inhibitor epoxomicin reduced distinctly the fluorescence of hemocytes, indicating inhibition of the proteasome within the cells (Fig. 1d).
Fig. 1.
Proteasome activities in shrimp hemocytes. Hemocytes from Crangon crangon showed fluorescence after incubation with specific substrates for a Try-, b Chy-, and c Cas-like activities. The bars represent 20 μm. d No fluorescence was detected when hemocytes were simultaneously incubated with the substrate for Chy-like activity and the inhibitor epoxomicin. The bar represents 20 μm
Native gel electrophoresis
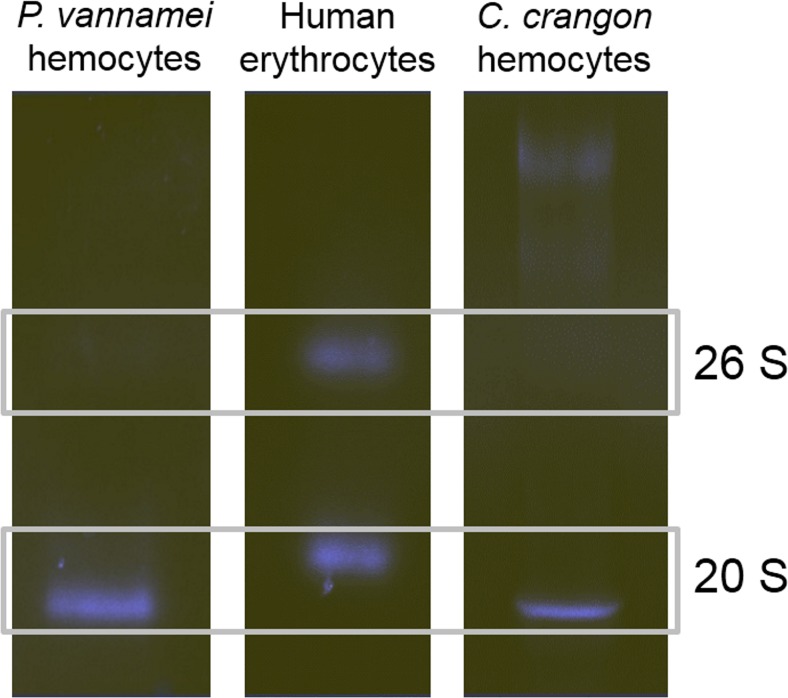
Both shrimp species showed active 20S proteasome bands (Fig. 2) with an approximate molecular mass of 600 kDa. Hence, they were slightly smaller than the 20S proteasome from human erythrocytes that accounted for 700 kDa. Neither crustacean species showed active 26S proteasome bands, which would be expected in the range of 2000 kDa.
Fig. 2.
Native electrophoresis of partly purified hemocyte proteasomes from Penaeus vannamei and Crangon crangon. Proteasome of human erythrocytes served as a positive control
Proteasome activities in hemocyte extracts
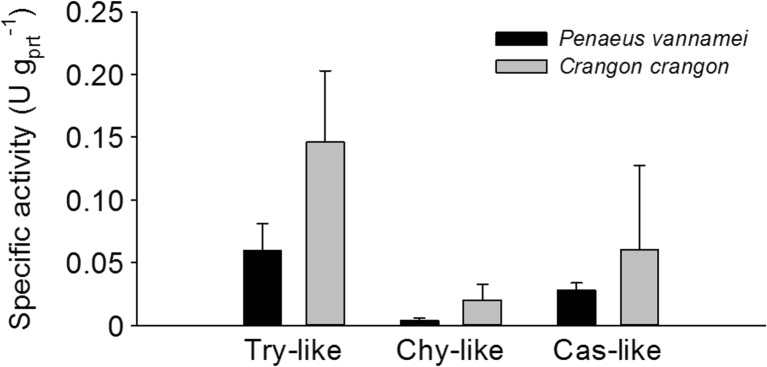
The activities of the proteasomal Try-like, Chy-like, and Cas-like sites were lower in P. vannamei than in C. crangon (Fig. 3). In P. vannamei, the average Try-like activity amounted to 0.060 ± 0.021 U gPrt −1 and in C. crangon 0.146 ± 0.057 U gPrt −1. Due to high variation among conspecifics, the differences were not statistically significant (p = 0.0717). The Chy-like activity in P. vannamei accounted for 0.004 ± 0.002 U gPrt −1 and in C. crangon (0.020 ± 0.012 U gPrt −1) (p = 0.0849). The Cas-like activity was 0.028 ± 0.005 U gPrt −1 in P. vannamei and 0.060 ± 0.067 U gPrt −1 in C. crangon (p = 0.4595).
Fig. 3.
Specific activities of the proteasome of hemocytes from Penaeus vannamei and Crangon crangon. Data are represented as the mean ± SD, n = 3
The ratios between the catalytic sites varied distinctly between species (Fig. 4). In P. vannamei, the Try-like activity was 15.4 ± 3.0 times higher than the Chy-like activity. In C. crangon, this ratio was significantly lower (p = 0.0177) than in P. vannamei showing 7.8 ± 1.7 times higher Try-like activity than Chy-like activity. The ratios between Try-like and Cas-like activities were more balanced, amounting to 2.2 ± 0.7 in P. vannamei and 2.5 ± 1.5 in C. crangon (p = 0.7045).The ratio between Cas-like and Chy-like activity was higher in P. vannamei (8.2 ± 4.8) than in C. crangon (2.3 ± 1.7), but differences were not statistically significant (p = 0.1192) due to the large scatter in data.
Fig. 4.
Ratios of proteasome activities in the hemocytes from Penaeus vannamei and Crangon crangon. The ratio values’ details are described in the text on the proteasome activities in hemocyte extracts. The asterisk (*) indicates significant difference between the activities of both organisms. Data are represented as the mean ± SD, n = 3
Stability assays
The stabilities of the Try-like and the Chy-like sites of the proteasome were investigated (Fig. 5a, b). Samples of both species were analyzed immediately after isolation, and subsamples of isolated hemocytes were immediately shock-frozen in liquid nitrogen. In both species, there were no significant differences in the Try-like and Chy-like activities between freshly extracted hemocytes and extracts from shock-frozen hemocytes. Repeated freezing and thawing, however, resulted in a strong loss of activity. The proteasome activity of P. vannamei was more susceptible to repeated freezing and thawing than that of C. crangon. P. vannamei extracts lost 90% of Try-like activity and the entire Chy-like activity after the first thawing cycle (Try-like activity: p < 0.0001, Chy-like activity: p < 0.0001). No activities were present after the second thawing cycle. Extracts of C. crangon maintained approximately 70–75% of Try-like activity and 55–60% of Chy-like activity after the first thawing cycle. After the second thawing cycle, extracts lost more than 90% of their original Try-like and Chy-like activities (Try-like activity: p < 0.0001, Chy-like activity: p < 0.0001).
Fig. 5.
Effects of repeated freezing and thawing on the proteasomal a trypsin-like and b chymotrypsin-like activities from hemocytes of Penaeus vannamei and Crangon crangon. Fresh (F) and shock-frozen (SF) hemocytes were extracted and analyzed immediately after isolation. Thereafter, extracts were frozen at −80 °C and thawed once (F1, SF1) and twice (F2, SF2) for activity determination. Means ± SD, n = 3. Treatments sharing same letters are not statistically significant from each other (p > 0.05)
Effects of inhibitors
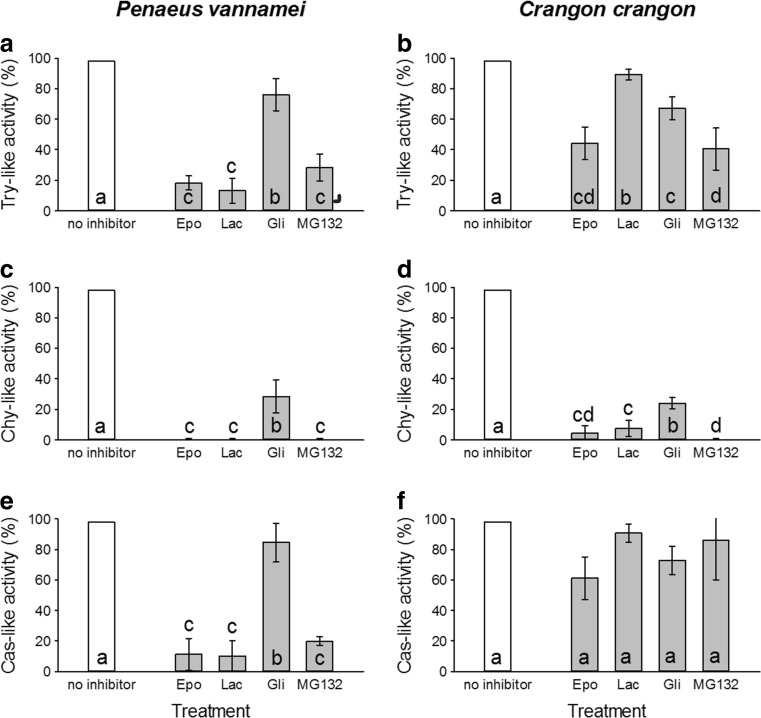
The proteasome activities of shrimp hemocytes were evaluated in the presence of four common proteasome inhibitors (Fig. 6a–f). In P. vannamei, the Try-like activity was reduced to less than 20% of its initial activity by epoxomicin and lactacystin. MG132 decreased the Try-like activity to 30%. Gliotoxin had the weakest effect on this catalytic site (Fig. 6a). The Try-like activity of C. crangon hemocytes was less affected by epoxomicin, MG132, and lactacystin than those of P. vannamei. Only gliotoxin showed a slightly higher inhibition in C. crangon than in P. vannamei (Fig. 6b). Epoxomicin, MG132, and lactacystin entirely vanished the Chy-like activity in P. vannamei hemocytes. A residual activity of 30% was observed after treatment with gliotoxin (Fig. 6c). The inhibitors had a similar effect on the Chy-like activity of C. crangon hemocytes. However, a residual activity of less than 10% remained after incubation with epoxomicin and lactacystin (Fig. 6d). The Cas-like activity of P. vannamei hemocytes was least affected by either inhibitor. The activity was reduced to 20% or less of the initial activity by epoxomicin, MG132, and lactacystin. Only low inhibition was observed with gliotoxin (Fig. 6e). The Cas-like activity of C. crangon hemocytes was less inhibited. Maximum inhibition, still leaving 60% of initial activity, was achieved with epoxomicin (Fig. 6f).
Fig. 6.
Effects of the inhibitors epoxomicin (epo), lactacystin (lac), gliotoxin (gli), and MG132 on the activities of proteasomal enzymes from hemocytes of Penaeus vannamei and Crangon crangon. a Trypsin-like activity of Penaeus vannamei (Epo, 17.83 ± 5.54%; Lac, 12.77 ± 8.92%; Gli, 76.63 ± 9.16%; MG132, 28.68 ± 7.47%), b trypsin-like activity of Crangon crangon (Epo, 43.37 ± 9.88%; Lac, 88.43 ± 4.10%; Gli, 66.75 ± 10.12%; MG132, 40.48 ± 12.53%), c chymotrypsin-like activity of Penaeus vannamei (Epo, 0.24 ± 0.48%; Lac, 0.39 ± 0.51%; Gli, 26.51 ± 12.50%; MG132, 0.39 ± 0.12%), d chymotrypsin-like activity of Crangon crangon (Epo, 4.34 ± 5.30%; Lac, 8.19 ± 7.71%; Gli, 26.51 ± 1.45%; MG132, 0.16 ± 0.06%), e caspase-like activity of Penaeus vannamei (Epo, 11.567 ± 10.12%; Lac, 10.12 ± 11.567%; Gli, 83.13 ± 13.25%; MG132, 19.28 ± 2.41%), and f caspase-like activity of Crangon crangon (Epo, 60.24 ± 16.63%; Lac, 89.16 ± 7.23%; Gli, 72.29 ± 21.21%; MG132, 84.34 ± 27.47%). Data represent means ± SD, n = 3. Treatments sharing same letters are not statistically significant from each other (p > 0.05)
pH profiles
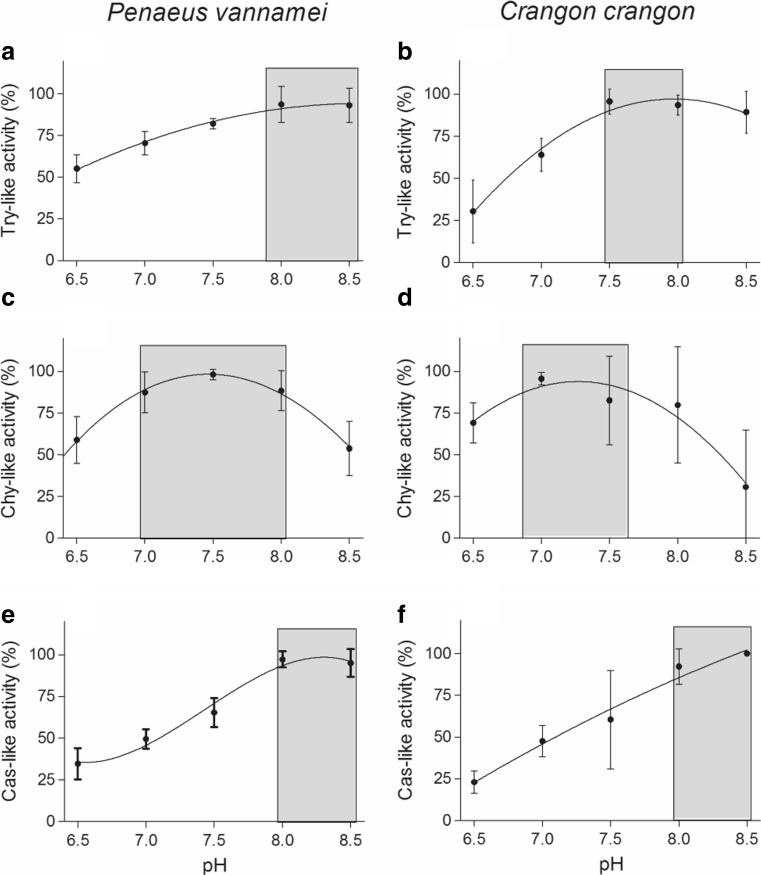
The pH profiles of the proteasome activities were similar in both species (Fig. 7a–f). The highest Try-like, Chy-like, and Cas-like activities were observed between pH 7.5–8.5, 7.0–8.0, and 8.0–8.5, respectively.
Fig. 7.
pH profiles of proteolytic enzymes from the hemocytes of Penaeus vannamei and Crangon crangon. Means ± SD, n = 3. a Trypsin-like activity of Penaeus vannamei, b trypsin-like activity of Crangon crangon, c chymotrypsin-like activity of Penaeus vannamei, d chymotrypsin-like activity of Crangon crangon, e caspase-like activity of Penaeus vannamei, and f caspase-like activity of Crangon crangon
Kinetics
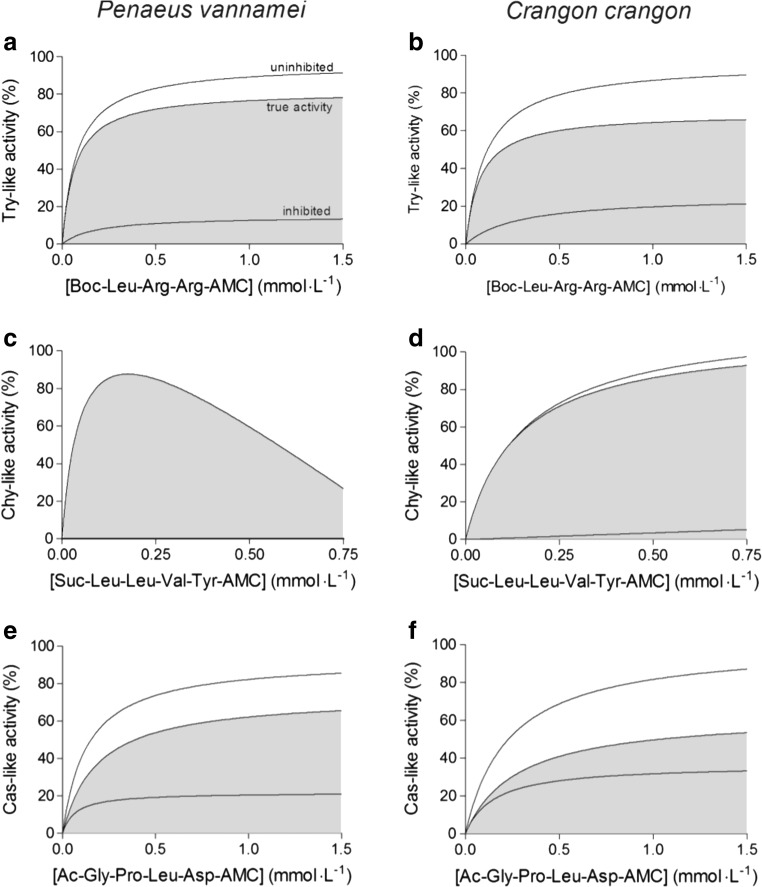
The Try-like and the Cas-like activities of the hemocytes of both species followed Michaelis-Menten kinetics (Fig. 8a, b, e, f). The same course of activity was shown by the Chy-like enzyme of C. crangon (Fig. 8d), but Chy-like activity of P. vannamei exhibited strong substrate or product inhibition at substrate concentrations higher than 0.25 mmol L−1 (Fig. 8c). The Michaelis-Menten constants (K M) of the Try-like activities did not differ significantly between species (Table 2). The K M value of the Chy-like reaction of P. vannamei could not be calculated for the entire range of the kinetic curve, but instead, it was calculated from the reaction velocities to a substrate concentration of up to 0.1 mmol L−1. It amounted to 0.309 ± 0.383 mmol L−1. In C. crangon, the K M value of the Chy-like activity was 0.167 mmol L−1. The average K M value of the Cas-like reaction was higher in C. crangon hemocytes. However, no statistically significant differences were found between both species (Table 2).
Fig. 8.
Reaction kinetics of proteasome activities of shrimp hemocytes. Means, n = 3. a Trypsin-like activity of Penaeus vannamei, b trypsin-like activity of Crangon crangon, c chymotrypsin-like activity of Penaeus vannamei, d chymotrypsin-like activity of Crangon crangon, e caspase-like activity of Penaeus vannamei, and f caspase-like activity of Crangon crangon
Table 2.
K M values for proteasome activities in the hemocytes of shrimps
| Activity | Penaeus vannamei | Crangon crangon |
|---|---|---|
| Trypsin-like | 0.072 ± 0.030 | 0.086 ± 0.009 |
| Chymotrypsin-like | 0.309 ± 0.383a | 0.167 ± 0.105 |
| Caspase-like | 0.196 ± 0.079 | 0.295 ± 0.262 |
aCalculated from the reaction velocity at substrate concentrations between 0 and 0.1 mmol L−1
Discussion
Despite its physiological and pathological relevance, the UPS is not sufficiently characterized in decapod crustaceans. Besides methodological reports (Götze and Saborowski 2011; Götze et al. 2013), almost no information is available about the catalytic properties of crustacean hemocyte proteasomes. However, it is crucial to understand such a cellular key process controlling several essential pathways (e.g., cell cycle, apoptosis, or differentiation) as this forms the basis for the investigation and interpretation of cellular stress responses or pathogen-mediated reactions.
Hemolymph sampling and hemocyte isolation were easily performed in P. vannamei because of the relatively large body size of the organisms (up to 20 cm length). In contrast, sampling of C. crangon specimens, which are smaller (5 cm length), was difficult. Low amounts of hemolymph were obtained, and thus, only a low amount of hemocytes could be isolated. However, due to the optimized assay procedure (Götze and Saborowski 2011), meaningful activity measurements were still feasible. Vital hemocytes of C. crangon showed hydrolysis of each of the three fluorogenic substrates, and particularly, the Chy-like activity was blocked by the specific proteasome inhibitor epoxomicin. Together, these results strongly show that crustacean hemocytes possess a functional proteasome complex.
During the process of hemolymph sampling, care must be taken to prevent contamination with other tissues as these residues may distort the determination of the hemocyte proteasome activities. In this study, no proteasome activity was detected in the hemolymph after sedimentation of the hemocytes (data not shown). Thus, it is concluded that the proteasome is located solely within the hemocytes, and no significant changes on the extracellular proteasome activity were observed. This may suggest that shrimp, and possibly crustaceans in general, lack extracellular circulating proteasomes (c-proteasomes/e-proteasomes) which are present in extracellular fluids and blood plasma of higher vertebrates (Sawada 2002; Sixt and Dahlmann 2008).
Native-PAGE showed a distinct 20S proteasome activity band, but no active 26S proteasome band. It seems unlikely that shrimp hemocytes do not possess 26S complexes. However, since it is known that this complex tends to be unstable (Hough et al. 1987; Waxman et al. 1987; Coux et al. 1996), it cannot be excluded that sampling, extraction, and storage of the hemocytes may have degraded the 26S complex so that only the more stable active 20S proteasome remained in both samples. Accordingly, all investigated shrimp proteasome characteristics may be solely attributed to the 20S proteasome. It should be noted, however, that from our perspective it seems improbable that the observed differences in activity of the 20S proteasome may change when complexed with the 26S proteasome activity. Previous studies have shown that in vivo, the 20S and 26S proteasome populations accomplish different tasks: the 26S proteasome degrades only ubiquitinated, primarily short-lived proteins, while the 20S core complex degrades oxidatively damaged proteins in an ubiquitin-independent manner (Grune 2000; Ciechanover 2005).
In contrast to the 20S proteasome of lobster or isolated 20S proteasome particles from other species (e.g. Mykles 1989), the 20S proteasomes of the shrimp hemocytes were fragile and quickly lost activity when subjected to repeated freezing and thawing. The low stability demands rapid processing and a strict adherence to focused experimental protocols. Particularly, repeated thawing and freezing should be strictly avoided since this can rapidly destroy proteasome activities and, thus, lead to incorrect analysis and misleading conclusions.
Our study showed that all three catalytic sites of the proteasome are active in a pH range between 6.5 and 8.5, and no differences appeared between species. The pH optimum of the three activities lies within the physiologically relevant intracellular pH range of around ~7.4. However, considerable differences in the biochemical properties of the proteasome were evident between P. vannamei and C. crangon, particularly in terms of inhibitor sensitivities and kinetics.
The selected inhibitors are commonly used in vertebrate proteasome research and are well characterized in their function and inhibitory capacity (Kisselev and Goldberg 2001; Meng et al. 1999; Dick et al. 1996; Kroll et al. 1999). These inhibitors are distinct in their modes of action, although all of them predominantly inhibit the Chy-like activity of the proteasome (Kisselev and Goldberg 2001). In vertebrate proteasome research, epoxomicin and lactacystin are known to be the most powerful inhibitors of the Chy-like site at very low concentrations in the micromolar to low millimolar range. Here, it was demonstrated for the first time that these two inhibitors, together with MG132, act most intensively on the Chy-like site of shrimp hemocyte proteasomes. Consistent with the binding mechanisms of these inhibitors, the Try-like and the Cas-like sites were less affected in both species. Remarkably, the Try-like and Cas-like sites of P. vannamei were significantly more affected by epoxomicin and lactacystin than those of C. crangon. This was also true for the inhibition of the Cas-like site in the presence of MG132. In contrast, gliotoxin was a less effective proteasome inhibitor for shrimp hemocyte proteasomes. These differences may indicate structural variation between the proteasomes of both species and demand further investigation.
Substrate kinetics for each catalytic site of the proteasome was performed to calculate the respective K M values. In both species, the Try-like and Cas-like activities apparently followed the Michaelis-Menten model while the Chy-like activity did so only in C. crangon. In P. vannamei, however, a substrate inhibition occurred above 0.1 mmol L−1 of the substrate. These results are partly in agreement with previous studies that reported that Try-like and Cas-like activities (the latter formerly denoted as PGPH-like activity) from bovine brain proteasomes followed Michaelis-Menten kinetics. Furthermore, and similar to our findings for P. vannamei, the Chy-like activity did not display Michaelis-Menten behavior but rather followed multibinding site kinetics with positive cooperativity (Piccinini et al. 2000). A complex kinetic was also reported for the Chy-like site of purified rabbit muscle 20S proteasome including substrate-induced hysteresis, substrate inhibition, and multiple substrate-binding sites (Stein et al. 1996). The authors suggested that the enzyme is subjected to substrate inhibition and that, in vivo, the enzyme requires conformational plasticity for its interactions with the regulatory complexes. A surprising result in our study is that the Chy-like activity displays different kinetics in P. vannamei and C. crangon. The fact that the Chy-like catalytic site of C. crangon follows a Michaelis-Menten model when the opposite is true for various vertebrate species, and for P. vannamei, demands further investigation.
The K M values of the catalytic sites from both shrimp hemocyte proteasomes were in the low to medium millimolar range. Shrimp proteasomes have a higher affinity for the Try-like substrate than the lobster proteasomes (K M < 0.09 mmol L−1 and K M 0.16 mmol L−1, respectively). The affinity for the Chy-like substrate is similar in either crustacean species (K M ~0.17 mmol L−1). Substrate affinities become lower when purified proteasome particles are analyzed. For example, purified rat liver 20S proteasome has a K M value of 0.06 mmol L−1 for the same Chy-like substrate as used in this study (Reidlinger et al. 1997). Various groups have investigated kinetic parameters of the proteasome (Djaballah and Rivett 1992; Cardozo et al. 1995; Kisselev et al. 2002). However, a comparison between species is not always possible since several artificial fluorogenic substrates with varying amino acid sequences were used, influencing degradation pattern and kinetic values (Luciani et al. 2005).
P. vannamei and C. crangon share similar ecological properties in terms of lifestyle, food preferences, salinity tolerance, and water temperature. Albeit these species are epibenthic inhabitants of estuaries and forage on various food sources (but are predominantly carnivorous), they occur in different climatic regions: P. vannamei is abundant in the tropical regions of the Pacific and Atlantic coasts of South and Middle America, whereas C. crangon dominates the temperate zones along the European coasts and estuaries. Though there may be a link between environmental conditions and the observed differences in the proteasome characteristics, it may be a very simplistic explanation for how climatic factors could be influencing the shape of the proteasome in both species. While our results cannot completely rule out the possibility of environmental influences, the possibility that such variations may reflect molecular and structural variation potentially due to phylogenetic separation should be also considered. Clearly, an ecological understanding of the functional role of the proteasome depends on a complete knowledge of its underlying biophysical and biochemical properties. Consequently, interpretation of activity data cannot be generalized among the phylum of crustacea but must be done in a species-specific manner. In turn, however, crustacean proteasomes may serve as a valuable model to study molecular function and evolution of the UPS in marine invertebrates.
Although there is little experimental evidence directly relating differences on proteasome activities with a precise systemic function, we can assume that the observed differences between the proteasomes of P. vannamei and C. crangon may contribute to differentially fulfill the specific physiological needs of both species. Consequently, such differences may be the key for the understanding of functional and evolutionary processes and deserve further investigation. Furthermore, as crustaceans inhabit almost all aquatic and some terrestrial ecological niches, and bear an extraordinary morphological variety, it seems obvious to predict an enormous plasticity of the proteasome system within members of this subphylum.
A limitation of our study may be the small sample size, as each analysis was run with only three specimens. This was due to the limited number of specific pathogen-free (WSSV and PstDV-1) shrimp from Sonora. Epidemiologic studies have demonstrated a high estimated prevalence for PstDV-1 in broodstock of P. vannamei from hatcheries on the northwest of Mexico (up to 63%) (Mendoza-Cano et al. 2016). Yet, we found significant differences among the properties of the proteasome of P. vannamei and C. crangon.
Organisms inhabiting aquatic environments frequently find a great variety of severe stress conditions, and establishing adequate responses to stressful stimuli is essential for their survival. A full understanding of such responses is important to provide the essential conditions for the health and survival of commercially important species. However, to understand the stress response, we must first completely comprehend its foundations. Thus, the characterization of the crustacean proteasome properties may help to enhance the knowledge of the fate of viral proteins and the specific roles of the proteasome individual components in the proteasome function. Furthermore, the biological significance of the interactions of the individual components of the crustacean proteasome remains to be explored, and it is still far from being fully understood.
Though the proteasomal capacity in both species has not been tested yet under challenging conditions, its differential functionality has been demonstrated in the current study by detailed in vitro assays and characterization of the three specific proteasome activities. Our study is the initial step for future research that may provide more resolved information about this topic as more evidence gradually accumulates.
Finally, this study identified proteasomal activities from two geographically distant decapod species and revealed their distinctive properties. Our results may be attractive for continuative basic research, and the proteasome may be exciting targets for potential future therapeutic agents against emerging pathogens affecting these species.
Acknowledgements
We are grateful to the crew of research vessel FK Uthörn for catching North Sea shrimp. Purified 26S proteasome from human erythrocytes was kindly provided by Dr. Burkhard Dahlmann (AG Kloetzel/Dahlmann, Charité, Berlin). Dr. Ulf Bickmeyer (AWI) assisted in fluorescence imaging. This work was partially supported by the grants MEX 10/011 of the International Bureau of the German Federal Ministry of Education and Research (IB-BMBF) and 102744 (to ASP) of the Consejo Nacional de Ciencia y Tecnología (CONACyT), México.
Ms. Sandra Götze and Ms. Oliviert Martinez Cruz received financial support from the International Bureau and from CONACyT for a research sojourns to Mexico and to Germany.
Contributor Information
Reinhard Saborowski, Phone: +49 (0)47148312220, Email: Reinhard.Saborowski@awi.de.
Arturo Sánchez-Paz, Phone: +52 (662) 213-15-93, Email: asanchez04@cibnor.mx.
References
- Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92(3):367–380. doi: 10.1016/S0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cardozo C, Eleuteri AM, Orlowski M. Differences in catalytic activities and subunit pattern of multicatalytic proteinase complexes (proteasome) isolated from bovine pituitary, lung, and liver. J Biol Chem. 1995;270(38):22645–22651. doi: 10.1074/jbc.270.38.22645. [DOI] [PubMed] [Google Scholar]
- Chongsatja PO, Bourchookarn A, Lo CF, Thongboonkerd V, Krittanai C. Proteomic analysis of differentially expressed proteins in Penaeus vannamei hemocytes upon Taura syndrome virus infection. Proteomics. 2007;7(19):3592–3601. doi: 10.1002/pmic.200700281. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6(1):79–86. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Cui H, Matsui K, Omura S, Schauer SL, Matulka RA, Sonenshein GE, Ju ST. Proteasome regulation of activation-induced T cell death. Proc Natl Acad Sci U S A. 1997;94(14):7515–7520. doi: 10.1073/pnas.94.14.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlmann B, Ruppert T, Kuehn L, Merforth S, Kloetzel PM. Different proteasome subtypes in a single tissue exhibit different enzymatic properties. J Mol Biol. 2000;303(5):643–653. doi: 10.1006/jmbi.2000.4185. [DOI] [PubMed] [Google Scholar]
- Dick LR, Cruikshank AA, Grenier L, Melandri FD, Nunes SL, Stein RL. Mechanistic studies on the inactivation of the proteasome by lactacystin. A central role for clasto-lactacystin β-lactone. J Biol Chem. 1996;271(13):7273–7276. doi: 10.1074/jbc.271.13.7273. [DOI] [PubMed] [Google Scholar]
- Djaballah H, Rivett AJ. Peptidylglutamyl-peptide hydrolase activity of the multicatalytic proteinase complex: evidence for a new high-affinity site, analysis of cooperative kinetics, and the effect of manganese ions. Biochemistry. 1992;31(16):4133–4141. doi: 10.1021/bi00131a033. [DOI] [PubMed] [Google Scholar]
- Drexler HCA. Activation of the cell death program by inhibition of proteasome function. Proc Natl Acad Sci U S A. 1997;94(3):855–860. doi: 10.1073/pnas.94.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D, Wahlster L, McLean PJ. Protein degradation pathways in Parkinson’s disease: curse or blessing. Acta Neuropathol. 2012;124(2):153–172. doi: 10.1007/s00401-012-1004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2012) The state of world fisheries and aquaculture 2012. Rome 209 pp
- Feng S, Zhan W, Xing J, Li J, Yang K, Wang J. Hematological changes in white spot syndrome virus-infected shrimp, Fenneropenaeus chinensis (Osbeck) J Ocean Univ China. 2008;7(3):287–293. doi: 10.1007/s11802-008-0287-7. [DOI] [Google Scholar]
- Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268(5211):726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366(6453):358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Glickmann MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Götze S, Saborowski R. NanoDrop fluorometry adopted for microassays of proteasomal enzyme activities. Anal Biochem. 2011;413(2):203–205. doi: 10.1016/j.ab.2011.02.023. [DOI] [PubMed] [Google Scholar]
- Götze S, Bose A, Abele D, Sokolova IM, Saborowski R. Pitfalls in invertebrate proteasome assays. J Exp Biol. 2013;216(8):1351–1354. doi: 10.1242/jeb.082792. [DOI] [PubMed] [Google Scholar]
- Groll M, Kim KB, Huber R, Crews CM. Crystal structure of epoxomicin:20S proteasome reveals a molecular basis for selectivity of α‘,β‘-epoxyketone proteasome inhibitors. J Am Chem Soc. 2000;122(6):1237–1238. doi: 10.1021/ja993588m. [DOI] [Google Scholar]
- Grune T. Oxidative stress, aging and the proteasomal system. Biogerontology. 2000;1(1):31–40. doi: 10.1023/A:1010037908060. [DOI] [PubMed] [Google Scholar]
- He F, Fenner BJ, Godwin AK, Kwang J. White spot syndrome virus open reading frame 222 encodes a viral E3 ligase and mediates degradation of a host tumor suppressor via ubiquitination. J Virol. 2006;80(8):3884–3892. doi: 10.1128/JVI.80.8.3884-3892.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hipp MS, Patel CN, Bersuker K, Riley BE, Kaiser SE, Shaler TA, Brandeis M, Kopito RR. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington’s disease. J Cell Biol. 2012;196(5):573–587. doi: 10.1083/jcb.201110093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough R, Pratt G, Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J Biol Chem. 1987;262(17):8303–8313. [PubMed] [Google Scholar]
- Huang L, Chen CH. Proteasome regulators: activators and inhibitors. Curr Med Chem. 2009;16(8):931–939. doi: 10.2174/092986709787581860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrousse AS, Gautier K, Apcher S, Badaoui S, Boissonnet G, Dadet MH, Henry L, Bureau JP, Schmid HP, Petit F. Relationships between proteasomes and viral gene products. Mol Biol Rep. 1999;26(1–2):113–117. doi: 10.1023/A:1006982023524. [DOI] [PubMed] [Google Scholar]
- Jiravanichpaisal P, Sricharoen S, Söderhäll I, Söderhäll K. White spot syndrome virus (WSSV) interaction with crayfish haemocytes. Fish Shellfish Immunol. 2006;20(5):718–727. doi: 10.1016/j.fsi.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Goldberg AL. Proteasome inhibitors: from research tools to drug candidates. Chem Biol. 2001;8(8):739–758. doi: 10.1016/S1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Akopian TN, Castillo V, Goldberg AL. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol Cell. 1999;4(3):395–402. doi: 10.1016/S1097-2765(00)80341-X. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Kaganovich D, Goldberg AL. Binding of hydrophobic peptides to several non-catalytic sites promotes peptide hydrolysis by all active sites of 20S proteasomes. Evidence for peptide induced channel opening in the α-rings. J Biol Chem. 2002;277(25):22260–22270. doi: 10.1074/jbc.M112360200. [DOI] [PubMed] [Google Scholar]
- Kroll M, Arenzana-Seisdedos F, Bachelerie F, Thomas D, Friguet B, Conconi M. The secondary fungal metabolite gliotoxin targets proteolytic activities of the proteasome. Chem Biol. 1999;6(10):689–698. doi: 10.1016/S1074-5521(00)80016-2. [DOI] [PubMed] [Google Scholar]
- Li F, Li M, Ke W, Ji Y, Bian X, Yan X. Identification of the immediate-early genes of white spot syndrome virus. Virology. 2009;385(1):267–274. doi: 10.1016/j.virol.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Luciani F, Kesmir C, Mishto M, Or-Guil M, de Boery RJ. A mathematical model of protein degradation by the proteasome. Biophys J. 2005;88(4):2422–2432. doi: 10.1529/biophysj.104.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker GD, Pica CM, Song J, Luker KE, Piwnica-Worms D. Imaging 26S proteasome activity and inhibition in living mice. Nat Med. 2003;9(7):969–973. doi: 10.1038/nm894. [DOI] [PubMed] [Google Scholar]
- Mendoza-Cano F, Enríquez-Espinoza T, Valenzuela-Castillo A, Encinas-García T, Sánchez-Paz A. Prevalence of the infectious hypodermal and hematopoietic necrosis virus in shrimp (Penaeus vannamei) broodstock in northwestern Mexico. Prev Vet Med. 2016;117(1):301–304. doi: 10.1016/j.prevetmed.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Meng L, Mohan R, Kwok BHB, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci U S A. 1999;96(18):10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo E, Romano A. Long-term memory consolidation depends on proteasome activity in the crab Chasmagnatus. Neuroscience. 2007;147(1):46–52. doi: 10.1016/j.neuroscience.2007.04.022. [DOI] [PubMed] [Google Scholar]
- Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4(3):192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- Mykles DL. Purification and characterization of a multicatalytic proteinase from crustacean muscle: comparison of latent and heat-activated forms. Arch Biochem Biophys. 1989;274(1):216–228. doi: 10.1016/0003-9861(89)90433-5. [DOI] [PubMed] [Google Scholar]
- Mykles DL. Structure and functions of arthropod proteasomes. Mol Biol Rep. 1999;26(1–2):103–111. doi: 10.1023/A:1006976524916. [DOI] [PubMed] [Google Scholar]
- Orian A, Whiteside S, Israël A, Stancovski I, Schwartz AL, Ciechanover A. Ubiquitin-mediated processing of NF-kB transcriptional activator precursor p105. Reconstitution of a free-cell system and identification of the uniquitin-carrier protein, E2, and a novel ubiquitin-protein ligase, E3, involved in conjugation. J Biol Chem. 1995;270(37):21707–21714. doi: 10.1074/jbc.270.37.21707. [DOI] [PubMed] [Google Scholar]
- Piccinini M, Tazartes O, Mostert M, Musso A, DeMarchi M, Rinaudo MT. Structural and functional characterization of 20S and 26S proteasomes from bovine brain. Mol Brain Res. 2000;76(1):103–114. doi: 10.1016/S0169-328X(99)00337-X. [DOI] [PubMed] [Google Scholar]
- Reidlinger J, Pike AM, Savory PJ, Murray RZ, Rivett AJ. Catalytic properties of 26S and 20S proteasomes and radiolabeling of MB1, LMP7, and C7 subunits associated with trypsin-like and chymotrypsin-like activities. J Biol Chem. 1997;272(40):24899–24905. doi: 10.1074/jbc.272.40.24899. [DOI] [PubMed] [Google Scholar]
- Sánchez-Paz A. White spot syndrome virus: an overview on an emergent concern. Vet Res. 2010;41(6):43. doi: 10.1051/vetres/2010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada H. Ascidian sperm lysin system. Zool Sci. 2002;19(2):139–151. doi: 10.2108/zsj.19.139. [DOI] [PubMed] [Google Scholar]
- Sixt SU, Dahlmann B. Extracellular, circulating proteasomes and ubiquitin—incidence and relevance. Biochim Biophys Acta. 2008;1782(12):817–823. doi: 10.1016/j.bbadis.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Stein RL, Melandri F, Dick L. Kinetic characterization of the chymotryptic activity of the 20S proteasome. Biochemistry. 1996;35(13):3899–3908. doi: 10.1021/bi952262x. [DOI] [PubMed] [Google Scholar]
- Stentiford GD, Lightner DV. Cases of white spot disease (WSD) in European shrimp farms. Aquaculture. 2011;319(1–2):302–306. doi: 10.1016/j.aquaculture.2011.06.032. [DOI] [Google Scholar]
- Tang KFJ, Navarro SA, Lightner DV. PCR assay for discriminating between infectious hypodermal and hematopoietic necrosis virus (IHHNV) and virus related sequences in the genome of Penaeus monodon. Dis Aquat Org. 2007;74(2):165–170. doi: 10.3354/dao074165. [DOI] [PubMed] [Google Scholar]
- Vargas-Albores F, Guzman MA, Ochoa JL. An anticoagulant solution for haemolymph collection and prophenoloxidase studies of penaeid shrimp (Penaeus californiensis) Comp Biochem Physiol. 1993;106A(2):299–303. doi: 10.1016/0300-9629(93)90516-7. [DOI] [Google Scholar]
- Waxman L, Fagan JM, Goldberg AL. Demonstration of two distinct high molecular weight proteases in rabbit reticulocytes, one of which degrades ubiquitin conjugates. J Biol Chem. 1987;262(2):2451–2457. [PubMed] [Google Scholar]
- Zhang S, Shi Y, Jin H, Liu Z, Zhang L, Zhang L. Covalent complexes of proteasome model with peptide aldehyde inhibitors MG132 and MG101: docking and molecular dynamics study. J Mol Model. 2009;15(12):1481–1490. doi: 10.1007/s00894-009-0515-0. [DOI] [PubMed] [Google Scholar]