Abstract
Human trisomies have recently been investigated using transcriptomics approaches to identify the gene expression (GE) signatures characteristic of each of these specific aneuploidy conditions. We hypothesized that the viability of cells with gross genomic imbalances might be associated with the activation of resilience mechanisms that are common to different trisomies and that are reflected by specific shared GE patterns. We report in this article our microarray GE analyses of amniocytes from fetuses with viable trisomy conditions, trisomy 21 or trisomy 18, to detect such common expression signatures. Comparative analysis of significantly differentially expressed genes in trisomies 18 and 21 revealed six dysregulated genes common to both: OTUD5, ADAMTSL1, TADA2A, PPID, PIAS2, and MAPRE2. These genes are involved in ubiquitination, protein folding, cell proliferation, and apoptosis. Pathway-based enrichment analyses demonstrated that both trisomies showed dysregulation of the PI3K/AKT pathway, cell cycle G2/M DNA damage checkpoint regulation, and cell death and survival, as well as inhibition of the upstream regulator TP53. Our data collectively suggest that trisomies 18 and 21 share common functional GE signatures, implying that common mechanisms of resilience might be activated in aneuploid cells to resist large genomic imbalances. To the best of our knowledge, this is the first study to use global GE profiling data to identify potential common mechanisms in fetal trisomies. Studies of other trisomies using transcriptomics and multiomics approaches might further clarify mechanisms activated in trisomy syndromes.
Keywords: : aneuploidy, transcriptomics, TP53, PI3K/AKT, cell-cycle, resilience
Background
Chromosomal aberrations are associated with significant morbidity and mortality in the prenatal period and in early infancy. Since the 1970s, trisomies of chromosomes 21, 18, and 13 have been known to be associated with well-defined syndromes, namely Down, Edwards, and Patau syndromes, respectively. The affected chromosomes in these potentially viable trisomies contain the fewest protein-coding genes of all chromosomes and represent the lowest net dosage imbalances that can be tolerated during in utero development (Torres et al., 2008).
Normal ontogenetic development is the result of complex regulatory mechanisms, which include adapted gene pool. Large genomic imbalances disturb these relationships and result in various clinically distinct conditions. Global gene expression (GE) analysis has the potential to identify the biological processes and pathways involved in numerous disorders in humans, including those in large genome imbalances.
Studies of the GE profiles in developing fetal or adult tissues in trisomies 21, 18, and other aneuploidies have demonstrated both the dysregulation of specific regions of chromosomes that are present in three copies and the widespread dysregulation of genes in regions of the euploid genome (Altug-Teber et al., 2007; FitzPatrick et al., 2002; Hui et al., 2012; Koide et al., 2011; Li et al., 2006; Lockstone et al., 2007; Mao et al., 2005; Slonim et al., 2009; Stingele et al., 2012; Volk et al., 2013). According to these studies, GE alterations cannot be attributed to dosage imbalances alone. Specifically, only few genes on the trisomic chromosomes show the expected 1.5-fold change in expression. Furthermore, several euploid genes show disrupted expression profiles (Disteche, 2013).
Previously performed studies focused on identifying specific GE signatures that are characteristic to individual trisomic syndromes in humans. However, none of the reports centered on scrutinizing the mechanisms or pathways associated with differentially expressed genes that were common to various trisomic syndromes. There are evidences that although the viable trisomies include different sets of disturbed genes, they share some similar phenotypic and molecular characteristics. Notably, trisomic cells, including stem and progenitor cells, have an impaired proliferation rate (Hibaoui et al., 2014; Liu et al., 2015), which might partially be associated with miscarriage in early pregnancy. About 50% of fetuses with trisomy 21 and up to 72% of those with trisomy 18 cases are miscarried (Morris and Savva, 2008; Witters et al., 2011).
Therefore, we hypothesized that in various trisomic syndromes, common GE signature might directly reflect gene dosage perturbation or might be associated with activation of homeostatic resilience mechanisms and that these would be detectable by differential gene expression (DGE) analysis. To address this issue, we analyzed the global transcriptome profiles of amniotic cells of fetuses with trisomy 21 and trisomy 18 with the aim of identifying common GE changes and biological mechanisms.
Materials and Methods
Ethics approval and consent to participate
This study was approved by the Slovenian National Ethics Committee (No. 169/04/13). All participants provided written informed consent before participation in the study, and all clinical investigations were conducted in accordance with the principles expressed in the Declaration of Helsinki.
Cell culture samples
Amniotic fluid samples were collected between the 16th and 18th week of gestation (average 16 5/7, median 16 4/7 and standard deviation of 3, 5 days) for routine cytogenetic analysis. Cell cultures of amniotic fluid were grown according to standard protocols using the tissue culture flask method and commercially available medium (AmnioMax, Invitrogen, CA) at 37°C in 5% CO2 environment. Following a routine diagnostic cytogenetic analysis, a second passage of amniotic cell culture was grown in the same condition as primary cell culture and used for total RNA extraction, 2–3 weeks after amniocentesis. The first sample set (sample set 1) included 19 samples, 10 that were derived from fetuses with trisomy 21 and 9 originating from normal euploid pregnancies. The second sample set (sample set 2) included 19 samples −9 that were derived from fetuses with trisomy 18 and 10 that were from normal euploid pregnancies. Clinical characteristics of the samples are presented in the Supplementary Tables S1 and S2.
RNA isolation
RNA was isolated from cultured amniocytes using the RNeasy mini kit (Qiagen) according to the manufacturer's instructions. The purity and yield of the isolated RNA samples were determined using the NanoDrop 2000c Spectrophotometer (Nanodrop Technologies, Wilmington, DE). The integrity of RNA samples selected for genome-wide profiling of GE was investigated on Agilent's Bioanalyzer using the RNA 6000 Nano Kit, and only samples with RNA integrity number values greater than 8.0 were used for the array experiments.
Global GE profiling
For the purposes of microarray analysis, we used the common reference design of the study: each experimental sample was separately hybridized against a common reference sample (Peng et al., 2003). Trisomy 21 samples (10 biological replicates) and control samples (9 biological replicates) from sample set 1 were hybridized against a common reference, obtained by pooling all samples in set 1. Similarly, trisomy 18 samples (9 biological replicates) and control samples (10 biological replicates) from sample set 2 were hybridized against a common reference, again obtained by pooling all samples from set 2.
In all cases, the reference RNA pool was labeled with Cy3 and samples were labeled with Cy5. We used Agilent 4 × 44 two-color Whole Human Genome Expression arrays (GPL6480), which contain 41,001 probes for interrogation of over 19,644 human genes, to estimate the extent of GE alterations in investigated cells with trisomy 18 and trisomy 21. RNA sample preparation, labeling, and hybridization were performed according to the manufacturer's instructions (Agilent Technologies).
After hybridization, the microarray slides were scanned using the Agilent High Resolution Microarray Scanner System using the manufacturer's recommended scanning settings. Subsequently, microarray features were extracted using Agilent Feature Extraction software v10.7.3.1. Further analysis steps were performed using the limma package from Bioconductor v2.8 in the R statistical environment version 2.13.1 (www.bioconductor.org). Probe annotations were obtained from the Agilent's eArray service (earray.chem.agilent.com) and appended to the initial datasets. Before and after normalization, the MA plots, boxplots, and expression value density plots were investigated for diagnostic purposes. Probe replicates were median-averaged before additional steps and the final results contained gene-level expression levels. In this way, results from two different platforms could be matched appropriately in downstream analyses.
To minimize the batch effects and other confounding factors, we performed the analyses for both datasets separately and the final gene- and pathway-based results were compared between the two trisomies. In brief, statistical comparison of the expression values was performed using a moderated t-test approach as implemented in limma (Smyth, 2003). Expression values were fitted to a linear model that accounted for the effects of trisomy status and differences between test samples and controls calculated by contrasting the two conditions in limma. Significance values and log fold-changes were calculated afterward, and p values were controlled for the issue of multiple testing by Benjamini and Hochberg method (Reiner et al., 2003). Power analyses for microarray experiments were performed using Microarray power analysis calculator (http://sph.umd.edu/department/epib/sample-size-and-power-calculations-microarray-studies), as previously described (Lee and Whitmore, 2002).
Enrichment analyses
Several different approaches such as GeneAnalytics, DAVID, ingenuity pathway analysis (IPA), and gene set enrichment analysis (GSEA) (Ben-Ari Fuchs et al., 2016; Huang da et al., 2009; Kramer et al., 2014; Subramanian et al., 2005) have been used to address the issue of gene set enrichment. We used IPA (Ingenuity System, Inc.) in addition to basic enrichment approaches, since IPA characterizes GE data in terms effects of effect direction (i.e., genes' up- and downregulation are taken into consideration), rather than mere associations as in GSEA. Given a GE dataset, our main goals were to identify the causal mechanistic networks and upstream biological causes (upstream regulators) and predicted impact on biological functions. IPA analysis also determines whether such regulators are activated or inhibited based on the observed GE pattern and detects which causal relationships previously reported in the literature are compatible with the signatures observed in the expression data (Kramer et al., 2014).
Datasets of the significantly differentially expressed genes in trisomy 21 and trisomy 18 (p value <0.05) were subjected to IPA Software version 26127183 (Release Date: November 30, 2015; Ingenuity Systems, Inc., Redwood City, CA) with general analysis settings: log ratio (range −4.14 to 5.17), p value <0.05. The software computes a Fisher's exact test to calculate statistical significance.
Results
To analyze the differences in GE and in key processes that underlie the molecular phenotypes of trisomic samples, the microarray data from cultivated amniocytes with trisomy 18 and trisomy 21 were used for DGE and functional enrichment analyses.
Amniocytes carrying an extra copy of chromosome 18 or 21 showed considerable differences in GE as shown by a comparison of their transcriptional profiles to those of euploid samples. We identified 1328 (5.7% of the total) unique genes with statistically significant differences in expression in trisomy 18 (adjusted p < 0.05) and 513 unique genes (2.4% of the total) in trisomy 21 (adjusted p < 0.05). In both trisomies approximately half of the significantly differentially expressed genes were downregulated and half of them upregulated.
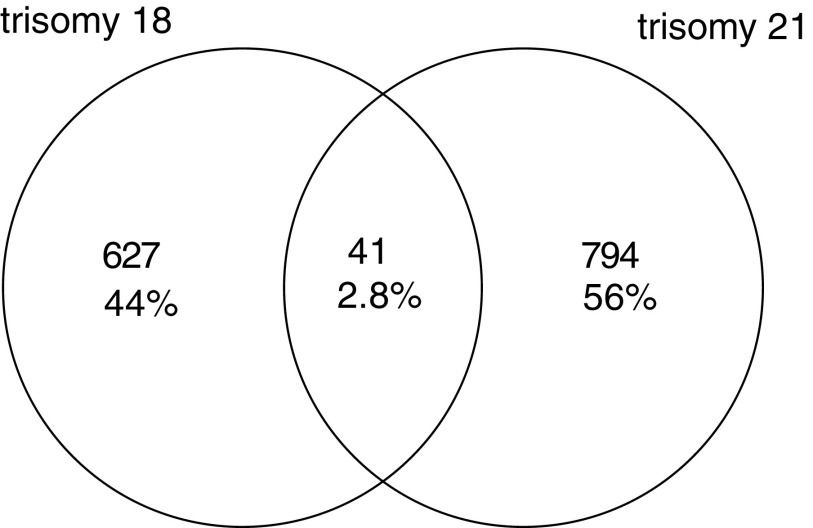
In addition, we compared top 1000 gene probes from the DGE data of trisomies 18 and 21 (adjusted p < 0.05) to detect genes that were common in both aneuploidies (Fig. 1).
FIG. 1.
A Venn diagram showing the number of differentially expressed gene probes that overlap between trisomies 18 and 21. All gene probes showed significant differences in expression (adj. p < 0.05).
We found 41 genes that were significantly differentially expressed in both trisomies compared with normal controls (Supplementary Table S1), yet, only six of them showed consistent direction of dysregulation (Table 1).
Table 1.
Symbols and GO Terms of Genes Showing Consistent Dysregulation in Trisomy 18 and Trisomy 21
| Gene | GO annotations | p value T18 | logFC T18 | p value T21 | logFC T21 |
|---|---|---|---|---|---|
| OTUD5 | GO:0004843(thiol-dependent ubiquitin-specific protease activity)|GO:0061578(Lys63-specific deubiquitinase activity)|GO:0101005 (ubiquitinyl hydrolase activity)|GO:1990380 (Lys48-specific deubiquitinase activity)|GO:0016579(protein deubiquitination)|GO:0032496(response to lipopolysaccharide) | 0.013 | −0.34 | 0.023 | −0.594 |
| ADAMTSL1 | (GO:0004222(metalloendopeptidase activity)|GO:0008233 (peptidase activity)|GO:0008270 (zinc ion binding)|GO:0006508(proteolysis) | 0.010 | 0.622 | 0.018 | 0.449 |
| TADA2A | (GO:0003677(DNA binding)|GO:0005634(nucleus)|GO:0006355 (regulation of transcription, DNA-templated)|GO:0035065 (regulation of histone acetylation) | 0.008 | 0.354 | 0.038 | 0.062 |
| PPID | GO:0003755(peptidyl-prolyl cis-trans isomerase activity)|GO:0000413(protein peptidy-prolyl isomerization)|GO:0006457 (protein folding) | 0.012 | 0.49 | 0.039 | 0.418 |
| PIAS2 | GO:0019789(SUMO transferase activity)|GO:0016925(protein sumoylation) | 0.007 | 0.942 | 0.019 | 0.445 |
| MAPRE2 | GO:0008017 (microtubule binding)|GO:0005874(microtubule) | 0.009 | 0.47 | 0.029 | 0.328 |
All genes were upregulated in both trisomies, except for OTUD5, which was downregulated. T18, trisomy 18 and T21, trisomy 21. The p values were adjusted using the Benjamini–Hochberg correction; logFC, log fold change. The GO annotations (function, process or cellular component) were searched using EMBL-EBI QuickGO (www.ebi.ac.uk).
GO.
Power analysis in our experiment has shown that we attained power of 89% to detect logFC (log fold change) values of at least 1.5 when analyzing 10 samples in each group, under assumptions that 10% of genes are differentially expressed in our experiment, permitting 10 false-positive results and sigma parameter of 1.0.
Datasets of differential expressed genes of trisomies 18 and 21 with adjusted p values <0.05 were used for further analysis. Lists of genes with adjusted p values and logFC values for trisomies 18 and 21 were uploaded into IPA software (IPA; Ingenuity System, Inc.). In this comparison, we discovered enrichment of genes in the PI3K/AKT signaling pathway and of genes involved in the cell cycle: G2/M DNA damage checkpoint regulation with p values <0.001 in both trisomies (Supplementary Tables S4 and S5).
The genes associated with PI3K/AKT pathway and cell cycle DNA damage checkpoint regulation were downregulated with negative activation scores (z = −0.9 in trisomy 21 and z = −1.1 in trisomy 18 and z scores of −1.8 in trisomy 21 and −1.2 in trisomy 18, respectively).
Upstream regulator analysis showed that the TP53 gene was predicted to be inhibited in both trisomies with p values <0.001 (Supplementary Tables S6 and S7). In terms of biological functions, IPA enrichment analyses of both trisomy 18 and trisomy 21 differentially expressed datasets identified overrepresentation of genes involved in cell cycle, cellular movement, and cell death and survival (p < 0.05). Differentially expressed genes involved in cellular movement and cell death and survival were downregulated, while cell cycle-associated genes showed a trend toward upregulation in both trisomies (Supplementary Tables S8 and S9).
In addition, in the group of genes associated with physiological system development and function, IPA enrichment analysis identified differentially expressed genes that were involved in organismal survival, tissue development, and connective tissue development and function (p < 0.05) (Supplementary Tables S6 and S7). Datasets of our studies are available at the Gene Expression Omnibus (GEO) public repository (www.ncbi.nlm.nih.gov/geo) under accession numbers GSE48051 for trisomy 21 and GSE89782 for trisomy 18.
Discussion
Using global GE analysis of cultivated fetal amniocytes with trisomy 18 and trisomy 21, we identified the genes, biological processes, and pathways that were common to both transcriptional profiles. To the best of our knowledge, this is the first study to use global GE profiling data to identify potential common mechanisms in fetal trisomies.
Even though there was no overall trend in global expression alterations in trisomy 18 and trisomy 21, comparative analysis of differentially expressed genes identified a subset of 6 differentially expressed genes, namely OTUD5, PIAS2, ADAMTSL1, TADA2A, PPID, and MAPRE2, involved in diverse biological processes, including protein deubiquitination and sumoylation of protein p53 (OTUD5, PIAS2), proteolysis and protein folding (ADAMTSL1, PPID), chromatin remodeling (TADA2A), and in addition, microtubule binding and the role in the proliferative control of normal cells (MAPRE2).
Processes involved in proliferative control and apoptosis have been widely associated with trisomies 21 and 18. Studies of trisomy 21 have consistently shown that the apoptosis is abnormal and that there is a decreased proliferation capacity in various human cell types with trisomy 21, that is, in induced pluripotent stem cells (Hibaoui et al., 2014), in fibroblasts and lymphoblastoid cells (Sullivan et al., 2016), human embryonic kidney cells (Wu et al., 2016). Abnormal apoptotic processes were also observed in trisomy 18. Notably, Koide et al. (2011) compared the global GE of cell-free fetal RNA in the amniotic fluid of mothers pregnant with trisomy 18 and euploid fetuses and found that genes that were relevant to the cell death network were downregulated in trisomy 18 samples. There is an increasing body of evidence that histone acetylation is involved in chromatin remodeling, which in turn influence biological processes, including apoptosis (Fullgrabe et al., 2010).
Using IPA enrichment analyses, we also found that genes that play a role in cell death and survival and in organismal survival were significantly overrepresented in differentially expressed datasets of studied trisomies, which is in agreement with earlier studies (Hibaoui et al., 2014; Koide et al., 2011; Sullivan et al., 2016; Zhao et al., 2016).
Our data showed downregulation of OTUD5, which acts as a cofactor in the activation of p53 in the DNA damage response (Luo et al., 2013; Park et al., 2015) and which might be related to the dysregulation noted above. In line with this, we found evidence of inhibition of the OTUD5 gene via its downstream targets, suggesting that the p53 pathway is downregulated. Inhibition of the p53 pathway might be a mechanism that is strongly associated with a tolerance for an aneuploid genome (Thompson and Compton, 2010). Inhibition of TP53 gene via dysregulation of the PI3K/AKT pathway, as observed in our study, was demonstrated previously in a postmortem analysis of brain tissue of subjects with trisomy 21 (Perluigi et al., 2014).
In this study, we provide evidence that several mechanisms that are involved in apoptosis and cell proliferation are dysregulated in both trisomies 21 and 18. Interestingly, these same mechanisms might also be involved in the pathogenesis of cancer, revealing a potential biological basis for the increased risk of cancer in patients with trisomies (Ganmore et al., 2009; Nizetic and Groet, 2012).
Among the differentially expressed genes, we identified genes involved in the ubiquitination process in both trisomies, highlighting the importance of the superior hierarchical biological processes, such as protein ubiquitination regulation, protein/cellular catabolic regulation, and protein modification regulation. Rozovski et al. (2007) demonstrated that protein ubiquitination was enriched for genes that were differentially expressed in trisomy 21. A related study showed that the level of protein ubiquitination is reduced in human trisomy 21 lymphoblastoid cells (Granese et al., 2013). In contrast, a recent study showed that fibroblasts in trisomy 21 exhibit increased protein ubiquitination and disruption of the proteostasis network and accumulation of misfolded proteins (Aivazidis et al., 2017).
However, there are no data on the association between protein ubiquitination and trisomy 18 in humans, and the role of ubiquitination in the trisomy 21 or 18 molecular phenotypes remains to be determined. A recent study suggested that even minor genetic changes, such as pathogenic variants associated with monogenic phenotypes, can impair protein–protein or protein–DNA interactions via increased chaperone–ubiquitin–proteasome system activity (Sahni et al., 2015). In addition, we detected overexpression of the PPID gene, which is involved in protein folding and apoptosis (Jandova et al., 2013).
Using enrichment analysis approach, we identified several mechanisms that were associated with trisomy 18 and trisomy 21 that might be implicated in the resilience mechanisms of the cells and help them resist genomic imbalances. A study by Gao and Barabasi (2016) showed that the behavior of the complex system might be modeled by the regulatory dynamics equation and that the system collapses upon introduction of a significant perturbation. This phenomenon is described by universal resilient function that predicts the behavior of the complex system and is independent of the network structure or the nature of the perturbation (Gao and Barabasi, 2016). Resilience mechanisms might provide insights into the pathogenic mechanisms in trisomy syndromes, which are crucial for designing new therapeutic approaches.
Nevertheless, while observing common mechanisms in two models, trisomy 18 and trisomy 21, it is difficult to discriminate between common expression profile related to shared phenotypic characteristics and expression profiles related to resilient mechanisms.
We used a conservative approach to investigate common differentially expressed genes in both trisomies to determine common dysregulated pathways. However, there are some limitations in our study: first, results are based on cultivated amniocyte samples. Cultivation process might influence the actual GE, although to culture the cells provides a more homogeneous environment, as the transcriptome profile is responding to the culture conditions and not to the natural challenging environment. Nevertheless, we have speculated that the impact of an extra chromosome, such as trisomy 18 or 21, on the DGE would greatly overcome the effects of the in vitro cell cultivation or type of the tissue investigated.
Second, exploring high-dimensional data poses a statistical challenge, since the number of samples is extremely lower than the number of testing genes, which can result in false-positive detection. Third, small effect sizes of genes associated with the disorder can remain undetected in the background or statistical noise. Fourth, the studies of the RNA expression patterns are very delicate and variations can be found between technical replicates and when samples are processed in multiple batches. The analyses for both two datasets were performed separately and might be confounded by batch effect.
In conclusion, we show in the present study that trisomies 18 and 21 share common expression signatures, implying that potential common mechanisms of resilience might be activated in aneuploid cells to resist large genomic imbalances. Studies comprising other trisomies might further clarify common mechanisms activated in trisomy syndromes.
Supplementary Material
Abbreviations Used
- Cy3
cyanine 3, greenish yellow fluorescent dye
- Cy5
cyanine 5, far-red fluorescent dye
- DGE
differential gene expression
- GE
gene expression
- GSEA
gene set enrichment analysis
- IPA
ingenuity pathway analysis
- log FC
log fold-change
- T18
trisomy of chromosome 18
- T21
trisomy of chromosome 21
- TP53
tumor protein 53
Acknowledgment
This study was supported by a grant P3-0326 from the Slovenian Research Agency.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Aivazidis S, Coughlan CM, Rauniyar AK, et al. (2017). The burden of trisomy 21 disrupts the proteostasis network in Down syndrome. PLoS One 12, e0176307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altug-Teber O, Bonin M, Walter M, et al. (2007). Specific transcriptional changes in human fetuses with autosomal trisomies. Cytogenet Genome Res 119, 171–184 [DOI] [PubMed] [Google Scholar]
- Ben-Ari Fuchs S, Lieder I, Stelzer G, et al. (2016). GeneAnalytics: an integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. Omics 20, 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche CM. (2013). How to correct chromosomal trisomy. Cell Res 23, 1345–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzPatrick DR, Ramsay J, McGill NI, Shade M, Carothers AD, and Hastie ND. (2002). Transcriptome analysis of human autosomal trisomy. Hum Mol Genet 11, 3249–3256 [DOI] [PubMed] [Google Scholar]
- Fullgrabe J, Hajji N, and Joseph B. (2010). Cracking the death code: Apoptosis-related histone modifications. Cell Death Differ 17, 1238–1243 [DOI] [PubMed] [Google Scholar]
- Ganmore I, Smooha G, and Izraeli S. (2009). Constitutional aneuploidy and cancer predisposition. Hum Mol Genet 18, R84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao JBB, and Barabasi AL. (2016). Universal resilience patterns in complex networks. Nature 530, 307–312 [DOI] [PubMed] [Google Scholar]
- Granese B, Scala I, Spatuzza C, et al. (2013). Validation of microarray data in human lymphoblasts shows a role of the ubiquitin-proteasome system and NF-kB in the pathogenesis of Down syndrome. BMC Med Genomics 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibaoui Y, Grad I, Letourneau A, et al. (2014). Modelling and rescuing neurodevelopmental defect of Down syndrome using induced pluripotent stem cells from monozygotic twins discordant for trisomy 21. EMBO Mol Med 6, 259–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 [DOI] [PubMed] [Google Scholar]
- Hui L, Slonim DK, Wick HC, Johnson KL, Koide K, and Bianchi DW. (2012). Novel neurodevelopmental information revealed in amniotic fluid supernatant transcripts from fetuses with trisomies 18 and 21. Hum Genet 131, 1751–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandova J, Janda J, and Sligh JE. (2013). Cyclophilin 40 alters UVA-induced apoptosis and mitochondrial ROS generation in keratinocytes. Exp Cell Res 319, 750–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide K, Slonim DK, Johnson KL, Tantravahi U, Cowan JM, and Bianchi DW. (2011). Transcriptomic analysis of cell-free fetal RNA suggests a specific molecular phenotype in trisomy 18. Hum Genet 129, 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J, Jr., and Tugendreich S. (2014). Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ML, and Whitmore GA. (2002). Power and sample size for DNA microarray studies. Stat Med 21, 3543–3570 [DOI] [PubMed] [Google Scholar]
- Li CM, Guo M, Salas M, et al. (2006). Cell type-specific over-expression of chromosome 21 genes in fibroblasts and fetal hearts with trisomy 21. BMC Med Genet 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Filippi S, Roy A, and Roberts I. (2015). Stem and progenitor cell dysfunction in human trisomies. EMBO Rep 16, 44–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockstone HE, Harris LW, Swatton JE, Wayland MT, Holland AJ, and Bahn S. (2007). Gene expression profiling in the adult Down syndrome brain. Genomics 90, 647–660 [DOI] [PubMed] [Google Scholar]
- Luo J, Lu Z, Lu X, et al. (2013). OTUD5 regulates p53 stability by deubiquitinating p53. PLoS One 8, e77682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Wang X, Spitznagel EL Jr., et al. (2005). Primary and secondary transcriptional effects in the developing human Down syndrome brain and heart. Genome Biol 6, R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JK, and Savva GM. (2008). The risk of fetal loss following a prenatal diagnosis of trisomy 13 or trisomy 18. Am J Med Genet A 146A, 827–832 [DOI] [PubMed] [Google Scholar]
- Nizetic D, and Groet J. (2012). Tumorigenesis in Down's syndrome: Big lessons from a small chromosome. Nat Rev Cancer 12, 721–732 [DOI] [PubMed] [Google Scholar]
- Park SY, Choi HK, Choi Y, Kwak S, Choi KC, and Yoon HG. (2015). Deubiquitinase OTUD5 mediates the sequential activation of PDCD5 and p53 in response to genotoxic stress. Cancer Lett 357, 419–427 [DOI] [PubMed] [Google Scholar]
- Peng X, Wood CL, Blalock EM, Chen KC, Landfield PW, and Stromberg AJ. (2003). Statistical implications of pooling RNA samples for microarray experiments. BMC Bioinformatics 4, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perluigi M, Pupo G, Tramutola A, et al. (2014). Neuropathological role of PI3K/Akt/mTOR axis in Down syndrome brain. Biochim Biophys Acta 1842, 1144–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, and Benjamini Y. (2003). Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375 [DOI] [PubMed] [Google Scholar]
- Rozovski U, Jonish-Grossman A, Bar-Shira A, Ochshorn Y, Goldstein M, and Yaron Y. (2007). Genome-wide expression analysis of cultured trophoblast with trisomy 21 karyotype. Hum Reprod 22, 2538–2545 [DOI] [PubMed] [Google Scholar]
- Sahni N, Yi S, Taipale M, Fuxman Bass JI, et al. (2015). Widespread macromolecular interaction perturbations in human genetic disorders. Cell 161, 647–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonim DK, Koide K, Johnson KL, et al. (2009). Functional genomic analysis of amniotic fluid cell-free mRNA suggests that oxidative stress is significant in Down syndrome fetuses. Proc Natl Acad Sci U S A 106, 9425–9429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2003). Normalisation of cDNA microarray data. Methods 31, 265–273 [DOI] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, and Storchova Z. (2012). Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol 8, 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Lewis HC, Hill AA, et al. (2016). Trisomy 21 consistently activates the interferon response. Elife 5, pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, and Compton DA. (2010). Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol 188, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Williams BR, and Amon A. (2008). Aneuploidy: Cells losing their balance. Genetics 179, 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk M, Maver A, Lovrecic L, Juvan P, and Peterlin B. (2013). Expression signature as a biomarker for prenatal diagnosis of trisomy 21. PLoS One 8, e74184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witters G, Van Robays J, Willekes C, et al. (2011). Trisomy 13, 18, 21, Triploidy and Turner syndrome: The 5T's. Look at the hands. Facts Views Vis Obgyn 3, 15–21 [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhang S, Xu Q, et al. (2016). Regulation of global gene expression and cell proliferation by APP. Sci Rep 6, 22460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang Z, Ren S, Zong Y, and Kong X. (2016). Co-expression network analysis of Down's syndrome based on microarray data. Exp Ther Med 12, 1503–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.