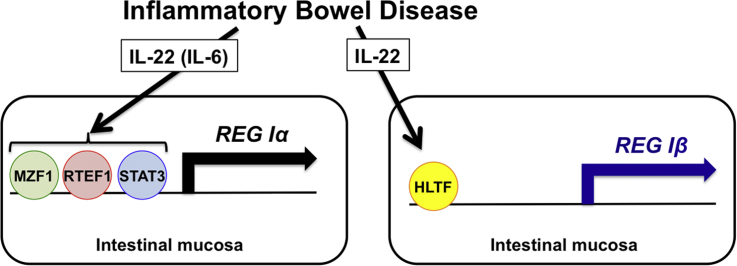
Abstract
The pathophysiology of inflammatory bowel disease (IBD) reflects a balance between mucosal injury and reparative mechanisms. Some regenerating gene (Reg) family members have been reported to be expressed in Crohn's disease (CD) and ulcerative colitis (UC) and to be involved as proliferative mucosal factors in IBD. However, expression of all REG family genes in IBD is still unclear. Here, we analyzed expression of all REG family genes (REG Iα, REG Iβ, REG III, HIP/PAP, and REG IV) in biopsy specimens of UC and CD by real-time RT-PCR. REG Iα, REG Iβ, and REG IV genes were overexpressed in CD samples. REG IV gene was also overexpressed in UC samples. We further analyzed the expression mechanisms of REG Iα, REG Iβ, and REG IV genes in human colon cells. The expression of REG Iα was significantly induced by IL-6 or IL-22, and REG Iβ was induced by IL-22. Deletion analyses revealed that three regions (− 220 to − 211, − 179 to − 156, and − 146 to − 130) in REG Iα and the region (− 274 to− 260) in REG Iβ promoter were responsible for the activation by IL-22/IL-6. The promoters contain consensus transcription factor binding sequences for MZF1, RTEF1/TEAD4, and STAT3 in REG Iα, and HLTF/FOXN2F in REG Iβ, respectively. The introduction of siRNAs for MZF1, RTEF1/TEAD4, STAT3, and HLTF/FOXN2F abolished the transcription of REG Iα and REG Iβ. The gene activation mechanisms of REG Iα/REG Iβ may play a role in colon mucosal regeneration in IBD.
Abbreviations: CD, Crohn's disease; CDX2, caudal-type homeobox transcription factor 2; FOXN2, forkhead box protein N2; GATA6, GATA DNA-binding protein 6; HLTF, helicase-like transcription factor; IBD, inflammatory bowel disease; IL, interleukin; MZF1, myeloid zinc finger 1; REG, regenerating gene; RTEF1, related transcriptional enhancer factor-1; siRNA, small interfering RNA; SOCS3, suppressors of the cytokine signaling 3; STAT3, signal transducer and activator of transcription 3; TEAD4, TEA Domain transcription Factor 4; UC, ulcerative colitis
Keywords: REG family genes, Crohn's disease, Ulcerative colitis, Transcription, Celiac disease
Graphical abstract
Highlights
-
•
REG Iα, REG Iβ, and REG IV were overexpressed in colon mucosa with ulcerative colitis.
-
•
REG IV was also overexpressed in colon mucosa with Crohn's disease.
-
•
REG Iα expression was induced by IL-22 and IL-6 via MZF1, RTEF1/TEAD4, and STAT3.
-
•
REG Iβ expression was induced by IL-22 via HLTF/FOXN2F.
-
•
REG IV expression was suppressed by TNFα via GATA6.
1. Introduction
Crohn's disease (CD) and ulcerative colitis (UC), the two primary forms of idiopathic human inflammatory bowel disease (IBD), are both characterized by chronic, destructive intestinal inflammation of unknown cause(s). Despite advances over the past decade in our understanding of cellular and molecular mechanisms underlying chronic inflammation, the precise etiopathogenic factors in IBD remain undefined. Current theories suggest that interplay between environmental, genetic, microbial, and immunologic factors results in the chronic gut inflammation that characterizes CD or UC.
Regenerating gene (REG) family proteins are structurally similar proteins belonging to the calcium-dependent (C-type) lectin superfamily. In humans, five REG family genes (i.e., REG Iα, REG Iβ, REG-related sequence (pseudogene), HIP/PAP (INGAP), and REG III) are tandemly ordered in the 95 kbp region of chromosome 2p12, whereas REG IV is located on chromosome 1q12-q21 [1], [2]. The first Reg gene was discovered in rat regenerating pancreatic islets and involved in β-cell regeneration [3], and Reg proteins have since been found in other physiological and pathophysiological processes [1], [2]. Their basic biological effects seem to be induction of cellular proliferation [4], [5], [6], [7]. Reg family proteins have been suggested to be involved in cellular proliferation in gastrointestinal cells [1], [2], [8]. Elsewhere in the gastrointestinal system, these proteins are found during tissue injury [1], [2]. They are also overexpressed in gastric and colorectal cancers [9], [10], [11], [12] and in colorectal cancer cell lines [13].
Concerning IBD, overexpression of REG Iα and REG Iβ mRNA in resected colonic tissue from CD and UC was first reported by Lawrance et al. [14]. We also showed the overexpression of HIP/PAP and REG III in IBD [15]. Subsequently, overexpression of REG Iα, REG Iβ, and REG III in IBD colon was reported [16], [17]. Overexpression of REG Iα mRNA and protein in UC, particularly in dysplasia or cancer, and a possible role for REG Iα as a marker for UC-associated neoplasia were also reported [18]. Recently, Granlund et al. analyzed four of REG family genes in five functional human REG family members, and found that the analyzed four genes (REG Iα, REG Iβ, HIP/PAP, and REG IV) were overexpressed in IBD samples [19].
Although we and others have previously suggested that REG family proteins have a trophic effect on mammalian epithelial cells and are involved in IBD, expression of all functional members of REG family genes (REG Iα, REG Iβ, REG III, HIP/PAP, and REG IV) in IBD is still unclear. In the present study, we examined all five REG family genes in IBD and found overexpression of REG Iα, REG Iβ, and REG IV mRNAs in CD and that of REG IV mRNA in UC. Reporter gene assays and siRNA-mediated knockdown experiments indicated that the overexpression of REG Iα, REG Iβ, and REG IV mRNA was mediated through several transcription factors, including myeloid zinc finger 1 (MZF1), related transcriptional enhancer factor-1 (RTEF1)/TEA Domain transcription Factor 4 (TEAD4), and signal transducer and activator of transcription 3 (STAT3) in REG Iα, helicase-like transcription factor (HLTF)/forkhead box protein N2 (FOXN2) in REG Iβ, and GATA DNA-binding protein 6 (GATA6) in REG IV gene expression.
2. Materials and methods
2.1. Tissue specimens and histological examination
Patients admitted to the Gastrointestinal Endoscopy Unit, Department of Gastroenterology, Saiseikai Nara Hospital (Nara, Japan) or Nara Medical University Hospital (Kashihara, Japan) for colonoscopy were included after informed consent. Colon biopsy samples were obtained from the rectum by endoscopy from 49 patients with CD (36 men and 13 women; mean age 33.21 ± 2.32 years, range 0.4–79) and 39 patients with UC (21 men and 18 women; mean age 43.07 ± 2.58, range 16–75). Normal control specimens were obtained from resected colon for cancer (44 patients, 26 men and 18 women; mean age 66.78 ± 1.65, range 43–91) and were used as normal control. The tissue specimens were fixed in 10% formalin solution, embedded in paraffin, and subjected to histopathological analyses.
This work was done with approval of the Review Board(s) of Saiseikai Nara Hospital and Nara Medical University Hospital. The diagnosis of UC was based on established endoscopic and histologic criteria [20].
2.2. Real-time RT-PCR
Total RNA was isolated from formalin-fixed, paraffin-embedded tissue specimens and from human colonic epithelial cells (HT-29 and LS-174T cells) using RNeasy FFPE Kit (Qiagen, Hilden, Germany) and RNeasy Protect Cell Mini Kit (Qiagen), respectively. The isolated RNA was reverse transcribed to the cDNA using High Capacity cDNA Synthesis Kit (Applied Biosystems, Foster City, CA) for the template of real-time PCR as described [21], [22], [23], [24], [25], [26], [27]. The cDNA was subjected to PCR with the following primers: β-actin (NM_001101) sense primer, 5’-GCGAGAAGATGACCCAGA-3’ and antisense primer, 5’-CAGAGGCGTACAGGGATA-3’; REG Iα (NM_002909) sense primer, 5’-AGGAGAGTGGCACTGATGACTT-3’ and antisense primer 5’-TAGGAGACCAGGGACCCACTG-3’; REG Iβ (NM_006507) sense primer, 5’-GCTGATCTCCTCCCTGATGTTC-3’ and antisense primer, 5’-TGTCAGTGATCTTGGTTTGAA-3’; REG III (AB161037) sense primer, 5’-GAATATTCTCCCCAAACTG-3’ and antisense primer, 5’-GAGAAAAGCCTGAAATGAAG-3’; HIP/PAP (NM_138937) sense primer, 5’-AGAGAATATTCGCTTAATTCC-3’ and antisense primer, 5’-AATGAAGAGACTGAAATGACA-3’; and REG IV (AY007243) sense primer, 5’-ATCCTGGTCTGGCAAGTC-3’ and antisense primer, 5’-CGTTGCTGCTCCAAGTTA-3’. All the PCR primers were synthesized by Nihon Gene Research Laboratories (Sendai, Japan). Real-time PCR was performed using KAPA SYBR® FAST qPCR Master Mix (Kapa Biosystems, Boston, MA) and Thermal Cycler Dice Real Time System (Takara Bio Inc, Kusatsu, Japan) as described [21], [22], [23], [24], [25], [26], [27]. PCR was performed with an initial step of 3 min at 95 °C followed by 40 cycles of 3 s at 95 °C and 20 s at 60 °C for β-actin, REG III, and HIP/PAP; 40 cycles of 3 s at 95 °C and 20 s at 64 °C for REG Iα, REG Iβ, and REG IV. Target cDNAs were cloned into pBluescript SK(-) plasmid (Stratagene, La Jolla, CA), and sequential 10-fold dilutions from 102 to 107 copies/µL were prepared. The serial dilutions were run to verify the specificity and to test the sensitivity of the SYBR Green-based real-time RT-PCR. Target mRNA value was normalized to that of β-actin mRNA, which was used to account for differences in the efficiency of reverse transcription between samples.
2.3. Cell culture and treatment
LS-174T and HT-29 human colonic epithelial cells were grown in RPMI 1640 medium (Nakarai Tesque, Inc., Kyoto, Japan) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 µg/mL streptomycin (Wako) [23]. Human interleukin (IL)− 6 and tumor necrosis factor (TNF)α were purchased from Roche Diagnostics (Indianapolis, IN), and human IL-8, IL-17A, IL-22, hepatocyte growth factor (HGF), basic fibroblast growth factor (bFGF), and epidermal growth factor (EGF) were from Wako. Cells were treated with 20 ng/mL IL-6, 10 nM IL-8, 1 µg/mL IL-17A, 20 ng/mL IL-22, 20 ng/mL TNFα, 50 ng/mL HGF, 10 nM bFGF, and 10 nM EGF for 24 h.
2.4. Promoter assay
The reporter constructs were prepared by inserting fragments of human REG Iα gene promoter (− 1190 to + 26, − 402 to + 26, − 370 to + 26, − 345 to + 26, − 301 to + 26, − 258 to + 26, − 247 to + 26, − 234 to + 26, − 226 to + 26, − 220 to + 26, − 211 to + 26, − 204 to + 26, − 179 to + 26, − 156 to + 26, − 146 to + 26, − 130 to + 26) [25], [26], human REG Iβ gene promoter (− 978 to + 30. − 806 to + 30, − 622 to + 30, − 562 to + 30, − 525 to + 30, − 478 to + 30, 441 to + 30, − 381 to + 30, − 326 to + 30, − 298 to + 30, − 274 to + 30, − 260 to + 30) [25] and human REG IV (− 1053 to + 22) upstream of a firefly luciferase reporter gene in pGL3-Basic vector (Promega, Madison, WI). The cells were grown in 24-well plates to 70–80% confluency and were transfected with reporter plasmids by lipofection. Briefly, 0.5 µg of each reporter plasmid and, as an internal control, 0.05 µg of pCMV-SPORT-β-galactosidase (Invitrogen, Carlsbad, CA) per well were mixed with Lipofectamine®3000 (Invitrogen) in a 24-well plate. After 24 h, the medium of each well was replaced with fresh medium containing stimulants and incubated further for 24 h. Cells were washed twice with PBS and extracts were prepared in extraction buffer (0.1 M potassium phosphate, pH 7.8/0.2% Triton X-100) as described [25], [26], [27]. Luciferase activity was measured using a PicaGene® Luciferase assay system (Toyo-ink, Tokyo, Japan) and was normalized by the β-galactosidase activity as described [25], [26], [27].
2.5. RNA interference (RNAi)
Small interfering RNA (siRNA)s directed against human MZF1, RTEF1/TEAD4, HLTF/FOXN2, STAT3, suppressors of the cytokine signaling 3 (SOCS3), caudal-type homeobox transcription factor 2 (CDX2), and GATA DNA-binding protein 6 (GATA6) were synthesized by Nihon Gene Research Laboratories. The sense sequences of siRNA for human MZF1, RTEF1/TEAD4, HLTF/FOXN2, STAT3, SOCS3, CDX2, and GATA6 were as follows: 5’-GAGGUCCUAUCAGAGAAGAtt-3’ for MZF1, 5’-GGGCAGACCUCAACACCAAtt-3’ for RTEF1/TESD4, 5’-GGAAUUUUAGCUGAUGAUAtt-3’ for HLTF/FOXN2, 5’-GCACCUUCCUGCUAAGAUUtt-3’ for STAT3, 5’-CCAAGAACCUGCGCAUCCAtt-3’ for SOCS3, 5’-AAGCCUCAGUGUCUGGCUCUGtt-3’ for CDX2, and 5’-GACAGAACGUGAUUCUCGUtt-3’ for GATA6. siRNA-scramble (Ambion®, Life Technologies) [25], [26], [27] was also used as a control. Transfection of siRNA to LS-174T cells was carried out using Lipofectamine® RNAiMAX (Thermo Fischer Scientific, Waltham, MA). Cells were transfected with 5 pmol of siRNA in a 24-well culture dish (4 × 105 cells/mL) as described [25], [26], [27].
2.6. Statistical analysis
All values for real-time RT-PCR were expressed as mean ± SE. The data were analyzed by unpaired two-tailed t-test using GraphPad Prism6 (GraphPad Software, La Jolla, CA). P value of < 0.05 was considered to be statistically significant.
3. Results
3.1. Expression of the REG family genes in IBD colon mucosa
REG family gene activation in the colon was evaluated in 88 patients with IBD (39 UC and 49 CD patients) and 44 controls (normal mucosa of surgical resected colon cancer specimens) from Saiseikai Nara Hospital and Nara Medical University Hospital. Patients with IBD were younger than control patients (mean ± SE ages of UC, CD, and control were 43.07 ± 2.58, 33.21 ± 2.32, and 66.78 ± 1.65 years old, respectively).
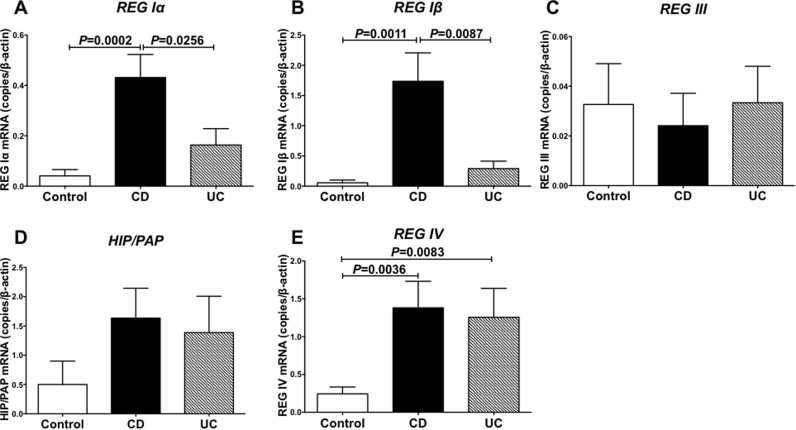
We extracted total RNA from the colon specimens and measured all REG family (REG Iα, REG Iβ, REG III, HIP/PAP, and REG IV) mRNA expression by real-time RT-PCR and found that REG Iα, REG Iβ, and REG IV genes were overexpressed in CD colon samples (Fig. 1A, B, and E). The REG IV gene was also overexpressed in UC colon samples (Fig. 1E). In contrast, the expression of type III REG genes (REG III and HIP/PAP) was not changed in IBD colon samples (Fig. 1C and D).
Fig. 1.
Expression of the REG family mRNAs in CD and UC colons. Expression of REG Iα (A), REG Iβ (B), REG III (C), HIP/PAP (D), and REG IV (E) was measured by real-time RT-PCR. The mRNA levels of REG Iα, REG Iβ, and REG IV in the CD colon were significantly higher than those of the control (P = 0.0002, 0.0011, and 0.0036, respectively). The mRNA level of REG IV in the UC colon was higher than that of the control (P = 0.0083). The mRNA levels of REG III and HIP/PAP were not different among control, CD, and UC. Data are expressed as mean ± SE for each group (n = 44 (Control), 49 (CD), and 39 (UC)). The statistical analyses were performed using Student's t-test.
3.2. Transcription of REG Iα is activated by IL-22 and IL-6 via MZF1, RTEF1/TEAD4, and STAT3
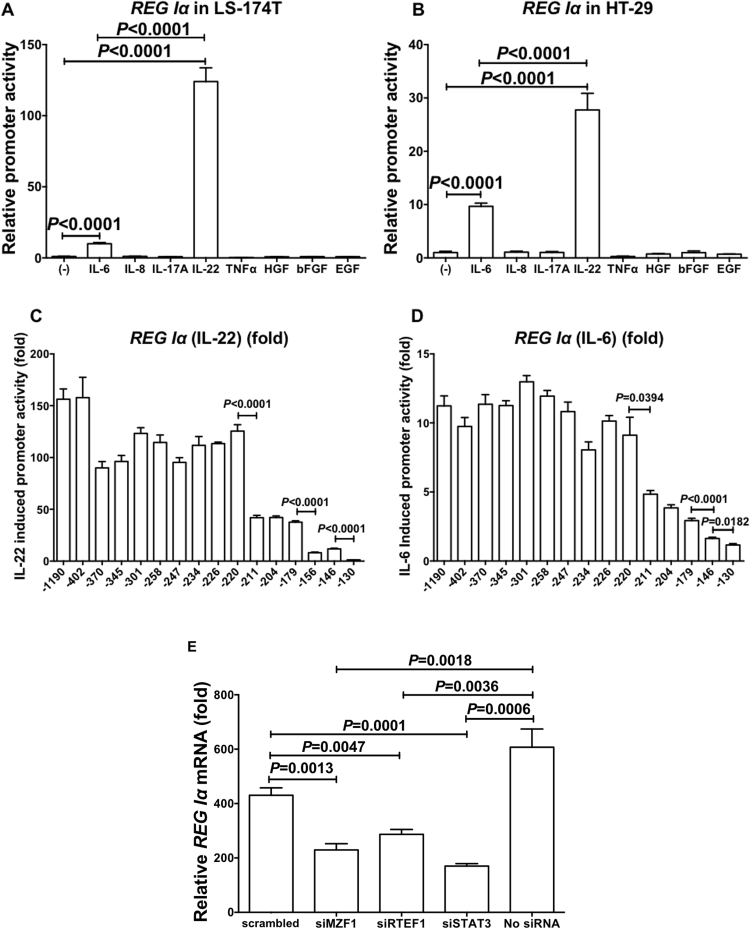
IL-6 and IL-8 are reported to induce REG family mRNA in vitro [8], [28], [29] and in vivo [22], [30], and IL-22 [31], TNFα [32], IL-17A [33], bFGF [34], HGF [35], and EGF [34] are also reported to induce REG family mRNA(s). To verify whether REG Iα gene expression is up-regulated by cytokine(s)/growth factor(s) in colon epithelial cells, we introduced luciferase reporter plasmid(s) containing REG Iα promoter (− 1190 to + 26) into colon epithelial cells (LS-174T and HT-29), stimulated by IL-6, IL-8, IL-17A, IL-22, TNFα, bFGF, HGF, or EGF, and measured transcriptional activities. As shown in Fig. 2A and B, REG Iα transcription was significantly increased by the addition of IL-6 and IL-22, and the IL-22-induced promoter activity of REG Iα showed greater increase than the IL-6-induced promoter activity in both LS-174T and HT-29 human colon cells.
Fig. 2.
Transcriptional activation of REG Iα by IL-22 and IL-6 via MZF1, RTEF1, and STAT3. Promoter activities of REG Iα in LS-174T (A) and HT-29 cells (B) stimulated with IL-6, IL-8, IL-17A, IL-22, TNFα, HGF, bFGF, or EGF were measured. The statistical analyses were performed using Student's t-test. Deletion analysis of REG Iα stimulated by IL-22 (C) and IL-6 (D). Human LS-174T cells were transfected with constructs containing various deletion mutants of REG Iα promoter. Constructs listed on ordinate are numbered according to their 5’ terminus in the REG Iα promoter. The transfected cells were stimulated with IL-22 (20 ng/mL) (C) or IL-6 (20 ng/mL) (D), after which the luciferase activities were measured. The diagram represents fold increase of luciferase activities to the untreated cells. All data are represented as the mean ± SE of the samples (n = 4). The statistical analyses were performed using Student's t-test against no addition. Effects of siRNA transfection on IL-22-induced increase of REG Iα mRNA in LS-174T cells (E). After siRNA was introduced, LS-174T human colon epithelial cells were stimulated with IL-22 (20 ng/mL). The expression of REG Iα (E) mRNA was measured by real-time RT-PCR using β-actin as an endogenous control. Data are expressed as mean fold (vs no addition) ± SE for each group (n = 4). The statistical analyses were performed using Student's t-test.
To map out the cis-element of REG Iα promoters that are responsible for REG Iα transcription in response to IL-22/IL-6 in colon epithelial cells, several lengths of REG Iα promoters were fused to the luciferase gene. After transfection of the reporter plasmid, cells were stimulated by IL-22 or IL-6 and transcriptional activity was measured. As shown in Fig. 2C, the deletion down to position − 220 in REG Iα promoter did not alter significantly the expression of the IL-22-induced reporter activation, but additional deletions to nucleotide − 211 (− 220 to − 211), − 179 to − 156, and − 146 to − 130 caused remarkable decreases of promoter activities. These results indicate that the regions of − 220 to − 211, − 179 to − 156, and − 146 to − 130 contain essential cis-elements for the IL-22 induced REG Iα promoter activities. A computer-aided search for sequences similar to known cis-acting element(s) revealed that each region has a possible binding site for MZF1, RTEF1/TEAD4, and STAT3, respectively.
We also tested nested deletion constructs of REG Iα reporter plasmids in the IL-6-induced REG Iα promoter activation. The deletion analyses showed that the essential regions for the IL-22-induced REG Iα promoter activation were also essential for the IL-6-induced REG Iα promoter (Fig. 2D), indicating that IL-6-induced REG Iα activation occurs in the same mechanism as IL-22-induced REG Iα activation.
To investigate the significance of MZF1, RTEF1/TEAD4, and STAT3 in IL-22-induced REG Iα expression in colon epithelial cells, we used RNA interference of MZF1, RTEF1/TEAD4, and STAT3 to identify whether these factors are essential for the IL-22-induced transcription of the REG Iα gene. As shown in Fig. 2E, the introduction of either MZF1, RTEF1/TEAD4, or STAT3 siRNA into the LS-174T cells significantly reduced the IL-22-induced expression of REG Iα mRNA as compared to no siRNA or scrambled siRNA introduction.
3.3. Transcription of REG Iβ is activated by IL-22 via HLTF/FOXN2
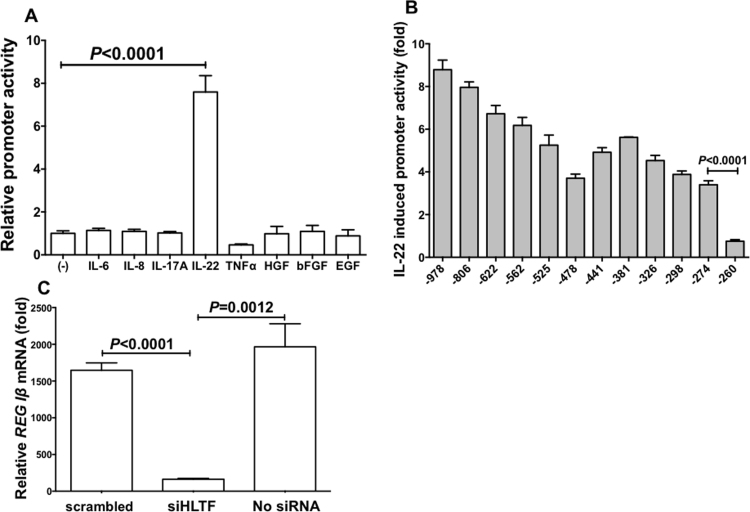
IL-22 showed significant increase in REG Iβ promoter activation, but the other cytokines/growth factors did not (Fig. 3A). Progressive deletions of the REG Iβ promoter gene were performed, the deleted constructs were transfected into LS-174T cells, and the IL-22-induced promoter activities were measured. The deletion analyses of REG Iβ promoter revealed that the − 274 to − 260 region was important for the IL-22-induced REG Iβ promoter activation (Fig. 3B), and that the region contains a possible HLTF/FOXN2 binding site.
Fig. 3.
Transcriptional activation of REG Iβ by IL-22 via HLTF. (A) Promoter activities of REG Iβ in LS-174T cells stimulated with IL-6, IL-8, IL-17A, IL-22, TNFα, HGF, bFGF, or EGF were measured. The statistical analyses were performed using Student's t-test. (B) Deletion analysis of REG Iβ promoter. Human LS-174T cells were transfected with constructs containing various deletion mutants of REG Iβ promoter. Constructs listed on ordinate are numbered according to their 5’ terminus in the REG Iβ promoter. The transfected cells were stimulated with IL-22 (20 ng/mL), after which the luciferase activities were measured. The diagram represents fold increase of luciferase activities to the untreated cells. All data are represented as the mean ± SE of the samples (n = 4). The statistical analyses were performed using Student's t-test against no addition. (C) Effects of siRNA transfection on IL-22-induced increase of REG Iβ mRNA in LS-174T cells. After siRNA was introduced, LS-174T human colon epithelial cells were stimulated with IL-22 (20 ng/mL). The expression of REG Iβ mRNA was measured by real-time RT-PCR using β-actin as an endogenous control. Data are expressed as mean fold (vs no addition) ± SE for each group (n = 4). The statistical analyses were performed using Student's t-test.
To investigate the significance of HLTF/FOXN2 for REG Iβ expression in colon epithelial cells, we used RNA interference of HLTF/FOXN2 to verify whether HLTF/FOXN2 is essential for the IL-22-induced transcription of REG Iβ gene. As shown in Fig. 3C, the introduction of HLTF/FOXN2 siRNA significantly reduced the IL-22-induced REG Iβ mRNA as compared to no siRNA or scrambled siRNA introduction.
3.4. GATA6 is required for REG IV expression
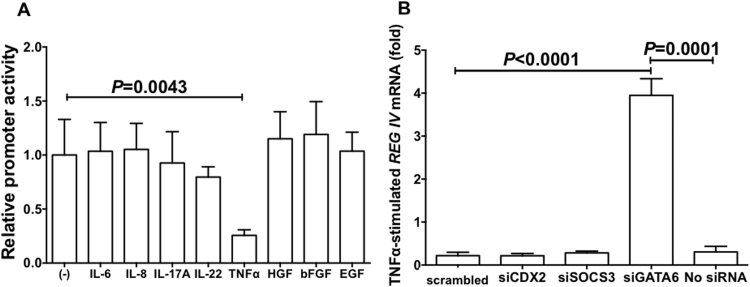
In REG IV promoter, no enhanced promoter activity was detected in all the cytokine/growth factor additions (IL-6, IL-8, IL-17A, IL-22, TNFα, HGF, bFGF, and EGF) in LS-174T cells. On the other hand, TNFα significantly decreased the REG IV promoter activity in LS-174T cells (Fig. 4A).
Fig. 4.
Requirement of GATA6 in REG IV expression. (A) Promoter activities of REG IV in LS-174T cells stimulated with IL-6, IL-8, IL-17A, IL-22, TNFα, HGF, bFGF, or EGF were measured. The statistical analyses were performed using Student's t-test. (B) Effects of siRNA transfection on TNFα-induced decrease of REG IV mRNA. After siRNA was introduced, LS-174T human colon epithelial cells were stimulated with TNFα (20 ng/mL). The expression of REG IV mRNA was measured by real-time RT-PCR using β-actin as an endogenous control. Data are expressed as mean fold (vs no addition) ± SE for each group (n = 4). The statistical analyses were performed using Student's t-test.
Concerning the TNFα-induced suppression of REG IV transcription, we tested siRNAs for CDX2, SOCS3, and GATA6 as CDX2 [36] and GATA6 [37] were reported to be involved in REG IV gene expression and TNFα was reported to control SOCS3 expression [38], and found that GATA6 siRNA abolished the TNFα-induced suppression of REG IV expression as compared to no siRNA or scrambled siRNA (Fig. 4B). This indicates that GATA6 but not CDX2 nor SOCS3 is an essential transcription factor for REG IV expression in colon epithelial cells and is responsible for the TNFα-induced suppression of REG IV expression.
4. Discussion
The pathophysiology of IBD such as UC and CD reflects a balance between mucosal injury related to an ongoing inflammatory process and mucosal reparative mechanisms. As proliferative mucosal factors may offer new therapeutic paradigms, Dieckgraefe et al. searched such candidate genes and found that REG family genes (REG Iα, REG Iβ, and HIP/PAP) were highly expressed in IBD colonic mucosa [17]. Ogawa et al. found that Reg IIIβ and Reg IIIγ were overexpressed in sodium dextran sulfate-induced mouse colitis as well as in a murine bacterial reconstitution model, and that HIP/PAP was overexpressed in human IBD colonic mucosa [15]. Numerous papers on expression analyses in one or more members of REG family genes in IBD mucosa followed these findings [16], [18], [19], [31], [39]. Despite these reports, expression of all members of REG family genes in IBD lesions has not been clarified. In the present study, we analyzed expression of all REG family mRNAs (REG Iα, REG Iβ, REG III, HIP/PAP, and REG IV) in CD and UC samples and found that REG family genes (REG Iα, REG Iβ, and REG IV) were overexpressed in CD colons and that the REG IV gene was also overexpressed in UC samples. In the present study, we used normal parts of resected colon cancer as normal ‘control’ and therefore the patients (CD and UC) were much younger than ‘control’. As Perfetti et al. showed that mouse pancreatic Reg I (mouse counterpart of REG Iα/REG Iβ) mRNA levels declined with age [40], we additionally analyzed 21 much younger control samples (mean age 15.6 years old) and found that mRNA levels of REG Iα, REG Iβ, and REG IV in the younger controls were not significantly different from those of ‘control’ in Fig. 1 (data not shown). In addition, we also analyzed REG family gene expression in another IBD, Celiac disease [41]. In subjects from Barcelona (12 Celiac disease patients before gluten-free diet and 10 age-related controls), we found that any of the REG family genes were not overexpressed in Celiac disease samples (Supplemental Fig. 1), suggesting that the REG family gene expression and its role(s) in disease progression are different among CD, UC, and Celiac disease, and the significance of the REG family gene expression as biomarkers is different. The autoimmune condition of Celiac disease and the role of some REG proteins as autoantigens [22], [42], [43] may contribute to explain these differences.
Originally, Reg was isolated as a gene specifically expressed in regenerating rat islets induced by 90% pancreatectomy with poly(ADP-ribose) polymerase inhibitor (such as nicotinamide and 3-aminobezamide) administrations [2], [3]. The Reg and Reg-related genes were isolated and were revealed to constitute a multigene family, the Reg gene family [1], [2]. Based on the primary structures of the Reg proteins, the members of the family are grouped into four subclasses; types I, II, III, and IV [1]. In humans, five REG family genes, i.e., REG Iα [3], [9], REG Iβ [44], REG-related sequence (pseudogene) (RS) [9], HIP/PAP [45], [46], and REG III [47] are tandemly ordered in the 95 kbp region of chromosome 2p12 [48], whereas REG IV locates on chromosome 1 [49]. In mouse genome, all Reg family genes except for Reg IV (i.e., Reg I, Reg II, Reg IIIα, Reg IIIβ, Reg IIIγ, and Reg IIIδ) were mapped to a contiguous 75 kbp region of chromosome 6 C [50], whereas Reg IV was mapped on chromosome 3. Type I (and type II) Reg proteins are expressed in regenerating islets [1] and are involved in β-cell regeneration [4], [5], [42], [51]. Reg family proteins have been suggested to be involved in cellular proliferation in gastrointestinal cells [52], hepatic cells [53], cardiovascular cells [30] and neuronal cells. Importantly, mouse Reg III was shown to be a Schwann cell mitogen that accompanies the regeneration of motor neurons [54], and Reg protein functions as a neurotrophic factor for motor neurons [54], [55]. Reg protein was also shown to mediate gastrointestinal epithelial cell proliferation in rats [56], [57]. Yonemura et al. showed that the expression of the REG Iα gene is closely related to the infiltrating property of gastric carcinoma, and may be a prognostic indicator of differentiated adenocarcinoma of the stomach [11]. In fact, following publication of this paper, correlations were reported between REG family gene expression and cancer prognosis [11], [12], [24], [58]. These observations suggest that the Reg gene family is involved in cell proliferation in a variety of cell types, including gastrointestinal cells.
In the present study, overexpression of REG Iα, REG Iβ, and REG IV in CD and of REG IV in UC colon mucosa was detected. It was reported previously that the REG family genes were expressed not only in various human inflammatory diseases such as gastritis [59], pancreatitis [60], salivary glanditis [22], [26], and colitis [15], but also in various experimental models of inflammation in animal tissues [30], [61]. REG Iα and REG Iβ were recently reported to be upregulated in human intestine during Entamoeba histolytica-infected acute colitis, and intestinal epithelia cells from Reg I (mouse counterpart of REG Iα/REG Iβ) knockout mice were found to be more susceptible to spontaneous, and parasite-induced, apoptosis [62], suggesting that REG Iα and REG Iβ could function to protect the intestinal epithelium from parasite-induced apoptosis. Thus, it is most likely that inflammation, regardless of whether it is autoimmune associated, is a key event that triggers REG family gene expression in many tissues. Therefore, whether REG family overexpression is associated directly with the immune disorder in patients with IBD is an interesting question. We performed real-time RT-PCR analyses of all REG family genes in IBD samples and found overexpression of REG Iα, REG Iβ, and REG IV in CD and of REG IV in UC colon mucosa (Fig. 1). These results support the idea that REG family mRNA overexpression is associated with inflammation triggered by autoimmune disorders such as IBD.
It was reported that Reg gene expression was regulated by several factors, such as nicotinamide [28], [30], glucocorticoids [28], nutrient factors [63], IL-6 [25], [26], [28], IL-8 [29], IL-17A [62], IL-22 [31], TNFα [32], HGF [35], bFGF [34], and EGF [34]. We tested the induction of REG Iα, REG Iβ, and REG IV by IL-6, IL-8, IL-17A, IL-22, TNFα, HGF, bFGF, and EGF and found that REG Iα was induced by IL-22/IL-6 via transcription factor(s) MZF1, RTEF1/TEAD4, and STAT3 (Fig. 1) and that REG Iβ was induced by IL-22 via HLTF/FOXN2 (Fig. 2). Recent reports have identified a crucial role for IL-22 in the regulation of both gut inflammation and epithelial barrier integrity. IL-22 gene delivery leads to rapid amelioration of dextran sodium sulfate-induced colitis [65], whereas IL-22 knockout mice show increased intestinal epithelial damage, along with systemic bacterial burden and significantly increased mortality [66]. Sekikawa et al. reported that IL-22 induced REG Iα expression via STAT3 in UC [31]. IL-6 signaling is thought to be of central importance for the maintenance of chronic intestinal inflammation in IBD such as CD and UC. IL-6 has also been implicated in the pathogenesis of colorectal cancer. In fact, IL-6 directly promotes tumor cell proliferation and survival through STAT3 activation. Due to its role in both types of diseases, IL-6 has been proposed as a missing link between inflammation and tumor development in the colon [67].
In this study, we showed the IL-22/IL-6-induced up-regulation of REG Iα and REG Iβ in LS-174T and HT-29 human colon cells and their molecular mechanism(s) in which several transcription factors (MZF1, RTEF1/TEAD4, and STAT3 in REG Iα, and HLTF/FOXN2 in REG Iβ) were involved. Recently, MZF1 was reported to be involved in colorectal cancer [68], and RTEF1/TEAD4 was associated with increased growth of gastric cancer [69]. Accumulating evidence indicates that IL-22/IL-6-STAT3 signaling is important in tumorigenesis and tumor growth of colitis-associated colorectal cancer [70]. HLTF/FOXN2 was reported to be associated with lower prognosis of glioblastoma patients [71]. These associations between transcription factors and cancer tumor growth/poor prognosis of cancer patients may be mediated via up-regulation of REG Iα/REG Iβ.
Up-regulation of REG IV gene in IBD has been reported previously [34], [39], [72]. However, the mechanism of REG IV expression in the colon is still controversial: Involvements of CDX2 [36] and GATA6 [37] were reported. In the present study, we found that REG IV mRNA was significantly up-regulated in UC and CD samples and down-regulated by the addition of TNFα in cultured colon cells. We therefore introduced siRNAs for CDX2, GATA6, and SOCS3 into LS-174T colon cells and evaluated the effect of the siRNAs on the TNFα-induced suppression. The introduction of GATA6 siRNA, but not CDX2 nor SOCS3 siRNA, was significantly attenuated the TNFα-induced REG IV suppression (Fig. 4), indicating that GATA6 is essential for REG IV expression and TNFα-induced REG IV suppression in colon epithelial cells.
Up-regulation of REG Iα, REG Iβ, and REG IV may have two conflicting sides like Dr. Jekyll and Mr. Hyde. The expression induces to protect/recover the intestinal epithelium from immune-mediated damages by proliferation of intestinal epithelial cells by REG family proteins in UC and CD. On the other hand, it may sometimes result in an increased risk of colon cancer. Therefore, long lasting IL-22/IL-6 production and the IL-22/IL-6-induced REG family gene up-regulation should be monitored and controlled for healthy recovery of intestinal epithelium. After recovery of damaged epithelium, anti-IL-22/anti-IL-6 antibodies or anti-REG family proteins may be a possible new preventive therapy to reduce colon cancer risk in UC and CD patients.
In the present study, we analyzed the expression of all REG family genes and found overexpression of REG Iα, REG Iβ, and REG IV in IBD. We tested cytokines that were reported to regulate the expression of the Reg family genes, regardless of whether they activate them, and found that IL-22/IL-6 up-regulated REG Iα via MZF1, RTEF1/TEAD4, and STAT3 and that IL-22 up-regulated REG Iβ via HLTF/FOXN2 in human colon cells. We also found that TNFα down-regulated REG IV expression via GATA6 in colon epithelial cells.
Acknowledgements
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, Japan Science and Technology Agency, and CERCA Programme/Generalitat de Catalunya, Catalan Government, and is in partial fulfillment by C. Tsuchida of the degree of Doctor of Medical Science at Nara Medical University. We would like to thank Drs. Masakazu Segawa, Michiyoshi Hisanaga, Teruhiko Imai, Shoji Teramoto in Saiseikai Nara Hospital for cooperating in collecting IBD patient samples. We are also grateful to Drs. Hiroyo Ota, Kiyomi Yoshimoto, Takanori Fujimura, Hiroki Tsujinaka, Akitaka Nonomura, and Shigeki Sugiura from Nara Medical University for encouragement.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2017.10.003.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2017.10.003.
Appendix A. Transparency document
Supplementary material
Appendix A. Supplementary material
Supplemental Fig. 1. Expression of the REG family mRNAs in Celiac disease colon samples. The mRNA levels of REG family (REG Iα, REG Iβ, REG III, HIP/PAP, and REG IV) in Celiac disease and control colon samples (12 Celiac disease patients before gluten-free diet and 10 age-related controls) in Barcelona were analyzed by real-time RT-PCR. The mRNA levels of REG Iα, REG Iβ, REG III, HIP/PAP, and REG IV normalized with β-actin mRNA level were not different between Celiac disease and the control. Data are expressed as mean ± SE for each group (n = 10 (Control: black circle), and 12 (Celiac disease: red square)). The statistical analyses were performed using Student's t-test. Supplementary material
References
- 1.Okamoto H., Takasawa S. Recent advances in the Okamoto model: the CD38-cyclic ADP-ribose signal system and the regenerating gene protein (Reg)-Reg receptor system in β-cells. Diabetes. 2002;51:S462–S473. doi: 10.2337/diabetes.51.2007.s462. [DOI] [PubMed] [Google Scholar]
- 2.Takasawa S. Regenerating gene (REG) product and its potential clinical usage. Expert Opin. Ther. Targets. 2016;20:541–550. doi: 10.1517/14728222.2016.1123691. [DOI] [PubMed] [Google Scholar]
- 3.Terazono K., Yamamoto H., Takasawa S., Shiga K., Yonemura Y., Tochino Y., Okamoto H. A novel gene activated in regenerating islets. J. Biol. Chem. 1988;263:2111–2114. [PubMed] [Google Scholar]
- 4.Watanabe T., Yonemura Y., Yonekura H., Suzuki Y., Miyashita H., Sugiyama K. Pancreatic beta-cell replication and amelioration of surgical diabetes by Reg protein. Proc. Natl. Acad. Sci. USA. 1994;91:3589–3592. doi: 10.1073/pnas.91.9.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takasawa S., Ikeda T., Akiyama T., Nata K., Nakagawa K., Shervani N.J. Cyclin D1 activation through ATF-2 in Reg-induced pancreatic β-cell regeneration. FEBS Lett. 2006;580:585–591. doi: 10.1016/j.febslet.2005.12.070. [DOI] [PubMed] [Google Scholar]
- 6.Ose T., Kadowaki Y., Fukuhara H., Kazumori H., Ishihara S., Udagawa J. Reg I-knockout mice reveal its role in regulation of cell growth that is required in generation and maintenance of the villous structure of small intestine. Oncogene. 2007;26:349–359. doi: 10.1038/sj.onc.1209799. [DOI] [PubMed] [Google Scholar]
- 7.Cui W., De Jesus K., Zhao H., Takasawa S., Shi B., Srikant C.B., Liu J.-L. Overexpression of Reg3α increases cell growth and the levels of cyclin D1 and CDK4 in insulinoma cells. Growth Factors. 2009;27:195–202. doi: 10.1080/08977190902863548. [DOI] [PubMed] [Google Scholar]
- 8.Sekikawa A., Fukui H., Fujii S., Takeda J., Nanakin A., Hisatsune H. REG Iα protein may function as a trophic and/or anti-apoptotic factor in the development of gastric cancer. Gastroenterology. 2005;128:642–653. doi: 10.1053/j.gastro.2004.12.045. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe T., Yonekura H., Terazono K., Yamamoto H., Okamoto H. Complete nucleotide sequence of human reg gene and its expression in normal and tumoral tissues. The reg protein, pancreatic stone protein, and pancreatic thread protein are one and the same product of the gene. J. Biol. Chem. 1990;265:7432–7439. [PubMed] [Google Scholar]
- 10.Zheng H.C., Sugawara A., Okamoto H., Takasawa S., Takahashi H., Masuda S., Takano Y. Expression profiling of the REG gene family in colorectal carcinoma. J. Histochem. Cytochem. 2011;59:106–115. doi: 10.1369/jhc.2010.956961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yonemura Y., Sakurai S., Yamamoto H., Endou Y., Kawamura T., Bandou E. REG gene expression is associated with the infiltrating growth of gastric carcinoma. Cancer. 2003;98:1394–1400. doi: 10.1002/cncr.11658. [DOI] [PubMed] [Google Scholar]
- 12.Dhar D.K., Udagawa J., Ishihara S., Otani H., Kinoshita Y., Takasawa S. Expression of Regenerating gene I in gastric adenocarcinomas: correlation with tumor differentiation status and patient survival. Cancer. 2004;100:1130–1136. doi: 10.1002/cncr.20097. [DOI] [PubMed] [Google Scholar]
- 13.Yamauchi A., Takahashi I., Takasawa S., Nata K., Noguchi N., Ikeda T. Thiazolidinediones inhibit REG Iα gene transcription in gastrointestinal cancer cells. Biochem. Biophys. Res. Commun. 2009;379:734–738. doi: 10.1016/j.bbrc.2008.12.113. [DOI] [PubMed] [Google Scholar]
- 14.Lawrance I.C., Fiocchi C., Chakravarti S. Ulcerative colitis and Crohn's disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum. Mol. Genet. 2001;10:445–456. doi: 10.1093/hmg/10.5.445. [DOI] [PubMed] [Google Scholar]
- 15.Ogawa H., Fukushima K., Naito H., Funayama Y., Unno M., Takahashi K. Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflamm. Bowel Dis. 2003;9:162–170. doi: 10.1097/00054725-200305000-00003. [DOI] [PubMed] [Google Scholar]
- 16.Wu F., Dassopoulos T., Cope L., Maitra A., Brant S.R., Harris M.L. Genome-wide gene expression differences in Crohn's disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm. Bowel Dis. 2007;13:807–821. doi: 10.1002/ibd.20110. [DOI] [PubMed] [Google Scholar]
- 17.Dieckgraefe B.K., Crimmins D.L., Landt V., Houchen C., Anant S., Porche-Sorbet R., Ladenson J.H. Expression of the regenerating gene family in inflammatory bowel disease mucosa: reg Iα upregulation, processing, and antiapoptotic activity. J. Investig. Med. 2002;50:421–434. doi: 10.1136/jim-50-06-02. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka H., Fukui H., Fujii S., Sekikawa A., Yamagishi H., Ichikawa K. Immunohistochemical analysis of REG Iα expression in ulcerative colitis-associated neoplastic lesions. Digestion. 2011;83:204–209. doi: 10.1159/000321808. [DOI] [PubMed] [Google Scholar]
- 19.Granlund Av, Beisvag V., Torp S.H., Flatberg A., Kleveland P.M., Ostvik A.E. Activation of REG family proteins in colitis. Scand. J. Gastroenterol. 2011;46:1316–1323. doi: 10.3109/00365521.2011.605463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matts S.G. The value of rectal biopsy in the diagnosisof ulcerative colitis, Quart. J. Med. 1961;30:393–407. [PubMed] [Google Scholar]
- 21.Masui T., Ota I., Itaya-Hironaka A., Takeda M., Kasai T., Yamauchi A. Expression of REG III and prognosis in head and neck cancer. Oncol. Rep. 2013;30:573–578. doi: 10.3892/or.2013.2521. [DOI] [PubMed] [Google Scholar]
- 22.Yoshimoto K., Fujimoto T., Itaya-Hironaka A., Miyaoka T., Sakuramoto-Tsuchida S., Yamauchi A. Involvement of autoimmunity to REG, a regeneration factor, in patients with primary Sjögren's syndrome. Clin. Exp. Immunol. 2013;174:1–9. doi: 10.1111/cei.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagawa K., Takasawa S., Nata K., Yamauchi A., Itaya-Hironaka A., Ota H. Prevention of Reg I-induced β-cell apoptosis by IL-6/dexamethasone through activation of HGF gene regulation. Biochim. Biophys. Acta. 1833;2013:2988–2995. doi: 10.1016/j.bbamcr.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Kimura M., Naito H., Tojo T., Itaya-Hironaka A., Dohi Y., Yoshimura M. REG Iα gene expression is linked with the poor prognosis of lung adenocarcinoma and squamous cell carcinoma patients via discrete mechanisms. Oncol. Rep. 2013;30:2625–2631. doi: 10.3892/or.2013.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamauchi A., Itaya-Hironaka A., Sakuramoto-Tsuchida S., Takeda M., Yoshimoto K., Miyaoka T. Synergistic activations of REG Iα and REG Iβ promoters by IL-6 and glucocorticoids through JAK/STAT pathway in human pancreatic β cells. J. Diabetes Res. 2015;2015:173058. doi: 10.1155/2015/173058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujimura T., Fujimoto T., Itaya-Hironaka A., Miyaoka T., Yoshimoto K., Yamauchi A. Interleukin-6/STAT pathway is responsible for the induction of gene expression of REG Iα, a new auto-antigen in Sjögren's syndrome patients, in salivary duct epithelial cells. Biochem. Biophy. Rep. 2015;2:69–74. doi: 10.1016/j.bbrep.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsujinaka H., Itaya-Hironaka A., Yamauchi A., Sakuramoto-Tsuchida S., Ota H., Takeda M. Human retinal pigment epithelial cell proliferation by the combined stimulation of hydroquinone and advanced glycation end-products via up-regulation of VEGF gene. Biochem. Biophys. Rep. 2015;2:123–131. doi: 10.1016/j.bbrep.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akiyama T., Takasawa S., Nata K., Kobayashi S., Abe M., Shervani N.J. Activation of Reg gene, a gene for insulin-producing β-cell regeneration: poly(adp-ribose) polymerase binds Reg promoter and regulates the transcription by autopoly(ADP-ribosyl)ation. Proc. Natl. Acad. Sci. USA. 2001;98:48–53. doi: 10.1073/pnas.240458597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshino N., Ishihara S., Rumi M.A.K., Ortega-Cava C.F., Yuki T., Kazumori H. Interleukin-8 regulates expression of Reg protein in Helicobacter pylori-infected gastric mucosa. Am. J. Gastroenterol. 2005;100:2157–2166. doi: 10.1111/j.1572-0241.2005.41915.x. [DOI] [PubMed] [Google Scholar]
- 30.Kiji T., Dohi Y., Takasawa S., Okamoto H., Nonomura A., Taniguchi S. Activation of regenerating gene Reg in rat and human hearts in response to acute stress. Am. J. Physiol. Heart Circ. Physiol. 2005;289:H277–H284. doi: 10.1152/ajpheart.01206.2004. [DOI] [PubMed] [Google Scholar]
- 31.Sekikawa A., Fukui H., Suzuki K., Karibe T., Fujii S., Ichikawa K. Involvement of the IL-22/ REG Iα axis in ulcerative colitis. Lab. Investig. 2010;90:496–505. doi: 10.1038/labinvest.2009.147. [DOI] [PubMed] [Google Scholar]
- 32.Fujishiro M., Nozawa K., Kawasaki M., Yamaguchi A., Iwabuchi K., Yanagida M. Regenerating gene (REG) 1 alpha promotes pannus progression in patients with rheumatoid arthritis. Mod. Rheumatol. 2012;22:228–237. doi: 10.1007/s10165-011-0564-y. [DOI] [PubMed] [Google Scholar]
- 33.Lai Y., Li D., Li C., Muehleisen B., Radek K.A., Park H.J. The antimicrobial protein REG3A regulates keratinocyte proliferation and differentiation after skin injury. Immunity. 2012;37:74–84. doi: 10.1016/j.immuni.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nanakin A., Fukui H., Fujii S., Sekikawa A., Kanda N., Hisatsune H. Expression of the REG IV gene in ulcerative colitis. Lab. Investig. 2007;87:304–314. doi: 10.1038/labinvest.3700507. [DOI] [PubMed] [Google Scholar]
- 35.Otonkoski T., Mally M.I., Hayek A. Opposite effects of β-cell differentiation and growth on Reg expression in human fetal pancreatic cells. Diabetes. 1994;43:1164–1166. doi: 10.2337/diab.43.9.1164. [DOI] [PubMed] [Google Scholar]
- 36.Naito Y., Oue N., Hinoi T., Sakamoto N., Sentani K., Ohdan H. Reg IV is a direct target of intestinal transcriptional factor CDX2 in gastric cancer. PLoS One. 2012;7:e47545. doi: 10.1371/journal.pone.0047545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawasaki Y., Matsumura K., Miyamoto M., Tsuji S., Okuno M., Suda S. REG4 is a transcriptional target of GATA6 and is essential for colorectal tumorigenesis. Sci. Rep. 2015;5:14291. doi: 10.1038/srep14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bode J.G., Nimmesgern A., Schmitz J., Schaper F., Schmitt M., Frisch W. LPS and TNFα induce SOCS3 mRNA and inhibit IL-6-induced activation of STAT3 in macrophages. FEBS Lett. 1999;463:365–370. doi: 10.1016/s0014-5793(99)01662-2. [DOI] [PubMed] [Google Scholar]
- 39.Kämäräinen M., Heiskala K., Knuutila S., Heiskala M., Winqvist O., Andersson L.C. RELP, a novel human REG-like protein with up-regulated expression in inflammatory and metaplastic gastrointestinal mucosa. Am. J. Pathol. 2003;163:11–20. doi: 10.1016/S0002-9440(10)63625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perfetti R., Egan J.M., Zenilman M.E., Shuldiner A.R. Differential expression of reg-I and reg-II genes during aging in the normal mouse. J. Gerontol. Biol. Sci. 1996;51A:B308–B315. doi: 10.1093/gerona/51a.5.b308. [DOI] [PubMed] [Google Scholar]
- 41.Vives-Pi M., Takasawa S., Pujol-Autonell I., Planas R., Cabré E., Ojanguren I. Biomarkers for diagnosis and monitoring of celiac disease. J. Clin. Gastroenterol. 2013;47:308–313. doi: 10.1097/MCG.0b013e31827874e3. [DOI] [PubMed] [Google Scholar]
- 42.Shervani N.J., Takasawa S., Uchigata Y., Akiyama T., Nakagawa K., Noguchi N. Autoantibodies to REG, a beta-cell regeneration factor, in diabetic patients. Eur. J. Clin. Investig. 2004;34:752–758. doi: 10.1111/j.1365-2362.2004.01419.x. [DOI] [PubMed] [Google Scholar]
- 43.Gurr W., Shaw M., Li Y., Sherwin R. RegII is a β-cell protein and autoantigen in diabetes of NOD mice. Diabetes. 2007;56:34–40. doi: 10.2337/db06-0669. [DOI] [PubMed] [Google Scholar]
- 44.Moriizumi S., Watanabe T., Unno M., Nakagawara K., Suzuki Y., Miyashita H. Isolation, structural determination and expression of a novel reg gene, human reg Iβ. Biochim. Biophys. Acta. 1994;1217:199–202. doi: 10.1016/0167-4781(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 45.Lasserre C., Christa L., Simon M.T., Vernier P., Bréchot C. A novel gene (HIP) activated in human primary liver cancer. Cancer Res. 1992;52:5089–8095. [PubMed] [Google Scholar]
- 46.Orelle B., Keim V., Masciotra L., Dagorn J.C., Iovanna J.L. Human pancreatitis-associated protein. Messenger RNA cloning and expression in pancreatic diseases. J. Clin. Investig. 1992;90:2284–2291. doi: 10.1172/JCI116115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nata K., Liu Y., Xu L., Ikeda T., Akiyama T., Noguchi N. Molecular cloning, expression and chromosomal localization of a novel human REG family gene, REG III. Gene. 2004;340:161–170. doi: 10.1016/j.gene.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Miyashita H., Nakagawara K., Mori M., Narushima Y., Noguchi N., Moriizumi S. Human REG family genes are tandemly ordered in a 95-kilobase region of chromosome 2p12. FEBS Lett. 1995;377:429–433. doi: 10.1016/0014-5793(95)01381-4. [DOI] [PubMed] [Google Scholar]
- 49.Hartupee J.C., Zhang H., Bonaldo M.F., Soares M.B., Dieckgraefe B.K. Isolation and characterization of a cDNA encoding a novel member of the human regenerating protein family: Reg IV. Biochim. Biophys. Acta. 2001;1518:287–293. doi: 10.1016/s0167-4781(00)00284-0. [DOI] [PubMed] [Google Scholar]
- 50.Abe M., Nata K., Akiyama T., Shervani N.J., Kobayashi S., Tomioka-Kumagai T. Identification of a novel Reg family gene, Reg IIIδ, and mapping of all three types of Reg family gene in a 75 kilobase mouse genomic region. Gene. 2000;246:111–122. doi: 10.1016/s0378-1119(00)00059-7. [DOI] [PubMed] [Google Scholar]
- 51.Planas R., Alba A., Carrillo J., Puertas M.C., Ampudia R., Pastor X. Reg (regenerating) gene overexpression in islets from non-obese diabetic mice with accelerated diabetes: role of IFNβ. Diabetologia. 2006;49:2379–2387. doi: 10.1007/s00125-006-0365-6. [DOI] [PubMed] [Google Scholar]
- 52.Kadowaki Y., Ishihara S., Miyaoka Y., Rumi M., Sato H., Kazumori H. Reg protein is overexpressed in gastric cancer cells, where it activates a signal transduction pathway that converges on ERK1/2 to stimulate growth. FEBS Lett. 2002;530:59–64. doi: 10.1016/s0014-5793(02)03398-7. [DOI] [PubMed] [Google Scholar]
- 53.Harada K., Zen Y., Kanemori Y., Chen T.C., Chen M.F., Yeh T.S. Human REG I gene is up-regulated in intrahepatic cholangiocarcinoma and its precursor lesions. Hepatology. 2001;33:1036–1042. doi: 10.1053/jhep.2001.24168. [DOI] [PubMed] [Google Scholar]
- 54.Livesey F.J., O'Brien J.A., Li M., Smith A.G., Murphy L.J., Hunt S.P. A Schwann cell mitogen accompanying regeneration of motor neurons. Nature. 1997;390:614–618. doi: 10.1038/37615. [DOI] [PubMed] [Google Scholar]
- 55.Namikawa K., Fukushima M., Murakami K., Suzuki A., Takasawa S., Okamoto H., Kiyama H. Expression of Reg/PAP family members during motor nerve regeneration in rat. Biochem. Biophys. Res. Commun. 2005;332:126–134. doi: 10.1016/j.bbrc.2005.04.105. [DOI] [PubMed] [Google Scholar]
- 56.Fukui H., Kinoshita Y., Maekawa T., Okada A., Waki S., Hassan S. Regenerating gene protein may mediate gastric mucosal proliferation induced by hypergastrinemia in rats. Gastroenterology. 1998;115:1483–1493. doi: 10.1016/s0016-5085(98)70027-7. [DOI] [PubMed] [Google Scholar]
- 57.Kazumori H., Ishihara S., Hoshino E., Kawashima K., Moriyama N., Suetsugu H. Neutrophil chemoattractant 2β regulates expression of the Reg gene in injured gastric mucosa in rats. Gastroenterology. 2000;119:1610–1622. doi: 10.1053/gast.2000.20262. [DOI] [PubMed] [Google Scholar]
- 58.Hayashi K., Motoyama S., Koyota S., Koizumi Y., Wang J., Takasawa S. REG I enhances chemo- and radiosensitivity in squamous cell esophageal cancer cells. Cancer Sci. 2008;99:2491–2495. doi: 10.1111/j.1349-7006.2008.00980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fukui H., Franceschi F., Penland R.L., Sakai T., Sepulveda A.R., Fujimori T. Effects of Helicobacter pylori infection on the link between regenerating gene expression and serum gastrin levels in Mongolian gerbils. Lab. Investig. 2003;83:1777–1786. doi: 10.1097/01.lab.0000106501.56339.ce. [DOI] [PubMed] [Google Scholar]
- 60.Satomura Y., Sawabu N., Ohta H., Watanabe H., Yamakawa O., Motoo Y. The immunohistochemical evaluation of PSP/reg-protein in normal and diseased human pancreatic tissues. Int. J. Pancreatol. 1993;13:59–67. doi: 10.1007/BF02795200. [DOI] [PubMed] [Google Scholar]
- 61.Asahara M., Mushiake S., Shimada S., Fukui H., Kinoshita Y., Kawakami C. Reg gene expression is increased in rat gastric enterochromaffin-like cells following water immersion stress. Gastroenterology. 1996;111:45–55. doi: 10.1053/gast.1996.v111.pm8698224. [DOI] [PubMed] [Google Scholar]
- 62.Peterson K.M., Guo X., Elkahloun A.G., Mondal D., Bardhan P.K., Sugawara A. The expression of REG 1A and REG 1B is increased during acute amebic colitis. Parasitol. Int. 2011;60:296–300. doi: 10.1016/j.parint.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiu L., List E.O., Kopchick J.J. Differentially expressed proteins in the pancreas of diet-induced diabetic mice. Mol. Cell. Proteomics. 2005;4:1311–1318. doi: 10.1074/mcp.M500016-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Loncle C., Bonjoch L., Folch-Puy E., Belen Lopez-Millan M., Lac S., Molejon M.I. IL17 functions through the novel REG3β-JAK2-STAT3 inflammatory pathway to promote the transition from chronic pancreatitis to pancreatic cancer. Cancer Res. 2015;75:4852–4862. doi: 10.1158/0008-5472.CAN-15-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sugimoto K., Ogawa A., Mizoguchi E., Shimomura Y., Andoh A., Bhan A.K. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J. Clin. Investig. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng Y., Veldez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 67.Waldner M.J., Neurath M.F. Master regulator of intestinal disease: il-6 in chronic inflammation and cancer development. Semin. Immunol. 2014;26:75–79. doi: 10.1016/j.smim.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 68.Chen G., Li H., Niu X., Li G., Han N., Li X. Identification of key genes associated with colorectal cancer based on the transcriptional network. Pathol. Oncol. Res. 2015;21:719–725. doi: 10.1007/s12253-014-9880-9. [DOI] [PubMed] [Google Scholar]
- 69.Lim B., Park J.L., Kim H.J., Park Y.K., Kim J.H., Sohn H.A. Integrative genomics analysis reveals the multilevel dysregulation and oncogenic characteristics of TEAD4 in gastric cancer. Carcinogenesis. 2014;35:1020–1027. doi: 10.1093/carcin/bgt409. [DOI] [PubMed] [Google Scholar]
- 70.Mao F., Xu M., Zuo X., Yu J., Xu W., Moussalli M.J. 15-Lipoxygenase-1 suppression of colitis-associated colon cancer through inhibition of the IL-6/STAT3 signaling pathway. FASEB J. 2015;29:2359–2370. doi: 10.1096/fj.14-264515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robertson E., Perry C., Doherty R., Madhusudan S. Transcriptomic profiling of forkhead box transcription factors in adult glioblastoma multiforme. Cancer Genomics Proteomics. 2015;12:103–112. [PubMed] [Google Scholar]
- 72.van Beelen Granlund A., Østvik A.E., Brenna Ø., Torp S.H., Gustafsson B.I., Sandvik A.K. REG gene expression in inflamed and healthy colon mucosa explored by in situ hybridisation. Cell Tissue Res. 2013;352:639–646. doi: 10.1007/s00441-013-1592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplemental Fig. 1. Expression of the REG family mRNAs in Celiac disease colon samples. The mRNA levels of REG family (REG Iα, REG Iβ, REG III, HIP/PAP, and REG IV) in Celiac disease and control colon samples (12 Celiac disease patients before gluten-free diet and 10 age-related controls) in Barcelona were analyzed by real-time RT-PCR. The mRNA levels of REG Iα, REG Iβ, REG III, HIP/PAP, and REG IV normalized with β-actin mRNA level were not different between Celiac disease and the control. Data are expressed as mean ± SE for each group (n = 10 (Control: black circle), and 12 (Celiac disease: red square)). The statistical analyses were performed using Student's t-test. Supplementary material