Abstract
Adeno-associated virus (AAV) inverted terminal repeats (ITRs) are key elements of AAV. These guanine-cytosine-rich structures are involved in the replication and encapsidation of the AAV genome, along with its integration in and excision from the host genome. These sequences are the only AAV-derived DNA sequences conserved in recombinant AAV (rAAV), as they allow its replication, encapsidation, and long-term maintenance and expression in target cells. Due to the original vector design, plasmids containing the gene of interest flanked by ITRs and used for rAAV production often present incomplete, truncated, or imperfect ITR sequences. For example, pSUB201 and its derivatives harbor a truncated (14 nt missing on the external part of the ITR), flop-orientated ITR plus 46 bp of non-ITR viral DNA at each end of the rAAV genome. It has been shown that rAAV genomes can be replicated, even with incomplete, truncated, or imperfect ITR sequences, leading to the production of rAAV vectors in transfection experiments. Nonetheless, it was hypothesized that unmodified wild-type (WT) ITR sequences could lead to a higher yield of rAAV, with less non-rAAV encapsidated DNA originating from the production cells and/or baculovirus shuttle vector genomes. This work studied the impact of imperfect ITRs on the level of encapsidated rAAV genomes and baculovirus-derived DNA sequences using the baculovirus/Sf9 cells production system. Replacement of truncated ITRs with WT and additional wtAAV2 sequences has an impact on the two major features of rAAV production: (1) a rise from 10% to 40% of full capsids obtained, and (2) up to a 10-fold reduction in non-rAAV encapsidated DNA. Furthermore, this study considered the impact on these major parameters of additional ITR elements and ITRs coupled with various regulatory elements of different origins. Implementation of the use of complete ITRs in the frame of the baculovirus-based rAAV expression system is one step that will be required to optimize the quality of rAAV-based gene therapy drugs.
Keywords: : rAAV, baculovirus, ITR, quality
Introduction
Adeno-associated virus (AAV) was first described in 19651 as a contaminant virus of purified adenovirus displaying a smaller capsid size (20–24 nm compared to the 80 nm capsid of adenovirus). Belonging to the Parvoviridae family, in the dependovirus genus, AAVs are dependent on a helper virus (herpesvirus or adenovirus) to replicate.2 The genetic organization of AAV is relatively simple, with three open reading frames (ORFs) coding for the regulatory proteins (the four Rep proteins) involved in all DNA-related activities, the capsid proteins (VP1, VP2, and VP3), and the assembly-activating protein (AAP), of importance for the intracellular trafficking and assembly of VP proteins. Furthermore, the AAV is framed on both sides by inverted terminal repeat (ITR) sequences. ITRs are the only cis-acting elements known to date. These 145 base T-shape structures found at both ends of AAV genomes are implicated in replication,3 second-strand synthesis, encapsidation, and insertion of the viral genome into human chromosome 19.4,5 One ITR is formed by two palindromic arms, called B–B′ and C–C′, embedded in a larger one, A–A′. The order of these palindromic sequences defines the flip or flop orientation of the ITR.6 Each wild-type (WT) AAV genome can contain either two flip-orientated or two flop-orientated ITRs, or one ITR of each orientation. Orientation of the nicked ITR changes with each round of replication. In addition to the palindromic sequences, a single-stranded remaining D sequence is present. This sequence is thought to play a role in genome encapsidation.7 The ITR also contains a Rep binding element (RBE)8 to which Rep78 or Rep68 proteins bind. This RBE consists of a quadruple repetition of a four nucleotide sequence 5′-GNGC-3′. Replication models suggest that Rep78/68, through their helicase activity, remodel the A–A′ sequence in a stem loop, exposing the terminal resolution site in a single-stranded conformation at its top.9–11
Production of recombinant AAV (rAAV) by transfection of mammalian cells uses two or three different plasmids, providing the AAV and adenoviral helper sequences12,13 (on two separated plasmids or altogether on one) as well as the vector plasmid with the genome of interest (GOI) flanked by the ITRs from AAV2.
One of the first constructs harboring the wtAAV2 genome and allowing production of rAAVs (pSUB201) was built by duplication of the 3′ end of the wtAAV2 genome (ITR + 46 bp of non-ITR viral DNA) onto the 5′ end.14,15 This duplication of one ITR had been performed in order to allow vector genome excision using a single restriction site (PvuII) from a double-stranded form in plasmids. Therefore, both AAV genome ends on this construct display an ITR in the same orientation plus 46 bp of non-ITR viral DNA directly flanking the GOI. In addition, pSUB201 presents a 14 bp deletion at each ITR extremity without affecting the ability to produce rAAV in HEK293 cells.
Although Samulski et al.16 and Wang et al.17 have published studies indicating that the use of truncated ITRs has no impact on rAAV vector production levels obtained in transfection, this issue has not been studied in the context of gene therapy vector manufacturing using the baculovirus/Sf9 cells production system. The present study considers the impact of ITR integrity and modification on the production of rAAV vectors encoding the gamma-sarcoglycan (γ-SGC) recombinant genome18 in the baculovirus/Sf9 production system. The study investigated rAAV titers, level of foreign DNA originating from baculovirus sequences, and full/empty particles ratio present in purified rAAV production.
Materials and Methods
ITR and constructs synthesis
The various constructs with ITR variants were generated by gene synthesis (Genewiz). ITR sequences had been confirmed by sequencing (SeqWright), and analysis performed with Snapgene software.
Bacmid generation
Bacmid DNA was generated through a modified Bac-to-Bac system, where chitinase and cathepsin genes were deleted using DH10Bac bacteria.
Baculovirus stock formation
Bacmid DNA and Cellfectin II (Invitrogen) were poured into two separate FACS tubes containing Sf900 III medium (Gibco), and were incubated for 15 min at room temperature. The two tubes were then mixed, incubated for a further 15 min, and added to one well of a six-well plate with one million Sf9 cells (Invitrogen). After 5 days, lysis plaques had been performed, and the stability of five clones was studied using quantitative polymerase chain reaction (qPCR) and Western blot. Two of the five clones were then amplified in a 125 mL shake flask (Corning) with a working volume of 75 mL. Baculovirus titrations were performed using the lysis plaque technique.19
Viral DNA extraction
Viral DNA extraction was performed in triplicate on 5 μL of cell pellet, cellular culture, supernatant, or purified rAAV. Five microliters of the sample was added to 45 μL of DNase I buffer (Tris HCl 1 M, CaCl2 0.1 M, and MgCl2 1 M) with 10 IU of DNase I (Invitrogen; 90083), and digestion was performed for 30 min at 37°C. Capsid degradation was performed with proteinase K digestion (Roche), and purification of viral genomes was performed using the MagNa Pure 96 DNA and Viral NA Small Volume Kit and the MagNa Pure 96 instrument (Roche) following the manufacturer's protocol. Elution volume was set to 50 μL.
qPCR
qPCR was performed by hydrolysis in LC480 (Roche). Baculovirus qPCR titration was performed on baculovirus DNA polymerase sequence with 5′-ATTAGCGTGGCGTGCTTTTAC-3′, 5′-GGGTCAGGCTCCTCTTTGC-3′ primers and 5′-CAAACACGCGCATTAACGAGAGCACC-3′ [5′]VIC[3′]TAMRA probe. For ITRconta titration, 5′-GCGGTACTTGGGTCGATATCA-3′, 5′-CCGCAGTGGCTCTCTATACAAA-3′ primers were used with 5′-AGTGCATCACTTCTTCCCGTATGCCCA-3′ [5′]6-FAM[3′]TAMRA probe. For γ-SGC titration, 5′-AAGTCGGTCCCAAAATGGTAGA-3′, 5′-TGCCGTCGTTGGAGTTGA-3′ primers with 5′-CAGAATCAACAGTTTCAG-3′ [5′]6-FAM[3′]MGB-NFQ probe.
rAAV production
A 125 mL shake flask (Corning) containing 70 mL of Sf900-III (Gibco) at one million Sf9 cells/mL was infected at a multiplicity of infection of 0.05 per baculovirus, and incubated for 96 h at 27°C under 170 rpm agitation.
rAAV purification
After a frost/defrost cycle, infected cells were incubated with 0.5% Triton X-100 for 2 h 30 min at 27°C under 170 rpm agitation. Clarification of the crude lysate was performed using a Pall Preflow Filter Capsule (0.45 μm; DFA3001UBC). The clarified product was then purified by affinity chromatography in a random order. AVB Sepharose gel (GE Healthcare) was used for the purification of rAAV2 and rAAV8. Poros9 resin (Thermo Fisher Scientific) was used for the purification of rAAV9. Collected fractions were concentrated with Amicon Ultra-15 (100 kDa; UFC910024) and re-suspended in 1 mL of phosphate-buffered saline (Gibco).
Analytical ultracentrifugation
Analytical ultracentrifugation (AUC) was performed in a ProteomeLab XL-1 centrifuge (Beckman) using an AN-60TI rotor (Beckman) at 20°C. Sample cell and counterbalance assembling were performed according to the manufacturer's instructions. Undiluted samples (400 μL) were loaded into each sample cell. A first set of runs (absorbance and interference measure) was performed at 3,000 rpm in order to set the wavelength of analysis and laser position properly. Absorbance and interference monitoring were performed at 16,000 rpm from 2 h to 2 h 30 min. Data were collected with ProteomeLab software (Beckman) and treated with Sedfit software. Runs were performed against a reference rAAV8 production produced without any transgene.
Western blot
Sf9 transfected cells were pelleted, and the supernatant was removed. Pellets were re-suspended in 100 μL of lysis buffer (Tris/phosphate 25 mM, pH 7.8, Glycerol 15%, DTT 1 mM, EDTA 1 mM, MgCl2 8 mM, Triton 0.2%) with Protease inhibitor cocktail (04693116001; Roche). Samples were kept on ice for 30 min before being centrifuged at 10,000 rpm for 10 min. The supernatant (20 μL) was mixed with LDS 4 × (NP0007; Invitrogen) and 10 × reducing agent (NP0004; Invitrogen). Samples were then heated for 15 min at 94°C and were run on a polyacrylamide gel (NuPAGE 4–12% Bis-Tris Gel; Invitrogen). After migration, proteins were transferred onto a nitrocellulose membrane using the iBlot transfer system (Invitrogen), following the manufacturer's protocol. Membranes were incubated for 1 h with blocking buffer (Odyssey). Membrane blotting was made with first antibody immunoglobulin B1 at 1/250 dilution (65158; Progen) and secondary antibody 680 LT (925-68020; Li-cor) diluted 1/20,000, following the manufacturer's instructions, and the image was acquired with an Odyssey device (Li-cor).
Ratio calculation
Specific baculoviral sequences present in purified rAAV particles are represented by the values obtained by qPCR for these sequences divided by the values obtained by qPCR for the rAAV vector γ-SGC.
Statistical analysis
Analysis and graphing were performed using GraphPad Prism software. Significance testing was performed using one-way analysis of variance followed by Dunnett's post test.
Results
As the only cis acting element, ITRs are crucial in AAV biology, particularly in genome replication and encapsidation. Sequencing of ITRs of the pSUB201 and derived plasmids used in the production of rAAV vectors has revealed deletions in the external parts of ITRs.
Additionally, both ITRs are flop orientated. Furthermore, a sequence of 46 nucleotides corresponding to wtAAV2 nt 4489–4534 originating from upstream of the 3′ ITR in the wtAAV2 genome has been identified just downstream of the 5′ ITR in the pSUB201. This repetition results from the duplication of the flop ITR along with the sequence of 46 bp during the original vector design in order to allow genome excision by restriction enzymes. Thus, the pSUB201 plasmid has a perfect repetition of a partially deleted flop ITR along with the described 46 nt sequence.
So far, influence of these modifications and wtAAV2-derived sequences has never been studied in the context of rAAV production using the baculovirus/Sf9 cells system. In order to do so, this study has investigated the impact on rAAV production using the baculovirus/Sf9 cells system of various forms of ITRs, including partially deleted ITRs, wild type ITRs, and ITRs coupled with various regulatory elements. Six constructs were evaluated for the rAAV production in a co-infection study with a baculovirus Rep2/Cap8.20 The γ-SGC recombinant gene was used as transgene for all evaluated variants.
The first construct, used as a reference, harbored the γ-SGC recombinant gene flanked by ITRs from the pSUB201 (SUB201-ITRs). On the 5′ side, both the flop ITR and 46 nt are conserved. On the 3′ side, the flop ITR is conserved, but instead of the 46 nt sequence, the same sequence is reduced to 37 nt following a SnaBI cloning reaction site (Fig. 1a). Thus, the remaining 37 nt correspond to wtAAV2 nt 4498–4534.
Figure 1.
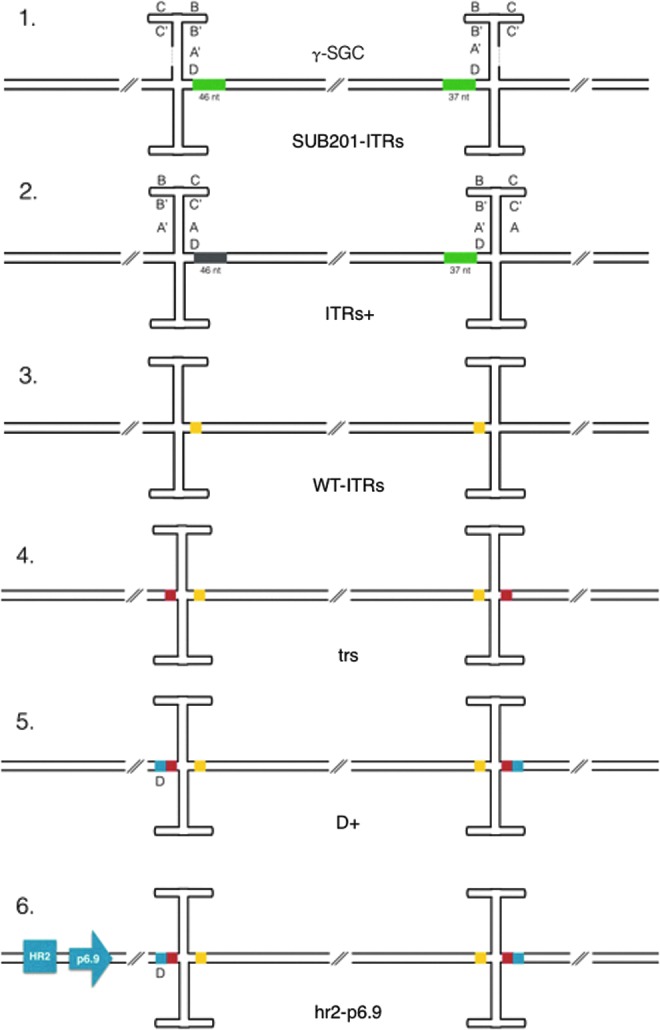
Genetic constructs assessed in this study (presented in the bacmid context). T-shape hairpin structures correspond to the inverted terminal repeat (ITR) double-stranded Holliday junction, with γ-SGG as the genetic construct between each ITR. 1. Schematization of truncated flop-flop ITRs (SUB201-ITRs), where the A sequence is missing, where green boxes correspond to the repeated sequence originated from 3′ end of the wild type (WT) adeno-associated virus serotype 2 (AAV2) sequence outside the ITRs, included in the γ-SGC sequence. 2. WT-ITR from wtAAV2 in correct orientation. Green and gray boxes represent the two sequences outside of the ITR included, coming respectively from the 3′ and the 5′ end of AAV2 genome. A–A′, B–B′, and C–C′ represent the ITR palindromic sequences, and D schematizes the localization of the D sequence (ITRs+). 3. WT-ITR. Yellow boxes represent the multiple cloning site inserted into the constructs to replace the two sequences flanking the ITRs (WT-ITRs). 4. Red boxes correspond to the trs +3 sequences inserted outside of the IT (trs). 5. Additional D sequence, represented by the blue boxes, inserted just before/after the trs +3 sequences (D+). 6. Modified version of the construct previously described26 in which baculoviral homologous region 2 and strong p6.9 promoter have been inserted (blues square and arrow) before the 5′ ITR (hr2-p6.9).
The second construct contained one ITR of each orientation combined with either the 46 or 37 bp sequences normally adjacent to it in the wtAAV2 genome, flip +46 bp (wtAAV2 nucleotides 146–191) on the 3′ end, and flop +37 bp on the 5′ end of the γ-SGC cassette (ITRs+; Fig. 1b).
In the third construct, the 46 bp and 37 bp sequences on each side of the transgene in ITRs+ have been replaced by a multiple cloning site (MCS; WT-ITRs; Fig. 1c) in order to facilitate transgene substitution.
For the fourth construct, it was hypothesized that rAAV genome rescue from the baculovirus shuttle vector could be a rate-limiting step in rAAV production using the baculovirus/Sf9 cells system. Rescue of the AAV genome occurs through Rep nicking activity. Efficient nicking requires three elements: the RBE,21 the small internal palindromes that comprise the tips of the hairpin terminal repeat (TR),21,22 and the terminal resolution site (trs).22 In order potentially to enhance rAAV genome rescue, the minimal trs sequence necessary for Rep cleavage (3′-CCGGT/TG-5′)10 has been re-created on the distal ends of ITRs by the addition of the three nucleotides normally following the trs (5′-GAG-3′; construct trs; Fig. 1d). Thus, the Rep nicking site was present four times instead of two (distal and proximal ends of both ITRs) on the double-stranded form of the rAAV genome in the baculovirus DNA.
The D sequence is the only non-palindromic ITR sequence. It is essential for recombinant genome packaging.23 Therefore, in the fifth construct, ITRs were extended on their distal end by the addition of both the three nucleotides and the D region (5′-AGGAACCCCTAGTGATG-3′) (D + l; Fig. 1e).
This construct was used to evaluate the impact of these additional D sequences on rescue and replication of the vector sequence,24 and potential packaging of ITR associated foreign DNA originating from the baculovirus genome bordering the rAAV genome.
Finally, the sixth construct was identical to “D+” but contained the baculoviral homologous repeats region 2 (hr2) and the strong baculoviral promoter p6.9 (hr2-p6.9; Fig. 1f) before the 5′-ITR. The aim of this construct was to assess the synergy between transcription and rAAV genome replication, as previously described for such enhancer elements.25,26
Three main characteristics were studied: the overall production level, the level of encapsidated DNA originating from the baculovirus genome, and the impact of these constructions on full/empty rAAV particle ratio using analytical ultra-centrifugation.
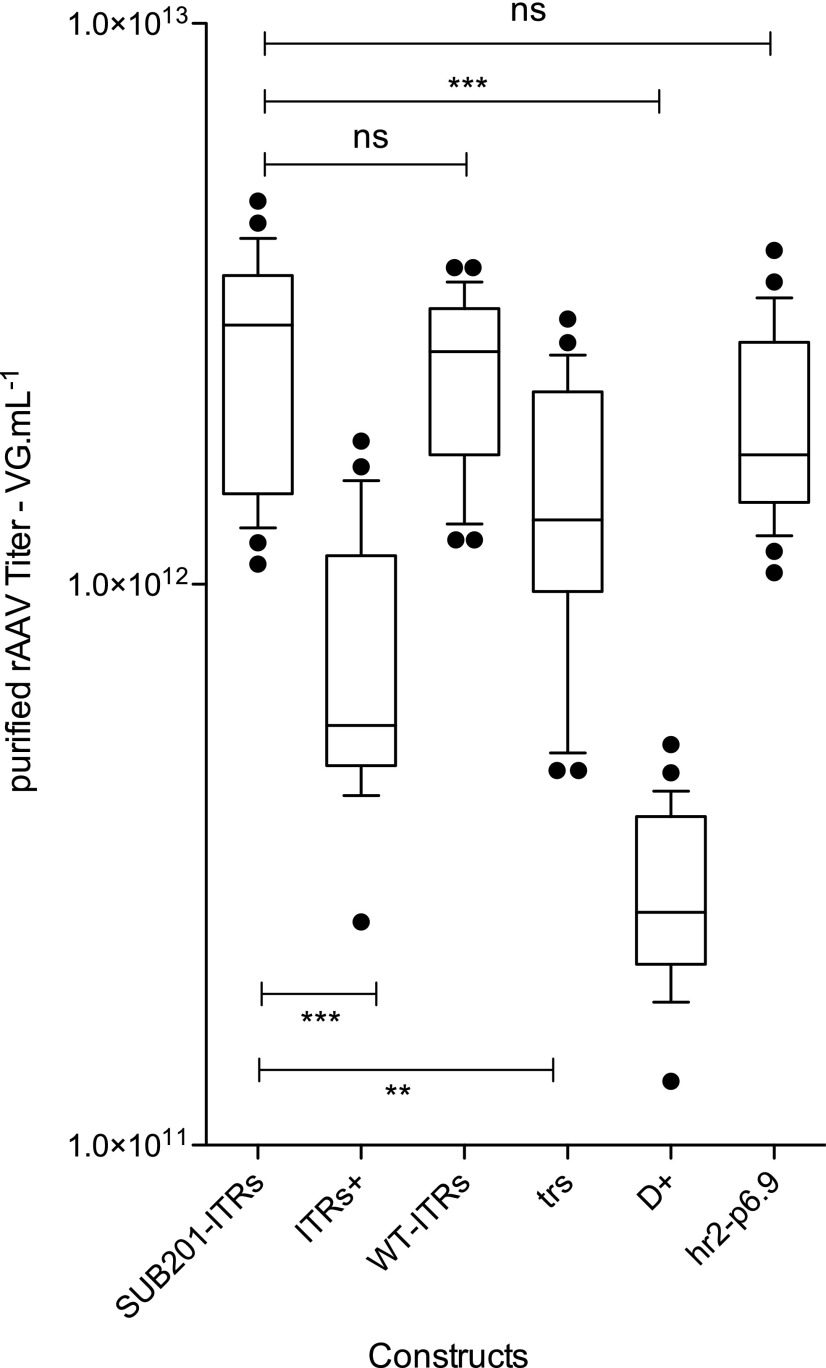
Three batches of rAAV per construct were produced. After affinity purification, the quantity of specific genome present in each production was determined using a γ-SGC-specific amplicon in a qPCR experiment. Figure 2 shows the level of specific viral genome encapsidated per construct. For the reference construct with the SUB201-ITRs, on average a rAAV titer of 2.6 × 1012 viral genomes (vg) per batch was obtained (Fig. 2). ITRs+ induced on average a threefold decrease of the overall rAAV titer compared to the SUB201 truncated ITRs, with an average of 8.0 × 1011 vg per batch (Fig. 2). The replacement of the two sequences originating from wtAAV2 by restriction endonuclease sites (WT-ITRs) led to an overall titer of 2.4 × 1012 vg, equivalent to that obtained with SUB201-ITRs (Fig. 2). Duplication of the trs site on the distal ends of ITRs led to a slight decrease of titer, with an average of 1.5 × 1012 vg per batch (Fig. 2).
Figure 2.
qPCR results on purified recombinant adeno-associated virus serotype 8 (rAAV8) produced using the different ITR variants. Three productions have been analyzed in triplicate with a probe specific to the genetic construct. Whiskers represent the 10th–90th percentile, and black dots represent points out of the 10th–90th percentile. Statistical analysis used one-way analysis of variance (ANOVA) and Dunnett's post test, with a significant p-value of <0.05.
The presence of the D sequence at both the 5′ and 3′ ends of each ITR led to a 10-fold reduction of rAAV genome encapsidation, with a yield of 3.0 × 1011 vg (Fig. 2). The addition of baculoviral elements hr2 p6.9 upstream to the D construction (hr2-p6.9) restored the overall yield of specific rAAV genome packaged (Fig. 2).
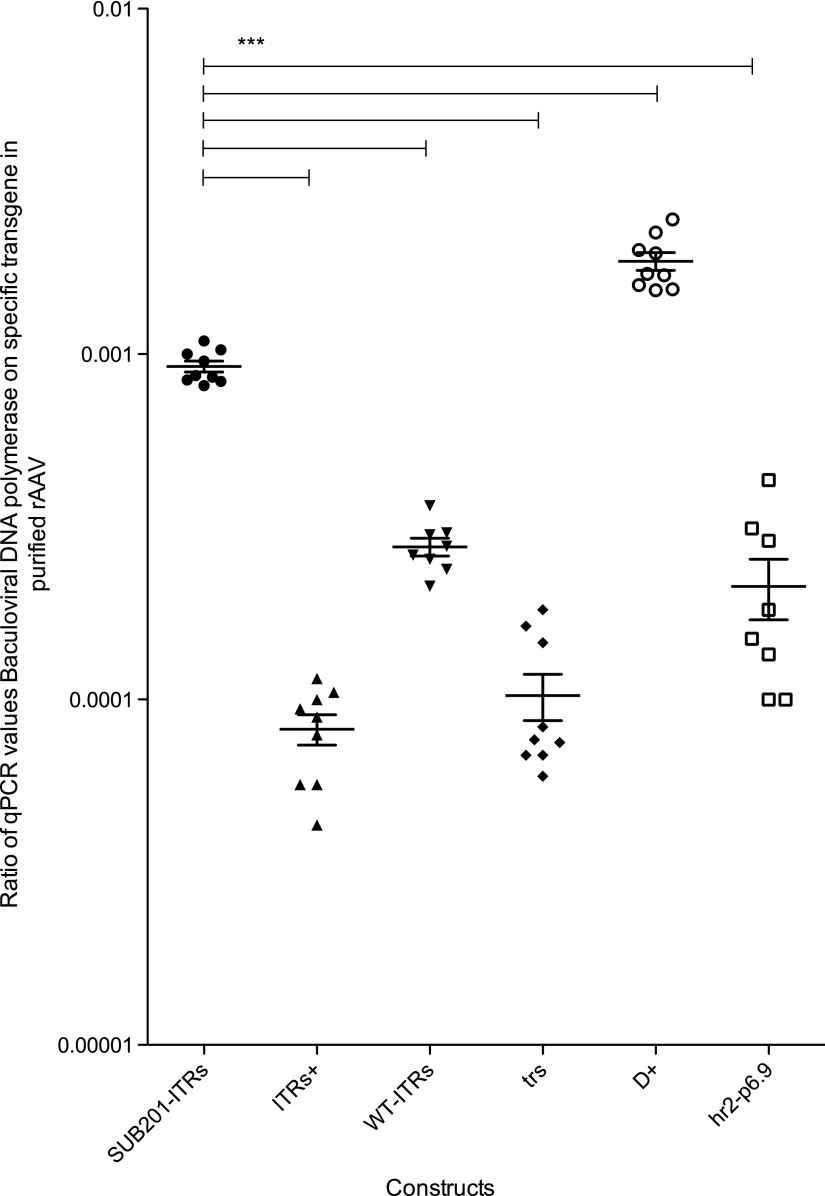
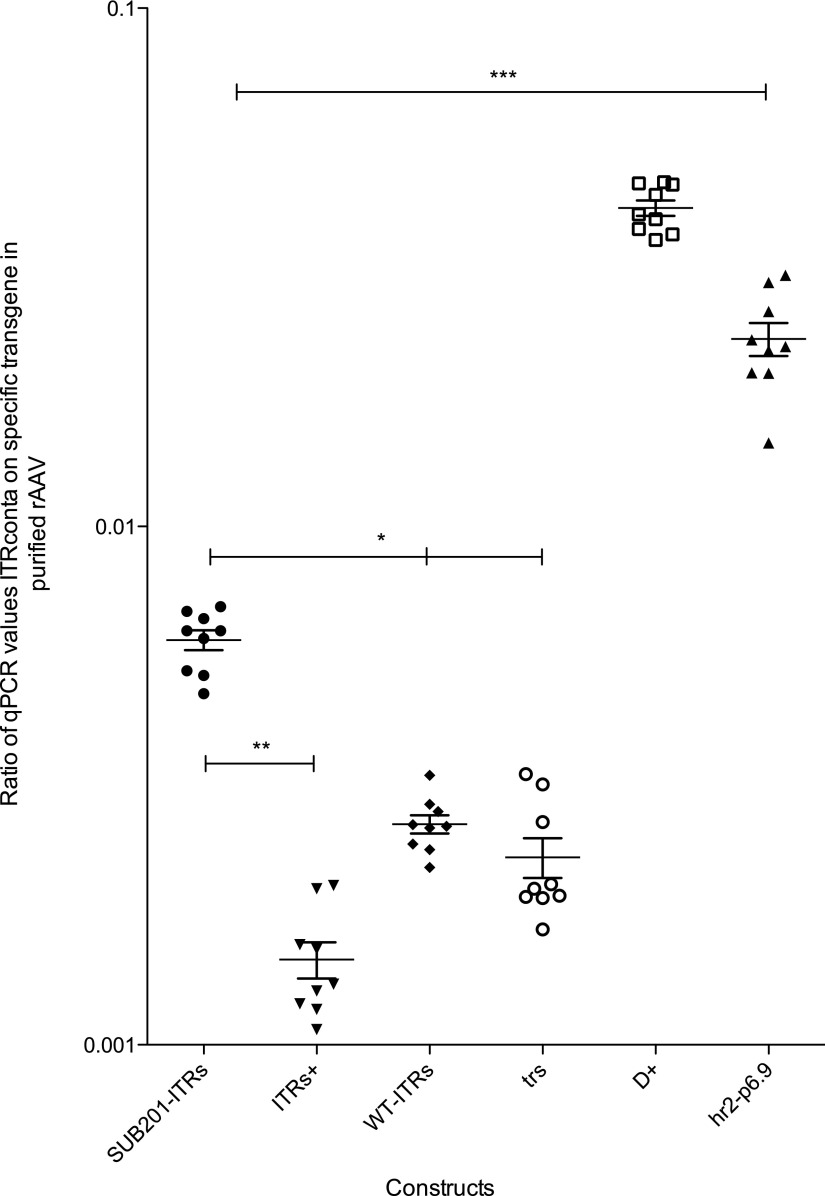
Next, the level of encapsidated DNA originating from the baculovirus backbone present in the purified particles was evaluated. To this purpose, the encapsidation of two DNA sequences originating from the baculovirus genome in the purified rAAV products was monitored. It was assumed that those sequences were representative of the global baculovirus DNA encapsidation. The first sequence (ITRconta) was located 1 kb downstream of the right ITR. The second studied sequence was located in the baculovirus DNA polymerase (BacDNApol amplicon) gene, 54 kb away from the right ITR. These two sequences were monitored to assess whether the distance from the ITR had an influence on the level of encapsidated baculovirus DNA in the rAAV8 particles. The ratios between the qPCR titer of BacDNApol and γ-SGC amplicons are presented in Fig. 3. Likewise, Fig. 4 shows the represented ratios between the qPCR titer of ITRconta and γ-SGC amplicons.
Figure 3.
Quantitative polymerase chain reaction (qPCR) results of baculoviral DNA polymerase sequence in purified recombinant adeno-associated virus (rAAV) particles. Dots represent the ratio of foreign DNA relative to specific construct. Plots represent means with the standard error to the mean. Statistical analysis used one-way ANOVA and Dunnett's post test, with a significant p-value of <0.05.
Figure 4.
qPCR results of baculoviral DNA sequence at 1 kb outside of ITR on encapsidated AAV particles. Dots represent the ratio of foreign DNA relative to specific construct. Plots represent means with the standard error to the mean. Statistical analysis used one-way ANOVA and Dunnett's post test, with a significant p-value of <0.05.
In the rAAV produced with the SUB201-ITRs, the qPCR results on the baculovirus DNA polymerase sequence gave a ratio of BacDNApol and γ-SGC qPCR titers (Fig. 3) representing 0.001 of the overall purified rAAV titer (Fig. 3). Restoration of the WT-ITRs with additional 46 (wtAAV2 nt 146–191) and 37 bp (wtAAV2 nt 4498–4534) sequences on ITRs+ construct resulted in a 10-fold decrease, with qPCR ratios going from 0.001 to 0.0001 of this specific amplicon in the purified rAAV particles (Fig. 3). Replacement of the wtAAV2 sequences flanking the GOI cassette by MCS _WT-ITRs) in combination with WT-ITRs also led to a reduced proportion of foreign baculovirus DNA polymerase sequence encapsidated in purified rAAV production compared to the SUB201-ITRs reference construct (Fig. 3). Nonetheless, this reduction in foreign DNA was two times smaller than the one brought with the ITRs+ construct (Fig. 3). This decrease in encapsidation specificity could be overcome by the duplication of the trs sequence on the distal end of ITRs. This could potentially be associated with a better rescue of the rAAV genome from the baculovirus genome.
The additional D motif outside the ITR led to a decrease in the overall γ-SGC titer. Furthermore, addition of this sequence dramatically increased the level of foreign DNA encapsidated in rAAV particles, with BacDNApol amplicon representing a ratio of 0.0018 of the purified rAAV (Fig. 3), 18 times higher than with the ITR+ construct.
Adding the hr2-p6.9 baculoviral genetic elements upstream of the left ITR partially restored the level of encapsidated foreign DNA at levels comparable to those monitored when using WT-ITRs (Fig. 3). Why addition of such baculoviral enhancer elements to the construct containing an additional D sequence allowed a 10-fold reduction of this baculovirus sequence encapsidation is not well understood. This could be either linked to transcriptional activity originating from the p6.9 promoter or replication activity originating from the hr2 baculovirus ori, limiting the access of the replication origin to the Rep proteins.
Due to their high guanine-cytosine-containing palindromic arms, ITRs fold into a T-shape conformation when genomes are single stranded. In a double-stranded sequence, however, as in plasmids or in the baculovirus used for rAAV production, these sequences form Holliday junctions.17,27 Several models have hypothesized how replicase proteins recognize RBE28,29 and initiate replication, and how they oligomerize to perform genome encapsidation.23,30 It remains unclear how repetition of motifs such as Holliday junctions influences replicase activities. Thus, this study considered how the vicinity of the ITRs influences the level of foreign DNA encapsidated in rAAV particles (ITRconta, a probe located 1 kb outside the right ITR) compared to DNA originating from a long distance (54 kb, BacDNApol; Figs. 3 and 4).
With a qPCR ratio (ITRconta on γ-SGC; Fig. 4) of 0.006 of the overall titer with SUB201-ITRs, the foreign sequence located near ITR represented a non-negligible proportion of encapsidated DNA (Fig. 4). The restoration of ITR integrity coupled with ITR-flanking sequences originating from the wtAAV2 in the ITRs+ construct greatly reduced the level of near-ITR DNA encapsidation (Fig. 4). Indeed, the obtained ratio was 0.0014—more than four times lower than with SUB201-ITRs. Deletion of the wtAAV2 sequences, and their replacement by a MCS (WT-ITRs; with or without the duplication of the trs sequence on the distal end of ITRs) did not seem to impact the specificity of DNA encapsidation, with ratios of 0.0026 and 0.0023, respectively (Fig. 4). Even if these values were two times higher than when using ITRs+, they remained lower than with SUB201-ITRs. Results obtained with constructs D+ and hr2-p6.9, which both contained an additional D sequence, tend to point to Ling's model,23 where this sequence is at least partially involved in the encapsidation process. The foreign sequence located 1 kb outside the right ITR represented a ratio of 0.0408 of the overall titer of the D+ purified rAAV (Fig. 4), a 20 times increase of such baculovirus sequences compared to the “trs” construct. The addition of the hr2-p6.9 elements to the D+ construct led to a two times decrease, with the ITRconta sequence representing a ratio of 0.0226 of the overall hr2-p6.9 purified rAAV (Fig. 4).
Considering how hr2-p6.9 baculoviral elements restore the overall rAAV titer and significantly decrease the baculovirus DNA polymerase levels (Fig. 2 and 3), this study synthesized a construct similar to hr2-p6.9 but without the additional D sequence. Unfortunately, this construct did not offer any improvement compared to ITRs+ or to the trs constructs, improving neither the overall rAAV titer nor the level of encapsidated baculoviral DNA (data not shown).
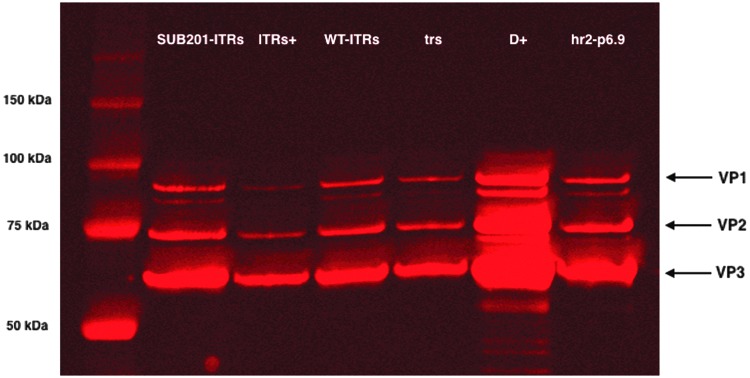
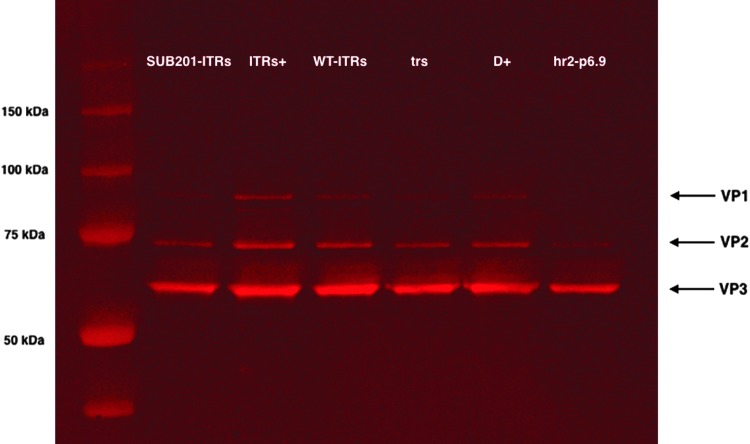
Based on qPCR titration of genome-containing viral particles, a Western blot anti-VP analysis on purified rAAV productions was performed on samples containing constant vg copies. A significant variation in the quantity of protein was noticed and could be explained by variable GOI encapsidation efficiencies (Fig. 5). Next, the amount of protein loaded into each well was extrapolated from these band intensities using the Image Studio software, and a new Western blot experiment was conducted with a theoretical constant amount of protein in each well (approximately 1 μg/sample) to assess the VP1:2:3 ratio in both full and empty rAAV capsids. At a constant protein level, VP1, VP2, and VP3 were detectable for all the constructs, except hr2-p6.9 where VP1 was not detectable. AAV capsids are comprised of 60 proteins, with VP1, VP2, and VP3 in a ratio of approximately 1:1:10, respectively.6 Nevertheless, due to the adaptation of AAV production to a heterologous production system (baculovirus/Sf9 cells), VP1 integration into formed capsids was limited in all constructs when compared to VP2 (Fig. 6). The absence of VP1 detection for the hr2-p6.9 construct can be explained by the approximately calculated protein amount, since VP1 could be detected in the Western blot performed at constant vg (Fig. 5).
Figure 5.
Western blot analysis of VP proteins on AVB chromatography purified rAAV8 preparations produced with ITR variants, separated on 8–12% SDS-PAGE. 5 × 109 genome particles loaded per lane.
Figure 6.
Western blot analysis of VP proteins on AVB chromatography purified rAAV8 preparations produced with ITR variants, separated on 8–12% SDS-PAGE. 1 μg of AAV capsids.
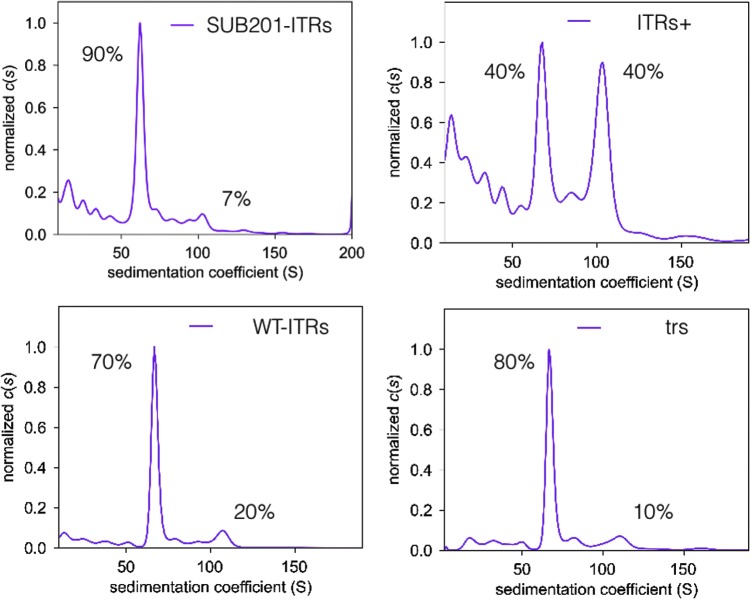
Another vital characteristic to look at in rAAV production is the proportion of full and empty capsids. In addition, it is important to discriminate different full capsid populations, since full-length genome, partial genome, or foreign DNA can be encapsidated. To assess the proportion of empty and full capsids, AUC was performed on ITRs+, WT-ITRs, and trs using the truncated SUB201-ITR γ-SGC as control. Constructs containing an additional D sequence (D+ and hr2-p6.9) were excluded from the AUC experiment, since they proved to have encapsidated significantly higher levels of foreign DNA (Fig. 4) based on qPCR experiments. As shown in Fig. 7, a full/empty particle ratio of around 10/90 was mainly obtained, comparable to other investigators.20,31 It is important to notice the significantly better encapsidation capacity observed with the ITRs+, with as high as 40% of full particles compared to 7% for the reference construct with truncated ITRs (SUB201-ITRs), 10% for the trs construct, and 20% of full capsids for the WT-ITRs construct. However, the AUC analysis of ITRs+ has detected slightly more contaminant species below 50S. This could be explained by the lower rAAV titer compared to the other constructs used for these experiments, putting the detection level closer to the background level.
Figure 7.
Analytical ultracentrifugation sedimentation coefficient results. Values have been normalized to c(s). Empty rAAV particles sedimentation are present between 64 and 66S, full rAAV8 particles are sedimentation between 99 and 103S. Values between 76 and 90S correspond to particles partially filled with a genome size between 1 and 3 kb. The values <60S correspond to impurities smaller than rAAV particles that could be free proteins or DNA.
Reconstitution of ITR integrity brings a slight improvement in the proportion of full particles. In addition to the importance of ITR integrity, this also suggests that the 46 bp (wtAAV2 nt 146–191) originating from wtAAV2 is of major importance for efficient encapsidation in cooperation with ITRs integrity, since WT-ITRs construction did not reach the specific encapsidation level of the ITRs+ construct. The 37 bp (wtAAV2 4498–4534) is probably not involved in the encapsidation process, as this element is also present in the SUB201 construct without any reduction of foreign DNA encapsidation when compared to WT-ITRs and ITR+ constructs.
Although the trs construct did not show a significant difference with the WT-ITRs construct regarding the reduction of baculoviral DNA polymerase sequence encapsidation (Fig. 2), the result obtained in the slight decrease of total capsid levels visible on the Western blot is encouraging (Fig. 5). It must be kept in mind that only short qPCR amplicons are used to monitor the foreign DNA encapsidation. Extrapolated to the totality of the AcMNPV baculovirus genome in the bacmid configuration (148 kb), the global reduction of baculovirus-derived DNA becomes highly significant.
In order to determine why ITRs+ allow a more selective encapsidation, new batches of rAAV were produced using SUB201-ITRs, ITRs+, WT-ITRs, and trs, and were monitored for replication and encapsidation kinetics of the rAAV genome over time. The hypothesis was that rAAV genomes with ITRs+ were replicated faster than other constructs, allowing earlier initiation of the encapsidation process.
However, this study did not yield any further information specific to full ITRs+ to explain why this construct led to a better encapsidation rate. Constructs with ITRs+ are not replicated faster than other constructs and are not encapsidated earlier (data not shown).
Regarding the particular interest of Généthon in muscular genetic diseases, this study mainly focused on rAAV8 production using the baculovirus/Sf9 cells system. Nevertheless, serotype 8 is not the only serotype of interest for clinical studies. Thus, after identifying ITRs+ as the most interesting construction regarding the quality of rAAV8, its influence on the quality of rAAVs of serotypes 2 and 9 has also been investigated (SUB201-ITRs was used as a reference). Serotypes 2 and 9 were chosen to assess the impact of this construction in a homologous system (ITR2, Rep2, Cap2) and in another serotype of clinical interest, respectively. Thus, rAAV2 (n = 1) and rAAV9 (n = 1) were produced using SUB201-ITRs or ITRs+ in co-infection with Rep2/Cap2 and Rep2/Cap9 encoding baculoviruses, respectively.
The level of γ-SGC transgene, along with DNA sequences originating from baculoviral DNA polymerase gene (BacDNApol amplicon) and from baculoviral DNA present downstream of ITR (ITRconta amplicon), were determined using qPCR titration. Then, BacDNApol and ITRconta titers were normalized to the γ-SGC titer for each rAAV production (BacDNApol/γ-SGC and ITRconta/γ-SGC ratios; Table 1), as previously performed for rAAV8. For both serotypes, the use of the ITRs+ construct instead of SUB201-ITRs led to a decrease in the level of encapsidated DNA originating from the baculovirus shuttle vector, as shown by the 1.5-fold difference between BacDNApol titers (normalized to γ-SGC titer) of SUB201-ITRs and ITRs+ for rAAV2 and the threefold difference for rAAV9. Once more, ITR vicinity was a major factor regarding the encapsidation of foreign DNA, as illustrated by the 2.5- (rAAV9, SUB201-ITRs) to 14-fold (rAAV2, ITRs+) difference between BacDNApol/γ-SGC and ITRconta/γ-SGC ratio (Table 1).
Table 1.
BacDNApol/γ-SGC and ITRconta/γ-SGC qPCR ratios for rAAV serotypes 2 and 9
| rAAV serotypes | Constructs | BacDNApol/γ-SGC | SUB201-ITRs/ITRs+ | ITRconta/γ-SGC | ITRconta/BacDNApol |
|---|---|---|---|---|---|
| rAAV2 | SUB201-ITRs | 0.000126 | 1.5 | 0.001129 | 8.9 |
| ITRs+ | 0.000084 | 0.001246 | 14.8 | ||
| rAAV9 | SUB201-ITRs | 0.000217 | 3.0 | 0.000547 | 2.5 |
| ITRs+ | 0.000073 | 0.000692 | 9.4 |
BacDNApol, baculovirus DNA polymerase; γ-SGC, gamma-sarcoglycan; rAAV, recombinant adeno-associated virus.
Regarding the proportions of empty and full particles in rAAV productions for the two additional serotypes and determined by AUC, SUB201-ITRs led to a much higher proportion of full particles when compared to rAAV8, with 64% and 67% of full particles for rAAV2 and rAAV9, respectively, compared to only 10% of full capsids obtained for rAAV8. Nevertheless, even more full particles were obtained using the ITRs+ construct, with 79% and 82% of full particles for rAAV2 and rAAV9, respectively (Table 2).
Table 2.
Percentages of empty/full particles in rAAV2 and rAAV9 production
| rAAV2 | rAAV9 | |||
|---|---|---|---|---|
| Construct | Empty particles (%) | Full particles (%) | Empty particles (%) | Full particles (%) |
| SUB201-ITRs | 29 | 64 | 24 | 67 |
| ITRs+ | 17 | 79 | 13 | 82 |
Proportions determined after analysis of analytical ultracentrifugation results.
Discussion
This study has demonstrated that replacement of the truncated ITRs originating from pSUB201 by ITRs+ (wtITR2 plus 46 bp [wtAAV2 nt146–191] and 37 bp [wtAAV2 4498–4534] sequences) in a baculovirus production context brings several improvements. The slight decrease in the overall rAAV titer is largely compensated by the decrease in the encapsidation of DNA originating from the baculovirus shuttle vector backbone in purified rAAV and by the higher level of full versus empty particles. In addition, it can also be concluded that the proximity of ITRs has a substantial influence on unspecific encapsidation, as demonstrated by higher titers obtained for the sequence located 1 kb outside the right ITR (ITRconta) when compared to the one located 54 kb away (BacDNApol). This was recently confirmed by Penaud-Budloo et al.34
In this work, encapsidation of DNA originating from the Sf9 cells was not studied, partly due to the difficulty of detecting low-level amplicons regarding the size of a cellular genome. However, those sequences must be kept under scrutiny, as demonstrated in the study of cell-derived DNA encapsidation during rAAV production by transfection of HEK293 cells published by Lecomte et al.32 With the observed reduction in contamination by major contaminants from the infected cells (i.e., the actively replicating baculovirus), one could think that reduction of foreign DNA encapsidation observed with ITR+ construct might also concern DNA originating from the Sf9 cells.
In addition to decreasing the encapsidated baculovirus DNA, significant variation in ratios of full/empty particles was noticed. Indeed, rAAVs produced with the construct associating WT-ITRs and their flanking sequences (ITRs+) had a full/empty particles ratio of 40/40 compared to only 7% of full particles with the truncated SUB201-ITRs.
These results indicate that while the number of replicated rAAV genomes is equal between the studied constructs, the 46 nt sequence of wtAAV2 (nt 146–191) and/or the 37 bp (wtAAV2 nt 4498–4534) enhance the specificity of rAAV packaging. These sequences have never previously been described as required in the encapsidation process. Whether these sequences are specifically required only in the context of rAAV production in the Sf9/baculovirus system or whether they could be of interest in other production systems remains to be established.
Replacement of the two wtAAV2 sequences present near each ITR by restriction sites (WT-ITRs) and duplication of the trs sequence on the distal end of ITRs (trs), even in the case of full-length ITRs, led to a full/empty ratio of only 10/90.
The results obtained with the construct containing additional D motifs validated the model where this sequence is partially implicated in the encapsidation process. In addition to increasing the encapsidation of near ITR sequences, it also increased the level of encapsidation of the baculoviral sequences 54 kb away and dramatically reduced the titer of purified rAAV. One explanation of such phenomena is the putative disruption of Rep78 replicase activities due to the presence of a supplementary D sequence on the distal ends of ITRs. As this sequence is recognized by the Rep proteins, its presence turns replicases away from vector genome encapsidation toward encapsidation of baculoviral sequences.
The addition of baculoviral elements hr2-p6.9 upstream of the flip ITR (hr2-p6.9) partially rescued the overall productivity and significantly reduced the foreign DNA encapsidation compared to the construct with additional D sequence (D+). Nevertheless, the same construct without additional D motifs did not bring any enhancement compared to ITRs+, WT-ITRs, or trs. These elements, in which hr2 is a transcriptional enhancer and p6.9 a very strong baculoviral promoter, could probably interact with the polyhedrin promoter of the rep gene sequence and modulate Rep expression, leading to the present results.
Understanding why these 46- and/or 37-base-long sequences flanking the internal part of ITRs improve the encapsidation of the rAAV vector genomes remains unclear. Further studies are required to obtain more insight on whether these sequences favor the interaction with the Rep protein complexes and stabilize them or whether these sequences are involved in folding into a particular conformation, leading to better presentation of the encapsidation motif. An indirect confirmation is the fact that neither the encapsidation rate nor the replication kinetics of the vector genome was significantly different for the ITR variants evaluated in this study.
Nonetheless, the ITRs+ construct improved the overall quality of rAAV particles by decreasing the encapsidated DNA originating from baculovirus, and greatly improved the full particles yield. Although rAAV9 and rAAV2 produced with SUB201-ITRs or ITRs+ as transgene have only been analyzed once compared to rAAV8, the aim was to know whether improvements observed in rAAV8 could be reproduced in other AAV serotypes. When using ITRs+ instead of reference SUB201-ITRs, a slight decrease of the rAAV titer was only observed in rAAV8. Reduction of encapsidated foreign DNA (baculoviral DNA) into purified rAAV was observed for all three serotypes, as depicted by the decrease of encapsidated DNA originating from 54 kb of the GOI. Reduction in the encapsidation of baculoviral DNA located near the ITR was only observed for rAAV8. Finally, even though the empty/full particle ratio is more related to the serotype considered (10%, 64%, and 67% of full particles using SUB201-ITRs for rAAV8, rAAV2, and rAAV9, respectively), an even better proportion of full capsids was observed for the three serotypes (40%, 79%, and 82% of full particles using ITRs+ for rAAV8, rAAV2, and rAAV9, respectively). Thus, the use of the ITRs+ construct has been proven conclusive on three different serotypes, especially regarding rAAV production quality.
ITRs are one of the key elements for any rAAV production, playing crucial roles for genome replication and encapsidation. The present study shows how ITR integrity could have an important impact on the quality of rAAV8 produced using the baculovirus/Sf9 system. As many different rAAV serotypes are currently used in several clinical trials, a better characterization of the encapsidated DNA that will be injected is a critical issue. Even if the ITR modifications did not bring any enhancements to the level of rAAV production, the 10-fold decrease in encapsidated foreign DNA will be of importance for regulatory reasons. Indeed, one of the major concerns of the European Medicines Agency (EMA) in relation to the delivery of the marketing authorization for uniQure's Glybera was the carry-over of baculovirus DNA because of the potential transfer of complete ORFs capable of being translated into functional baculovirus proteins (EMA Assessment report EMA/882900/2011). Thus, improvements of the biological system either via optimization of ITRs in the baculovirus context (this paper) or modification of the rep sequences33 or any further modification should be performed and implemented if proven useful.
In conclusion, ITRs have been previously demonstrated as a crucial element in AAV biology, and this study shows that in the context of the baculovirus, correct ITR sequences associated with sequences flanking them in the wtAAV2 genome have a positive impact, especially on the quality of the rAAV8 vectors. In particular, ITRs+ allow up to 40% of full particles to be obtained when other ITR constructs never exceeded 20%. In addition, such ITRs+ construct yields a 10-fold decrease in baculovirus DNA in rAAV particles compared to the γ-SGC with deleted ITR, thus improving the overall rAAV quality. Even though this development represents an important advance for rendering the Sf9/baculovirus more efficient for the production of AAV vectors of high quality, further improvements are required, in particular with respect to vector quantity, quality, and potency.
Acknowledgments
The authors are grateful to Dr. Christopher Binny for his corrections and proofreading. We would like to thank Cyril Luc, Florence Lacoste, and Christine Le Bec for performing the analytical ultracentrifugation analysis.
Author Disclosure
There is no conflict of interest.
References
- 1.Atchison RW, Casto BC, Hammon WM. Adenovirus-associated defective virus particles. Science 1965;149:754–756 [DOI] [PubMed] [Google Scholar]
- 2.Geoffroy M-C, Salvetti A. Helper functions required for wild type and recombinant adeno-associated virus growth. Curr Gene Ther 2005;5:265–271 [DOI] [PubMed] [Google Scholar]
- 3.Berns KI. Parvovirus replication. Microbiol Rev 1990;54:316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samulski RJ, Zhu W, Xiao X, et al. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J 1991;10:3941–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samulski RJ. Adeno-associated virus: integration at a specific chromosomal locus. Curr Opin Genetics Dev 1993;3:74–80 [DOI] [PubMed] [Google Scholar]
- 6.Gonçalves MAFV. Adeno-associated virus: from defective virus to effective vector. Virol J 2005;2:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang XS, Srivastava A. A novel terminal resolution-like site in the adeno-associated virus type 2 genome. J Virol 1997;71:1140–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryan JH, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol 1996;70:1542–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder RO, Im DS, Ni T, et al. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol 1993;67:6096–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brister JR, Muzyczka N. Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and trans esterification. J Virol 1999;73:9325–9336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brister JR, Muzyczka N. Mechanism of Rep-mediated adeno-associated virus origin nicking. J Virol 2000;74:7762–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol 1998;72:2224–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimm D, Kern A, Rittner K, et al. Novel tools for production and purification of recombinant adeno-associated virus vectors. Hum Gene Ther 1998;9:2745–2760 [DOI] [PubMed] [Google Scholar]
- 14.Samulski RJ, Chang LS, Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J Virol 1987;61:3096–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samulski RJ, Berns KI, Tan M, et al. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci U S A 1982;79:2077–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samulsi RJ, Berns KI, Srivastave A, et al. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell 1983;33:135–143 [DOI] [PubMed] [Google Scholar]
- 17.Wang XS, Ponnazhagan S, Srivastava A. Rescue and replication of adeno-associated virus type 2 as well as vector DNA sequences from recombinant plasmids containing deletions in the viral inverted terminal repeats: selective encapsidation of viral genomes in progeny virions. J Virol 1996;70:1668–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herson S, Faycal Hentati F, Rigolet A, et al. A phase I trial of adeno-associated virus serotype 1-γ-sarcoglycan gene therapy for limb girdle muscular dystrophy type 2C. Brain 2012;135:483–492 [DOI] [PubMed] [Google Scholar]
- 19.Wood HA. An agar overlay plaque assay method for Autographa californica nuclear polyhedrosis virus. J Invertebr Pathol 1977;29:304–307 [Google Scholar]
- 20.Smith RH, Levy JR, Kotin RM. A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol Ther 2009;17:1888–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarty DM, Pereira DJ, Zolotukhin I, et al. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J Virol 1994;68:4988–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snyder RO, Im DS, Ni T, et al. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol 1993;67:6096–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling C, Wang Y, Lu Y, et al. The adeno-associated virus genome packaging puzzle. J Mol Genet Med 2015;9:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao X, Xiao W, Li J, et al. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J Virol 1996;71:941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aslanidi G, Lamb K, Zolotukhin S. An inducible system for highly efficient production of recombinant adeno-associated virus (rAAV) vectors in insect Sf9 cells. Proc Natl Acad Sci U S A 2009;106:5059–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gómez-Sebastián S, López-Vidal J, Escribano JM. Significant productivity improvement of the baculovirus expression vector system by engineering a novel expression cassette. PLoS One 2014;9:96562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holliday R. A mechanism for gene conversion in fungi. Genet Res 2007;89:285–307 [DOI] [PubMed] [Google Scholar]
- 28.Tullis GE, Shenk T. Efficient replication of adeno-associated virus type 2 vectors: a cis-acting element outside of the terminal repeats and a minimal size. J Virol 2000;74:11511–11521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Musatov S, Roberts J, Pfaff D, et al. A cis-acting element that directs circular adeno-associated virus replication and packaging. J Virol 2002;76:12792–12802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zarate-Perez F, Mansilla-Soto J, Bardelli M, et al. Oligomeric properties of adeno-associated virus Rep68 reflect its multifunctionality. J Virol 2013;87:1232–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Urabe M, Ding C, Kotin RM. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum Gene Ther 2002;13:1935–1943 [DOI] [PubMed] [Google Scholar]
- 32.Lecomte E, Benoît Tournaire B, Cogné B, et al. Advanced characterization of DNA molecules in rAAV vector preparations by single-stranded virus next-generation sequencing. Mol Ther Nucleic Acids 2015;4:e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noordman Y, Lubelski J, Bakker AC. Mutated rep encoding sequences for use in AAV production. US 2013/0023034
- 34.Penaud-Budloo M, Lecomte E, Guy-Duché A, et al. Accurate identification and quantification of DNA species by next-generation sequencing in adeno-associated viral vectors produced in insect cells. Hum Gene Ther Methods 2017;28:148–162 [DOI] [PubMed] [Google Scholar]