Abstract
cis-Prenyltransferases (cis-PTs) constitute a large family of enzymes conserved during evolution and present in all domains of life. In eukaryotes and archaea, cis-PT is the first enzyme committed to the synthesis of dolichyl phosphate, an obligate lipid carrier in protein glycosylation reactions. The homodimeric bacterial enzyme, undecaprenyl diphosphate synthase, generates 11 isoprene units and has been structurally and mechanistically characterized in great detail. Recently, we discovered that unlike undecaprenyl diphosphate synthase, mammalian cis-PT is a heteromer consisting of NgBR (Nus1) and hCIT (dehydrodolichol diphosphate synthase) subunits, and this composition has been confirmed in plants and fungal cis-PTs. Here, we establish the first purification system for heteromeric cis-PT and show that both NgBR and hCIT subunits function in catalysis and substrate binding. Finally, we identified a critical RXG sequence in the C-terminal tail of NgBR that is conserved and essential for enzyme activity across phyla. In summary, our findings show that eukaryotic cis-PT is composed of the NgBR and hCIT subunits. The strong conservation of the RXG motif among NgBR orthologs indicates that this subunit is critical for the synthesis of polyprenol diphosphates and cellular function.
Keywords: enzyme, enzyme catalysis, glycosylation, isoprenoid, lipid, cis-prenyltransferase, dolichol, prenyl
Introduction
Dolichyl phosphate is an obligate lipid carrier for protein N-glycosylation, O-mannosylation, C-mannosylation, and glycosylphosphatidylinositol-anchor synthesis in eukaryotic cells and undecaprenyl phosphate is essential for peptidoglycan biosynthesis in bacteria. cis-Prenyltransferase (cis-PT)2 is the rate-limiting enzyme committed to dolichyl phosphate biosynthesis in Eukaryotes and Archaea, as well as undecaprenyl phosphate biosynthesis in Eubacteria (1, 2). Both eukaryotic and prokaryotic cis-PTs belong to a large protein family, well conserved during evolution (1), and the cis-PT family was identified among the 355 protein families that trace to the last universal common ancestor of all cells (LUCA, or the progenote) by phylogenetic criteria (3).
The bacterial enzyme, undecaprenyl diphosphate synthase (UPPS) is a homodimeric enzyme that catalyzes chain elongation of farnesyl diphosphate (FPP) by sequential reactions with eight isopentenyl diphosphate (IPP) molecules. UPPS has been structurally and mechanistically characterized in great detail (4, 5); however, the eukaryotic enzyme has not been purified to date. Our group discovered that unlike UPPS, mammalian and fungal cis-PT is heteromeric complex consisting of NgBR (Nus1) and hCIT (dehydrodolichol diphosphate synthase) subunits in human cells (6, 7), and these findings were confirmed for a number of plant cis-PTs (8–12), demonstrating a major difference in the composition of cis-PT activity in prokaryotes and eukaryotes. Moreover, loss of function mutations identified in patients via exome sequencing in either NgBR or hCIT causes a congenital glycosylation disorder and results in severe clinical manifestations including cognitive defects and retinitis pigmentosa (6, 13–16), and microdeletions within NgBR locus are linked to pediatric epilepsy (17, 18). Further phylogenetic analysis of NgBR and UPPS suggests that a C-terminal motif important for mammalian and fungal cis-PT is shared by the single-subunit cis-PT UPPS (1).
In the rubber plant (Hevea brasiliensis), a primary source of natural rubber, an NgBR ortholog is essential for rubber synthase activity (8), confirming previous work in rubber producing lettuce (10) and dandelions (11). In these papers, the enzymatic activity of the rubber synthase complex is inferred to be solely attributable to the hCIT component of complex. Moreover, the NgBR ortholog is proposed to serve as a scaffold necessary for targeting the hCIT ortholog to the rubber particle monolayer but not critical for catalysis. In the present study, we have purified and characterized for the first time a human heteromeric cis-PT complex composed of NgBR and hCIT subunits. Biochemical characterization of purified WT and various mutants of the NgBR–hCIT complex shows that both subunits contribute to catalytic activity. Furthermore, we provide evidence that a conserved C-terminal motif RXG motif is critical for enzyme catalysis in both two-component and single-subunit cis-PTs.
Results
Protein sequence alignment of cis-prenyltransferase homology domain bearing proteins reveals similarity between prokaryotic and eukaryotic cis-PTs
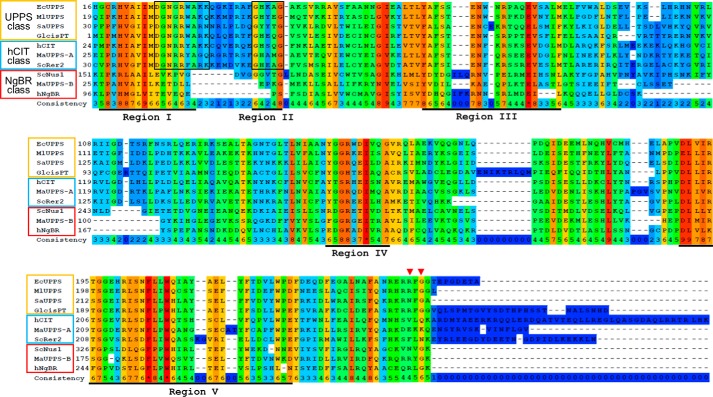
Sequential alignment of nonredundant homomeric cis-PTs from all three domains of life together with heteromeric orthologs define the position of cis-PT homology domain among NgBR orthologs (Fig. 1). Analysis of the architecture of representative cis-PT homology domains revealed a number of characteristic features for each of the group (single-subunit orthologs, hCIT orthologs, and NgBR orthologs) separately or shared between two groups (1, 4, 7, 10, 19). Only four of the previously identified five conserved regions (4) are present in all three groups. The NgBR group lacks region II and has highly degenerate regions I and III. Among other conserved residues, the NgBR class is missing a highly conserved stretch of the six amino acid residues involved in FPP binding and catalysis in region I (boxed region) found in UPPS and hCIT. Regions IV and V, including the predicted dimer interface based on the structural data obtain for the UPPS of Micrococcus luteus and Escherichia coli (20, 21), are well conserved among all three groups. Finally, the NgBR group shares with homomeric enzymes, but not with the hCIT group, an RXG C-terminal conserved motif (see red arrowheads) (1, 6), with Gly being absolutely conserved in EcUPPS and NgBR classes. The Arg residue is substituted with Asn in SaUPPS and a majority of plant and fungal NgBR orthologs (6, 8–12, 22). In most cases, X is any nonpolar amino acid however putative Hypocreales NgBR orthologs, as well as UPPS of some Trypanosomatidae have positively charged His or Arg at this position. Meta-analysis of available experimental data concerning cis-PTs suggests that both the catalytic motif of region I and the C-terminal RXG motif are indispensable for enzymatic activity being part of the same subunit or separated between hCIT (catalytic motif) and NgBR (RXG) class of cis-PTs proteins. To directly examine the contribution of each subunit to enzymatic activity of the heteromeric hCIT–NgBR complex, a purification scheme of the complex was undertaken.
Figure 1.
Multiple alignment of the proteins bearing cis-PT homology domain. Proteins represented in the figure are single subunit cis-PTs: EcUPPS (E. coli, GenBankTM accession no. P60472), MlUPPS (M. luteus; GenBankTM accession no. BAA31993.1), SaUPPS (Sulfolobus acidocaldarius; GenBankTM accession no. WP_011277635.1), and Glcis-PT (G. lamblia; GenBankTM accession no. XP_001709868.1); orthologs of hCIT cis-PT subunit: hCIT (human, GenBankTM accession no. BAB14439), ScRer2 (S. cerevisiae, GenBankTM accession no. P35196), and MaUPPS-A (M. acetovorans, GenBankTM accession no. AAM07075.1); and orthologs of NgBR cis-PT subunit: NgBR (human, GenBankTM accession no. NP_612468), ScNus1 (S. cerevisiae, GenBankTM accession no. NP_010088), and MaUPPS-B (M. acetovorans, GenBankTM accession no. WP_011024281.1). The conservation scoring is performed by PRALINE. The scoring scheme works from 0 for the least conserved alignment position, up to 10 (*) for the most conserved alignment position. Positions of five conserved regions originally described among cis-PTs are underlined. Residues involved in catalysis and substrate binding conserved between hCIT orthologs and single subunit cis-PTs are boxed. Red triangles indicate the positions of conserved arginine/asparagine and glycine in the C-terminal RXG motif of NgBR orthologs and homomeric cis-PTs.
Purification and biochemical characterization of human cis-PT
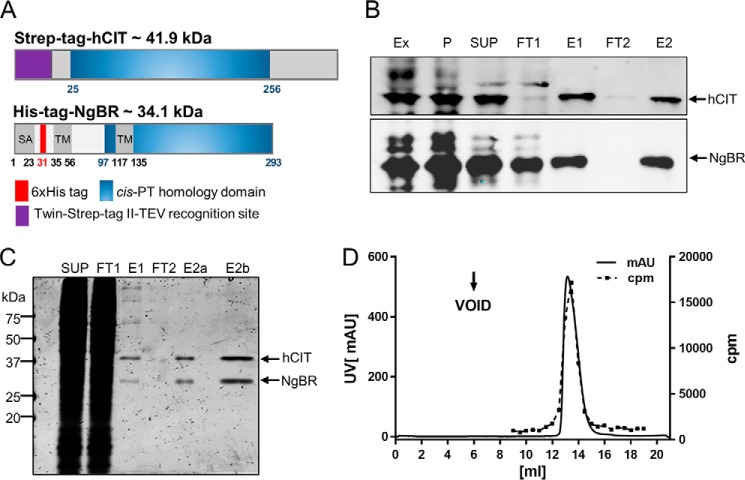
Optimal expression and tagging strategy was first established using a previously described expression system in a triple knock-out strain lacking nus1Δ, rer2Δ, srt1Δ in Saccharomyces cerevisiae (6). Cell survival was ensured because of expression of Glcis-PT on the URA3 plasmid, and cells were co-transformed with the LEU2 and MET15 plasmids bearing WT or epitope-tagged versions of hCIT and NgBR, respectively. The yeast cells were streaked onto complete plates or synthetic complete medium containing 1% 5-fluoroorotic acid (FOA). Because Ura3 protein, which is expressed from the URA3 marker present in the plasmids, converts FOA to toxic 5-fluorouracil, the survival of the yeast cells on the FOA plate depends on the functionality of the expressed NgBR–hCIT complex. hCIT isoform 1 (UniProtKB no. Q86SQ9-1) and isoform 2, (UniProtKB no. Q86SQ9-2), but not isoform 3 (UniProtKB no. Q86SQ9-3), supported the growth of yeast when co-expressed with NgBR in the triple delete strain (data not shown). N-terminal tagging, but not C-terminal tagging, of hCIT generated a construct indistinguishable from WT, non-tagged hCIT in yeast. Both N- and C-terminal tagging of NgBR reduced its stability and activity when expressed in yeast; therefore a His6 tag was placed internally after Gly31 between the putative signal anchor and TM1 (Fig. 2A). This tag did not affect the growth phenotype and cis-PT activity in yeast co-expressing N-terminally Strep-tagged hCIT.
Figure 2.
Purification of NgBR–hCIT cis-PT complex. A, epitope-tagged NgBR and hCIT were transiently expressed in Expi293 cells from bicistronic vector and purified as described in the text. SA, putative signal anchor; TM, putative transmembrane domain. B, representative Western blot monitoring purification of NgBR–hCIT complex. Ex, total crude extract, P, 200,000 × g membrane pellet; SUP, 200,000 × g supernatant after Triton X-100 solubilization of the membrane fraction; FT1 and FT2, flow-through from Strep-Tactin and nickel column, respectively; E1, elution from the Strep-Tactin; E2, final elution from the nickel column. C, Coomassie-stained SDS/PAGE showing SUP, FT1, E1, two (E2a), and eight times concentrated final fraction (E2b). D, size-exclusion chromatography profile of purified NgBR–hCIT cis-PT complex. UV absorption at 254 nm was measured as a readout for protein elution (left y axis, black line). Incorporation of 14C-labeled IPP into organic fraction was measured as a readout for cis-PT activity (right y axis, red line).
Next, the cDNAs encoding His6-NgBR and Strep-tagged-hCIT were cloned into an internal ribosome entry site (IRES)-containing bicistronic vector, allowing the simultaneous expression of two proteins. This was necessary based on our previous work demonstrating that co-translation of both subunits was required for activity in an in vitro translation system (6). To generate protein for purification, the construct was transiently transfected into Expi293F cells and cells collected 72 h later by centrifugation. The pellet was washed with PBS and lysed in detergent free buffer (Ex). To prefractionate cell extracts prior to affinity chromatography, crude membrane extracts were subjected to ultracentrifugation, pellet (P) was solubilized by Dounce homogenization in the presence with 0.5% Triton X-100, and solubilized protein was cleared by a second ultracentrifugation step (SUP). N-terminally Strep-tagged hCIT and internally tagged His6–NgBR complex were purified using a dual affinity purification scheme (Fig. 2B; see Table 1 for enrichment). Each step of purification was monitored by Western blotting and measurement of cis-PT activity, and this scheme enriched the specific activity of the enzyme 1,700-fold over the starting material. As judged by SDS-PAGE and Coomassie staining of each fraction, the hCIT–NgBR complex was ∼95% pure (Fig. 2C), and the purified heteromeric complex ran as monodispersed peak on size-exclusion chromatography (Fig. 2D, solid line) that tracked with cis-PT activity (Fig. 2D, dashed line).
Table 1.
Purification of human cis-PT from Expi 293 cells
Mean ± S.D. values of three replicates are shown for specific activity.
| Step | Total protein | Total activity | Specific activity | Yield | Purification fold |
|---|---|---|---|---|---|
| mg | μmol | μmol/h/mg protein | % | ||
| Crude extract | 240.24 | 6.32 | 0.023 (±0.003) | 100 | |
| Membrane pellet | 140.7 | 2.20 | 0.016 (±0.002) | 34.8 | 0.7 |
| SUP | 71.45 | 2.14 | 0.030 (±0.001) | 33.9 | 1.3 |
| S-Tactin | 0.06 | 1.45 | 24.13 (±4.91) | 22.9 | 1049 |
| Nickel column | 0.024 | 0.98 | 40.97 (±9.39) | 15.5 | 1781 |
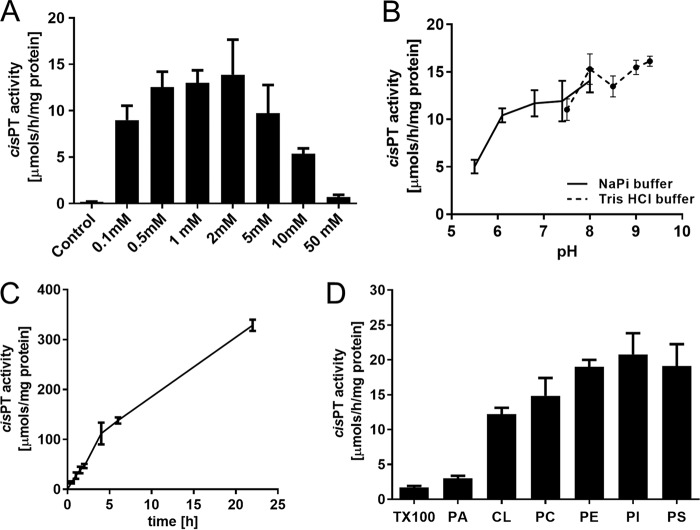
The purified hCIT–NgBR complex required Mg2+ for its activity consistent with previous studies in bacterial and eukaryotic enzymes (23–29) with maximum activity attained at 0.5–2.0 mm MgCl2. (Fig. 3A). The purified enzyme had a broad pH optimum (Fig. 3B) (24) and was highly stable, demonstrating that linear cis-PT activity occurred over 24 h at 37 °C (Fig. 3C).
Figure 3.
Biochemical properties of purified human cis-PT. cis-PT activity was measured using purified hcis-PT complex as described under “Experimental Procedures” unless otherwise stated. A, optimal concentration of MgCl2. cis-PT activity was measured in the presence of 5 mm EDTA (to chelate MgCl2 in the enzyme storage buffer) or 0.1, 0.5, 1, 2, 5, 10, and 50 mm MgCl2. B, pH dependence of cis-PT activity. cis-PT activity was measure in the 50 mm sodium phosphate buffer (pH 5.5, 6.1, 7.4, or 8.0) or 50 mm Tris-HCl (pH 7.5, 8.0, 8.5, 9.0, or 9.3). C, time dependence of cis-PT activity. D, phospholipids stimulate cis-PT activity. cis-PT activity was measured in the buffer containing 0.35% (w/v) Triton X-100, 1% (w/v) phosphatidic acid (PA), 1% (w/v) cardiolipin (CL), 1% (w/v) phosphatidylcholine (PC), 1% (w/v) phosphatidylethanolamine (PE), 1% (w/v) phosphatidylinositol (PI), or 1% (w/v) phosphatidylserine (PS). The data are means ± S.D. values of three technical replicates.
It was previously reported that exogenous lipids stimulate the activity of semipurified M. luteus UPPS (30) and phospholipids may modulate the functional properties of several membrane proteins (31). To test the influence of lipids on human cis-PT activity, Triton X-100 was removed from the enzyme preparation by buffer exchange, and activity was measured upon the addition of Triton X-100 or different phospholipids at concentration above their critical micellar concentration. As seen in Fig. 3D, phosphatidic acid modestly increased enzymatic activity (1.8-fold), whereas other phospholipids had a marked effect in activating the enzyme. Cardiolipin increased cis-PT activity 7-fold, and phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, and phosphatidylserine increased the activity by ∼8–12-fold, suggesting that the lipid environment strongly impacts cis-PT activity.
Phenotypic analysis and biochemical characterization of the purified proteins support importance of the RXG motif
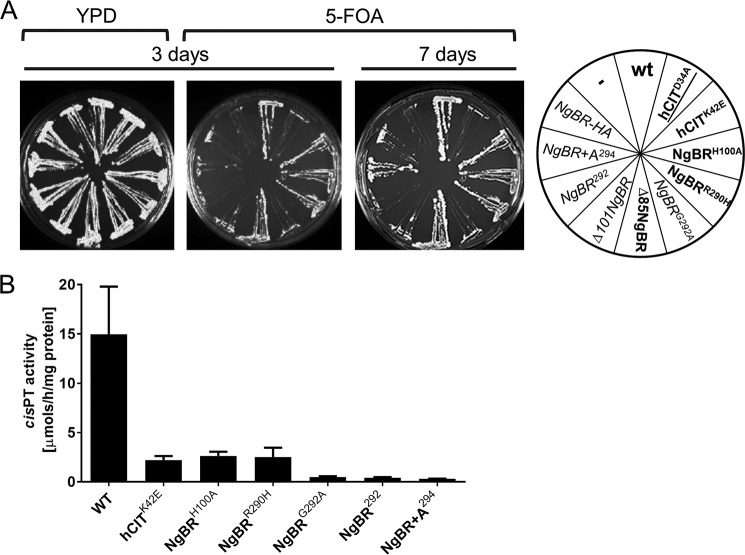
To examine the role of the conserved RXG motif in hcis-PTenzymatic activity and to confirm the predicted position of the cis-PT homology domain in NgBR, we performed phenotypic analysis of a series of truncation and point mutants of NgBR or hCIT using the yeast triple deletion strain (rer2Δ, srt1Δ, and nus1Δ) mentioned above. As seen in Fig. 4A, no growth defects were observed with complexes containing the following mutations compared with WT hcis-PT–NgBRH100A (His is the first amino acid in region I and is highly conserved among proteins bearing cis-PT homology domain), NgBRR290H (a human mutation causing a glycosylation disorder), Δ85-NgBR (a truncated protein lacking the first 84 amino acids including TM domain conserved across eukaryotic orthologs of NgBR), and hCITK42E (a human mutation of hCIT that causes retinitis pigmentosa). Based on our previous work (6), this results suggest that at least 20% residual enzymatic activity of above mutants is necessary to generate sufficient amount of dolichol to support intact glycosylation and survival in yeast. In addition to the above variants, we sought to test the essentiality of the conserved Asp34 in hCIT, which corresponds to the strictly conserved Asp26 of EcUPPS involved in catalysis (20, 23, 32). As seen in Fig. 4A, the hCITD34A–NgBR complex did not support the growth of the triple knock-out yeast cells, confirming its predicted role in enzymatic activity of hcis-PT. Furthermore, cells expressing a truncated Δ101-NgBR (lacking the first 100 amino acids) had a severe growth defect, suggesting that the first conserved region in NgBR, despite its degeneration, is important for function. This finding is in line with the recent structural analysis of the plant Z,Z-farnesyl diphosphate synthase (zFPPS), suggesting that the N terminus of the enzyme might extend to the active site of the neighboring monomer near the C terminus (33).
Figure 4.
Characterization of human cis-PT mutants. A, the rer2Δ, srt1Δ, nus1Δ triple deletion strain expressing G. lamblia cis-PT from URA3 plasmid was co-transformed with the LEU2 and MET15 plasmids bearing wild-type or mutated variant of hCIT and NgBR as indicated. The cells were streaked onto complete plates (YPD) or synthetic complete medium containing 1% FOA. The Ura3 protein, which is expressed from the URA3 marker present in the plasmids, converts FOA to toxic 5-fluorouracil. The growth of cells was monitored over time to assess phenotypic differences. The combination of alleles affecting the growth is indicated in italic, and the combination not supporting the growth is marked in red. B, cis-PT activity was measured using purified WT or mutant hcis-PT complex as described under “Experimental Procedures.” The values are the means ± S.D. of three to nine independent measurements from at least two independent isolations of enzyme except for hCIT–NgBR+A294 and hCIT–NgBR292 performed on one batch.
Finally, the conservation of the RXG motif was confirmed by a number of modifications at the C terminus of NgBR. Deletion of the C-terminal K293 (NgBR292), the addition of Ala to NgBR (NgBR+A294), HA tagging (NgBR-HA), or NgBRG292A substitutions all reduced growth documenting the importance of the last four residues in NgBR for function (Fig. 4A). Next, we compared the steady-state activities of purified mutants able to support the growth of yeast compared with the purified WT enzyme. We did not include the N-terminal truncation variants of NgBR because introducing the epitope tag after the predicted TM1 (35–56 amino acids) rendered the enzyme inactive in yeast. As shown in Fig. 4B, all mutants analyzed displayed lower enzymatic activity compared with WT enzyme. In addition, mutations that strongly impacted growth in yeast (NgBRG292A, NgBR292, and NgBR+A294) had the lowest enzymatic activity under varying concentrations of co-substrates FPP and IPP (Fig. 5 and Table 2). These data support the important role of C terminus, in particular Gly292 in cis-PT activity.
Figure 5.
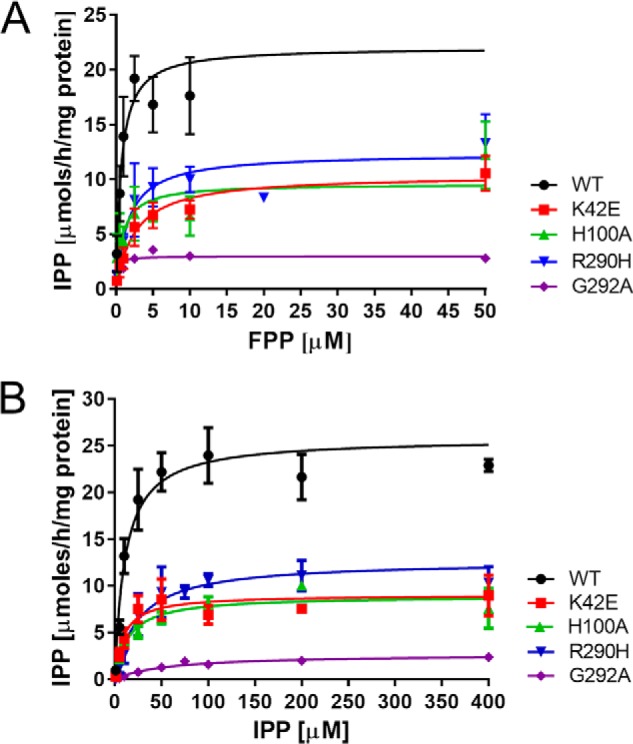
Michaelis–Menten plot of hcis-PT kinetics. A and B, the measurements of Km for FPP (A) and IPP (B). Each data points represent three to eight independent measurements (n = 1 for measurement of Km for FPP of hCIT–NgBR G292A enzyme, and n = 2 for other samples). The details are provided under “Experimental Procedures.”
Table 2.
Kinetic parameters of human cis-PTs
Mean ± S.D. values of three replicates are shown.
|
Km |
kcat | ||
|---|---|---|---|
| IPP | FPP | ||
| μm | μm | 1/s | |
| hcis-PT, WT | 11.1 (±1.3) | 0.68 (±0.14) | 0.58 (±0.02) |
| hcis-PT, hCIT K42E | 9.4 (±2.1) | 2.59 (±0.43) | 0.12 (±0.03) |
| hcis-PT, NgBR H100A | 14.9 (±2.0) | 0.89 (±0.32) | 0.16 (±0.01) |
| hcis-PT, NgBR R290H | 24.8 (±4.4) | 1.64 (±0.32) | 0.12 (±0.01) |
| hcis-PT, NgBR G292A | 57.7 (±7.4) | 0.19 (±0.06) | 0.05 (±0.002) |
To further characterize hcis-PT and to elucidate the impact of mutations on catalysis and substrate binding, steady-state kinetic parameters were measured for the complexes containing the WT subunits, hCIT–NgBR, hCITK42E/NgBR, hCIT–NgBRH100A, hCIT–NgBRR290H, and hCIT–NgBRG292A (Table 2 and Fig. 5, A and B). The K42E substitution of hCIT reduced kcat and increased the Km for FPP but not the affinity for IPP. NgBRH100A and NgBRR290H decreased the kcat ∼4-fold compared with WT enzyme. However, NgBRH100A did not exhibit marked changes in affinity for IPP or FPP, whereas NgBRR290H had moderately but significantly reduced affinities for IPP. Substitution of G292A in NgBR, which markedly reduced the growth phenotype in yeast, decreased the catalytic activity of the enzyme to an even greater extent with a kcat that is 10-fold lower than that of the WT enzyme. In this case, there were modest (5-fold) but significant differences in the Km for IPP and a slight decrease in the Km for FPP implying that C-terminal mutations in NgBR markedly affected enzymatic activity and IPP binding.
The RXG motif in homomeric EcUPPS and Glcis-PT is critical for cis-PT function and enzymatic activity
Comparison of the primary amino acid sequences of single- and two-component enzymes reveals that both classes share a conserved RXG C-terminal motif (Fig. 1). Based on the crystal structure of UPPS, Arg242 in the RXG motif is involved in the binding of the diphosphate group of IPP (32). Furthermore, the role of C terminus in IPP binding and catalysis is supported by structural information obtained for decaprenyl diphosphate synthase of Mycobacterium tuberculosis and recent studies on zFPPS of Solanum habrochaites (33, 34), implicating the C terminus in IPP binding. To verify the importance of the RXG motif in homomeric cis-PTs, we assayed growth of the triple deletion yeast strain, transformed with WT or mutant forms of homomeric cis-PT, namely EcUPPS (E. coli UPPS) and Glcis-PT (Giardia lamblia UPPS). The cells were transformed with LEU2 plasmids bearing WT, R242H, and G244A of EcUPPS or WT, R236H, and G238A of Glcis-PT, and the growth was monitored over 7 days. As seen in Fig. 6A, WT E. coli and G. lamblia cis-PTs exhibited normal growth phenotypes on YPD and FOA plates and Arg to His mutants because each construct had similar phenotypes compared with WT transformed cells. In contrast, UPPSG244A and Glcis-PT G238A mutants failed to grow on FOA plates, demonstrating an essential role of G in the RXG motif in homomeric cis-PT function. In addition, cis-PT activity was measured using purified WT, R236H, and G238A mutants of Glcis-PT. Mutation of both residues reduced activity, with Glcis-PTR236H having ∼5 times lower activity and Glcis-PTG238A being virtually inactive (Fig. 6B) compared with WT.
Figure 6.
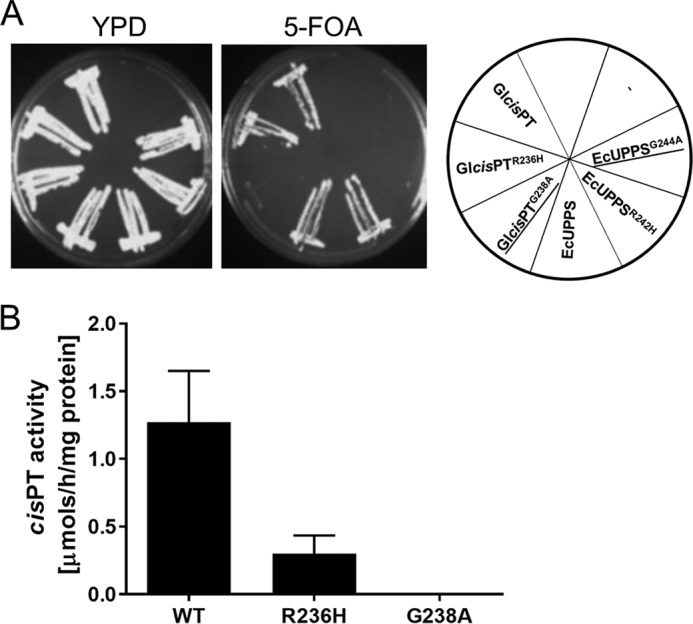
The RXG motif of EcUPPS and Glcis-PT is essential for growth and activity. A, the rer2Δ, srt1Δ, nus1Δ triple deletion strain expressing G. lamblia cis-PT from URA3 plasmid was transformed with the LEU2 plasmid bearing WT or mutants ORFs for Glcis-PT or EcUPPS. As a negative control, cells were transformed with empty vector. The mutated genes G244A and G238A did not supporting the growth and are underlined. B, cis-PT activity was measured using purified wild-type or mutant Glcis-PT. The data are means ± S.D. values of three technical replicates.
Presence of heteromeric cis-PT in Methanosarcina acetovorans supports the importance of the RXG motif
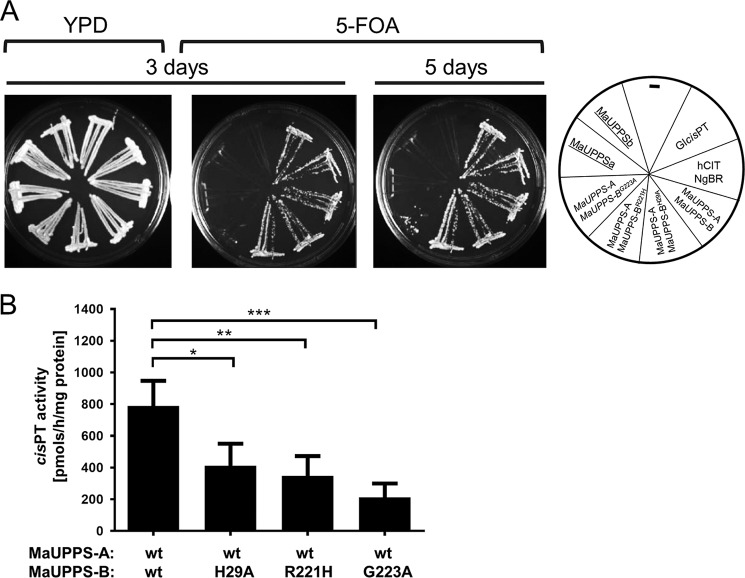
Heteromeric cis-PTs was predicted in subgroup of Euryarchaeota, based on the fact that Halomebacteria and Archaeoglobaceae have at least two UPPS orthologs: a putative cis-PT that is closely related to single subunit Archea enzymes but lacks the C-terminal RXG motif and a NgBR/Nus1-like protein (1, 35) (Fig. 1). To test the conservation of this motif, we cloned putative undecaprenyl diphosphate synthase of M. acetovorans consisting of MaUPPS-A (MA3723, hCIT group) and MaUPPS-B (MA4402, NgBR/Nus1 group). The triple deletion strain bearing Glcis-PT on the URA3 plasmid was co-transformed with the LEU2 and MET15 plasmids expressing wild-type or mutated variants of MaUPPS-A and MaUPPS-B, respectively. The growth of yeast cells on the FOA plates was monitored over 5 days. As seen in Fig. 7A, co-expression of MaUPPS-A and MaUPPS-A is indispensable to support cell growth, but neither MaUPPS-A nor MaUPPS-B is sufficient. Similarly, as it was observed in case of NgBR, mutation of His29 (corresponding to His100 of NgBR) and Arg221 (corresponding to Arg290 of NgBR) in MaUPPS-B does not impair growth, but MaUPPS-BG223A substitution caused a severe growth delay. Quantification of the cis-PT activity reveals that each of the analyzed mutation inhibits enzyme activity with most profound effect of MaUPPS-B G223A substitution (Fig. 7B).
Figure 7.
Phenotypical and biochemical analysis of heteromeric MaUPPS supports predicted length of cis-PT domain of NgBR and importance of C-terminal RXG motif. A, the rer2Δ, srt1Δ, nus1Δ triple deletion strain expressing G. lamblia cis-PT from URA3 plasmid was co-transformed with the LEU2 and MET15 plasmids bearing wild-type or mutated variant of MaUPPS-A (hCIT/Rer2/Srt1 ortholog) and MaUPPS-B (NgBR/Nus1 ortholog) as indicated. Cells transformed with empty plasmid were used as negative control, and cells transformed with LEU2 Glcis-PT or co-transformed with hCIT (LEU2) and NgBR (MET15) were used as positive control. The cells were streaked onto YPD or synthetic complete medium containing 1% FOA. The growth of cells was monitored overtime to assess phenotypic differences. The combination of alleles affecting the growth is indicated in italics, and the combination not supporting the growth is underlined. B, cis-PT activity was measured using S. cerevisiae rer2Δ, srt1Δ, nus1Δ membrane fractions expressing WT or mutated MaUPPs subunits as indicated. The data are means ± S.D. values of four or five replicates in two independent experiments.
Discussion
Here we demonstrate that two subunits of the human cis-PT, NgBR and hCIT, are required to form a functional enzyme. The purification of the cis-PT complex supports prior work showing that co-translation of both subunits is required for polyprenol synthesis in vitro and for survival in yeast lacking orthologs of each component (6). Moreover, the catalytic Asp34 in hCIT and RXG motif in NgBR, both conserved in homomeric cis-PT such as UPPS, are critical for catalytic activity of the complex. These data highlight the evolutionary conservation of essential elements required for cis-PT function throughout all walks of life.
Eukaryotic cis-PTs were initially presumed to be homomeric based on detailed studies in undecaprenyl diphosphate synthases of E. coli and M. luteus (4, 5, 36–38). Recent work by us (6, 7, 19) and others have shown the essential role of NgBR (and its orthologs including Nus1 in S. cerevisiae) and hCIT (and its orthologs including Rer2 and Srt1 in S. cerevisiae) as both being required for cis-PT activity and polyprenol synthesis (6, 8, 10–12). The expression of only hCIT or only NgBR does not support growth in the nus1Δ, rer2Δ, srt1Δ strain of S. cerevisiae, and in vitro translation of either subunit is not catalytically active (6). At first glance, this may appear to be in conflict with previous reports showing that eukaryotic hCIT orthologs heterologously expressed in S. cerevisiae were active without co-expression of NgBR/Nus1 orthologs. However, this can be explained by the presence of the endogenous Nus1 in S. cerevisiae. Furthermore, a number of hCIT orthologs in plants and protists that were studied in yeast are single-subunit enzymes that have been acquired through horizontal gene transfer and are more closely related to EcUPPS than to hCIT (1, 28, 39–43). This complexity can be overcome and simplified by complementation experiments in the nus1Δ, rer2Δ, srt1Δ strain as performed previously (6, 11), where it is clear that both NgBR and hCIT or a single-subunit enzyme is required.
Previous experiments have shown that co-expression of NgBR with hCIT results in their interaction and stabilization based on co-immunoprecipitation experiments (6, 7). Whether NgBR/Nus1 serves as an accessory subunit for docking the complex to the ER membrane or is a structural component contributing to catalysis and substrate binding is still a matter of debate. Experiments showing that 1) the loss of NgBR does not lead to changes in the relative amount of hCIT associated with ER membrane fractions (7) and 2) a mutation in the C terminus of NgBR causes a congenital disorder of glycosylation syndrome because of defective dolichol synthesis (6) support the role of NgBR as a structural component of hcis-PT involved in catalysis. The idea of NgBR being solely a docking component was recently reported by Yamashita et al. (8), who identified a three-component system composed of HRBP (a NgBR ortholog), HRT (a hCIT ortholog) and rubber elongation factor (44), all of which were required for long-chain prenol or natural rubber synthesis when reconstituted into washed rubber particles. Sequential alignment of nonredundant proteins bearing cis-PT homology domains presented in this paper enabled the observation of a highly conserved C-terminal RXG motif shared between NgBR/Nus1 and the UPPS group of enzymes. Furthermore, meta-analysis of available experimental data concerning cis-PT suggests that the C-terminal RXG motif is indispensable for enzymatic activity. To further elucidate the role of NgBR in hcis-PT activity, we successfully purified the WT NgBR–hCIT complex and its mutated versions to homogeneity and biochemically characterized the proteins. Mutations in either subunit of hcis-PT reduces enzymatic activity in comparison to the WT enzyme, and these results are in agreement with the predicted function of the analyzed residue in homomeric cis-PTs.
The first observation supporting a functional role of the extreme C terminus of NgBR regulating enzyme activity is derived from patients harboring an R290H mutation in NgBR, which reduces activity in vivo and in vitro (6) and now seen with the purified complex (Table 2) by 70–80%. In addition, epitope tagging the C terminus of NgBR reduces activity (Fig. 4), and this has been previously shown in the cis-prenyltransferase (CPT)-like NgBR ortholog of Lactuca sativa (10). Crystallographic data obtained for homomeric EcUPPS show that Arg242 in the RXG motif is involved in Mg+2-IPP binding (32). In addition, the role of the C terminus in IPP binding and catalysis is supported by structural information obtained for decaprenyl diphosphate synthase of M. tuberculosis (34) and zFPPS (33). In the structure of diphosphate synthase, the extreme C terminus of one monomer interacts with the active site of the other subunit. Based on our biochemical data and the available structural information for homomeric cis-PTs, it is likely that Arg290 in the RXG motif of NgBR is involved in IPP binding, and the flexible nature of glycine in the motif may permit conformational changes to seal the active site cavity. Collectively, these data imply that both subunits contribute to enzymatic activity, and the C-terminal tail of NgBR regulates aspects of cis-PT catalytic activity.
The general function of NgBR orthologs as a cis-PT subunit is further supported by the heteromeric cis-PT from M. acetovorans. MaUPPS-B (NgBR ortholog) is missing any predicted TM domains, and yet the MaUPPS-A–MaUPPS-B complex is indispensable for supporting the growth of triple deletion strain of S. cerevisiae. Because the mutations in the conserved region of MaUPPS-B have similar impact on MaUPPS activity as those in distantly related hcis-PT, we postulate a common mechanism for both enzymes.
In summary, this study strongly advances the concept that eukaryotic cis-PT is composed of two subunits, NgBR and hCIT, and this two component system is conserved in M. acetovorans. Interestingly, conservation of the RXG motif across phyla implicates a catalytic role of NgBR orthologs in the synthesis of polyprenol diphosphates critical for cellular function. Future structural studies on the heteromeric NgBR–hCIT complex will permit a deeper mechanistic understanding of how the C terminus regulates substrate binding and catalysis.
Experimental procedures
Materials
Unless otherwise stated, all reagents were of analytical grade and purchased from Sigma-Aldrich, Thermo Fisher Scientific, and Zymo Research (Irvine, CA). Restriction enzymes were from New England Biolabs (Ipswich, MA). [1-14C]IPP (50 mCi/mmol) was purchased from American Radiolabeled Chemicals (St. Luis, MO). Reverse phase thin layer chromatography (RP18-HTLC) plates were from Merck. Undecaprenol was obtained from the isoprenoids collection from the Institute of Biochemistry and Biophysics (Polish Academy of Science). Primary antibodies used in this study include α-NgBR (Abcam, ab168351), α-dehydrodolichol diphosphate synthase (Sigma, SAB2100572), and α-Strep-tag (IBA Solutions for Life Sciences, StrepMAB-Classic). Site-directed mutagenesis was performed using TagMaster site-directed mutagenesis kit (GM Biosciences). The E. coli UPPS was amplified from genomic DNA. M. acetovorans ORFs encoding MaUPPS-A (MA3723) and MaUPPS-B (MA4402) were amplified from synthetically synthetized codon optimized for expression in S. cerevisiae gBlocks Gene Fragments (ITD Integrated Technologies, Coralville, IA). Glcis-PT, NgBR, and hCIT isoform 1 were amplified from previous plasmids. The cassette containing synthetic intron and IRES was amplified from pIRESneo1 (Clontech). The Invitrogen Gateway cloning strategy was used to insert cDNA into yeast expression vectors. All PCR products were cloned into pCR8/GW/TOPO TA (Invitrogen), sequenced, and subcloned into final yeast expression vector pKG-GW1 or pKG-GW2. To express His-Strep tag II–Glcis-PT in bacteria, the Strep tag II–Glcis-PT was amplified with restriction enzyme recognition sites, and the Strep tag sequence was inserted in the primers. The PCR product was ligated in frame with the internal His6 tag of the pRSF-DUET1 plasmid.
To express both subunits of hcis-PT from single mRNA in mammalian cells, the IRES surrounded by two multicloning sites was introduced into pCEP4 (Invitrogen) using NEBUilder HiFi DNA Assembly (NEB) to obtain the pKGmDUET vector. His6 internally tagged NgBR was subcloned into BamHI/NotI sites of pKGmDUET from pKG-GW2-(31-HIS)NgBR plasmid, and SmaI/EcoRI/Klenow-treated Strep-hCIT was subcloned into PmeI site from pKG-GW1-Strep-hCIT plasmid.
Yeast complementation assay
For yeast complementation analysis of cis-PTs, S. cerevisiae strains KG405 (nus1Δ rer2Δ srt1Δ), carrying the Glcis-PT gene on a plasmid with a URA3 marker was used (45). To analyze homomeric Glcis-PT and EcUPPS mutants, strain KG405 was transformed with vector pKG-GW1 (leucine selection) carrying WT or mutated versions of corresponding genes or empty vector as negative control. To phenotypically analyze hcis-PT, strain KG405 was transformed with vectors pKG-GW1 carrying hCIT variants (leucine selection) and pKG-GW2 carrying NgBR variants (methionine selection) in combination or empty vectors as negative control. To analyze putative heteromeric UPPS of M. acetovorans (MaUPPS), strain KG405 was transformed with vector pKG-GW1 carrying MaUPPS-A (MA3723, hCIT ortholog) and vector pKG-GW2 carrying WT and mutated variants of MaUPPS-B (MA4402, NgBR ortholog) in combination or with the corresponding empty vectors as negative control. Transformed yeast cells were grown overnight at 30 °C in synthetic defined medium lacking uracil and leucine or lacking uracil, methionine, and leucine were streaked onto synthetic defined medium containing all amino acids, nucleotide supplements, and 1% (w/v) 5-FOA (Zymo Research) and onto YPD plates. The plates were incubated for up to 7 days at 30 °C. Colonies growing on the 5-FOA plates were streaked on synthetic defined medium lacking uracil and incubated at 30 °C for 3 days to verify the loss of the pNEV-Glcis-PT plasmid. Yeast strain KG405 and its derivative carrying MaUPPS complex, hcis-PT complex or single subunits enzymes expressed from pKG-GW1 plasmid instead of pNEV-Glcis-PT were cultured in 2% (w/v) Bacto peptone and 1% (w/v) yeast extract supplemented with 2% glucose (w/v) (YPD). Synthetic minimal media were made of 0.67% (w/v) yeast nitrogen base and 2% (w/v) supplemented with auxotrophic requirements. For solid medium, agar (BD Biosciences, Sparks, MD) was added at a 2% (w/v) final concentration. Yeast cells were transformed using the Frozen-EZ yeast transformation II kit (Zymo Research).
Purification of human cis-PT
To purify the human hcis-PT complex, constructs containing internally tagged His6-NgBR and N-terminally tagged Strep-hCIT(pKGmDUET-hCIT–NgBR) were transiently transfected in 200 ml of culture of Expi293F cells according to the manufacturer's protocol (Invitrogen). The cells were harvested 72 h post-transfection by centrifugation and washed with PBS. Each gram of the cells was resuspended in S-Tactin buffer (100 mm Tris-HCl, pH 8, 300 mm NaCl, 2 mm 2-mercaptoethanol, 1 mm MgCl2), and cells were disrupted by sonication on ice. Unbroken material was cleared with a 1,000 × g centrifugation, and the supernatant from this spin was recentrifuged at 200,000 × g for 30 min at 4 °C to obtain total membrane fractions. Total membrane fraction was homogenized in S-Tactin buffer supplemented with 0.5% Triton X-100 to solubilize membrane proteins. Solubilization was followed by additional 200,000 × g centrifugation. Strep-hCIT was purified from 200,000 × g supernatant using the Strep-Tactin XT system (IBA GmbH). The Strep-hCIT–His6-NgBR complex was eluted from Strep-Tactin XT resin with 100 mm Tris-HCl, pH 8, 300 mm NaCl, 2 mm 2-mercaptoethanol, 1 mm MgCl2, 0.1% Triton X-100, 5% glycerol, 50 mm biotin elution buffer. The Strep-Tactin purification of Strep-hCIT was followed by nickel column purification of His6-NgBR using HisPur nickel-nitrilotriacetic acid resin (Thermo Fisher Scientific). Sephadex G-25 in PD-10 desalting columns (GE Healthcare Life Sciences) were used to exchange buffer for 20 mm NaPi, pH 8, 300 mm NaCl, 2 mm 2-mercaptoethanol, 1 mm MgCl2, 0.1% Triton X-100, 20% glycerol. Purification efficiency was tracked by Western blot analysis of each fraction, Coomassie staining of SDS-PAGE of final eluate, and measurement of specific cis-PT activity (Fig. 1 and Table 1).
Purification of Glcis-PT
To purify Glcis-PT, the pRSF-DUET1–Glcis-PT plasmid was transformed into E. coli Rosetta 2 cells (Novagen). E. coli was grown in autoinduction medium (46) until the logarithmic growth phase at 37 °C flowed 24 h of incubation at 17 °C to express heterologous protein. The cells were harvested by centrifugation, washed with PBS, and stored −80 °C. The proteins were extracted using B-PER reagent (Thermo Fisher Scientific) supplemented with 1 mm MgCl2 and 2 mm 2-mercaptoethanol. The Glcis-PT was first purified using nickel column HisPur nickel-nitrilotriacetic acid resin (Thermo Fisher Scientific) followed by the Strep-Tactin XF (IBA GmbH) purification. Sephadex G-25 in PD-10 desalting columns (GE Healthcare) were used to exchange buffer for 20 mm NaPi, pH 8, 300 mm NaCl, 2 mm 2-mercaptoethanol, 1 mm MgCl2, 0.1% Triton X-100, 20% glycerol. Purification efficiency was tracked by Western blot analysis and Coomassie staining of SDS-PAGE of final eluate.
Size-exclusion chromatography
The size-exclusion chromatography was carried out using an ÄKTA purifier (GE Healthcare) with the size-exclusion column Superdex 200 10/300 GL (GE Healthcare) at flow rate of 0.5 ml/min. The UV absorption at 254 nm was measured as a readout for protein elution. The used buffer contained 50 mm Tris-HCl buffer, pH 8.0, and 300 mm NaCl, 2 mm 2-mercaptoethanol, 1 mm MgCl2, 0.1% Triton X-100, 5% (v/v) glycerol. The column was calibrated by using the low molecular weight and high molecular weight calibration kit (GE Healthcare). For determination of the void, volume blue dextran (2,000 kDa; GE Healthcare) was used. Size determination was calculated based on the standard linear equation based on the calibration of the column.
cis-PT enzymatic activity of hcis-PT
Standard incubation mixture contained, in a final volume of 50 μl, 50 μm FPP, 100 μm [1-14C]IPP (55 mCi/mmol), 50 mm Tris-HCl, pH 8, 1 mm MgCI2, 20 mm 2-mercaptoethanol, 1 mg/ml BSA, 10 mm KF, and 1% (w/v) phosphatidylinositol. Membrane or crude protein (100 μg) or 20–100 ng of purified protein was used for activity assays. In some experiments with crude membranes, zaragozic acid A (10 μm) was added. Reactions were incubated for 60 min at 37 °C and terminated by the addition of 1 ml of chloroform-methanol 3:2. The protein pellet was removed by centrifugation, and the supernatant was washed three times with 1/5 volume of 10 mm EDTA in 0.9% NaCl. The incorporation of [14C]IPP into organic fractions containing polyprenyl diphosphate was measured by scintillation counting.
Kinetic parameters
Standard 25 or 100 μl (hCIT–NgBRG292 mutant) reaction mixture containing 9–25 nm hcis-PT was used. To measure kinetic parameters for FPP, 0.1–50 μm FPP was use along with 100 μm IPP for wild-type enzyme, hCITK42E and NgBRH100A mutants, 200 μm IPP for NgBRR292H mutant, and 400 μm IPP for NgBRG292 mutant. To measure kinetic parameters for IPP, 0.1–400 μm IPP was used along with 50 μm FPP. The initial velocity data were fitted to the Michaelis–Menten equation using the GraphPad Prism 7.02 computer program (GraphPad Software, Inc.) to obtain Km values. kcat values were obtained from the Michaelis–Menten equation for IPP.
cis-PT enzymatic activity of Glcis-PT
Glcis-PT activity was measured as before (39) with minor modifications. Briefly the incubation mixture contained, in a final volume of 50 μl, 45 μm FPP, 100 μm [1-14C]IPP (55 mCi/mmol), 25 mm Tris-HCl, pH 7.4, 1 mm MgCI2, 20 mm 2-mercaptoethanol, 10 mm KF, 0.1% Triton X-100, 1 mg/ml BSA, 2 μg of purified enzyme. After 60 min of incubation at 37 °C, the reaction was terminated by the addition of 1 ml of chloroform-methanol (3:2). The protein pellet was removed by centrifugation, and the supernatant was washed three times with volume of 10 mm EDTA in 0.9% NaCl. The incorporation of [14C]IPP into organic fractions containing polyprenyl diphosphate was measured by scintillation counting.
cis-PT enzymatic activity of MaUPPS
For S. cerevisiae expressing MaUPPS complex membrane fractions were prepared as described (47), and cis-PT activity measured (47, 48) with minor modifications. Briefly the incubation mixture contained, in a final volume of 100 μl, 45 μm FPP, 100 μm [1-14C]IPP (55 mCi/mmol) 25 mm Tris-HCl, pH 7.4, 1 mm MgCI2, 20 mm 2-mercaptoethanol, 10 mm KF, 10 μm zaragozic acid A, and 250 μg of membranes protein. After 90 min of incubation at 30 °C, the reaction was terminated by the addition of 4 ml of chloroform-methanol (3:2). The protein pellet was removed by centrifugation, and the supernatant was washed three times with volume of 10 mm EDTA in 0.9% NaCl. The organic phase was concentrated under a stream of nitrogen. Then organic solvent was evaporated, and lipids were loaded onto HPTLC RP-18 precoated plates with a concentrating zone and run in acetone containing 50 mm H3PO4. The plates were exposed to film to visualize the products of IPP incorporation. To measure incorporation of radioactive IPP into polyprenol fraction, the gel from the zone containing radiolabeled polyprenols was scraped and subjected to liquid scintillation counting. To analyze the length of polyprenol diphosphates before running the TLC analysis lipids were subjected to mild acid hydrolysis. Lipids were loaded onto HPTLC RP-18 together with undecaprenol as internal standard and subjected to radioautography as was described above.
Author contributions
K. A. G. designed all the expression systems for NgBR–hCIT, generated yeast strains, performed enzymatic assays on the NgBR–hCIT complex, and wrote the manuscript; B. H. E. cloned, expressed, purified, and characterized Glcis-PT WT and mutant enzymes and edited the manuscript; E. J. P. cloned and characterized NgBR constructs and participated in experimental design and writing; J. R. K. assisted with purification strategies and gel filtration chromatography; and W. C. S. supervised overall project and wrote manuscript.
Acknowledgment
We acknowledge the gift of undecaprenol from Dr. Ewa Kula-Swiezewska (Polish Academy of Science).
This work was supported by National Institutes of Health Grants R01 HL64793, R01 HL61371, and HL133018; a grant from the Leducq Fondation (MIRVAD network; to W. C. S.); and an American Heart Association Scientist Development Grant (to E. J. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- cis-PT
- cis-prenyltransferases
- UPPS
- undecaprenyl diphosphate synthase
- FPP
- farnesyl diphosphate
- IPP
- isopentenyl diphosphate
- FOA
- 5-fluoroorotic acid
- Strep
- streptomycin
- IRES
- internal ribosome entry site
- zFPPS
- Z,Z-farnesyl diphosphate synthase
- SUP
- supernatant..
References
- 1. Grabińska K. A., Park E. J., and Sessa W. C. (2016) cis-Prenyltransferase: new insights into protein glycosylation, rubber synthesis, and human diseases. J. Biol. Chem. 291, 18582–18590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hartley M. D., and Imperiali B. (2012) At the membrane frontier: a prospectus on the remarkable evolutionary conservation of polyprenols and polyprenyl-phosphates. Arch. Biochem. Biophys. 517, 83–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weiss M. C., Sousa F. L., Mrnjavac N., Neukirchen S., Roettger M., Nelson-Sathi S., and Martin W. F. (2016) The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1, 16116. [DOI] [PubMed] [Google Scholar]
- 4. Takahashi S., and Koyama T. (2006) Structure and function of cis-prenyl chain elongating enzymes. Chem. Rec. 6, 194–205 [DOI] [PubMed] [Google Scholar]
- 5. Teng K.-H., and Liang P.-H. (2012) Structures, mechanisms and inhibitors of undecaprenyl diphosphate synthase: a cis-prenyltransferase for bacterial peptidoglycan biosynthesis. Bioorg. Chem. 43, 51–57 [DOI] [PubMed] [Google Scholar]
- 6. Park E. J., Grabińska K. A., Guan Z., Stránecký V., Hartmannová H., Hodaňová K., Barešová V., Sovová J., Jozsef L., and Ondrušková N. (2014) Mutation of Nogo-B receptor, a subunit of cis-prenyltransferase, causes a congenital disorder of glycosylation. Cell Metab. 20, 448–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harrison K. D., Park E. J., Gao N., Kuo A., Rush J. S., Waechter C. J., Lehrman M. A., and Sessa W. C. (2011) Nogo-B receptor is necessary for cellular dolichol biosynthesis and protein N-glycosylation. EMBO J. 30, 2490–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamashita S., Yamaguchi H., Waki T., Aoki Y., Mizuno M., Yanbe F., Ishii T., Funaki A., Tozawa Y., Miyagi-Inoue Y., Fushihara K., Nakayama T., and Takahashi S. (2016) Identification and reconstitution of the rubber biosynthetic machinery on rubber particles from Hevea brasiliensis. eLife 5, e19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kwon M., Kwon E.-J., and Ro D. K. (2016) cis-Prenyltransferase and polymer analysis from a natural rubber perspective. Methods Enzymol. 576, 121–145 [DOI] [PubMed] [Google Scholar]
- 10. Qu Y., Chakrabarty R., Tran H. T., Kwon E.-J., Kwon M., Nguyen T.-D., and Ro D.-K. (2015) A lettuce (Lactuca sativa) homolog of human Nogo-B receptor interacts with cis-prenyltransferase and is necessary for natural rubber biosynthesis. J. Biol. Chem. 290, 1898–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Epping J., van Deenen N., Niephaus E., Stolze A., Fricke J., Huber C., Eisenreich W., Twyman R. M., Prüfer D., and Gronover C. S. (2015) A rubber transferase activator is necessary for natural rubber biosynthesis in dandelion. Nat. Plants 1, 10.1038/nplants.2015.48 [DOI] [Google Scholar]
- 12. Brasher M. I., Surmacz L., Leong B., Pitcher J., Swiezewska E., Pichersky E., and Akhtar T. A. (2015) A two-component enzyme complex is required for dolichol biosynthesis in tomato. Plant J. 82, 903–914 [DOI] [PubMed] [Google Scholar]
- 13. Sabry S., Vuillaumier-Barrot S., Mintet E., Fasseu M., Valayannopoulos V., Héron D., Dorison N., Mignot C., Seta N., Chantret I., Dupré T., and Moore S. E. (2016) A case of fatal type I congenital disorders of glycosylation (CDG I) associated with low dehydrodolichol diphosphate synthase (DHDDS) activity. Orphanet J. Rare Dis. 11, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Venturini G., Koskiniemi-Kuendig H., Harper S., Berson E. L., and Rivolta C. (2015) Two specific mutations are prevalent causes of recessive retinitis pigmentosa in North American patients of Jewish ancestry. Genet. Med. 17, 285–290 [DOI] [PubMed] [Google Scholar]
- 15. Züchner S., Dallman J., Wen R., Beecham G., Naj A., Farooq A., Kohli M. A., Whitehead P. L., Hulme W., Konidari I., Edwards Y. J., Cai G., Peter I., Seo D., Buxbaum J. D., et al. (2011) Whole-exome sequencing links a variant in DHDDS to retinitis pigmentosa. Am. J. Hum. Genet. 88, 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zelinger L., Banin E., Obolensky A., Mizrahi-Meissonnier L., Beryozkin A., Bandah-Rozenfeld D., Frenkel S., Ben-Yosef T., Merin S., Schwartz S. B., Cideciyan A. V., Jacobson S. G., and Sharon D. (2011) A missense mutation in DHDDS, encoding dehydrodolichyl diphosphate synthase, is associated with autosomal-recessive retinitis pigmentosa in Ashkenazi Jews. Am. J. Hum. Genet. 88, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Milani D., Cagnoli G. A., Baccarin M., Alfei E., Guerneri S., and Esposito S. (2016) Insights into 6q21-q22: refinement of the critical region for acro-cardio-facial syndrome. Congenit. Anom. (Kyoto) 56, 187–189 [DOI] [PubMed] [Google Scholar]
- 18. Szafranski P., Von Allmen G. K., Graham B. H., Wilfong A. A., Kang S.-H., Ferreira J. A., Upton S. J., Moeschler J. B., Bi W., Rosenfeld J. A., Shaffer L. G., Wai Cheung S., Stankiewicz P., and Lalani S. R. (2015) 6q22. 1 microdeletion and susceptibility to pediatric epilepsy. Eur. J. Hum. Genet. 23, 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miao R. Q., Gao Y., Harrison K. D., Prendergast J., Acevedo L. M., Yu J., Hu F., Strittmatter S. M., and Sessa W. C. (2006) Identification of a receptor necessary for Nogo-B stimulated chemotaxis and morphogenesis of endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 103, 10997–11002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ko T.-P., Chen Y.-K., Robinson H., Tsai P.-C., Gao Y.-G., Chen A. P.-C., Wang A. H.-J., and Liang P.-H. (2001) Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis. J. Biol. Chem. 276, 47474–47482 [DOI] [PubMed] [Google Scholar]
- 21. Fujihashi M., Zhang Y.-W., Higuchi Y., Li X.-Y., Koyama T., and Miki K. (2001) Crystal structure of cis-prenyl chain elongating enzyme, undecaprenyl diphosphate synthase. Proc. Natl. Acad. Sci. U.S.A. 98, 4337–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang H., Ohyama K., Boudet J., Chen Z., Yang J., Zhang M., Muranaka T., Maurel C., Zhu J.-K., and Gong Z. (2008) Dolichol biosynthesis and its effects on the unfolded protein response and abiotic stress resistance in Arabidopsis. Plant Cell 20, 1879–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo R.-T., Ko T.-P., Chen A. P.-C., Kuo C.-J., Wang A. H.-J., and Liang P.-H. (2005) Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis. J. Biol. Chem. 280, 20762–20774 [DOI] [PubMed] [Google Scholar]
- 24. Kurisaki A., Sagami H., and Ogura K. (1996) Proteolytic release of dehydrodolichyl diphosphate synthase from pig testis microsomes. Biosci. Biotechnol. Biochem. 60, 1109–1114 [DOI] [PubMed] [Google Scholar]
- 25. Adair W. L. Jr., and Cafmeyer N. (1987) Characterization of the Saccharomyces cerevisiae cis-prenyltransferase required for dolichyl phosphate biosynthesis. Arch. Biochem. Biophys. 259, 589–596 [DOI] [PubMed] [Google Scholar]
- 26. Adair W. L. Jr., Cafmeyer N., and Keller R. K. (1984) Solubilization and characterization of the long chain prenyltransferase involved in dolichyl phosphate biosynthesis. J. Biol. Chem. 259, 4441–4446 [PubMed] [Google Scholar]
- 27. Takahashi I., and Ogura K. (1982) Prenyltransferases of Bacillus subtilis: undecaprenyl pyrophosphate synthetase and geranylgeranyl pyrophosphate synthetase. J. Biochem. 92, 1527–1537 [DOI] [PubMed] [Google Scholar]
- 28. Oh S. K., Han K. H., Ryu S. B., and Kang H. (2000) Molecular cloning, expression, and functional analysis of a cis-prenyltransferase from Arabidopsis thaliana: implications in rubber biosynthesis. J. Biol. Chem. 275, 18482–18488 [DOI] [PubMed] [Google Scholar]
- 29. Apfel C. M., Takács B., Fountoulakis M., Stieger M., and Keck W. (1999) Use of genomics to identify bacterial undecaprenyl pyrophosphate synthetase: cloning, expression, and characterization of the essential uppS gene. J. Bacteriol. 181, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koyama T., Yoshida I., and Ogura K. (1988) Undecaprenyl diphosphate synthase from Micrococcus luteus BP 26: essential factors for the enzymatic activity. J. Biochem. 103, 867–871 [DOI] [PubMed] [Google Scholar]
- 31. Dowhan W. (2017) Understanding phospholipid function: why are there so many lipids? J . Biol. Chem. 292, 10755–10766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang S.-Y., Ko T.-P., Liang P.-H., and Wang A. H.-J. (2003) Catalytic mechanism revealed by the crystal structure of undecaprenyl pyrophosphate synthase in complex with sulfate, magnesium, and triton. J. Biol. Chem. 278, 29298–29307 [DOI] [PubMed] [Google Scholar]
- 33. Chan Y.-T., Ko T.-P., Yao S.-H., Chen Y.-W., Lee C.-C., and Wang A. H.-J. (2017) Crystal structure and potential head-to-middle condensation function of a Z,Z-farnesyl diphosphate synthase. ACS Omega 2, 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang W., Dong C., McNeil M., Kaur D., Mahapatra S., Crick D. C., and Naismith J. H. (2008) The structural basis of chain length control in Rv1086. J. Mol. Biol. 381, 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ogawa T., Emi K., Koga K., Yoshimura T., and Hemmi H. (2016) A cis-prenyltransferase from Methanosarcina acetivorans catalyzes both head-to-tail and nonhead-to-tail prenyl condensation. FEBS J. 283, 2369–2383 [DOI] [PubMed] [Google Scholar]
- 36. Shridas P., Rush J. S., and Waechter C. J. (2003) Identification and characterization of a cDNA encoding a long-chain cis-isoprenyltranferase involved in dolichyl monophosphate biosynthesis in the ER of brain cells. Biochem. Biophys. Res. Commun. 312, 1349–1356 [DOI] [PubMed] [Google Scholar]
- 37. Endo S., Zhang Y.-W., Takahashi S., and Koyama T. (2003) Identification of human dehydrodolichyl diphosphate synthase gene. Biochim. Biophys. Acta 1625, 291–295 [DOI] [PubMed] [Google Scholar]
- 38. Sato M., Sato K., Nishikawa S., Hirata A., Kato J., and Nakano A. (1999) The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodescis-prenyltransferase, a key enzyme in dolichol synthesis. Mol. Cell. Biol. 19, 471–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grabińska K. A., Cui J., Chatterjee A., Guan Z., Raetz C. R., Robbins P. W., and Samuelson J. (2010) Molecular characterization of the cis-prenyltransferase of Giardia lamblia. Glycobiology 20, 824–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Surmacz L., Plochocka D., Kania M., Danikiewicz W., and Swiezewska E. (2014) cis-Prenyltransferase AtCPT6 produces a family of very short-chain polyisoprenoids in planta. Biochim. Biophys. Acta 1841, 240–250 [DOI] [PubMed] [Google Scholar]
- 41. Akhtar T. A., Matsuba Y., Schauvinhold I., Yu G., Lees H. A., Klein S. E., and Pichersky E. (2013) The tomato cis-prenyltransferase gene family. Plant J. 73, 640–652 [DOI] [PubMed] [Google Scholar]
- 42. Kera K., Takahashi S., Sutoh T., Koyama T., and Nakayama T. (2012) Identification and characterization of a cis, trans-mixed heptaprenyl diphosphate synthase from Arabidopsis thaliana. FEBS J. 279, 3813–3827 [DOI] [PubMed] [Google Scholar]
- 43. Cunillera N., Arró M., Forés O., Manzano D., and Ferrer A. (2000) Characterization of dehydrodolichyl diphosphate synthase of Arabidopsis thaliana, a key enzyme in dolichol biosynthesis. FEBS Lett. 477, 170–174 [DOI] [PubMed] [Google Scholar]
- 44. Dennis M. S., Henzel W. J., Bell J., Kohr W., and Light D. R. (1989) Amino acid sequence of rubber elongation factor protein associated with rubber particles in Hevea latex. J. Biol. Chem. 264, 18618–18626 [PubMed] [Google Scholar]
- 45. Park E. J., Grabińska K. A., Guan Z., and Sessa W. C. (2016) NgBR is essential for endothelial cell glycosylation and vascular development. EMBO Reports 17, 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Studier F. W. (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 47. Szkopińska A., Grabińska K., Delourme D., Karst F., Rytka J., and Palamarczyk G. (1997) Polyprenol formation in the yeast Saccharomyces cerevisiae: effect of farnesyl diphosphate synthase overexpression. J. Lipid Res. 38, 962–968 [PubMed] [Google Scholar]
- 48. Grabińska K., Sosińska G., Orłowski J., Swiezewska E., Berges T., Karst F., and Palamarczyk G. (2005) Functional relationships between the Saccharomyces cerevisiae cis-prenyltransferases required for dolichol biosynthesis. Acta Biochim. Pol. 52, 221–232 [PubMed] [Google Scholar]