Abstract
Genotyping-by-sequencing (GBS) was conducted on 333 Cercospora isolates collected from Beta vulgaris (sugar beet, table beet and swiss chard) in the USA and Europe. Cercospora beticola was confirmed as the species predominantly isolated from leaves with Cercospora leaf spot (CLS) symptoms. However, C. cf. flagellaris also was detected at a frequency of 3% in two table beet fields in New York. Resolution of the spatial structure and identification of clonal lineages in C. beticola populations using genome-wide single nucleotide polymorphisms (SNPs) obtained from GBS was compared to genotyping using microsatellites. Varying distance thresholds (bitwise distance = 0, 1.854599 × 10−4, and 1.298 × 10−3) were used for delineation of clonal lineages in C. beticola populations. Results supported previous reports of long distance dispersal of C. beticola through genotype flow. The GBS-SNP data set provided higher resolution in discriminating clonal lineages; however, genotype identification was impacted by filtering parameters and the distance threshold at which the multi-locus genotypes (MLGs) were contracted to multi-locus lineages. The type of marker or different filtering strategies did not impact estimates of population differentiation and structure. Results emphasize the importance of robust filtering strategies and designation of distance thresholds for delineating clonal lineages in population genomics analyses that depend on individual assignment and identification of clonal lineages. Detection of recurrent clonal lineages shared between the USA and Europe, even in the relaxed-filtered SNP data set and with a conservative distance threshold for contraction of MLGs, provided strong evidence for global genotype flow in C. beticola populations. The implications of intercontinental migration in C. beticola populations for CLS management are discussed.
Introduction
Understanding the genetic structure and evolutionary trajectory of pathogen populations in agroecosystems is fundamental to sustainable disease management [1]. Population genetics studies provide insights into pathogen biology, epidemiology, and co-evolutionary interactions with plants, which, in turn, help define management units and dynamic management strategies in response to ever-changing pathogen populations [1,2]. Recently, microsatellites (also known as simple sequence repeats) have been the markers of choice for population genetics studies, and have improved our knowledge of the biology and epidemiology of plant pathogens [3–8]. Some of the characteristics that make microsatellites popular (high mutation rate and variability) also complicate data analysis due to homoplasy, null alleles and complex mutation patterns that may violate assumptions of mutation-migration-drift equilibrium [9,10]. Moreover, ascertainment bias, caused by selection of the most polymorphic microsatellite loci after screening a limited number of individuals, may introduce a systematic bias in the estimates of population variation and structure [11,12]. Some studies also have proposed that microsatellites do not accurately reflect genome-wide patterns of diversity [13,14].
Alternatively, single nucleotide polymorphisms (SNPs) offer genome-wide coverage; and confer stable inheritance and analytical simplicity [15]. However, the biallelic nature and lower information content of SNPs demand a higher number of loci to be analyzed, which has traditionally hindered the use of SNPs in population genetics studies of non-model organisms [16–18]. Moreover, ascertainment bias persists as a substantial complication in methods of SNP discovery that use a non-representative sample of individuals for discovery of loci that are subsequently used for genotyping a broader set of individuals [16].
The advent of reduced-representation sequencing (RRS) methods [19] that facilitate genome-wide discovery of a large number of SNPs at lower costs has enabled the transition from population genetics to population genomics [20,21]. RRS approaches such as restriction site associated DNA sequencing (RAD-seq) [22] and genotyping-by-sequencing (GBS) [23] use restriction enzymes to reduce genome complexity before sequencing. These methods enable sequencing of a targeted genome fraction for a large number of individuals thereby reducing ascertainment bias through simultaneous marker discovery and genotyping [12,14]. Population genomics approaches can improve population genetics studies through generating multitudes of polymorphic markers that better reflect the genome-wide genetic diversity of populations [13,24]; and enhance resolution to identify fine-scale genetic variation [25–27] or detect rare recombination events [28].
An essential step in population genomics analyses using RRS genotyping techniques is filtering of poor quality reads and loci with low read depth, low minor allele frequency and a high proportion of missing data [20]. However, best filtering practices are poorly defined for population genomics studies [20,21]. Another potential area of confusion when using RRS genotyping techniques, especially for population genomics analyses of clonal populations, is the identification of genotypes and clones [20,29]. When using only a handful of microsatellite loci, a clone is defined as a unique multi-locus genotype (MLG) [30,31]. However, sequencing and SNP calling errors and missing data in RRS genotyping techniques result in identification of genetically identical individuals as different MLGs [25,29,32]. Moreover, in populations where clonal reproduction predominates, accumulation of somatic mutations over generations results in genetically non-identical individuals within clones, which may be more readily differentiated through high throughput genotyping methods with improved resolution. Thus, assignment of clones based only on MLG will result in highly inflated estimates of clonal diversity [12,20]. Presence of genotyping errors or somatic mutations at high frequencies may be detected through a peak at very low distance magnitudes in the frequency distribution of genetic distances. This may be used to define a distance threshold below which distinct MLGs are assembled into multi-locus lineages (MLL) [32]. Use of analytical tools that depict clonal boundaries based on defined genetic distance thresholds is, therefore, indispensable for identification of MLLs in genome-wide SNP data sets [20,29,32].
Cercospora beticola Sacc. is a haploid fungus and the cause of Cercospora leaf spot (CLS) on Beta vulgaris L. (sugar beet, table beet and swiss chard) worldwide. Previous population genetic studies using microsatellite markers uncovered cryptic recombination and high genetic diversity [33–38] in C. beticola populations, yet also revealed the predominance of multiple clones in New York and Hawaii [37–38]. Discovery of low population differentiation and distribution of recurrent genotypes across table beet fields separated by kilometers was interpreted as evidence for long distance dispersal of C. beticola in its asexual form, i.e., genotype flow [38]. Since asexual spores of C. beticola are reported to be dispersed by water or wind over short distances [39,40], long distance dispersal of clones may be mediated by other mechanisms such as agricultural machinery [41,42], seedborne inocula [34,43,44], or insects [39,43]. The relative roles of these mechanisms in initiation of CLS epidemics are not yet fully understood.
Detection of genotype flow in microsatellite population genetics studies of C. beticola may also have been a function of the low power of the 12, microsatellite loci to discriminate non-identical genotypes. In addition, due to the high mutation rate of microsatellite loci, identical multi-locus genotypes may arise through convergent evolution (homoplasy), and not be identical by descent [12]. The use of molecular markers with lower mutation rates, e.g., SNPs, and more refined genotyping methods such as GBS that provide thousands of markers and greater resolution of clonal structure may improve our understanding of genotype flow in C. beticola populations.
The primary objective of this study was to investigate the occurrence of global genotype flow in C. beticola populations using microsatellites [29,33] and GBS [23]. We used the R package poppr [29,45] for defining clonal boundaries in a GBS-SNP data set of C. beticola populations, and assessed the impact of various filtering strategies and distance thresholds on the estimation of clonal diversity and genotype flow. A complementary objective was to compare measures of clonal diversity, differentiation and structure of C. beticola populations obtained from both genotyping approaches.
Materials and methods
Fungal isolates and species identification
In total, 333 Cercospora isolates sampled from B. vulgaris (sugar beet, table beet and swiss chard) in the USA and Europe were included in this study (Table 1). The populations from Hawaii (Diamond Head community garden) and New York (Farms 1 and 2, Fields 3 and 5) were described in a previous study [38]. In brief, the Hawaiian population consisted of 67 isolates collected from swiss chard and table beet growing in sympatry in a community garden in Honolulu, Hawaii. The New York populations were collected from two mixed-cropping farms (Farms 1 and 2) and two monoculture table beet fields (Fields 3 and 5 planted to cultivars ‘Ruby Queen’ and ‘Red Ace’, respectively). The mixed-cropping farms in New York consisted of small-scale organic vegetable gardens that produce table beet and swiss chard, intermixed with other fresh market vegetables, on an annual basis. The monoculture table beet fields consisted of broad-acre (> 0.2 km2) table beet fields with at least 2- to 3-year rotations with non-host crops. The C. beticola isolates from Michigan, North Dakota and Europe were obtained from personal collections (LEH and GAS). The identity of all isolates was confirmed as C. beticola using PCR primers CercoCal-beta and CercoCal-R [46].
Table 1. Cercospora isolates collected from Beta vulgaris in the United States of America and Europe, and characterized through genotyping-by-sequencing and microsatellites.
| Population | Location | Year | Host (Variety) | N |
|---|---|---|---|---|
| Europe | Denmark (n = 5), England (n = 2), Germany (n = 5), Italy (n = 6), Sweden (n = 2), Turkey (n = 5) | 2009–11 | Sugar beet | 25 |
| Hawaii | Diamond Head community garden, Honolulu | 2015 | swiss chard | 34 |
| Table beet | 33 | |||
| Michigan | Michigan State University Research Field | 2011 | Sugar beet | 4 |
| New York | Farm 1, Hector | 2015 | Table beet | 16 |
| Farm 2, Phelps | 2015 | swiss chard | 27 | |
| Table beet (Detroit) | 39 | |||
| Table beet (Touchstone Gold) | 38 | |||
| Field 3, Batavia | 2015 | Table beet (Ruby Queen) | 54 | |
| Field 5, Mt Morris | 2015 | Table beet (Red Ace) | 51 | |
| North Dakota | USDA Research Field | 2014 | Sugar beet | 12 |
| Total | 333 |
Microsatellite genotyping
Amplification of 12 SSR loci in the isolates from Michigan (n = 4), North Dakota (n = 12), and Europe (n = 25), fragment analysis, and microsatellite allele calling was conducted as described by Vaghefi et al. [37]. Fragment analysis was conducted at the Cornell University Institute of Biotechnology Genomic Diversity Facility, using a GeneScan-500 LIZ size standard (Applied Biosystems) on an ABI 3730xl DNA Analyzer. To reduce the effect of genotyping error and missing data on the results, the data were filtered in poppr [29,45] using a filtering threshold estimated by the function cutoff_predict (0.02083333). These data were combined with previously published data for the C. beticola isolates collected from New York (n = 225) and Hawaii (n = 67) [37].
Genotyping-by-sequencing
DNA extraction of single-conidium-derived isolates was conducted on lyophilized mycelial tissue as described by Vaghefi et al. [36]. DNA integrity was evaluated by gel electrophoresis and quantification conducted using a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). DNA from each isolate (40–100 μl; >20 ng/μl) was submitted to the Cornell University Institute for Genomic Diversity (IGD) for DNA clean-up and GBS [23]. In brief, a reduced representation library was created by digesting genomic DNA with the restriction enzyme Pst1; oligonucleotide adapters were ligated onto restriction fragments; samples were pooled, enriched by PCR, and sequenced (100 bp single-end) on an Illumina Hi-Seq2500 sequencer (Illumina, San Diego, CA, USA). Due to its dependence on digestion with restriction enzymes, the GBS method developed by Elshire et al. [23] is highly sensitive to DNA sample quality as impurities may prevent complete digestion and result in lower read numbers. Therefore, a commercial kit (ZR-96 Genomic DNA Clean & Concentrator, Zymo Research, CA, USA) was used by the IGD to purify the DNA samples before library preparation.
To assess the reproducibility of GBS, three Cercospora isolates were genotyped six to eight times (Table 2). The same DNA sample for each of these isolates was included in each plate and sequencing run to evaluate the contribution of sequencing and SNP calling errors to polymorphism.
Table 2. Replicated DNA samples in genotyping-by-sequencing of Cercospora isolates, and genetic distance among the replicates.
| Isolatea | Replicate | DNA plate | Sequencing run |
|---|---|---|---|
| Tb15-092 | 1 | 1 | 1 |
| 2 | 1 | 1 | |
| 3 | 2 | 1 | |
| 4 | 2 | 1 | |
| 5 | 3 | 2 | |
| 6 | 3 | 2 | |
| 7 | 4 | 2 | |
| 8 | 4 | 2 | |
| Average genetic distance among replicatesb | 0.000283 (0.001855) | ||
| Tb15-169 | 1 | 1 | 1 |
| 2 | 1 | 1 | |
| 3 | 2 | 1 | |
| 4 | 2 | 1 | |
| 5 | 3 | 2 | |
| 6 | 4 | 2 | |
| Average genetic distance among replicates | 0.001298 (0.004451) | ||
| Tb15-547 | 1 | 1 | 1 |
| 2 | 2 | 1 | |
| 3 | 3 | 2 | |
| 4 | 3 | 2 | |
| 5 | 4 | 2 | |
| 6 | 4 | 2 | |
| Average genetic distance among replicates | 0.000101 (0.000538) | ||
a Isolates Tb15-092 and Tb15-169 are C. beticola. Tb15-547 was later identified as C. cf. flagellaris.
b Bitwise distance as estimated in poppr v 2.0 [45] for the SNP data set with relaxed filtering parameters. The maximum distance among replicates is given in parentheses. For the strictly filtered SNP data set, the bitwise distance among the DNA replicates was zero.
SNP calling and strict vs. relaxed quality filtering
Genotype calling was performed by IGD using the TASSEL-GBS pipeline implemented in Tassel v. 3.0.174 [47]. In this method, only sequences that align to the reference genome (74% of the sequence tags) yield SNPs. However, SNPs are identified based on differences among the isolates and not relative to the reference genome. In brief, sequence tags (unique sequences trimmed of ambiguous nucleotides and barcodes to 64 bp) were aligned to the draft genome of C. beticola isolate Tb14-085 (collected from table beet in Batavia, New York [36]) using the Burrows-Wheeler Aligner (BWA; [48]). Only sequence tags present at least three times (pooled sample depth of three) were used to identify SNPs. The resulting variant call format (VCF) file was filtered to include only SNPs with minor allele frequency greater than 0.01 and maximum missing data of 90% by IGD. The data set obtained from the IGD was further filtered for more stringent parameters using TASSEL v. 5.2.33 [49] and Vcftools v. 0.1.14 [50] on the Linux cluster at the Cornell University BioHPC Computing Lab, Ithaca, New York, USA.
A preliminary exploratory analysis was conducted on the entire data set using principal component analysis (PCA) in TASSEL. Genotypes were converted to numeric scores and missing data was imputed to the mean score for each site. The resulting eigenvalues were visualized within a scatter plot.
As C. beticola is haploid, all heterozygous sites derived from sequencing or SNP calling errors were recoded as missing. The data set was further filtered using two approaches; i) relaxed filtering for a minimum minor allele frequency of 0.01 and a maximum of 0.25 missing data for each locus; and ii) stricter filtering for a minimum locus-by-individual (genotype) read depth of three, minimum minor allele frequency of 0.01, and a maximum of 0.10 missing data for each locus, i.e., only loci with at least 90% coverage in all isolates were retained. These two SNP data sets are referred to as the “relaxed-filtered” (S1 File) and “strictly filtered” (S2 File) data sets herein. In both data sets, genotypes with > 20% missing data were removed from the analyses. Variable sites produced by each method were converted into VCF files to enable importation to R, using the software vcfR [51]. Bitwise genetic distance, which calculates the fraction of different loci among samples, counting missing data as equivalent in comparison, was estimated among the replicated DNA samples for the relaxed-filtered and strictly filtered SNP data sets [29,45].
The replicated DNA samples were removed from data sets for all subsequent analyses. Removal of the genotypes with more than 20% missing data from the relaxed-filtered and strictly filtered SNP data sets resulted in a total of 307 and 310 individuals, respectively. To enable meaningful comparisons of clonal diversity indices among the data sets, the microsatellite and the strictly filtered SNP data sets were reduced to include only the 307 individuals in the relaxed-filtered SNP data set.
Data analysis
Delineation of multi-locus lineages (MLLs)
Sequencing and scoring errors, somatic mutations, and missing data may inflate the number of clones by assigning individuals belonging to the same clone to separate MLGs [32]. To reduce the effect of such phenomena on identification of clones, the microsatellite and SNP data sets were contracted using distance thresholds identified in poppr v. 2.0 [45]. The microsatellite data was contracted using Bruvo’s distance [52] “farthest neighbour” algorithm, and a filtering threshold of 0.02083333, estimated by the cutoff_predict function in poppr. For the SNP data sets, two approaches were taken. In the first approach, the cutoff_predict function in poppr was used to estimate the distance threshold for MLL boundaries (mlg.filter threshold = 1.854599 × 10−4 and 0 for the relaxed-filtered and strictly filtered data sets, respectively), and all genotypes with the estimated distance threshold were collapsed into the same multi-locus lineage (MLL). In the second approach, the average bitwise distance among the replicated DNA samples was used (1.298 × 10−3 and 0, for the relaxed-filtered and strictly filtered data sets, respectively; Table 2) to collapse all genotypes with the calculated distance threshold into the same MLL. This resulted in three SNP data sets; 1) relaxed-filtered contracted using the more conservative distance threshold estimated by poppr (1.854599 × 10−4), herein referred to as the “relaxed-filtered data set 1”; 2) relaxed-filtered contracted using a larger distance threshold (1.298 × 10−3) based on the average distance among replicated DNA samples, herein referred to as the “relaxed-filtered data set 2”; and 3) strictly filtered data set contracted with the distance threshold of zero. All subsequent analyses were conducted on the contracted data sets.
For the SNP and microsatellite data sets, Nei’s measure of allelic diversity (He) [53], the number of multi-locus lineages (MLLs), clonal fraction (CF), Simpson’s complement index of genotypic diversity (λ) [54] corrected for sample size, and recurrent MLLs (MLLs that occurred more than once) were obtained using poppr. For the microsatellite data set, allelic richness (Ra) was estimated with rarefaction in ADZE v. 1.0 [55]. Pearson’s correlation coefficient was used to assess the association between indices of clonal diversity estimated from the microsatellite and the SNP data sets. For the microsatellite data set, the probability that recurrent MLGs (MLGs that occurred more than once) could have arisen through sexual reproduction was estimated through Psex in GenClone 2.0 [32], and statistical significance was computed by 999 randomizations.
Population structure and differentiation
Jost’s measure of population differentiation (D) [56], pairwise Nei’s GST [57] and pairwise FST [58] were estimated using the package mmod [59] implemented in adegenet [60], and hierfstat [61]. The Mantel test [62] was performed using ade4 [63] implemented in mmod, with 999 permutations, to quantify associations between values of D, GST and FST obtained from the microsatellite and SNP data sets. Discriminant analysis of principal components (DAPC) among C. beticola populations was conducted using adegenet [60]. The optimal number of principal components (PCs) for each data set was determined using the xvalDapc function.
The number of genetic clusters (K) and assignment of individuals to each cluster without a priori assumption of populations were estimated using the program, STRUCTURE [64]. Assignment of MLLs to clusters was inferred for K = 1–10. Each model was simulated five times with 100,000 iterations and a burn-in period of 10,000 Monte Carlo Markov Chains. The optimal number of clusters was chosen by computing Evanno’s ΔK [65] through STRUCTURE HARVESTER v.0.6.94 [66]. The replicated runs for the optimal K were combined using CLUMPAK [67] and a single graphical output was generated.
Results
GBS data summary
A total of 349 genotyped DNA samples had an average of 1,168,032 ± 512,427 reads that passed quality filtering. One DNA sample with less than 10,000 reads failed the quality filtering. The unfiltered data set containing all the 350 DNA samples representing 333 individuals (17 replicates of three individuals) included 27,838 SNPs, which reduced to 19,126 SNPs after initial filtering by the IGD. Further filtering of the entire data set for minor allele frequency of at least 0.01, and minimum 80% coverage of loci resulted in 7,431 SNPs.
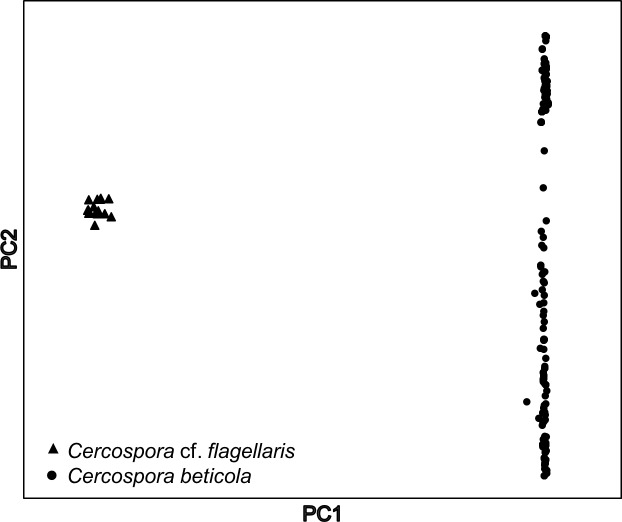
PCA using the entire data set (333 individuals and 7,431 SNPs) detected two distinct clusters of individuals (Fig 1), separating 18 DNA samples (from Fields 3 and 5) into a distinct cluster. Microsatellite loci failed to amplify in these same samples. Subsequent multi-locus phylogenetic analyses (ITS, actin, calmodulin, histone H3, and translation elongation factor 1-α) revealed these samples were a different species; Cercospora cf. flagellaris, and were not included in subsequent analyses.
Fig 1. Principal component analysis of 333 Cercospora spp. isolates collected from Beta vulgaris genotyped through genotyping-by-sequencing (GBS).
SNPs (n = 7,431) obtained through GBS detected two distinct clusters later identified as C. cf. flagellaris (triangles) and C. beticola (circles) using multi-locus sequence typing.
Relaxed vs. strict-filtering of SNP data set
For the relaxed-filtered data set, 2,696 SNPs were retained in 319 DNA samples (307 individuals); and 4.89% of the data set was missing. Strict filtering parameters retained 1,631 SNPs in 322 DNA samples (310 individuals), resulting in 1.35% missing data.
Replicated samples
For the relaxed-filtered SNP data set, none of individuals had the same MLG, including the replicated samples. The bitwise distance among the replicated samples ranged from 4.1 × 10−4 to 1.855 × 10−3 (Table 2). For the strictly filtered SNP data set, the bitwise distance among the replicated DNA samples was zero. However, 37–50% of the replicated samples were identified as different MLGs, which were attributed to loci with missing data.
Delineation of clonal lineages
Genotyping of the 307 C. beticola isolates using microsatellites resulted in detection of 130 MLGs. Contracting the microsatellite data set using the threshold estimated by the cutoff_predict function in poppr resulted in collapsing of 10 genotypes and retention of 120 MLLs (Table 3).
Table 3. Indices of multi-locus diversity for Cercospora beticola populations from Hawaii (HI), New York (NY) and Europe (EUR) after genotyping using 12 microsatellite loci [31,33] and genotyping-by-sequencing (GBS) [23].
The relaxed-filtered GBS data set included minimum minor allele frequency of 0.01 and a maximum of 25% missing data for each locus. The strictly filtered data set included a minimum locus-by-individual read depth of three, minimum minor allele frequency of 0.01, and a maximum of 10% missing data for each locus.
| Populationa | Nb | Microsatellite loci | GBS–Relaxed-Filtered (2,696 SNPs) | GBS–Strictly Filtered (1,631 SNPs) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Data set 1 (mlg.filter threshold = 1.855 × 10−4) | Data set 2 (mlg.filter threshold = 1.298 × 10−3) | mlg.filter threshold = 0 | |||||||||||
| MLLc | λd | CFe | MLLc | λd | CFe | MLLc | λd | CFe | MLLc | λd | CFe | ||
| HI | 65 | 7 | 0.42 | 0.89 | 30 | 0.96 | 0.53 | 12 | 0.90 | 0.81 | 6 | 0.68 | 0.91 |
| ND | 12 | 8 | 0.89 | 0.33 | 11 | 0.98 | 0.08 | 10 | 0.97 | 0.17 | 8 | 0.89 | 0.33 |
| NY–Farm 1 | 15 | 6 | 0.70 | 0.60 | 13 | 0.98 | 0.13 | 7 | 0.78 | 0.53 | 4 | 0.73 | 0.73 |
| NY–Farm 2 | 98 | 35 | 0.96 | 0.64 | 82 | 0.99 | 0.16 | 60 | 0.98 | 0.38 | 34 | 0.96 | 0.65 |
| NY–Field 3 | 47 | 32 | 0.98 | 0.32 | 41 | 0.99 | 0.13 | 35 | 0.98 | 0.25 | 29 | 0.97 | 0.38 |
| NY–Field 5 | 43 | 17 | 0.89 | 0.61 | 36 | 0.99 | 0.16 | 24 | 0.94 | 0.44 | 15 | 0.88 | 0.65 |
| EUR | 23 | 21 | 0.99 | 0.09 | 22 | 0.99 | 0.04 | 22 | 0.95 | 0.04 | 20 | 0.98 | 0.13 |
a Due to the small number of individuals from Michigan (n = 4), the indices of clonal diversity were not estimated for this population
b N = population size
c MLL = Number of multi-locus lineages after contracting the data set using the mlg.filter function in poppr [45]
d λ = Simpson’s complement index of genotypic diversity defined as the probability that two genotypes randomly chosen from the population are different
e CF = clonal fraction = (N–number of MLLs)/N.
The relaxed-filtered SNP data set (2,696 SNPs in 307 individuals) included 307 MLGs. Collapsing the data set using the more conserved threshold estimated by cutoff_predict resulted in 235 contracted MLLs (relaxed-filtered SNP data set 1). Using the average bitwise distance among the replicated DNA samples as the threshold resulted in 166 MLLs within the 307 individuals (relaxed-filtered SNP data set 2; Table 3).
The strictly filtered SNP data set (1,631 SNPs in 307 individuals) included 275 MLGs, i.e., 32 individuals had the same genotype at all 1,631 SNPs loci. The distance threshold estimated by the cutoff_predict function and maximum distance among the replicated DNA samples were estimated as zero. Therefore, the strictly filtered SNP data set was only contracted once, collapsing all the individuals with a bitwise distance of zero to the same MLL, resulting in 111 MLLs (Table 3).
Recurrent lineages and genotype flow
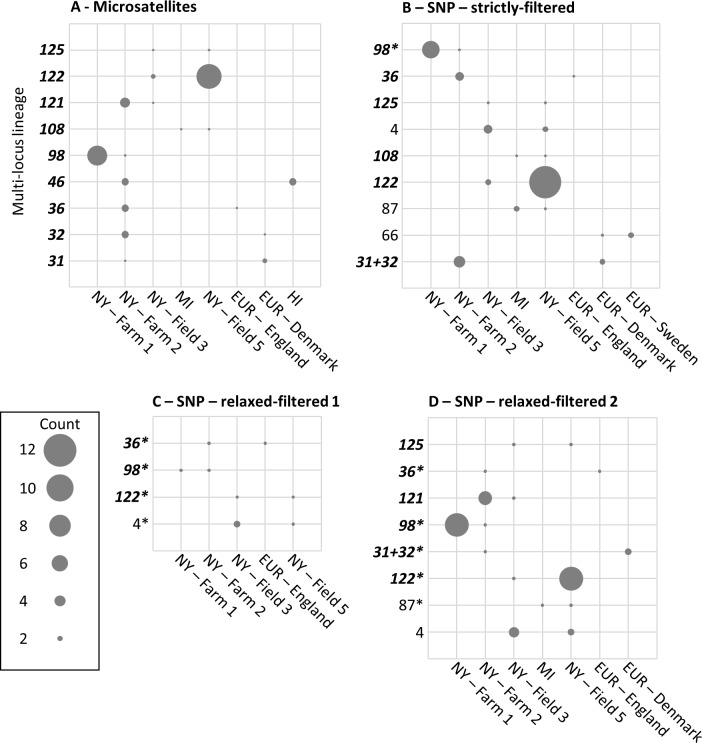
Of the 120 MLLs identified using microsatellites, 43 were recurrent and nine were shared among populations (Fig 2A). MLLs 98, 121, 122 and 125 were shared among the C. beticola populations from New York. MLLs 46 and 108 were shared between different states within the USA. Three MLLs were shared between New York and Europe, all of which occurred in Farm 2 (MLLs 31, 32 and 36). The probability of the recurrent MLLs having originated from independent sexual events was ≤ 0.003 for all microsatellite MLLs.
Fig 2. Recurrent multi-locus lineages (MLLs) shared among Cercospora beticola populations.
Circles represent MLLs shared among Hawaii (HI), Michigan (MI), New York (NY), and Europe (EUR), with circle sizes proportional to MLL frequencies. The vertical axes show the MLLs detected in the microsatellite (A) and single nucleotide polymorphism (SNP) data sets generated through genotyping-by-sequencing (B [strictly filtered], C [relaxed-filtered data set 1], and D [relaxed-filtered data set 2]). MLLs detected using microsatellites are indicated in bold and italic font. When the same MLL was detected in a SNP data set, the original MLL number was replaced with the microsatellite MLL number to allow comparisons between markers. SNP MLLs that included some, but not all, of the individuals in a microsatellite MLLs are indicated with an asterisk.
The strictly filtered SNP data set (111 MLLs) included 46 recurrent MLLs and nine were shared among populations from different locations (Fig 2B). Four MLLs were shared among the populations within New York, and two MLLs occurred in Michigan and New York. The microsatellite MLL36 also was detected in the strictly filtered SNP data set as shared between Farm 2 (New York) and England. Microsatellite MLLs 31 and 32 were shared between Farm 2 and Denmark but not differentiated from each other in the strictly filtered SNP data set and were identified as a single clonal lineage. An additional MLL (66) was only detected in the strictly filtered SNP data set as shared between Denmark and Sweden.
Of the 235 MLLs detected in the relaxed-filtered data set 1, 46 were recurrent and four were shared among populations. Three MLLs were shared among various farms and fields within New York, while one MLL occurred in both Farm 2 and England (Fig 2C). Of the 166 MLLs in relaxed-filtered data set 2, 52 were recurrent, and eight were shared among populations; five MLLs among table beet fields and farms within New York; one MLL between New York and Michigan; and two MLLs between Farm 2 and Europe (Fig 2D).
Indices of genetic diversity
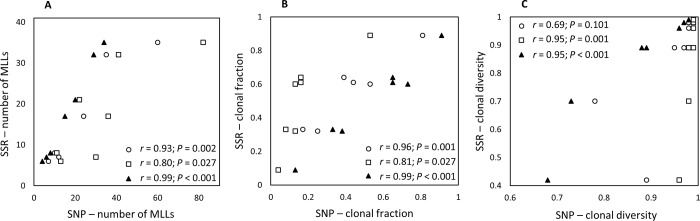
The indices of allelic diversity (He) and richness estimated using microsatellite loci were not significantly correlated with the He obtained from the SNP data sets (r < 0.69; P > 0.085) (S1 Table). A significant positive correlation was detected between the number of MLLs and clonal fraction estimated from the microsatellites and SNP data sets (Fig 3A and 3B). Simpson’s complement index of genotypic diversity (λ) obtained with the microsatellite and one of the relaxed-filtered SNP data sets was not significantly correlated (Fig 3C).
Fig 3.
Relationships between the (A) Number of multi-locus lineages (MLLs); (B) Clonal fraction; and (C) Simpson’s complement index of genotypic diversity for Cercospora beticola populations estimated using 12 microsatellites (SSR) and single nucleotide polymorphisms (SNPs) generated using genotyping-by-sequencing. Values were estimated using the strictly filtered SNP data set (filled triangles), relaxed-filtered SNP data set 1 (open circles) and relaxed-filtered SNP data set 2 (open squares).
Population structure and differentiation
Pairwise indices of differentiation obtained using microsatellites showed low differentiation among the two table beet monoculture fields (Fields 3 and 5) within New York (D = 0.06, GST = 0.05, FST = 0.06) while the populations from the mixed-cropping farms were more differentiated from each other (D = 0.46, GST = 0.22, FST = 0.12) and the other two fields (D > 0.20, GST > 0.13, FST > 0.12; S2–S4 Tables). The C. beticola population from Europe showed low to moderate differentiation compared to populations from Farm 2 (New York) (D = 0.07, GST = 0.03, FST = 0.03) and North Dakota (D = 0.20, GST = 0.08, FST = 0.09), but higher differentiation when compared to other populations (D > 0.46, GST > 0.21, FST > 0.21; S2–S4 Tables).
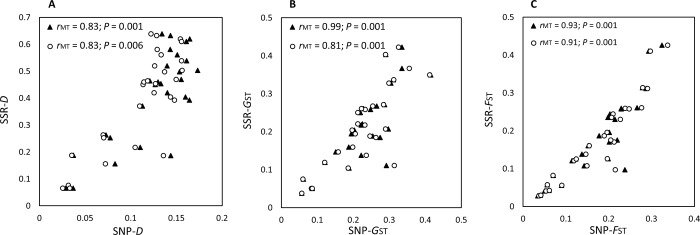
Patterns of population differentiation obtained from SNP data sets were similar to those obtained using microsatellites (S2–S4 Tables). Mantel tests revealed strong and significant correlations between the values of pairwise D, GST, and FST estimated by microsatellite markers and SNP data sets (Fig 4).
Fig 4. Relationships between indices of population differentiation.
(A) Jost’s D, (B) Pairwise Nei’s GST, and (C) Pairwise FST between Cercospora beticola populations estimated using 12 microsatellites (SSR) and single nucleotide polymorphisms (SNPs) identified using genotyping-by-sequencing. Values were estimated using the strict SNP data set (filled triangles) and relaxed-filtered SNP data set 1 (open circles). Values estimated using the relaxed-filtered SNP data set 2 were almost identical to those obtained from data set 1.
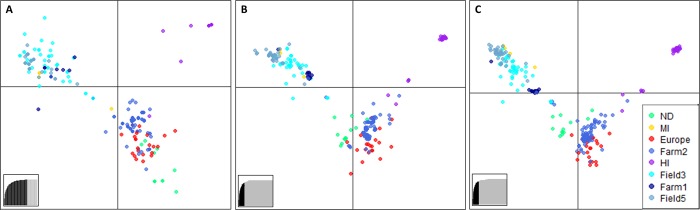
Patterns of population structure were not affected by marker or SNP filtering parameters (Fig 5). DAPC analysis demonstrated that C. beticola isolates from the two, monoculture table beet fields (Fields 3 and 5) and Farm 1 in New York clustered together, and individuals from Farm 2 clustered with isolates from Europe and North Dakota (Fig 5). The majority of the Hawaiian isolates formed a distinct cluster.
Fig 5.
Discriminant analysis of principal components (DAPC) for Cercospora beticola populations from Hawaii (HI), Michigan (MI), New York (Farms 1 and 2; Fields 3 and 5), North Dakota (ND), and Europe using (A) microsatellite, (B) strictly filtered and (C) relaxed-filtered SNP data sets generated using genotyping-by-sequencing.
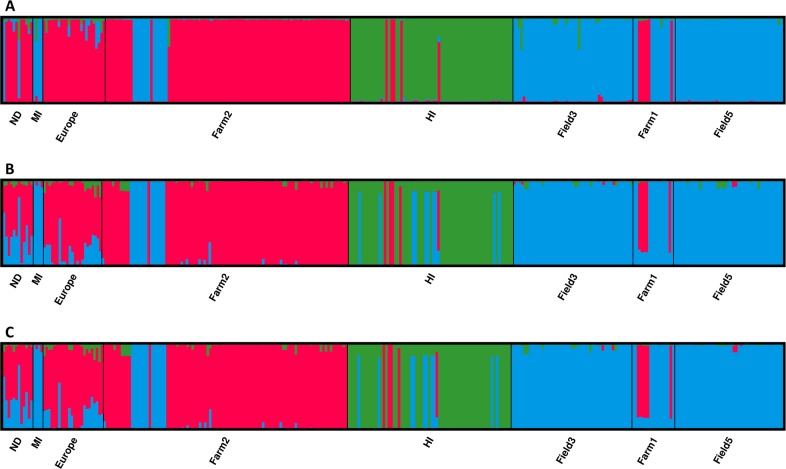
Population structure analysis of microsatellite and SNP data sets resulted in three distinct clusters (Fig 6). The number of clusters was not affected by marker or SNP data set filtering parameters. However, assignment of some individuals to populations differed between data sets generated from the two markers (Fig 6). This could be due to hypervariability of the microsatellite loci and resultant homoplasy in MLGs that are not identical by descent depicted by state. Therefore, individuals with different SNP profiles may be assigned to the same clonal lineage when only analyzed at 12 microsatellite loci.
Fig 6.
Assignment of Cercospora beticola isolates from Hawaii (HI), Michigan (MI), New York (Farms 1 and 2; Fields 3 and 5), North Dakota (ND) and Europe to three clusters detected through Bayesian clustering analysis of (A) microsatellite, and single nucleotide polymorphism (SNP) data sets generated by genotyping-by-sequencing using (B) strict filtering and (C) relaxed filtering. Each bar represents one individual and the bar height indicates estimated membership fraction of each individual in the inferred clusters.
The three clusters detected in the microsatellite data set consisted of 60 (cluster 1; green in Fig 6A), 120 (cluster 2; blue in Fig 6A) and 127 (cluster 3; red in Fig 6A) individuals. The most distinct of the clusters was composed entirely of individuals from Hawaii (cluster 1). Five individuals from Hawaii were assigned to cluster 3 using microsatellites and SNP data sets. Analysis of the SNP data sets assigned an additional nine isolates from Hawaii to cluster 2 (Fig 6B and 6C). Cluster 2 almost exclusively included individuals from New York and Michigan, in addition to two individuals from North Dakota (Fig 6A). The SNP data sets also assigned two individuals from North Dakota to cluster 2, but their identity was not consistent between the microsatellite and the SNP data sets. Unique to the SNP data sets, was the assignment of an individual from Europe (Germany) to cluster 2. Cluster 3, was the most diverse in all data sets, and included individuals from Europe, Hawaii, New York and North Dakota.
Discussion
Advances in high throughput SNP genotyping approaches have facilitated the identification of multitudes of SNPs in non-model organisms [19], which offer potential advantages over microsatellites for population genetics analyses due to genome-wide coverage, enhanced resolution of population diversity and structure, and stable inheritance facilitating simpler data analysis [14–18]. Our comparative analyses revealed major differences in the estimates of genetic diversity obtained from GBS and microsatellite data sets. However, both marker types and filtering parameters for the GBS data set revealed similar patterns of structure and differentiation in C. beticola populations from the USA and Europe.
The SNP data sets provided higher resolution in identification of MLGs compared to microsatellites but also greater error rates [68]. A previous study revealed that the error rates for the 12 microsatellite markers used in the current work for genotyping of C. beticola populations were zero, except for a single hypervariable locus (CbSSR3) with an error rate of 0.01 [37]. Here, we assessed GBS genotyping error through including replicated samples and estimating the mismatch rate [69]. Genetic distance among replicated DNA samples varied among isolates and was attributed to a combination of error sources including sensitivity of GBS results to DNA sample quality and quantity, sequencing error and locus drop out. Relaxed filtering of the SNP data set resulted in a bitwise distance of 4.1 × 10−4 to 1.855 × 10−3 among replicated samples. In contrast, strict filtering parameters resulted in an error rate of zero. It has been suggested that higher mismatch rates among replicates are associated with lower read depth [70]. For diploid organisms, filtering for minimum read depth from four to seven has been used to reduce error rates [25,70]. For the haploid C. beticola, filtering the SNP data set for a minimum locus-by-individual read depth of three was sufficient to reduce the mismatch rate of replicated samples to zero. Stringent filtering approaches are especially critical for population genomics analyses that rely on individual identification and delineation of clonal lineages [69–71].
Higher error rates and resolution of GBS approaches can substantially inflate the number of MLGs [12,20]. Even when the SNP data set was filtered using stringent parameters, some of the replicated samples with a mismatch rate of zero were assigned to different MLGs due to missing data. Thus, a more biologically relevant representation of clonality was obtained by collapsing MLGs to MLLs. Contraction of MLGs using a threshold that was too low resulted in inflation of the number of MLLs, and genotypic diversity in all populations was greater than 0.96 (relaxed-filtered data set 1). When the relaxed-filtered data set was contracted using an arbitrary threshold based on the distance among replicated samples, the index of genotypic diversity decreased for some populations but did not correlate with the microsatellite data or the strictly filtered SNP data set. More stringent filtering parameters reduced the number of SNPs from 2,696 to 1,361, and the indices of genotypic diversity were strongly correlated with those obtained from the microsatellite data (Fig 3). There is no “standard” way of filtering and contracting genome-wide SNP data sets, and such parameters are selected based on the type of data, objectives, and biology of the organism in question [20,71]. As filtering parameters and distance thresholds for contracting MLGs substantially impacted the number of retained loci and MLLs, we suggest that population genomics studies based on GBS use variable filtering parameters to critically assess the sensitivity of the results to the filtering approaches.
Detection of repeated MLLs of C. beticola across two continents strengthens the argument for genotype flow [36,38]. Even with relaxed-filtering of the SNP data set and a conservative distance threshold for contraction of MLGs to MLLs, which was even lower than the average error rate, one recurrent clonal lineage was shared between New York and England (Fig 2C) and multiple MLLs were shared among table beet fields in New York. The microsatellite data set (Fig 2A) and strictly filtered SNP data set (Fig 2B) detected many more MLLs that were shared between continents, and among different states in the USA.
A potential means of long distance migration of clonal lineages is contaminated seed. Cercospora beticola has been associated with raw sugar beet seed (‘beet balls’) [43] and population genetics studies in Europe have suggested infested seed as a potential source of inoculum [34,44]. However, other studies have not found C. beticola on sugar beet seed [72,73]; and the presence of C. beticola on table beet seed lots in New York is also unknown. Commercially available table beet seed is not polished to remove the exterior corky layer as is done in sugar beet seed to enhance germination and reduce pathogen contamination. The seed planted in the monoculture fields in New York (Fields 3 and 5) originated from Skagit Co., Washington State, USA, where occurrence of C. beticola in seed crops is rare [74]. If the high number of shared MLLs and low population differentiation of C. beticola populations from table beet monoculture fields in New York is not a result of contaminated seed, transfer by agricultural machinery [41,42] or insects [39,43] may also be involved. The shared clonal lineages between New York and Europe were collected from table beet at Farm 2, which is a mixed-cropping organic production enterprise. The table beet seed used at Farm 2 is obtained annually from various organic certified seed providers, while the seed planted at Farm 1 had been produced at that location for many years.
The predominant source of C. beticola inoculum in table beet and sugar beet fields is most likely to be alternative weedy hosts or volunteers [43,75], infested plant debris [76,77] or soilborne inoculum [78]. Cercospora beticola is reported to persist for 22 months to over three years on infested plant debris [76,77], and also remain virulent for 27 and 20 months in sterilized and C. beticola-infested field soil, respectively [78]. CLS epidemics then spread rapidly within a few weeks through short-range rain splash of asexual spores, which are unlikely to be involved in long-distance dispersal [39,40]. The presence of a sexual form remains unknown but multiple studies have postulated cryptic sex [79–81]. Even if C. beticola is capable of sexual reproduction, due to its heterothallic nature [79], ascospores are unlikely to be of the same clonal lineage and will most likely have recombinant genotypes [82]. Thus, ascospores are unlikely to be the source of long distance dispersal of MLLs in this study. Other plausible routes for inter-continental transmission of C. beticola is international trade of diseased alternative hosts. For example, Groenewald et al. [83] reported C. beticola from Chrysanthemum spp. suggesting that trade in cut flowers and ornamentals could be involved. Evidence for long distance migration of C. beticola genotypes provided here warrants further investigations on potential seedborne inoculum as a means of dispersal.
Although measures of genetic diversity (He and λ) were not associated between different marker systems, there was significant correlation among the number of clonal lineages and clonal fraction estimated from microsatellite and SNP data sets, which led to similar clonality rankings of populations across data sets. The population in Hawaii was the most clonal while the European population had the highest diversity. This is most likely an artefact of sampling strategies as the population from Hawaii was isolated from one community garden while the isolates from Europe were collected from a broad geographical area. Lack of significant correlation between expected heterozygosity obtained from microsatellites (SSR-He) and SNPs (SNP-He) has been reported, and may be due to the restricted genome coverage of microsatellites [13,84].
Absolute values of population differentiation (D, GST and FST) obtained from the microsatellite and SNP data sets varied, however, there was a strong, significant correlation between the indices of differentiation, supporting results of other studies [13,84–87]. Analyses based on allele frequencies, such as estimates of genetic differentiation, are not as affected by genotyping errors as those based on individual identification [69]. Although shared recurrent MLLs were indicative of long distance dispersal of inoculum, pairwise population genetic differentiation and clustering of individuals based on geographic location indicated that the C. beticola population is not panmictic within New York, nor worldwide, providing evidence for restricted dispersal of inoculum.
Spatial patterns in C. beticola population structure were not affected by markers or filtering parameters for the SNP data set. However, minor differences were observed in the assignment of individuals to inferred clusters. Other studies have also found no or little impact of marker system on biological conclusions concerning the broad-scale structure of populations [70,88,89]. In general, due to the lower information of SNPs, accurate estimation of the number of populations has been reported to require more SNPs than microsatellites [11,24,85,89,90].
In conclusion, different marker systems, filtering approaches and distance thresholds used to collapse MLGs to MLLs, strongly affected clone identification in C. beticola. However, general patterns of variation or population structure were not affected by marker type or filtering parameters used to interrogate SNP data sets. For analyses based on allele frequencies, maximizing the number of SNPs may be more beneficial. In contrast, for analyses that require confidence in genotype of individuals, more stringent filtering for locus-by-individual read depth and clear definition of clonal boundaries based on genotyping error rate is necessary. The results also emphasize the need for development of species-specific molecular markers for rapid and reliable detection of C. beticola. The PCR markers based on the calmodulin region reliably differentiate C. beticola from C. apii [46], but failed to differentiate C. cf. flagellaris isolates collected from CLS symptoms on table beet. An earlier study also demonstrated these primers did not differentiate C. chenopodii from C. beticola [38]. The GBS-SNP data produced here may be useful for discovery of informative SNPs for diagnostic assay development [90,91].
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(VCF)
(VCF)
Acknowledgments
Thanks to Carol Bowden, Elizabeth Maloney, David Strickland and Viviana Rivera-Varas for excellent technical support, and growers and gardeners for access to their fields and plots for sample collection. The authors declare no competing financial interests.
Funding
This research was supported by the United States Department of Agriculture, National Institute of Food and Agriculture Hatch project NYG-625424, and the Federal Capacity Funds Initiative (2015-16-118) managed by the New York State Agricultural Experiment Station (NYSAES), Cornell University, Geneva, New York, USA, and the NYSAES Director’s Controlled Endowment Fund.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the United States Department of Agriculture, National Institute of Food and Agriculture Hatch project NYG-625424, and the Federal Capacity Funds Initiative (2015-16-118) managed by the New York State Agricultural Experiment Station (NYSAES), Cornell University, Geneva, New York, USA, and the NYSAES Director’s Controlled Endowment Fund.
References
- 1.McDonald BA. How can research on pathogen population biology suggest disease management strategies? The example of barley scald (Rhynchosporium commune). Plant Pathol. 2015; 64:1005–1013. [Google Scholar]
- 2.Milgroom MG, Peever TL. Population biology of plant pathogens: the synthesis of plant disease epidemiology and population genetics. Plant Dis. 2003; 87:608–617. [DOI] [PubMed] [Google Scholar]
- 3.Biasi A, Martin FN, Cacciola SO, di San Lio GM, Grünwald NJ, Schena L. Genetic analysis of Phytophthora nicotianae populations from different hosts using microsatellite markers. Phytopathology 2016; 106:1006–1014. doi: 10.1094/PHYTO-11-15-0299-R [DOI] [PubMed] [Google Scholar]
- 4.Lehner MS, de Paula Júnior TJ, Del Ponte EM, Mizubuti ESG, Pethybridge SJ. Independently founded populations of Sclerotinia sclerotiorum from a tropical and a temperate region have similar genetic structure. PloS One 2017; 12:e0173915 doi: 10.1371/journal.pone.0173915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mascheretti S, Croucher PJP, Vettraino A, Prospero S, Garbelotto M. Reconstruction of the Sudden Oak Death epidemic in California through microsatellite analysis of the pathogen Phytophthora ramorum. Mol. Ecol. 2008; 17:2755–2768. doi: 10.1111/j.1365-294X.2008.03773.x [DOI] [PubMed] [Google Scholar]
- 6.Schoebel CN, Stewart J, Grunwald NJ, Rigling D, Prospero S. Population history and pathways of spread of the plant pathogen Phytophthora plurivora. PLoS One 2014; 9:e85368 doi: 10.1371/journal.pone.0085368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sommerhalder RJ, McDonald BA, Mascher F, Zhan J. Sexual recombinants make a significant contribution to epidemics caused by the wheat pathogen Phaeosphaeria nodorum. Phytopathology 2010; 100:855–862. doi: 10.1094/PHYTO-100-9-0855 [DOI] [PubMed] [Google Scholar]
- 8.Vercauteren A, De Dobbelaere I, Grünwald NJ, Bonants P, Van Bockstaele E, Maes M, et al. Clonal expansion of the Belgian Phytophthora ramorum populations based on new microsatellite markers. Mol. Ecol. 2010; 19:92–107. doi: 10.1111/j.1365-294X.2009.04443.x [DOI] [PubMed] [Google Scholar]
- 9.Hedrick PW. Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution 1999; 53:313–318. doi: 10.1111/j.1558-5646.1999.tb03767.x [DOI] [PubMed] [Google Scholar]
- 10.Putman AI, Carbone I. Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecol. Evol. 2014; 4:4399–4428. doi: 10.1002/ece3.1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haasl RJ, Payseur BA. Multi-locus inference of population structure: a comparison between single nucleotide polymorphisms and microsatellites. Heredity 2011; 106:158–171. doi: 10.1038/hdy.2010.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milgroom MG. Population Biology of Plant Pathogens: Genetics, Ecology, and Evolution. St. Paul, MN: APS Press; 2015. 399 p. [Google Scholar]
- 13.Fischer MC, Rellstab C, Leuzinger M, Roumet M, Gugerli F, Shimizu KK, et al. Estimating genomic diversity and population differentiation–an empirical comparison of microsatellite and SNP variation in Arabidopsis halleri. BMC Genomics 2017; 18:69 doi: 10.1186/s12864-016-3459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Väli Ü, Einarsson A, Waits L, Ellegren H. To what extent do microsatellite markers reflect genome wide genetic diversity in natural populations? Mol. Ecol. 2008; 17:3808–3817. doi: 10.1111/j.1365-294X.2008.03876.x [DOI] [PubMed] [Google Scholar]
- 15.Landegren U, Nilsson M, Kwok P. Reading bits of genetic information: methods for single-nucleotide polymorphism analysis. Genome Res. 1998; 8:769–776. [DOI] [PubMed] [Google Scholar]
- 16.Morin PA, Luikart G, Wayne RK. SNPs in ecology, evolution and conservation. Trends Ecol. Evolut. 2004; 19:208–216. [Google Scholar]
- 17.Seeb JE, Carvalho G, Hauser L, Naish K, Roberts S, Seeb LW. Single nucleotide polymorphism (SNP) discovery and applications of SNP genotyping in non-model organisms. Mol. Ecol. Res. 2011; 11:1–8. [DOI] [PubMed] [Google Scholar]
- 18.Garvin MR, Saitoh K, Gharrett AJ. Application of single nucleotide polymorphisms to non-model species: a technical review. Mol. Ecol. Res. 2010; 10:915–934. [DOI] [PubMed] [Google Scholar]
- 19.Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter, ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011; 12:499–510. doi: 10.1038/nrg3012 [DOI] [PubMed] [Google Scholar]
- 20.Grünwald NJ, McDonald BA, Milgroom MG. Population genomics of fungal and oomycete pathogens. Annu. Rev. Phytopathol. 2016; 54:323–346. doi: 10.1146/annurev-phyto-080614-115913 [DOI] [PubMed] [Google Scholar]
- 21.Luikart G, England FR, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nature Rev. Genet. 2003; 4:981–994. doi: 10.1038/nrg1226 [DOI] [PubMed] [Google Scholar]
- 22.Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 2008; 3:e3376 doi: 10.1371/journal.pone.0003376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A robust, simple Genotyping-by-Sequencing (GBS) approach for high diversity species. PLoS One 2011; 6:e19379 doi: 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narum SR, Buerkle CA, Davey JW, Miller MR, Hohenlohe PA. Genotyping- by -sequencing in ecological and conservation genomics. Mol. Ecol. 2013; 22:2841–2847. doi: 10.1111/mec.12350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen ZR, Everts KL, Fry WE, Gevens AJ, Grünwald NJ, Gugino BK, et al. Genetic variation within clonal lineages of Phytophthora infestans revealed through genotyping-by-sequencing, and implications for Late Blight epidemiology. PLoS One 2016; 11:e0165690 doi: 10.1371/journal.pone.0165690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milgroom MG, Jiménez-Gasco M, Olivares-García C, Jiménez-Díaz RM. Clonal expansion and migration of a highly virulent, defoliating lineage of Verticillium dahliae. Phytopathology 2016; 106:1038–1046. doi: 10.1094/PHYTO-11-15-0300-R [DOI] [PubMed] [Google Scholar]
- 27.Summers CF, Gulliford CM, Carlson CH, Lillis JA, Carlson MO, Cadle-Davidson L, et al. Identification of genetic variation between obligate plant pathogens Pseudoperonospora cubensis and P. humuli using RNA sequencing and genotyping-by-sequencing. PLoS One 2015; 10:e0143665 doi: 10.1371/journal.pone.0143665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milgroom MG, Jiménez-Gasco M, García CO, Drott MT, Jiménez-Díaz RM. Recombination between clonal lineages of the asexual fungus Verticillium dahliae detected by genotyping by sequencing. PLoS One, 2014; 9:e106740 doi: 10.1371/journal.pone.0106740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamvar ZN, Brooks JC, Grünwald NJ. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015; 6:208 doi: 10.3389/fgene.2015.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grünwald NJ, Hoheisel GA. Hierarchical analysis of diversity, selfing, and genetic differentiation in populations of the oomycete Aphanomyces euteiches. Phytopathology 2006; 96:1134–1141. doi: 10.1094/PHYTO-96-1134 [DOI] [PubMed] [Google Scholar]
- 31.Cooke DEL, Cano LM, Raffaele S, Bain RA, Cooke LR, Etherington GJ, et al. Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLoS Pathog. 2012; 8:e1002940 doi: 10.1371/journal.ppat.1002940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnaud-Haond S, Duarte CM, Alberto F, Serrao EA. Standardizing methods to address clonality in population studies. Mol. Ecol. 2007; 16:51155139. [DOI] [PubMed] [Google Scholar]
- 33.Groenewald M, Groenewald JZ, Linde CC, Crous PW. Development of polymorphic microsatellite and single nucleotide polymorphism markers for Cercospora beticola (Mycosphaerellaceae). Mol. Ecol. Notes 2007; 890–892. [Google Scholar]
- 34.Moretti M, Karaoglanidis G, Saracchi M, Fontana A, Farina G. Analysis of genotypic diversity in Cercospora beticola Sacc. field isolates. Ann. Microbiol. 2006; 56:215–221. [Google Scholar]
- 35.Weiland J, Eide J, Rivera-Varas V, Secor G. Genetic diversity of Cercospora beticola in the U.S. and association of molecular markers with tolerance to the fungicide triphenyltin hydroxide (TPTH). Phytopathology, 2001; 91:94. [Google Scholar]
- 36.Vaghefi N, Hay FS, Kikkert JR, Pethybridge SJ. Genotypic diversity and resistance to azoxystrobin of Cercospora beticola on processing table beet in New York. Plant Dis. 2016; 100:1466–1473. [DOI] [PubMed] [Google Scholar]
- 37.Vaghefi N, Kikkert JR, Bolton MD, Hanson LE, Secor GA, Pethybridge SJ. De novo assembly of Cercospora beticola for microsatellite development and validation. Fung. Ecol. 2017; 26:125–134. [Google Scholar]
- 38.Vaghefi N, Nelson SC, Kikkert JR, Pethybridge SJ. Genetic structure of Cercospora beticola populations on Beta vulgaris in New York and Hawaii. Sci. Rep. 2017; 7:1726 doi: 10.1038/s41598-017-01929-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meredith DS. Conidium release and dispersal in Cercospora beticola. Phytopathology 1967; 57:889–893. [Google Scholar]
- 40.Lawrence JS, Meredith DS. Wind dispersal of conidia of Cercospora beticola. Phytopathology 1970; 60:1076–1078. [Google Scholar]
- 41.Vereijssen J, Schneider JHM, Stein A, Jeger MJ. Spatial pattern of Cercospora leaf spot of sugar beet in fields in long-and recently established areas. Eur. J. Plant Pathol. 2006; 116:187–198. [Google Scholar]
- 42.Vereijssen J, Schneider JHM, Jeger MJ. Epidemiology of Cercospora leaf spot on sugar beet: modeling disease dynamics within and between individual plants. Phytopathology 2007; 97:1550–1557. doi: 10.1094/PHYTO-97-12-1550 [DOI] [PubMed] [Google Scholar]
- 43.McKay MB, Pool VW. Field studies of Cercospora beticola. Phytopathology 1918; 8:119–136. [Google Scholar]
- 44.Turgay EB, Bakir M, Ozeren P, Katircioglu YZ, Maden S. Detection of pathotypes and genetic diversity of Cercospora beticola. Plant Pathol. J. 2010; 26:306–312. [Google Scholar]
- 45.Kamvar ZN, Tabima JF, Grünwald NJ. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. Peer J. 2014; 2:e281 doi: 10.7717/peerj.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groenewald M, Groenewald JZ, Crous PW. Distinct species exist within the Cercospora apii morphotype. Phytopathology 2005; 95:951–959. doi: 10.1094/PHYTO-95-0951 [DOI] [PubMed] [Google Scholar]
- 47.Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire R, Sun Q, et al. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 2014; 9:e90346 doi: 10.1371/journal.pone.0090346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009; 25:1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 2007; 23:2633–2635. doi: 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- 50.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, De Pristo MA, et al. The variant call format and VCFtools. Bioinformatics 2011; 27:2156–2158. doi: 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knaus BJ, Grunwald NJ. VcfR: an R package to manipulate and visualize VCF format data. BioRxiv 2016; 041277. [DOI] [PubMed]
- 52.Bruvo R, Michiels NK, D’Souza TG, Schulenburg H. A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol. Ecol. 2004; 13:2101–2106. doi: 10.1111/j.1365-294X.2004.02209.x [DOI] [PubMed] [Google Scholar]
- 53.Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978; 89:583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simpson EH. Measurement of diversity. Nature 1949; 163:688. [Google Scholar]
- 55.Szpiech ZA, Jakobsson M, Rosenberg NA. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 2008; 24:2498–2504. doi: 10.1093/bioinformatics/btn478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jost L. GST and its relatives do not measure differentiation. Mol. Ecol. 2008; 17:4015–4026. [DOI] [PubMed] [Google Scholar]
- 57.Nei M, Chesser RK. Estimation of fixation indices and gene diversities. Ann. Hum. Genet. 1983; 47:253–259. [DOI] [PubMed] [Google Scholar]
- 58.Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad. Sci. USA 1973; 70:3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winter DJ. MMOD: an R library for the calculation of population differentiation statistics. Mol. Ecol. Res. 2012; 12:1158–1160. [DOI] [PubMed] [Google Scholar]
- 60.Jombart T. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 2008. 24:1403–1405. doi: 10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- 61.Goudet J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 2005; 5:184–186. [Google Scholar]
- 62.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967; 27:209–220. [PubMed] [Google Scholar]
- 63.Dray S, Dufour AB. The ade4 package: implementing the duality diagram for ecologists. J. Stat. Softw. 2007; 22:1–20. [Google Scholar]
- 64.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000; 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005; 14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- 66.Earl DA, von Holdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Res. 2012; 4:359–361. [Google Scholar]
- 67.Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Res. 2015; 15:1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pompanon F, Bonin A, Bellemain E, Taberlet P. Genotyping errors: causes, consequences and solutions. Nat. Rev. Genet. 2005; 6:847–846. doi: 10.1038/nrg1707 [DOI] [PubMed] [Google Scholar]
- 69.Schilling MP, Wolf PG, Duffy AM, Rai HS, Rowe CA, Richardson BA, et al. Genotyping-by-sequencing for Populus population genomics: an assessment of genome sampling patterns and filtering approaches. PLoS One 2014; 9: e95292 doi: 10.1371/journal.pone.0095292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mesak F, Tatarenkov A, Earley RL, Avise JC. Hundreds of SNPs vs. dozens of SSRs: which dataset better characterizes natural clonal lineages in a self-fertilizing fish? Front. Ecol. Evol. 2014; 2:74. [Google Scholar]
- 71.Andrews KR, Good JM, Miller MR, Luikart G, Hohenlohe PA. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat. Rev. Genet. 2016; 17:81–92. doi: 10.1038/nrg.2015.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agarwal PC, Singh B, Dev U, Rani I, Chand D, Khetarpal RK. Seed-borne fungi detected in sugar beet seeds imported into India during last three decades. Plant Health Prog. 2006; doi: 10.1094/PHP-2006-1211-01-RS [Google Scholar]
- 73.Byford WJ, Gambogi P. Phoma and other fungi on beet seed. Trans. Br. Mycol. Soc. 1985; 84:21–28. [Google Scholar]
- 74.Du Toit LJ. Crop Profile for Table Beet Seed in Washington. United States Department of Agriculture Pest Management Centers; December 2007. Available from: http://www.ipmcenters.org/CropProfiles/docs/WAbeetseed.pdf. Cited 20 April 2017. [Google Scholar]
- 75.Vestal EF. Pathogenicity, host response and control of Cercospora leaf-spot of sugar beets Agricultural Experiment Station, Iowa State College of Agriculture and Mechanic Arts, Research Bulletin; 1933; 168. [Google Scholar]
- 76.Khan J. del Rio LE, Nelson R, Rivera-Varas V, Secor GA, Khan MFR. Survival, dispersal, and primary infection site for Cercospora beticola in sugar beet. Plant Dis. 2008; 92:741–745. [DOI] [PubMed] [Google Scholar]
- 77.Solel Z. Survival of Cercospora beticola, the causal agent of sugar beet leaf spot, in Israel. Trans. Br. Mycol. Soc. 1970; 54:504–506. [Google Scholar]
- 78.Nagel CM. The longevity of Cercospora beticola in soil. Phytopathology 1938; 28:342–350. [Google Scholar]
- 79.Groenewald M, Groenewald JZ, Harrington TC, Abeln EC, Crous PW. Mating type gene analysis in apparently asexual Cercospora species is suggestive of cryptic sex. Fung. Genet. Biol. 2006; 43:813–825. [DOI] [PubMed] [Google Scholar]
- 80.Groenewald M, Linde CC, Groenewald JZ, Crous PW. Indirect evidence for sexual reproduction in Cercospora beticola populations from sugar beet. Plant Pathol. 2008; 57: 25–32. [Google Scholar]
- 81.Bolton MD, Secor GA, Rivera V, Weiland JJ, Rudolph K, Birla K, et al. Evaluation of the potential for sexual reproduction in field populations of Cercospora beticola from USA. Fungal Biol. 2012; 116:511–521. doi: 10.1016/j.funbio.2012.01.011 [DOI] [PubMed] [Google Scholar]
- 82.Ni M, Feretzaki M, Sun S, Wang X, Heitman J. Sex in fungi. Annu. Rev. Genet. 2011; 45:405–430. doi: 10.1146/annurev-genet-110410-132536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Groenewald JZ, Nakashima C, Nishikawa J, Shin H-D, Park J-H, Jama AN, Groenewald M, Braun U, Crous PW. Species concepts in Cercospora: spotting the weeds among the roses. Stud. Mycol. 2013; 75:115–170. doi: 10.3114/sim0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.DeFaveri J, Viitaniemi H, Leder E, Merilä J. Characterizing genic and nongenic molecular markers: comparison of microsatellites and SNPs. Mol. Ecol. Res. 2013; 13:377–92. [DOI] [PubMed] [Google Scholar]
- 85.Ryynänen HJ, Tonteri A, Vasemägi A, Primmer CR. A comparison of biallelic markers and microsatellites for the estimation of population and conservation genetic parameters in Atlantic salmon (Salmo salar). J. Hered. 2007; 98:692–704. doi: 10.1093/jhered/esm093 [DOI] [PubMed] [Google Scholar]
- 86.Ozerov M, Vasemägi A, Wennevik V, Diaz-Fernandez R, Kent M, Gilbey J, et al. Finding markers that make a difference: DNA pooling and SNP-arrays identify population informative markers for genetic stock identification. PLoS One 2013; 8:e82434 doi: 10.1371/journal.pone.0082434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Glover KA, Hansen MM, Lien S, Als TD, Høyheim B, Skaala Ø. A comparison of SNP and STR loci for delineating population structure and performing individual genetic assignment. BMC Genetics 2010; 11:2 doi: 10.1186/1471-2156-11-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jeffries DL, Copp GH, Lawson Handley L, Olsén KH, Sayer CD, Hänfling B. Comparing RADseq and microsatellites to infer complex phylogeographic patterns, an empirical perspective in the Crucian carp, Carassius carassius, L. Mol. Ecol. 2016; 25: 2997–3018. doi: 10.1111/mec.13613 [DOI] [PubMed] [Google Scholar]
- 89.Granevitze Z, David L, Twito T, Weigend S, Feldman M, Hillel J. Phylogenetic resolution power of microsatellites and various single nucleotide polymorphism types assessed in 10 divergent chicken populations. Anim. Genet. 2014; 45:87–95. doi: 10.1111/age.12088 [DOI] [PubMed] [Google Scholar]
- 90.Summers CF, Adair NL, Gent DH, McGrath MT, Smart CD. Pseudoperonospora cubensis and P. humuli detection using species-specific probes and high definition melt curve analysis. Can. J. Plant Pathol. 2015; 37:315–330. [Google Scholar]
- 91.Paschou P, Ziv E, Burchard EG, Choudhry S, Rodriguez-Cintron W, Mahoney MW, Drineas P. PCA-correlated SNPs for structure identification in worldwide human populations. PLoS Genet. 2007; 3: e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(VCF)
(VCF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.