Abstract
The inhibition of host innate immunity pathways is essential for the persistence of attaching and effacing pathogens such as enteropathogenic Escherichia coli (EPEC) and Citrobacter rodentium during mammalian infections. To subvert these pathways and suppress the antimicrobial response, attaching and effacing pathogens use type III secretion systems to introduce effectors targeting key signaling pathways in host cells. One such effector is the arginine glycosyltransferase NleB1 (NleBCR in C. rodentium) that modifies conserved arginine residues in death domain-containing host proteins with N-acetylglucosamine (GlcNAc), thereby blocking extrinsic apoptosis signaling. Ectopically expressed NleB1 modifies the host proteins Fas-associated via death domain (FADD), TNFRSF1A-associated via death domain (TRADD), and receptor-interacting serine/threonine protein kinase 1 (RIPK1). However, the full repertoire of arginine GlcNAcylation induced by pathogen-delivered NleB1 is unknown. Using an affinity proteomic approach for measuring arginine-GlcNAcylated glycopeptides, we assessed the global profile of arginine GlcNAcylation during ectopic expression of NleB1, EPEC infection in vitro, or C. rodentium infection in vivo. NleB overexpression resulted in arginine GlcNAcylation of multiple host proteins. However, NleB delivery during EPEC and C. rodentium infection caused rapid and preferential modification of Arg117 in FADD. This FADD modification was extremely stable and insensitive to physiological temperatures, glycosidases, or host cell degradation. Despite its stability and effect on the inhibition of apoptosis, arginine GlcNAcylation did not elicit any proteomic changes, even in response to prolonged NleB1 expression. We conclude that, at normal levels of expression during bacterial infection, NleB1/NleBCR antagonizes death receptor-induced apoptosis of infected cells by modifying FADD in an irreversible manner.
Keywords: Escherichia coli (E. coli), glycosylation, innate immunity, pathogenesis, post-translational modification (PTM), bacterial effectors
Introduction
Enteropathogenic Escherichia coli (EPEC)5 is one of the most common causes of diarrheagenic disease in infants and young children in low income countries (1, 2). Upon ingestion, EPEC rapidly colonizes the mucosa of the small intestine, forming a tight association with the apical surface of host enterocytes leading to the destruction of brush-border microvilli and the formation of actin-rich pedestal-like structures (3). This distinct intestinal histopathology is known as the attaching and effacing (A/E) lesion and is the hallmark of A/E pathogen infection by EPEC, enterohemorrhagic E. coli, and the murine pathogen Citrobacter rodentium (4). A/E lesion formation is the result of the activities of the locus of enterocyte effacement (LEE)-encoded type III secretion system (5) and its associated effector proteins such as the translocated intimin receptor (6, 7) and WXXXE effector mitochondria-associated protein Map (8–10). Although these LEE effectors are essential for enabling the establishment of infection for A/E pathogens, additional effectors located outside the LEE, known as non-LEE (nle) effectors (11), also play an important role in the augmentation of host signaling and subversion of the host innate immune response (4). For example, NleE from EPEC inhibits NF-κB signaling via methylation of key cysteine residues in TAB2 and TAB3, thereby blocking their binding to ubiquitylated tumor necrosis factor (TNF) receptor-associated factors (12). Similarly, cell death pathways may be modulated by A/E pathogens through the cooperative action of nle effectors such as EspL, which degrades receptor-interacting protein homotypic interaction motif -containing proteins, thereby blocking receptor-interacting protein homotypic interaction motif-dependent inflammatory and necroptotic signaling pathways (13), and NleF, which directly inhibits caspase-4, -8, and -9 activation (14–16).
Another nle effector, NleB1, is the prototypic member of a novel family of bacterial glycosyltransferase enzymes that mediate the glycosylation of arginine residues, an atypical post-translational modification (PTM) not observed in eukaryotic cells (17, 18). NleB1 inhibits Fas ligand and TNF-mediated apoptosis by blocking death domain interactions in the corresponding receptor complexes (17, 18). To date, multiple death domain-containing host targets of NleB1 have been reported, including the Fas-associated death domain protein (FADD), TNFRSF1A-associated via death domain (TRADD), and receptor-interacting serine/threonine protein kinase 1 (17, 18). Modification of these substrates occurs at a conserved arginine residue within the death domain corresponding to Arg117 of FADD and Arg235 of TRADD (17, 18). In addition, NleB from C. rodentium (here referred to as NleBCR) was reported to inhibit NF-κB activation (19) and to reduce type I Interferon production (20) through modification of GAPDH. Although GAPDH can be glycosylated in vitro by NleBCR (19, 21), whether glycosylation occurs in vivo has yet to be determined (18). EPEC NleB1 was also recently shown to interact with Ensconsin and block vesicular movement along microtubules, although this did not require glycosyltransferase activity (22). Importantly, none of these studies investigated the extent of arginine glycosylation during wild-type EPEC infection in vitro or C. rodentium infection in vivo. Hence, some of the target modifications observed may be the result of NleB overexpression and may not occur when native levels of NleB are delivered by the wild-type pathogen.
Using a recently developed antibody specific for Arg-GlcNAc linkages (23), we established an Arg-GlcNAc-specific enrichment method coupled with mass spectrometry (MS) to provide a robust means to monitor arginine GlcNAcylation during A/E pathogen infection. Here, we applied this to identify the endogenous targets modified by NleB1/NleBCR during wild-type EPEC and C. rodentium infection. We observed that human and mouse FADD were preferentially targeted by both these enzymes at Arg117 under wild-type infection conditions and that overexpression of NleB1 led to indiscriminate Arg GlcNAcylation of non-authentic targets. The resulting modification of Arg117 in FADD was stable and resistant to both environmental and enzymatic activities. However, despite the permanence of the Arg-GlcNAc modification, its presence did not elicit any changes within the host cell proteome even after prolonged expression of NleB1. Thus, these findings expand our understanding of NleB1/NleBCR-mediated Arg GlcNAcylation as an irreversible and silent modification and highlight the promiscuous nature of NleB1 under non-wild-type infection conditions.
Results
Development of an Arg-GlcNAc immunoenrichment method
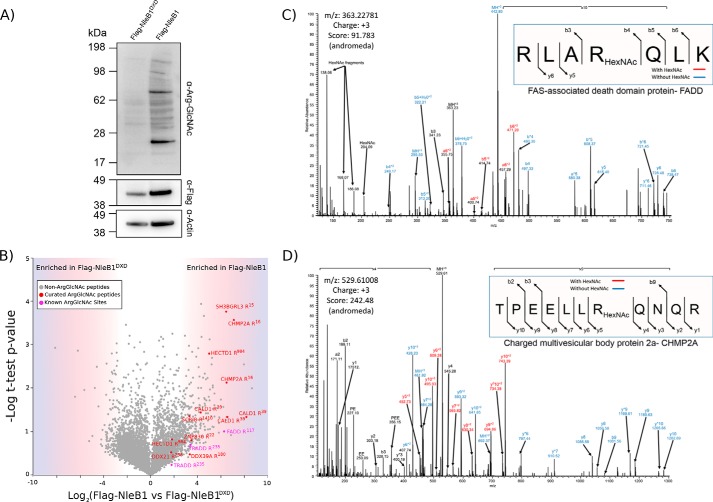
The addition of GlcNAc to arginine by NleB1 is thought to target a limited subset of death domain-containing proteins during infection (17, 18). However, until now, no assessment of the kinetics or repertoire of endogenous targets has been undertaken. As with other glycosylation events, the tendency of glycopeptides to undergo ion competition/suppression in the presence of abundant non-glycosylated peptides (24, 25) suggests that enrichment would be required for a proteome-wide investigation of arginine GlcNAcylation, similar to approaches used for other PTMs (26–28). With the recent development of a specific Arg-GlcNAc antibody (23), we assessed the use of antibody-based capture of arginine-GlcNAcylated peptide for proteome-wide assessment of arginine GlcNAcylation mediated by ectopically expressed NleB1. Using HeLa cell lines stably expressing doxycycline-inducible FLAG-NleB1, we have previously shown that Fas signaling can be inhibited (14). Consistent with the mechanism of inhibition, robust levels of arginine GlcNAcylation could be observed after 24 h compared with HeLa cells stably expressing catalytically inactive FLAG-NleB1DXD (Fig. 1A). To identify arginine GlcNAcylation events during stable expression of FLAG-NleB1, proteins were digested, and glycosylated peptides were enriched using Arg-GlcNAc antibody (supplemental Fig. 1A). Using this approach, 42 unique arginine-GlcNAcylated peptides were identified (supplemental Table 3). Using manual curation, we identified 15 Arg-GlcNAc glycopeptides corresponding to 10 unique sites with high confidence enriched within FLAG-NleB1 compared with cells stably expressing FLAG-NleB1DXD (Fig. 1B and supplemental Annotations). Although glycosylation events are typically labile under collision-based fragmentation, we observed Arg-GlcNAc to be partially stable, consistent with previous observations of Arg rhamnosylation (29, 30). This stability enabled the localization of Arg-GlcNAc to the previously reported sites for FADD (Arg117; Fig. 1C) and TRADD (Arg235; supplemental Annotations) (17, 18) while also enabling the assessment of novel arginine GlcNAcylation events, including of the charged multivesicular body protein 2a (Arg16; Fig. 1D). To further complement these assignments, electron transfer dissociation/EThcD fragmentation was undertaken, resulting in the validation of five arginine GlcNAcylation sites within HeLa cells stably expressing FLAG-NleB1 (supplemental Annotations). Thus, our glycopeptide immunoenrichment method enabled the detection of arginine GlcNAcylation events on endogenous protein substrates after ectopic expression of FLAG-NleB1.
Figure 1.
Establishing a peptide Arg-GlcNAc enrichment approach to identify endogenously modified targets. A, stable expression of FLAG-NleB1 and FLAG-NleB1DXD for 24 h results in arginine GlcNAcylation of multiple host proteins only within NleB1. B, label-free quantification of Arg-GlcNAc peptide pulldown results from induction of NleB1 versus NleB1DXD. The scatter plot shows the mean ion intensity peptide ratios of NleB1 versus NleB1DXD plotted against negative logarithmic t test p values from biological triplicate experiments. Within biological replicates of NleB1-expressing cells, multiple putative arginine-glycosylated proteins were identified with manually curated Arg-GlcNAc peptides shown in red and Arg-GlcNAc peptides containing previously known Arg-GlcNAc modifications shown in purple. C and D, manually curated MS-MS spectra showing the previously known Arg-GlcNAc site of FADD Arg117 and the novel site Arg16 within charged multivesicular body protein 2a. HexNAc, N-acetylhexosamine.
Arg117 in FADD is the preferred target for NleB1 during wild-type EPEC infection
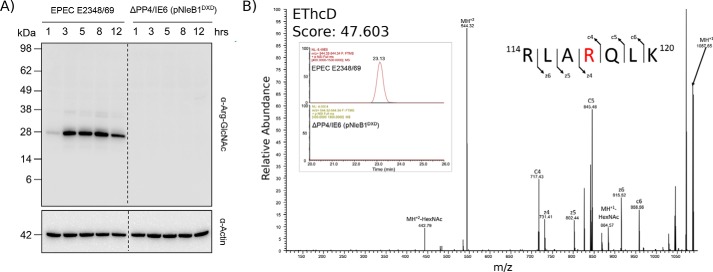
The ability to enrich Arg-GlcNAc glycopeptides using the Arg-GlcNAc-specific antibody provided an ideal means to assess the extent of arginine GlcNAcylation during EPEC E2348/69 infection. In contrast to stable expression of FLAG-NleB1, which resulted in the modification of multiple host proteins, only a single protein of ∼25 kDa was detected by immunoblotting during EPEC E2348/69 infection (Fig. 2A). This dominant band appeared rapidly and became saturated within 3 h. Arginine-GlcNAc enrichment of glycopeptides from EPEC E2348/69 infection at 3 h confirmed arginine GlcNAcylation of Arg117 in FADD as the dominant modification (Fig. 2B). Surprisingly, multiple bacterially derived proteins, including glutathione synthetase (B7UHZ4) and 30S ribosomal protein S4 (B7UK20), were also identified (supplemental Table 4).
Figure 2.
The Arg-GlcNAcylated host protein repertoire during EPEC infection is predominantly restricted to FADD. A, time course of EPEC E2348/69 infection over 12 h shows the appearance of a single dominant band consistent with the size of FADD (∼25 kDa). B, extracted ion chromatograms showing the presence of modified FADD peptides at 3 h postinfection with wild-type EPEC E2348/69 that are absent in the Arg-GlcNAc enrichment of EPEC E2348/69 ΔPP4/IE6 (pNleB1DXD)-infected cells 3 h postinfection. EThcD fragmentation confirms that the site of modification in FADD corresponds to Arg117. HexNAc, N-acetylhexosamine.
As additional faint bands were observed at later time points of infection, we also assessed the targets of arginine GlcNAcylation 8 h postinfection. Similar to the 3-h time point, Arg117 in FADD and the bacterial targets were readily detectable (supplemental Fig. 2 and supplemental Table 4). However, at this time point, arginine GlcNAcylation of at Arg235 in TRADD could also be observed, albeit inconsistently across the biological replicates (supplemental Fig. 2 and supplemental Table 4). No arginine GlcNAcylation was detected during infection with EPEC expressing catalytically inactive NleB1, strain EPEC E2348/69 ΔPP4/IE6 (pNleB1DXD) (Fig. 2, A and B). Proteome analysis of the input sample confirmed the presence of both TRADD and FADD within HT-29 cells during EPEC infection with comparative proteomics using intensity-based absolute quantification supporting the presence of FADD at higher relative levels of abundance than TRADD (supplemental Fig. 3 and supplemental Table 5). Taken together, these results demonstrate that Arg117 in FADD was the preferred target of NleB1 during EPEC E2348/69 infection.
Extent of Arg GlcNAcylation upon overexpression of NleB1 during infection
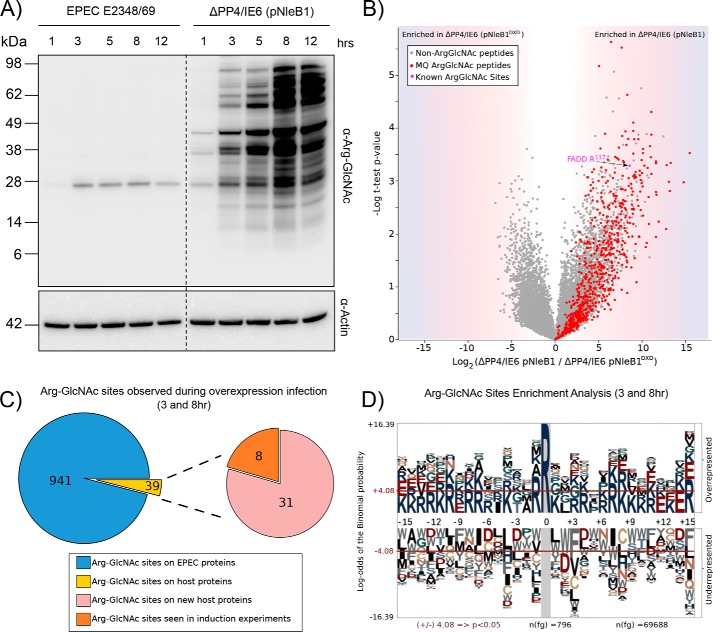
Given the discrepancy in levels of arginine GlcNAcylation between HeLa cells stably expressing FLAG-NleB1 and NleB1 delivered by wild-type EPEC E2348/69 during infection, we assessed whether overexpression of NleB1 during EPEC E2348/69 infection altered the range of arginine-GlcNAcylated targets of NleB1. As two potential Arg glycosyltransferases exist within EPEC (NleB1, located within the IE6 genomic island, and NleB2, located within the PP4 genomic island), complementation was undertaken within the ΔPP4/IE6 background. NleB1 expressed from a multicopy plasmid during EPEC infection resulted in extensive Arg GlcNAcylation compared with that observed under wild-type infection conditions (Fig. 3A). The enrichment of arginine-GlcNAcylated glycopeptides at 3 and 8 h after infection revealed extensive modification of both bacterial proteins and host proteins, including FADD (Fig. 3B and supplemental Table 6). In total, 1154 arginine-GlcNAcylated glycopeptides, corresponding to 980 unique sites with a localization of >0.75, were identified. The majority of the observed glycosylation sites were of bacterial origin (941 sites; Fig. 3C), whereas 39 sites were identified in host proteins. Modification of Arg117 in FADD was readily detected, whereas modification of TRADD was undetectable at 3 or 8 h (Fig. 3B). We detected modification of a further eight proteins, which were also seen under conditions of stable FLAG-NleB1 expression (Fig. 3C). The presence of 941 sites of arginine GlcNAcylation within EPEC E2348/69 proteins enabled an analysis of the composition of targeted sites, which revealed that arginine GlcNAcylation events appeared to target sites rich in basic amino acids, similar to death domains (Fig. 3D). However, this was not an absolute requirement as host Arg-GlcNAcylated proteins did not demonstrate the same extent of enrichment in flanking basicity (supplemental Fig. 4). To correlate the appearance of Arg-GlcNAcylated substrates with NleB1 levels, we also attempted to monitor NleB1 levels using both an NleB1-specific antibody and proteomic analysis. However, because of its low abundance (31), NleB1 is undetectable by both methodologies during wild-type infections compared with the ΔPP4/IE6 background expressing NleB1 from a multicopy plasmid despite the detection of Arg GlcNAcylation of FADD (supplemental Fig. 5 and supplemental Table 5). Taken together, this supports that increased levels of NleB1 expression result in indiscriminate arginine GlcNAcylation of both bacterial and host proteins.
Figure 3.
Overexpression of NleB1 results in non-authentic modifications of both bacterial and host substrates. A, time course of EPEC E2348/69 infection over 12 h shows extensive arginine GlcNAcylation when NleB1 is overexpressed compared with wild-type EPEC E2348/69 infections. B, label-free quantification of Arg-GlcNAc peptide pulldown results from 8-h infection of HT-29 cells with EPEC E2348/69 ΔPP4/IE6 (pNleB1) versus EPEC E2348/69 ΔPP4/IE6 (pNleB1DXD). The scatter plot shows the mean ion intensity peptide ratios of E232489/69 ΔPP4/IE6 (pNleB1) versus E2348/69 ΔPP4/IE6 (pNleB1DXD) plotted against negative logarithmic t test p values from biological triplicate experiments. Within biological replicates of EPEC E2348/69 ΔPP4/IE6 (pNleB1) infections, multiple putative arginine-GlcNAcylated peptides were enriched from both bacterial and host origin (colored in red) with Arg-GlcNAc peptides containing previously known Arg-GlcNAc modifications shown in purple. MQ, MaxQuant. C, Arg-GlcNAc sites observed within EPEC E2348/69 ΔPP4/IE6 (pNleB1) infections are predominantly derived from bacterial origin with the host proteins modified during infection identical to those seen during stable NleB1 induction. D, pLogo motif analysis of Arg-GlcNAc sites observed within EPEC E2348/69-derived proteins. Arg-GlcNAc sites are flanked by basic residues.
Preferred target of NleB during C. rodentium infection
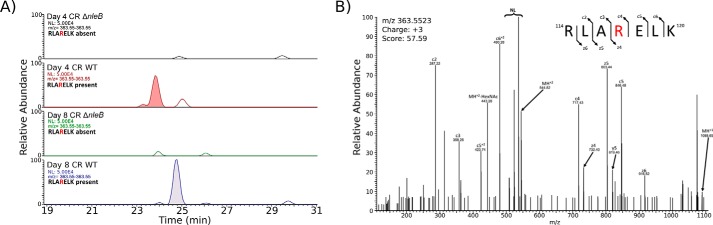
The identification of Arg117 in FADD as the preferred target of arginine GlcNAcylation during EPEC infection and the emergence of non-authentic targets upon overexpression of NleB1 prompted us to investigate arginine GlcNAcylation during mouse infection with C. rodentium. Previously, Li et al. (18) showed that, upon ectopic expression, NleBCR glycosylated FADD but was unable to modify TRADD. To verify the target of NleB during C. rodentium infection in vivo, we performed immunoenrichment and parallel reaction monitoring (32) to selectively monitor the modification of Arg117 in FADD and Arg235 in TRADD at days 4 and 8 after wild-type C. rodentium or ΔnleBCR mouse infection. Consistent with NleB1 from EPEC E2348/69, arginine GlcNAcylation of Arg117 in FADD was readily observable upon wild-type C. rodentium infection at days 4 and 8, whereas no modified substrates were observed during infection with ΔnleBCR (Fig. 4, A and B). Indeed, Arg117 in FADD was the only modification that could be observed. Data-independent analysis of enriched arginine-glycosylated samples also failed to identify any additional substrates of arginine GlcNAcylation during mouse infection with wild-type C. rodentium (data not shown). Proteome analysis of the input sample confirmed the presence of TRADD and FADD as well as C. rodentium proteins within samples (supplemental Fig. 6 and supplemental Table 7). Thus, similar to EPEC E2348/69 infection, Arg117 in FADD was modified by NleBCR during C. rodentium infection in vivo.
Figure 4.
FADD is the preferred target of NleB during C. rodentium infection. A, extracted ion chromatograms showing the presence of modified FADD peptides at days 4 and 8 postinfection with wild-type C. rodentium ICC169 that are absent in the Arg GlcNAcylation enrichment of C. rodentium ΔnleB-infected samples. NL corresponds to neutral loss associated with ETD fragmentation. B, EThcD fragmentation confirms that the site of modification in FADD corresponds to Arg117 during C. rodentium infection. HexNAc, N-acetylhexosamine.
Stability and permanence of the Arg117 FADD modification by NleB1
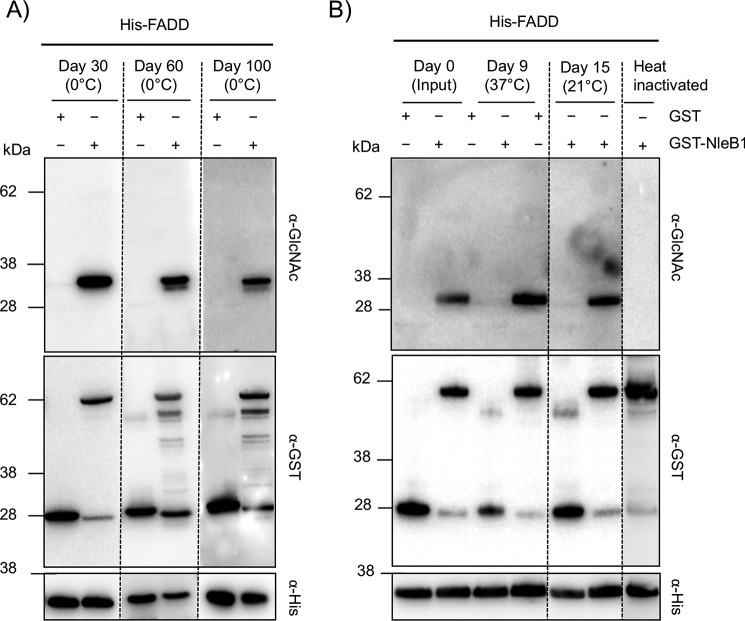
We postulated that the observed saturation of Arg117 arginine GlcNAcylation in FADD during EPEC E2348/69 infection (Fig. 2A) supported arginine GlcNAcylation being a stable and irreversible modification given that NleB1 is not translocated to high levels during wild-type infection (31). To assess stability of the Arg-GlcNAc modification, we performed in vitro experiments to track changes in arginine GlcNAcylation over time in response to environmental conditions and host enzymatic activity. Purified His-FADD was incubated with GST-NleB1 and UDP-GlcNAc for 3 h at 37 °C and then further incubated at 37, 21, or 4 °C for extended periods of time before being subjected to immunoblot analysis. When kept at 4 °C, arginine GlcNAcylation of FADD was still detectable after 100 days of incubation (Fig. 5A) even though some protein degradation was observed as indicated by the presence of additional bands when probing for GST-NleB1 (Fig. 5A). To exclude the possibility that over time the Arg-GlcNAc modification of FADD was lost and then reincorporated onto FADD by active GST-NleB1, heat treatment was used to inactivate GST-NleB1 after initial incubation. Arg-GlcNAc modification of FADD was still detected when the heat-inactivated mixtures were kept at 37 °C for 9 days or at 21 °C for 15 days (Fig. 5B). Thus, under physiological temperatures, the arginine GlcNAcylation of FADD was highly stable.
Figure 5.
Arg-GlcNAcylated FADD is stable and irreversible in response to physiological temperatures. Immunoblots of in vitro generated Arg-GlcNAcylated His-FADD incubated at various temperatures are shown. A, time course of in vitro NleB1-modified His-FADD stored at 4 °C for up to 100 days. B, time course of in vitro NleB1-modified His-FADD stored at 37 °C for 9 days and 21 °C for 15 days. Heat inactivation of NleB1 prior to incubation with His-FADD resulted in the inhibition of Arg GlcNAcylation. Incubated proteins were separated by SDS-PAGE, immunoblotted, and probed with anti-GlcNAc antibody to detect modified FADD. Anti-His, anti-GST, and anti-β-actin antibodies were used for control immunoblots.
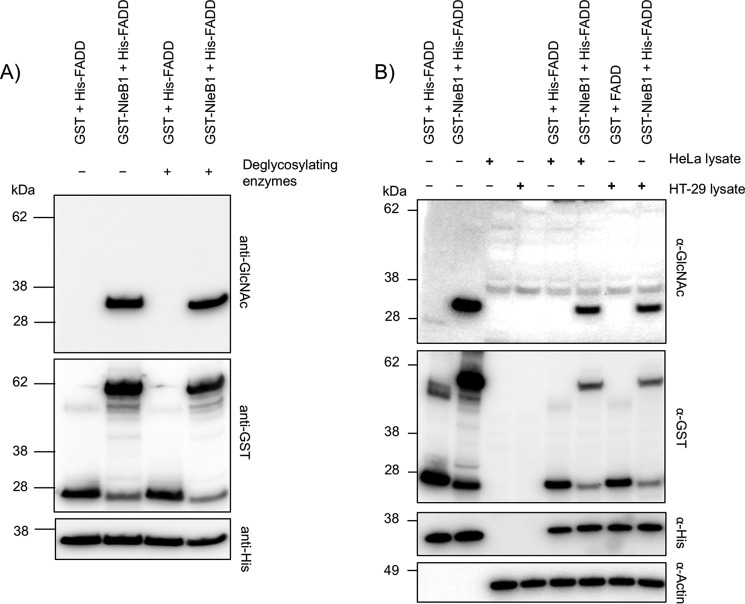
The enzymatic removal of glycosylation is frequently utilized for proteomic analysis of glycoproteins. Most asparagine N-linked glycosylations can be removed by the enzyme peptide:N-glycosidase F (PNGase F), whereas a variety of enzymes are required to remove O-linked glycosylation (33). These enzymes have varying specificities, and their ability to recognize Arg-linked glycosylation was unknown. Hence, we tested commonly used glycosidases to assess whether they could hydrolyze Arg GlcNAcylation. Using denatured bovine fetuin, we found that PNGase F, sialidase A, and endo-α-N-acetylgalactosaminidase (O-glycanase) were functional under the conditions used as shown by a decrease in molecular weight of fetuin upon their coincubation (supplemental Fig. 7). However, when the glycosidases were incubated with NleB1-modified FADD, there was no loss of GlcNAc from FADD (Fig. 6A), suggesting these enzymes are unable to recognize and hydrolyze the Arg-GlcNAc glycosidic bond.
Figure 6.
Arg-GlcNAcylated FADD is stable and irreversible in response to glycosidase and cellular enzymatic activities. Immunoblots of in vitro generated Arg-GlcNAcylated His-FADD incubated with glycosidases and mammalian cell lysates are shown. A, immunoblots of His-FADD incubated with either GST or GST-NleB1 at 37 °C for 3 h, then heat-inactivated, and left at 37 °C overnight with or without the glycosidases PNGase F, sialidase A, and O-glycanase. B, in vitro NleB1-modified His-FADD was then heat-inactivated and incubated with and without HeLa or HT-29 cell lysate overnight. Incubated proteins were separated by SDS-PAGE, immunoblotted, and probed with anti-GlcNAc antibody to detect modified FADD. Anti-His, anti-GST, and anti-β-actin antibodies were used for control immunoblots.
To further probe the sensitivity of Arg-GlcNAcylated FADD to enzymatic activities within the host cell, we investigated the stability of this modification in the presence of cellular lysates. NleB1-modified His-FADD carrying the Arg-GlcNAc modification was incubated with HeLa and HT-29 cell lysates and then subjected to immunoblot analysis. Upon overnight incubation, we observed a small decrease in the levels of FADD Arg GlcNAcylation (Fig. 6B); however, this decrease was proportional to changes in the protein levels of His-FADD and was consistent with protein degradation rather than loss of the modification (Fig. 6B). Therefore, there did not appear to be enzymes within HeLa or HT-29 cell lysates capable of removing the Arg-GlcNAc modification. Thus, once FADD is modified by Arg GlcNAcylation, it is irreversible within the host cell.
Host cell response to arginine GlcNAcylation
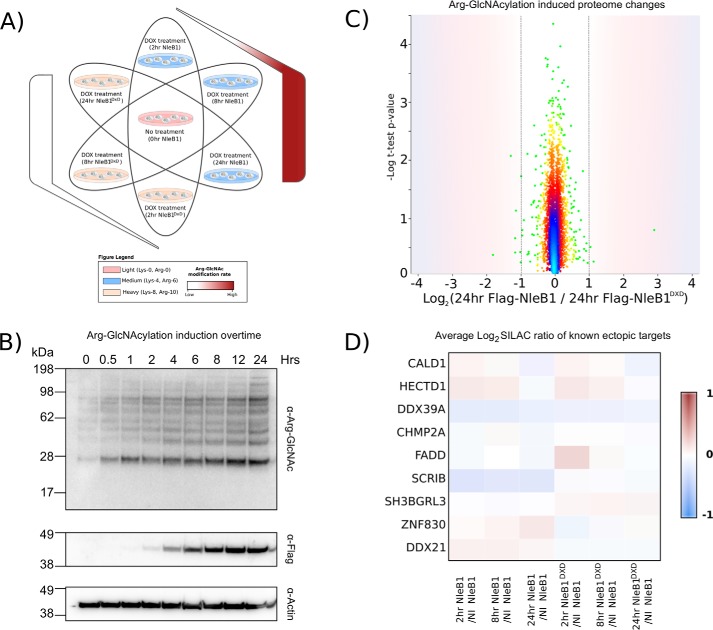
The identification of arginine GlcNAcylation as an irreversible modification in mammalian cells prompted us to determine whether the modification was detected by host innate immune sensing mechanisms. To exclude the contribution of bacterially induced proteome changes, we utilized the stable inducible FLAG-NleB1 and FLAG-NleB1DXD cell lines to assess proteome changes in response to NleB1 activity after 2, 8, and 24 h of FLAG-NleB1 expression using SILAC-based quantitative proteomics (Fig. 7A). Using this approach, 6532 proteins were identified with 3502 quantified at all time points, including all previously observed arginine glycosylation targets with the exception of TRADD (supplemental Fig. 8 and supplemental Table 8). Strikingly, the induction of Arg GlcNAcylation had no significant impact on the cell proteome despite robust arginine GlcNAcylation induced upon expression of FLAG-NleB1 (Fig. 7, B and C, and supplemental Table 8). Similar to the total proteome, levels of Arg-GlcNAcylated targets were largely unaffected by modification (Fig. 7D). Thus, even upon expression of FLAG-NleB1, the proteome and arginine-GlcNAcylated targets appeared unaffected by NleB1 activity. Together these experiments support the notion that host protein arginine GlcNAcylation by NleB1 is a silent and irreversible modification.
Figure 7.
Arginine GlcNAcylation does not elicit proteome changes within the host. A, experimental design of SILAC experiment to investigate the effect of Arg GlcNAcylation on the host proteome. FLAG-NleB1 and FLAG-NleB1DXD stable cell lines were induced with doxycycline (DOX) for 2, 8, or 24 h and compared with uninduced NleB1 controls. B, a time course of Arg GlcNAcylation demonstrates a temporal dependence of Arg GlcNAcylation that coincides with the expression of FLAG-NleB1. C, SILA-based quantitation comparing the changes in the proteome between cells stably expressing NleB1 or NleB1DXD after 24 h of doxycycline induction. The scatter plot shows the mean SILAC protein ratio of cells stably expressing FLAG-NleB1 versus FLAG-NleB1DXD plotted against negative logarithmic t test p values from biological triplicate experiments. Density-based coloring demonstrates that the vast majority of proteins are unaffected by the induction of Arg GlcNAcylation with no statistically significant protein changes observed within the proteome. D, known arginine GlcNAcylation targets show no changes in protein abundance in response to NleB1 induction in all time points examined.
Discussion
During EPEC and C. rodentium infection, several studies have demonstrated that the NleB effectors target multiple host proteins for arginine GlcNAcylation (17, 18, 21). These studies were all performed under conditions of NleB overexpression and/or ectopic expression. Here, we also found that ectopic expression of NleB or infection with EPEC E2348/69 overexpressing NleB1 lead to Arg GlcNAcylation of multiple host targets. This confirmed that the NleB effectors modify a greater range of proteins, including bacterial proteins, than previously reported. Arginine GlcNAcylation in bacterial targets typically occurred in regions rich in basic residues akin to the sequences observed within the death domains of mammalian proteins (Fig. 3C). However, this preference in modification site was not absolute with numerous host arginine GlcNAcylation events observed within regions lacking flanking basic residues, such as Arg16 of charge multivesicular body protein 2a (Fig. 1D and supplemental Fig. 4).
In contrast to the results observed upon overexpression of NleB1, wild-type EPEC E2348/69 infection led to arginine GlcNAcylation of a single target, which was confirmed as Arg117 in the death domain of FADD (Fig. 2 and supplemental Table 4). This observation was replicated in vivo during wild-type C. rodentium infection (Fig. 4), suggesting that FADD is the dominant and preferred target of NleB1/NleBCR. The preferential modification of FADD under wild-type conditions supports our previous finding of the importance of Fas signaling in limiting the duration of C. rodentium infection (17). As FADD is the sole adapter required for caspase-8 activation during Fas signaling (34, 35), blocking this key protein prevents the initiation of Fas ligand-mediated apoptosis and the subsequent elimination of infected enterocytes (17).
The observation of a narrower host target range of NleB1/NleBCR during wild-type infection compared with ectopic and overexpression during infection has been noted for other bacterial effector studies (36, 37). For example, the type III Shigella protease IpaJ has been shown to target only the ADP-ribosylation factor and ADP-ribosylation factor-like GTPases during infection compared with the majority of the N-myristoylome during ectopic expression (37). Interestingly, in this work, we did observe modification of TRADD when infections were extended to 8 h, albeit inconsistently within replicates (one of three biological replicates; supplemental Fig. 2 andsupplemental Table 4). Although this observation supports the finding that TRADD can be targeted by wild-type levels of NleB1, the contribution of a later and weaker modification of TRADD to the inhibition of TNF signaling is unclear as this function is redundant with several other EPEC effector proteins (12, 13, 36, 38, 39). Furthermore, we were unable to detect Arg GlcNAcylation of TRADD during C. rodentium infection either 4 or 8 days postinfection despite the detection of TRADD protein in the input material used for Arg GlcNAcylation enrichment (supplemental Table 7 and supplemental Fig. 6). This supports the previous finding that NleBCR is unable to modify the death domain of TRADD (18).
The observation that overexpression of NleB1 leads to widespread non-authentic arginine GlcNAcylation of substrates sheds light on the range of targets reported for NleB effectors. A previous study used in vitro labeling coupled to Western blotting-based analysis to suggest that NleBCR glycosylated GAPDH (19). However, MS and radiolabeling experiments performed in another study later refuted this target (18). In this study, at no point did we detect arginine GlcNAcylation of GAPDH during wild-type infections or upon overexpression of NleB1 during infection or ectopic expression, although the modification of arginine residues within GAPDH was recently demonstrated (21). It should be noted that an important nuance of the identification of modified peptides is that the identification alone does not directly provide information about the occupancy rate of the site. As MS instrumentation improves, even low-occupancy sites of modification may be able to be detected, but these may not contribute to any observable phenotype. This feature coupled to the complication of substrate promiscuity means that great care needs to be taken when assigning the targets of Arg GlcNAcylation under non-infection conditions as these targets may be artifactual in nature. Recently, it was suggested that the Salmonella NleB homologues SSeK1 and SseK3 arginine-GlcNAcylated FADD and TRADD, but again these assignments are based on ectopic expression showing markedly different arginine GlcNAcylation profiles compared with wild-type Salmonella infection (40).
In addition to the modification of host proteins by NleB1, we also observed arginine GlcNAcylation of EPEC proteins even when grown under laboratory conditions (supplemental Fig. 9 and supplemental Table 9). Bacterial glycosylation has been noted in multiple pathogens (41, 42), and although we believe this is the first example of Arg GlcNAcylation of bacterial proteins, the functional consequences of these modifications, if any, are still unclear.
Finally, as FADD is the predominant target of NleB1/NleBCR-mediated arginine GlcNAcylation, we directly assessed the stability of the modification in response to environmental conditions, including incubation with host enzymes and the effect of NleB1 activity on the proteome (Figs. 5–7). We found that arginine GlcNAcylation of FADD is highly stable and unaffected by glycosidases/host enzymatic activities or physiological temperatures. This suggests that similar to other bacterially mediated PTMs, such as threonine eliminylation (43), arginine GlcNAcylation leads to a permanent and irreversible modification within the host cell. Despite the fact that arginine GlcNAcylation appears insensitive to removal by host factors, the modification does not induce detectable changes in the host proteome even when NleB1 is overexpressed, suggesting that, unlike bacterial glycosylation mediated by the clostridial toxin TcdA/B (44, 45), NleB1-mediated arginine GlcNAcylation is not sensed by cell-intrinsic defense pathways (46) (Fig. 7).
In summary, we conclude that, when overexpressed, NleB1 can modify a far wider repertoire of proteins than previously appreciated. However, during wild-type pathogen infection, Arg117 of FADD is the only detectable target of biological significance both in vitro and in vivo.
Experimental procedures
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in supplemental Table 1. Strains of E. coli and C. rodentium were grown at 37 °C in Luria-Bertani (LB) broth with shaking or in Dulbecco's modified Eagle's medium (DMEM) without shaking. When required, the following antibiotics were added at the indicated concentrations: ampicillin, 100 μg/ml; kanamycin, 50 or 100 μg/ml; nalidixic acid, 50 μg/ml; chloramphenicol, 25 μg/ml.
Analysis of Arg glycosylation in stable inducible cell lines expressing FLAG-NleB1 and FLAG-NleB1DXD
HeLa cell lines expressing FLAG-NleB1 and catalytically inactive FLAG-NleB1DXD were constructed as described previously (14) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS). For immunoblotting studies, cells were seeded into 6-well plates at 3 × 105 cells/well 1 day prior to induction with doxycycline (20 ng/ml) for 24, 16, 8, 4, 2, 1, 0.5, and 0 h. At the desired time points, medium was removed, and cells were collected in Kal B buffer (50 mm Tris, pH 7.4, 1.0% Triton X-100, 150 mm NaCl, 1 mm EDTA with Complete protease inhibitor mixture (Roche Applied Science) added immediately prior to use). Lysates were subjected to SDS-PAGE using precast 4–12% gels (Invitrogen) and transferred to nitrocellulose membranes. Membranes were blocked for 1 h in 5% skim milk in TBS-Tween 20 and then incubated for at least 16 h at 4 °C with either anti-Arg-GlcNAc antibody (1:2,000; ab195033, Abcam), anti-FLAG-horseradish peroxidase (HRP) M2 antibody (1:2,000; A8592, Sigma-Aldrich), or mouse monoclonal anti-β-actin antibody (1:5,000; catalogue number AC-15, Sigma-Aldrich). Proteins were detected using anti-rabbit or anti-mouse IgG HRP-conjugated secondary antibody (1:3,000; catalogue numbers NEF812001EA and NEF822001EA, respectively, PerkinElmer Life Sciences) and developed with ECL Prime Western blotting reagent (GE Healthcare). All antibodies were diluted in TBS with 1% bovine serum albumin (BSA; Sigma-Aldrich) and 0.1% Tween 20. Images were visualized using an MFChemiBis imaging station (DNR Bio-Imaging Systems). For proteomic analysis, 100-mm dishes were seeded at 2 × 106 cells/well the day prior to induction. Effector protein expression was induced with doxycycline (20 ng/ml) for 24 h, and cells were washed three times in ice-cold PBS to remove media-related proteins prior to lysis with ice-cold guanidinium chloride lysis buffer (6 m guanidinium chloride, 100 mm Tris, pH 8.5, 10 mm tris(2-carboxyethyl)phosphine, 40 mm 2-chloroacetamide) according to the protocol of Humphrey et al. (47). All experiments were performed in biological triplicate.
SILAC analysis of inducible cell lines expressing FLAG-NleB1 and FLAG-NleB1DXD
SILAC labeling of HeLa derivatives was accomplished as described previously (48, 49). Briefly, DMEM lacking Lys and Arg was supplemented with 10% dialyzed FBS (Invitrogen), 1× penicillin/streptomycin (Sigma), and combinations of the following lysine and arginine isotopologues: for “light” labeled cells, l-arginine (34 mg/liter) and l-lysine (73 mg/liter) (Sigma-Aldrich); for “medium” labeled cells, l-[13C6]arginine (35 mg/liter) and d4-l-lysine (74.8 mg/liter); and for “heavy” labeled cells, l-[13C6,15N4]arginine (35.8 mg/liter) and l-[13C6,15N2]lysine (76.6 mg/liter) (Cambridge Isotope Laboratories, Andover, MA). Cells were split 1:4 into the three SILAC media formulations and passaged five times for complete replacement of labeled amino acids. For each condition within a biological replicate, one confluent 100-mm dish was used with cells washed three times in ice-cold PBS prior to being lysed with ice-cold guanidinium chloride lysis buffer and mixing 1:1:1 prior to sample preparation. All experiments were performed in biological triplicate.
Infection of HT-29 cells with EPEC for Arg glycosylation analysis
HT-29 cells were infected with EPEC E2348/69 (50) and ΔPP4/IE6 derivatives (17, 39) to induce arginine glycosylation of host proteins and analyzed by either immunoblotting or Arg glycosylation peptide affinity purification of cell lysates. For immunoblotting, 1 day prior to infection, HT-29 cells were seeded at 2.5 × 105 cells/ml in 24-well plates (Greiner Bio One), and various EPEC derivatives were cultured in 10 ml of LB broth overnight at 37 °C. The following day, bacterial cultures were subinoculated 1:75 in DMEM and incubated at 37 °C with 5% CO2 for 2.5 h prior to infection. HT-29 cells were infected with a multiplicity of infection of 1 with various EPEC strains for 1, 3, 5, 8, or 12 h. At the required time point, HT-29 cells were lysed in Kal B buffer and subjected to immunoblotting as outlined above. For Arg glycosylation peptide affinity purification, HT-29 cells were seeded at 4 × 106 cells in 100-mm dishes the day prior to infection, and various EPEC strains were cultured in 10 ml LB broth overnight at 37 °C. The following day, bacterial cultures were subinoculated 1:75 in DMEM and incubated at 37 °C with 5% CO2 for 2.5 h prior to infection. HT-29 cells were infected with a multiplicity of infection of 1 with EPEC derivatives for either 3 or 8 h. At the required time, cells were washed three times in ice-cold PBS to remove media-related proteins and lysed with ice-cold guanidinium chloride lysis buffer. All experiments were performed in biological triplicate. Western blot analyses of biological were performed as above using anti-Arg-GlcNAc antibody (1:2,000), rabbit polyclonal anti-NleB1 (1:500; produced by the Walter and Eliza Hall Institute antibody facility from recombinant NleB1), and mouse monoclonal anti-β-actin antibody (1:5,000).
Infection of mice with C. rodentium
Animal infections were performed according to protocol of Pearson et al. (17) and Wong Fok Lung et al. (51). All animal experiments were approved by the University of Melbourne Animal Ethics Committee. C. rodentium ICC169 (52) and C. rodentium ICC169 ΔnleBCR (17) were cultured in LB broth containing antibiotics, as required, overnight at 37 °C with shaking. On the following day, bacterial cells were harvested by centrifugation at 3,220 × g for 10 min at room temperature, and the bacterial pellet was resuspended in PBS. Unanesthetized 5- 8-week-old female C57BL/6 mice were each given 200 μl of a bacterial suspension containing ∼1 × 109 CFU in PBS by oral gavage. The viable count of the inoculum was determined by retrospective serial dilution and plating on Luria agar containing the required antibiotic. Five mice were used per infection group, and colonic epithelial cells were harvested at days 4 and 8.
Colon epithelial cell isolation for Arg glycosylation peptide affinity purification
For isolation of colonic epithelial cells, colons were removed, cut longitudinally, and rinsed in ice-cold Hanks' balanced salt solution (137 mm NaCl, 5.4 mm KCl, 0.25 mm Na2HPO4, 0.1 g glucose, 0.44 mm KH2PO4, 1.3 mm CaCl2, 1.0 mm MgSO4, 4.2 mm NaHCO3) to remove fecal material. Prior to epithelial cell release, colonic tissue was washed in ice-cold 0.5 mm DTT, RPMI 1640 medium and then dissected into 0.25-cm2 sections. Tissue sections were transferred into 3 mm EDTA, 0.5 mm DTT in Ca2+/Mg2+-free Hanks' balanced salt solution and incubated for 15 min at 37 °C with shaking. Epithelial cells were released by vortexing and isolated by straining through a 100-μm cell strainer. Tissue sections were subjected to two rounds of epithelial cell release with 3 mm EDTA, 0.5 mm DTT in Ca2+/Mg2+-free Hanks' balanced salt solution and straining through a 100-μm cell strainer. Isolated epithelial cells were washed twice with ice-cold PBS and snap frozen prior to lysis with ice-cold guanidinium chloride lysis buffer.
Recombinant protein production
Recombinant protein was produced as described previously (17). Briefly, plasmids for the expression of His6-tagged FADD and GST-tagged NleB1 were transformed into BL21 C43(DE3) E. coli. LB overnight cultures of BL21 containing the appropriate expression vector were used to inoculate 200 ml of LB broth at 1:100 and grown at 37 °C with shaking to an optical density (A600) of 0.6. Cultures were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside and grown for a further 2.5 h before being pelleted by centrifugation. Before purification, bacterial pellets were resuspended in the appropriate binding buffer from Novagen His-Bind and GST-Bind kits. Bacterial suspensions were lysed using an EmulsiFlex-C3 high-pressure homogenizer (Avestin) according to the manufacturer's instructions. Purification of proteins was performed according to the manufacturer's protocols (Novagen). Proteins were dialyzed using Cellu-Sep T3 regenerated cellulose tubing (Fisher Biotec), and protein concentrations were determined using a bicinchoninic acid (BCA) kit (Thermo Scientific).
In vitro arginine GlcNAcylation stability assays
Purified GST, GST-NleB, and His-FADD were used for in vitro N-acetylglycosylation assays, which involved incubation of 2 μg of proteins either alone or in combination in the presence of 1 mm UDP-GlcNAc (Sigma) at 37 °C for 3 h in 150 mm NaCl, 20 mm Tris, pH 8. The mixtures were then either incubated at 4 °C or heat-inactivated at 80 °C for 10 min and then incubated at 4 °C, room temperature (∼21 °C), or 37 °C for extended periods of time. At various time points following the initial incubation, some of the mixture was taken for immunoblotting.
To assess stability in response to glycosidases, in vitro glycosylated FADD was incubated with and without glycosidases according to the ProZyme Enzymatic Deglycosylation kit protocol (ProZyme, Hayward, CA). Briefly, proteins were denatured at 100 °C in the presence of 0.1% SDS and 60 mm mercaptoethanol for 5 min and allowed to cool before Nonidet P-40 was added to a final concentration of 0.8%. PNGase F, sialidase A, and endo-α-N-acetylgalactosaminidase were then added, and the reaction was allowed to proceed overnight at 37 °C before adding sample buffer, boiling, and subjecting the samples to SDS-PAGE and immunoblotting as above. Bovine fetuin control reactions were performed according to the manufacturer's protocol and subjected to SDS-PAGE. Detection of proteins for the control reaction was performed by colloidal Coomassie staining of the polyacrylamide gels.
To assess stability in response to cellular lysates, HeLa and HT-29 were prepared from confluent 100-mm dishes by scraping in PBS with Complete protease inhibitor mixture. The HeLa or HT-29 cell suspensions were passed through a 26-gauge needle 50 times to lyse the cells before pelleting to remove cell debris. In vitro glycosylated protein was added directly to lysates and incubated for 16 h at 37 °C. Sample buffer was added to incubated samples before they were boiled, subjected to SDS-PAGE, and transferred to nitrocellulose membranes. Membranes were probed with mouse monoclonal anti-GlcNAc (1:2,000; CTD110.6, Cell Signaling Technology), mouse monoclonal anti-His (1:2,000; AD1.1.10, AbD Serotech), rabbit polyclonal anti-GST (1:2,000; 26H1, Cell Signaling Technology), or mouse monoclonal anti-β-actin (1:5,000) primary antibody and HRP-conjugated anti-mouse or anti-rabbit secondary antibody and developed as described above.
Isolation of proteins for proteome analysis and Arg glycosylation peptide affinity purification
Cells lysed in ice-cold guanidinium chloride lysis buffer were collected and boiled at 95 °C for 10 min with shaking at 2000 rpm to shear DNA and inactivate protease activity. Lysates were then cooled for 10 min on ice and boiled again at 95 °C for 10 min with shaking at 2000 rpm. Lysates were cooled, and protein concentration was determined using a BCA assay. 2 mg of protein from each sample was precipitated by mixing 4 volumes of ice-cold acetone with 1 volume of sample. Samples were precipitated overnight at −20 °C and then centrifuged at 4,000 × g for 10 min at 4 °C. The precipitated protein pellets were resuspended with 80% ice-cold acetone and precipitated for an additional 4 h at −20 °C. Samples were centrifuged at 17,000 × g for 10 min at 4 °C to collect precipitated protein, supernatant was discarded, and excess acetone was driven off at 65 °C for 5 min.
Digestion of complex protein lysates
Dried protein pellets were resuspended in 6 m urea, 2 m thiourea, 40 mm NH4HCO3 and reduced/alkylated prior to digestion with Lys-C (1:200, w/w) and then trypsin (1:50, w/w) overnight as described previously (53). Digested samples were acidified to a final concentration of 0.5% formic acid and desalted with 50 mg of tC18 Sep-Pak (Waters Corp.) according to the manufacturer's instructions. Briefly, tC18 Sep-Pak was conditioned with Buffer B (0.1% formic acid (FA), 80% acetonitrile (ACN)), washed with 10 volumes of Buffer A* (0.1% trifluoroacetic acid (TFA), 2% ACN), the sample was loaded, the column was washed with 10 volumes of Buffer A*, and bound peptides were eluted with Buffer B and then dried.
Arg glycosylation affinity purification
Peptide affinity purification was accomplished according to the protocol of Udeshi et al. (54) but modified to allow for Arg-GlcNAc enrichment. Briefly, aliquots of 100 μl of Protein A/G Plus-agarose beads (Santa Cruz Biotechnology, Santa Cruz CA) were washed three times with 1 ml of immunoaffinity purification (IAP) buffer (10 mm Na3PO4, 50 mm NaCl, 50 mm MOPS, pH 7.2) and tumbled overnight with 10 μg of anti-Arg-GlcNAc antibody at 4 °C. Beads coupled to anti-Arg-GlcNAc were then washed three times with 1 ml of 100 mm sodium borate, pH 9, to remove non-bound proteins and cross-linked for 30 min with tumbling using 20 mm dimethyl pimelimidate (Thermo Scientific) in 100 mm HEPES, pH 8.0. Cross-linking was quenched by washing beads with 200 mm ethanolamine, pH 8.0, three times and then tumbling beads in an additional 1 ml of 200 mm ethanolamine, pH 8.0, for 2 h at 4 °C. Beads were washed three times with IAP buffer and used immediately.
Purified peptides were resuspended in 1 ml of IAP buffer, and the pH was checked to ensure compatibility with affinity conditions. Peptide lysates were then added to the prepared cross-linked anti-Arg-GlcNAc antibody beads and tumbled for 3 h at 4 °C. Upon completion, antibody beads were centrifuged at 3,000 × g for 2 min at 4 °C, and the unbound peptide lysates were collected. Antibody beads were then washed six times with 1 ml of ice-cold IAP buffer, and Arg-GlcNAc peptides were eluted using two rounds of acid elution. For each elution round, 100 μl of 0.2% TFA was added, and antibody beads were allowed to stand at room temperature with gentle shaking every minute for 10 min. Peptide supernatants were collected and desalted using C18 StageTips (55, 56) before analysis by LC-MS.
High-pH fractionation of SILAC proteomes
Fractionation of SILAC-labeled samples was achieved by basic reversed-phase chromatography according to the protocol of Udeshi et al. (54) with minor modifications. Briefly, peptides were resuspend in Buffer A (5 mm ammonium formate, 2% ACN, pH 10) and separated using a 1100 series high-performance liquid chromatography instrument (Agilent Technologies, Santa Clara, CA) with an XBridge C18 column (1.0 × 150 mm, 3.5 μm; Waters) and a flow rate of 100 μl/min. Separation was accomplished using a 90-min gradient. The concentration of Buffer B (5 mm ammonium formate, 90% ACN, pH 10) was ramped from 0 to 6% Buffer B over 5 min and then to 8% over 2 min followed by an increase to 27% Buffer B in 38 min, to 31% Buffer B in 4 min, to 39% Buffer B in 4 min, and to 60% Buffer B in 7 min and completed with a 4-min run at 100% Buffer B and a 26-min gradient back to 100% Buffer A. 100-μl fractions were collected in a 96-well plate with every sixth fraction combined to generate a total of six fractions, which were concentrated by vacuum centrifugation, desalted using C18 StageTips, and subjected to mass spectrometric analysis.
HCD identification of SILAC proteomics- and FLAG-NleB1 ectopic expression-generated Arg-GlcNAc affinity-enriched peptide using reversed-phase LC-MS
Purified peptides were resuspended in Buffer A* and separated using an in-house packaged 25-cm, 75-μm-inner diameter, 360-μm-outer diameter, 1.7-μm 130-Å CSH C18 (Waters) reversed-phase analytical column with an integrated HF-etched nanoelectrospray ionization tip. Samples were loaded directly onto the column using an ACQUITY UPLC M-Class System (Waters) at 600 nl/min for 20 min with Buffer A (0.1% FA) and eluted at 300 nl/min using a gradient altering the concentration of Buffer B (99.9% ACN, 0.1% FA) from 0 to 32% Buffer B over 90 min, then from 32 to 40% Buffer B in the next 10 min, then increased to 80% Buffer B over an 8-min period, held at 100% Buffer B for 2 min, and then dropped to 0% Buffer B for another 20 min. Reversed phase-separated peptides were infused into a Q-Exactive (Thermo Scientific) mass spectrometer, and data were acquired using data-dependent acquisition. One full precursor scan (resolution, 70,000; 350–2,000 m/z; AGC target of 3 × 106) followed by 10 data-dependent HCD MS-MS events (resolution, 17,500; AGC target of 1 × 105 with a maximum injection time of 200 ms; normalized collision energy of 28 with 20% stepping) were allowed with 35 s dynamic exclusion enabled.
EThcD identification of Arg-GlcNAc affinity-enriched peptides derived from infections using reversed-phase LC-MS
Purified peptides were resuspended in Buffer A* and separated using a two-column chromatography setup comprising a PepMap100 C18 20-mm × 75-μm trap and a PepMap C18 500-mm × 75-μm analytical column (Thermo Scientific). Samples were concentrated onto the trap column at 5 μl/min for 5 min and infused into an Orbitrap FusionTM LumosTM TribridTM mass spectrometer (Thermo Scientific) at 300 nl/min via the analytical column using a Dionex Ultimate 3000 UPLC system (Thermo Scientific). 125-min gradients were run, altering the buffer composition from 1 to 28% Buffer B over 90 min, from 28 to 40% Buffer B over 10 min, and from 40 to 100% Buffer B over 2 min, and then the composition was held at 100% Buffer B for 3 min, dropped to 3% Buffer B over 5 min, and held at 3% Buffer B for another 15 min. The Lumos mass spectrometer was operated in data-dependent mode, automatically switching between the acquisition of a single Orbitrap MS scan (resolution, 120,000) every 3 s and Orbitrap EThcD for each selected precursor (maximum fill time, 100 ms; AGC of 5 × 104 with a resolution of 30,000 for Orbitrap MS-MS scans). For parallel reaction monitoring (32) experiments, the known tryptic Arg-modified sites of TRADD (18) and FADD (17) (UniProt accession numbers Q3U0V2/B2RRZ7 and Q3U0V2, respectively) were monitored using the predicted m/z for the +2 and +3 charge states (supplemental Table 2). Data-independent acquisition was performed by switching between the acquisition of a single Orbitrap MS scan (resolution, 120,000; m/z 300–1500) every 3 s and Orbitrap EThcD for each PRM precursor (maximum fill time, 100 ms; AGC of 5 × 104 with a resolution of 60,000 for Orbitrap MS-MS scans).
Data analysis
Identification of proteins and Arg-glycosylated peptides was accomplished using MaxQuant (v1.5.3.1) (57). Searches were performed against the mouse (UniProt proteome ID UP000000589; Mus musculus; downloaded May 18, 2016; 50,306 entries), E. coli O127:H6 strain E2348/69 (UniProt proteome ID UP000008205; E. coli O127:H6 strain E2348/69/EPEC; downloaded July 28, 2014; 4,595 entries), C. rodentium ICC168 (UniProt proteome ID UP000001889; C. rodentium strain ICC168; downloaded December 12, 2016), or human (UniProt proteome ID UP000005640; Homo sapiens; downloaded October 24, 2013; 84,843 entries) proteome depending on the samples with carbamidomethylation of cysteine set as a fixed modification. Searches were performed with trypsin cleavage specificity allowing two miscleavage events and variable modifications of oxidation of methionine, N-acetylhexosamine addition to arginine (Arg-GlcNAc), and acetylation of protein N termini. The precursor mass tolerance was set to 20 ppm for the first search and 10 ppm for the main search with a maximum false discovery rate of 1.0% set for protein and peptide identifications. To enhance the identification of peptides between samples, the match between runs option was enabled with the precursor match window set to 2 min and an alignment window of 10 min. For label-free quantitation, the MaxLFQ option within MaxQuant (58) was enabled in addition to the requantification module. The resulting outputs were processed within the Perseus (v1.4.0.6) (59) analysis environment to remove reverse matches and common protein contaminants prior to further analysis. For comparisons of relative protein levels within samples, intensity-based absolute quantification was used (60). Glycopeptides identified using HCD fragmentation were manually assessed according to the guidelines of Chen et al. (61) and are provided within the supplemental figures. For EThcD-identified glycopeptides, only Class I (localization >0.75) sites were considered for motif analysis with pLogo (62). For label-free quantification comparisons, missing values were imputed using Perseus with the resulting Pearson correlations and heat maps visualized using R. All mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (63) partner repository with the data set identifier PXD006810.
Author contributions
N. E. S. conceived and designed all MS-based experiments, performed MS sample preparations/enrichments, analyzed the results, and wrote most of the paper. C. G. performed in vitro arginine GlcNAcylation stability studies and aided in the preparation of the manuscript. G. L. P. generated the inducible NleB1 cell lines and aided in the preparation of the manuscript. C. L. K. performed murine infections. A. I. W. and N. A. W. provided mass spectrometry access. J. S. P. performed EPEC infections and aided in the preparation of the manuscript. E. L. H. aided in the design of experiments, financially facilitated the work, and aided in the preparation of the manuscript.
Supplementary Material
This work was supported in part by National Health and Medical Research Council of Australia (NHMRC) Project Grants APP1098826 (to E. L. H.) and APP1100164 (to N. E. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Annotations, Figs. 1–9, and Tables 1–9.
The mass spectrometric raw data and spectral libraries associated with this manuscript are available from ProteomeXchange with the accession number PXD006810.
- EPEC
- enteropathogenic E. coli
- A/E
- attaching and effacing
- LEE
- locus of enterocyte effacement
- nle
- non-LEE
- PTM
- post-translational modification
- FADD
- Fas-associated via death domain
- TRADD
- TNFRSF1A-associated via death domain
- EThcD
- electron-transfer/higher-energy collision dissociation
- PNGase F
- peptide:N-glycosidase F
- SILAC
- stable isotope labeling with amino acids in cell culture
- FA
- formic acid
- ACN
- acetonitrile
- IAP
- immunoaffinity purification
- AGC
- automatic gain control
- HCD
- higher-energy collision dissociation.
References
- 1. Ochoa T. J., and Contreras C. A. (2011) Enteropathogenic Escherichia coli infection in children. Curr. Opin. Infect. Dis. 24, 478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kotloff K. L., Nataro J. P., Blackwelder W. C., Nasrin D., Farag T. H., Panchalingam S., Wu Y., Sow S. O., Sur D., Breiman R. F., Faruque A. S., Zaidi A. K., Saha D., Alonso P. L., Tamboura B., et al. (2013) Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382, 209–222 [DOI] [PubMed] [Google Scholar]
- 3. Moon H. W., Whipp S. C., Argenzio R. A., Levine M. M., and Giannella R. A. (1983) Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41, 1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pearson J. S., Giogha C., Wong Fok Lung T., and Hartland E. L. (2016) The genetics of enteropathogenic Escherichia coli virulence. Annu. Rev. Genet. 50, 493–513 [DOI] [PubMed] [Google Scholar]
- 5. McDaniel T. K., Jarvis K. G., Donnenberg M. S., and Kaper J. B. (1995) A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 92, 1664–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hartland E. L., Batchelor M., Delahay R. M., Hale C., Matthews S., Dougan G., Knutton S., Connerton I., and Frankel G. (1999) Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol. Microbiol. 32, 151–158 [DOI] [PubMed] [Google Scholar]
- 7. Kenny B., DeVinney R., Stein M., Reinscheid D. J., Frey E. A., and Finlay B. B. (1997) Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91, 511–520 [DOI] [PubMed] [Google Scholar]
- 8. Alto N. M., Shao F., Lazar C. S., Brost R. L., Chua G., Mattoo S., McMahon S. A., Ghosh P., Hughes T. R., Boone C., and Dixon J. E. (2006) Identification of a bacterial type III effector family with G protein mimicry functions. Cell 124, 133–145 [DOI] [PubMed] [Google Scholar]
- 9. Simpson N., Shaw R., Crepin V. F., Mundy R., FitzGerald A. J., Cummings N., Straatman-Iwanowska A., Connerton I., Knutton S., and Frankel G. (2006) The enteropathogenic Escherichia coli type III secretion system effector Map binds EBP50/NHERF1: implication for cell signalling and diarrhoea. Mol. Microbiol. 60, 349–363 [DOI] [PubMed] [Google Scholar]
- 10. Kenny B., and Jepson M. (2000) Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell. Microbiol. 2, 579–590 [DOI] [PubMed] [Google Scholar]
- 11. Dean P., and Kenny B. (2009) The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr. Opin. Microbiol. 12, 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang L., Ding X., Cui J., Xu H., Chen J., Gong Y. N., Hu L., Zhou Y., Ge J., Lu Q., Liu L., Chen S., and Shao F. (2011) Cysteine methylation disrupts ubiquitin-chain sensing in NF-κB activation. Nature 481, 204–208 [DOI] [PubMed] [Google Scholar]
- 13. Pearson J. S., Giogha C., Mühlen S., Nachbur U., Pham C. L., Zhang Y., Hildebrand J. M., Oates C. V., Lung T. W., Ingle D., Dagley L. F., Bankovacki A., Petrie E. J., Schroeder G. N., Crepin V. F., et al. (2017) EspL is a bacterial cysteine protease effector that cleaves RHIM proteins to block necroptosis and inflammation. Nat. Microbiol. 2, 16258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pollock G. L., Oates C. V., Giogha C., Wong Fok Lung T., Ong S. Y., Pearson J. S., and Hartland E. L. (2017) Distinct roles of the anti-apoptotic effectors NleB and NleF from enteropathogenic Escherichia coli. Infect. Immun. 85, e01071–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blasche S., Mörtl M., Steuber H., Siszler G., Nisa S., Schwarz F., Lavrik I., Gronewold T. M., Maskos K., Donnenberg M. S., Ullmann D., Uetz P., and Kögl M. (2013) The E. coli effector protein NleF is a caspase inhibitor. PLoS One 8, e58937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pallett M. A., Crepin V. F., Serafini N., Habibzay M., Kotik O., Sanchez-Garrido J., Di Santo J. P., Shenoy A. R., Berger C. N., and Frankel G. (2017) Bacterial virulence factor inhibits caspase-4/11 activation in intestinal epithelial cells. Mucosal Immunol. 10, 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pearson J. S., Giogha C., Ong S. Y., Kennedy C. L., Kelly M., Robinson K. S., Lung T. W., Mansell A., Riedmaier P., Oates C. V., Zaid A., Mühlen S., Crepin V. F., Marches O., Ang C. S., et al. (2013) A type III effector antagonizes death receptor signalling during bacterial gut infection. Nature 501, 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li S., Zhang L., Yao Q., Li L., Dong N., Rong J., Gao W., Ding X., Sun L., Chen X., Chen S., and Shao F. (2013) Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nature 501, 242–246 [DOI] [PubMed] [Google Scholar]
- 19. Gao X., Wang X., Pham T. H., Feuerbacher L. A., Lubos M. L., Huang M., Olsen R., Mushegian A., Slawson C., and Hardwidge P. R. (2013) NleB, a bacterial effector with glycosyltransferase activity, targets GAPDH function to inhibit NF-κB activation. Cell Host Microbe 13, 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao X., Pham T. H., Feuerbacher L. A., Chen K., Hays M. P., Singh G., Rueter C., Hurtado-Guerrero R., and Hardwidge P. R. (2016) Citrobacter rodentium NleB protein inhibits tumor necrosis factor (TNF) receptor-associated factor 3 (TRAF3) ubiquitination to reduce host type I interferon production. J. Biol. Chem. 291, 18232–18238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. El Qaidi S., Chen K., Halim A., Siukstaite L., Rueter C., Hurtado-Guerrero R., Clausen H., and Hardwidge P. R. (2017) NleB/SseK effectors from Citrobacter rodentium, Escherichia coli, and Salmonella enterica display distinct differences in host substrate specificity. J. Biol. Chem. 292, 11423–11430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Law R. J., Law H. T., Scurll J. M., Scholz R., Santos A. S., Shames S. R., Deng W., Croxen M. A., Li Y., de Hoog C. L., van der Heijden J., Foster L. J., Guttman J. A., and Finlay B. B. (2016) Quantitative mass spectrometry identifies novel host binding partners for pathogenic Escherichia coli type III secretion system effectors. J. Proteome Res. 15, 1613–1622 [DOI] [PubMed] [Google Scholar]
- 23. Pan M., Li S., Li X., Shao F., Liu L., and Hu H. G. (2014) Synthesis of and specific antibody generation for glycopeptides with arginine N-GlcNAcylation. Angew. Chem. Int. Ed. Engl. 53, 14517–14521 [DOI] [PubMed] [Google Scholar]
- 24. Wang Z., Udeshi N. D., O'Malley M., Shabanowitz J., Hunt D. F., and Hart G. W. (2010) Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 9, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Halim A., Larsen I. S., Neubert P., Joshi H. J., Petersen B. L., Vakhrushev S. Y., Strahl S., and Clausen H. (2015) Discovery of a nucleocytoplasmic O-mannose glycoproteome in yeast. Proc. Natl. Acad. Sci. U.S.A. 112, 15648–15653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carlson S. M., Moore K. E., Green E. M., Martín G. M., and Gozani O. (2014) Proteome-wide enrichment of proteins modified by lysine methylation. Nat. Protoc. 9, 37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guan K. L., Yu W., Lin Y., Xiong Y., and Zhao S. (2010) Generation of acetyllysine antibodies and affinity enrichment of acetylated peptides. Nat. Protoc. 5, 1583–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Udeshi N. D., Mertins P., Svinkina T., and Carr S. A. (2013) Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Protoc. 8, 1950–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lassak J., Keilhauer E. C., Fürst M., Wuichet K., Gödeke J., Starosta A. L., Chen J. M., Søgaard-Andersen L., Rohr J., Wilson D. N., Häussler S., Mann M., and Jung K. (2015) Arginine-rhamnosylation as new strategy to activate translation elongation factor P. Nat. Chem. Biol. 11, 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang S., Corcilius L., Sharp P. P., Rajkovic A., Ibba M., Parker B. L., and Payne R. J. (2017) Synthesis of rhamnosylated arginine glycopeptides and determination of the glycosidic linkage in bacterial elongation factor P. Chem. Sci. 8, 2296–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mills E., Baruch K., Aviv G., Nitzan M., and Rosenshine I. (2013) Dynamics of the type III secretion system activity of enteropathogenic Escherichia coli. MBio 4, e00303–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peterson A. C., Russell J. D., Bailey D. J., Westphall M. S., and Coon J. J. (2012) Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics 11, 1475–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aquino-Gil M., Pierce A., Perez-Cervera Y., Zenteno E., and Lefebvre T. (2017) OGT: a short overview of an enzyme standing out from usual glycosyltransferases. Biochem. Soc. Trans. 45, 365–370 [DOI] [PubMed] [Google Scholar]
- 34. Strasser A., Jost P. J., and Nagata S. (2009) The many roles of FAS receptor signaling in the immune system. Immunity 30, 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lavrik I. N., and Krammer P. H. (2012) Regulation of CD95/Fas signaling at the DISC. Cell Death Differ. 19, 36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pearson J. S., Riedmaier P., Marchès O., Frankel G., and Hartland E. L. (2011) A type III effector protease NleC from enteropathogenic Escherichia coli targets NF-κB for degradation. Mol. Microbiol. 80, 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burnaevskiy N., Peng T., Reddick L. E., Hang H. C., and Alto N. M. (2015) Myristoylome profiling reveals a concerted mechanism of ARF GTPase deacylation by the bacterial protease IpaJ. Mol. Cell 58, 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yen H., Ooka T., Iguchi A., Hayashi T., Sugimoto N., and Tobe T. (2010) NleC, a type III secretion protease, compromises NF-κB activation by targeting p65/RelA. PLoS Pathog. 6, e1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Newton H. J., Pearson J. S., Badea L., Kelly M., Lucas M., Holloway G., Wagstaff K. M., Dunstone M. A., Sloan J., Whisstock J. C., Kaper J. B., Robins-Browne R. M., Jans D. A., Frankel G., Phillips A. D., et al. (2010) The type III effectors NleE and NleB from enteropathogenic E. coli and OspZ from Shigella block nuclear translocation of NF-κB p65. PLoS Pathog. 6, e1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Günster R. A., Matthews S. A., Holden D. W., and Thurston T. L. (2017) SseK1 and SseK3 T3SS effectors inhibit NF-κB signalling and necroptotic cell death in Salmonella-infected macrophages. Infect. Immun. 85, e00010–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nothaft H., and Szymanski C. M. (2010) Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 [DOI] [PubMed] [Google Scholar]
- 42. Valguarnera E., Kinsella R. L., and Feldman M. F. (2016) Sugar and spice make bacteria not nice: protein glycosylation and its influence in pathogenesis. J. Mol. Biol. 428, 3206–3220 [DOI] [PubMed] [Google Scholar]
- 43. Meijer B. M., Jang S. M., Guerrera I. C., Chhuon C., Lipecka J., Reisacher C., Baleux F., Sansonetti P. J., Muchardt C., and Arbibe L. (2017) Threonine eliminylation by bacterial phosphothreonine lyases rapidly causes cross-linking of mitogen-activated protein kinase (MAPK) in live cells. J. Biol. Chem. 292, 7784–7794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jochim N., Gerhard R., Just I., and Pich A. (2014) Time-resolved cellular effects induced by TcdA from Clostridium difficile. Rapid Commun. Mass Spectrom. 28, 1089–1100 [DOI] [PubMed] [Google Scholar]
- 45. Zeiser J., Gerhard R., Just I., and Pich A. (2013) Substrate specificity of clostridial glucosylating toxins and their function on colonocytes analyzed by proteomics techniques. J. Proteome Res. 12, 1604–1618 [DOI] [PubMed] [Google Scholar]
- 46. Xu H., Yang J., Gao W., Li L., Li P., Zhang L., Gong Y. N., Peng X., Xi J. J., Chen S., Wang F., and Shao F. (2014) Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature 513, 237–241 [DOI] [PubMed] [Google Scholar]
- 47. Humphrey S. J., Azimifar S. B., and Mann M. (2015) High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 33, 990–995 [DOI] [PubMed] [Google Scholar]
- 48. Stoehr G., Schaab C., Graumann J., and Mann M. (2013) A SILAC-based approach identifies substrates of caspase-dependent cleavage upon TRAIL-induced apoptosis. Mol. Cell. Proteomics 12, 1436–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scott N. E., Rogers L. D., Prudova A., Brown N. F., Fortelny N., Overall C. M., and Foster L. J. (2017) Interactome disassembly during apoptosis occurs independent of caspase cleavage. Mol. Syst. Biol. 13, 906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Levine M. M., Bergquist E. J., Nalin D. R., Waterman D. H., Hornick R. B., Young C. R., and Sotman S. (1978) Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet 1, 1119–1122 [DOI] [PubMed] [Google Scholar]
- 51. Wong Fok Lung T., Giogha C., Creuzburg K., Ong S. Y., Pollock G. L., Zhang Y., Fung K. Y., Pearson J. S., and Hartland E. L. (2016) Mutagenesis and functional analysis of the bacterial arginine glycosyltransferase effector NleB1 from enteropathogenic Escherichia coli. Infect. Immun. 84, 1346–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mundy R., Pickard D., Wilson R. K., Simmons C. P., Dougan G., and Frankel G. (2003) Identification of a novel type IV pilus gene cluster required for gastrointestinal colonization of Citrobacter rodentium. Mol. Microbiol. 48, 795–809 [DOI] [PubMed] [Google Scholar]
- 53. Scott N. E., Parker B. L., Connolly A. M., Paulech J., Edwards A. V., Crossett B., Falconer L., Kolarich D., Djordjevic S. P., Højrup P., Packer N. H., Larsen M. R., and Cordwell S. J. (2011) Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10, M000031–MCP000201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Udeshi N. D., Svinkina T., Mertins P., Kuhn E., Mani D. R., Qiao J. W., and Carr S. A. (2013) Refined preparation and use of anti-diglycine remnant (K-ϵ-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics 12, 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ishihama Y., Rappsilber J., and Mann M. (2006) Modular stop and go extraction tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J. Proteome Res. 5, 988–994 [DOI] [PubMed] [Google Scholar]
- 56. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 57. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 58. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 60. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 61. Chen Y., Zhang J., Xing G., and Zhao Y. (2009) Mascot-derived false positive peptide identifications revealed by manual analysis of tandem mass spectra. J. Proteome Res. 8, 3141–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. O'Shea J. P., Chou M. F., Quader S. A., Ryan J. K., Church G. M., and Schwartz D. (2013) pLogo: a probabilistic approach to visualizing sequence motifs. Nat. Methods 10, 1211–1212 [DOI] [PubMed] [Google Scholar]
- 63. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.